Abstract
Epigenetic changes in chromatin through histone post-translational modifications are essential for altering gene transcription in response to environmental cues. How histone modifications are regulated by environmental stimuli remains poorly understood yet this process is critical for delineating how epigenetic pathways are influenced by the cellular environment. We have used the target of rapamycin (TOR) pathway, which transmits environmental nutrient signals to control cell growth, as a model to delineate mechanisms underlying this phenomenon. A chemical genomics screen using the TOR inhibitor rapamycin against a histone H3/H4 mutant library identified histone H3 lysine 56 acetylation (H3K56ac) as a chromatin modification regulated by TOR signaling. We demonstrate this acetylation pathway functions in TOR-dependent cell growth in part by contributing directly to ribosomal RNA (rRNA) biogenesis. Specifically, H3K56ac creates a chromatin environment permissive to RNA polymerase I transcription and nascent rRNA processing by regulating binding of the high mobility group protein Hmo1 and the small ribosomal subunit (SSU) processome complex. Overall, these studies identify a novel chromatin regulatory role for TOR signaling and support a specific function for H3K56ac in ribosomal DNA (rDNA) gene transcription and nascent rRNA processing essential for cell growth.
INTRODUCTION
The extracellular and intracellular environment induces chromatin alterations to regulate gene expression yet the mechanisms underlying such interactions remain poorly understood (1). Since environmental signals such as nutrient availability influence gene expression and epigenetic processes affecting cell development (2), delineating these mechanisms has profound importance for many complex human diseases. The target of rapamycin (TOR)-signaling pathway transmits nutrient (i.e. growth factor and amino acid) information to regulate cell growth and proliferation, and this pathway is deregulated in many diseases, including cancer, diabetes and cardiovascular disease (3). TOR was originally identified in the budding yeast Saccharomyces cerevisiae but is conserved in all eukaryotes (4–6). The TOR pathway consists of two signaling branches. The TORC1 branch controls transcriptional and translational processes necessary for growth and proliferation and is inhibited by the drug rapamycin, while the TORC2 complex controls the cytoskeletal changes necessary for growth and is rapamycin insensitive (7). The yeast TORC1 complex consists of either the Tor1 or Tor2 kinases, Lst8, Kog1 and Tco89 (7). Increases in intravacuolar amino acid concentration leads to TORC1 activation through association with the vacuole-localized EGO complex, consisting of the Ego1 and Ego3 proteins as well as the small GTPases Gtr1 and Gtr2 (8). TORC1 activation can then lead to direct phosphorylation of the AGC kinase family member, Sch9, to mediate some of TORC1’s effect on cell growth (9,10). However, TORC1 signaling also has Sch9-independent effects. In particular, Tor kinases are recruited to the promoter regions of many downstream target genes, including the ribosomal DNA (11) (rDNA) transcribed by RNA Polymerase I (Pol I) in yeast and to RNA Pol I, Pol II and Pol III transcribed genes in mammalian cells (12–14).
Although TORC1 signaling is critical for controlling gene expression essential for cell growth, how it regulates chromatin structure to control transcription is not well understood. Previous studies in yeast have linked the RSC chromatin remodeling complex (15), the Rpd3 histone deacetylase complex (16,17) and the Esa1 histone acetyltransferase (18) to TORC1-dependent gene expression, but whether TORC1 signaling directly controls these chromatin modifiers has not been addressed. Acetylation of histones plays a key role in decompacting chromatin to permit transcriptional activity (19). In particular, histone H3 lysine 56 acetylation (H3K56ac) promotes nucleosome disassembly at promoter regions to facilitate transcription initiation by disrupting the histone H3–DNA interactions that occur close to where DNA enters and exits the nucleosome (20–23). H3K56ac is regulated by the combined actions of the histone chaperone Asf1 and the acetyltransferase Rtt109 (24–27) and this pathway contributes not only to gene transcription but also to DNA repair and replication (21,28). While H3K56ac levels may peak during S-phase to facilitate nascent chromatin formation (29), recent studies suggest that H3K56ac is also expressed throughout the entire cell cycle (30,31), suggesting this histone mark has cell-cycle independent roles as well. However, the mechanisms regulating H3K56ac levels and the role this histone mark plays in cell function still remain poorly understood.
To elucidate mechanisms by which TORC1 regulates chromatin, we have completed a systematic rapamycin-based chemical genomics screen of a histone H3/H4 library (32) to identify histone residues involved in TORC1-regulated growth, since mutations in many TORC1 pathway components cause rapamycin sensitivity (manuscript in preparation). Our screen identified acetylation at histone H3 lysine 56 (H3K56ac), controlled by the histone chaperone Asf1 and the acetyltransferase Rtt109 (24–27,33), as a chromatin pathway regulated by TORC1 signaling and necessary for optimal rDNA transcription and ribosomal RNA (rRNA) processing.
MATERIALS AND METHODS
Yeast media and cell growth conditions
Yeast deletion mutants and derivative strains were derived from the BY4741 genetic background. The histone H3/H4 mutant library was purchased from Open Biosystems and used to engineer the specific genomically-tagged alleles described herein using standard yeast molecular biology approaches (34). A full listing of the yeast strains and plasmids used in this study are found in Supplementary Table S1. Unless otherwise stated, yeast strains were cultured either in nutrient rich media consisting of 1% yeast extract, 2% peptone and 2% glucose (YPD), nutrient defined media (SC), or SC media lacking uracil (SC-Ura) to select for plasmid maintenance. All yeast growth media and reagents were purchased from either Research Products International or US Biologicals. Rapamycin was purchased from Fisher Scientific and reconstituted in methanol. Cells were typically grown to an OD600 = 0.8–1.2 unless otherwise stated.
Whole-cell extract preparation, ChIP and antibodies used
Whole-cell extracts (WCE) were prepared and coimmunoprecipitations were performed as previously described using 750 µg of WCE (35). ChIP experiments were performed as described (35) with minor modifications. The sonicated chromatin extract was clarified using a low speed centrifugation step (4000g) to maintain soluble chromatin fragments containing large protein complexes bound (36). For the immunoprecipitation (IP) step, 500 µg of soluble chromatin was immunoprecipitated overnight and immune complexes isolated using Protein A agarose beads (Santa Cruz Biotechnology) after extensive washing and cross-link reversal. Purified DNA was isolated using DNA binding columns (Genesee, Co.) and DNA was eluted in 50 µl of water for the immunoprecipitated material. An amount of 50 µg of input material was also purified similarly and eluted in 100 µl of water. For qPCR, the IP material was diluted 1:5 and the input material diluted 1:50 and qPCR performed using a Roche SYBR Green master mix on a Roche 480 Lightcycler real-time PCR machine. IP specific signals were first normalized to input levels via the ΔΔCt method then further normalized to an intergenic region on Chromosome V to control for immunoprecipitation and DNA purification efficiency. Primer sequences are listed in Supplementary Table S2. The α-Myc and α-HA antibodies were purchased from Millipore, the α-G6PDH antibody purchased from Sigma-Aldrich, and the α-H3K56ac and α-H3 antibodies purchased from Active Motif.
RNA extraction and cDNA synthesis
Total RNA was isolated from cells using TriReagent (Sigma-Aldrich, Inc.) and bead beating. RNA was digested to completion with DNase I (Promega, Inc.) and then 1 µg of RNA was used in a cDNA synthesis reaction with random hexamer primers using the Im-Prom II cDNA synthesis kit (Promega, Inc.) per the manufacturer’s protocol. cDNAs were resuspended to a final volume of 100 µl and then diluted 1:5 before performing qPCR as described above. rDNA specific signals were normalized to the levels of yeast actin mRNA via the ΔΔCt method.
Flow cytometry analysis
Yeast cells were stained with SYTOX Green (Invitrogen, Inc.) as previously described (37) and samples analyzed on a Becton Dickinson LSR II flow cytometer and data analyzed with Modfit software at the UT Flow Cytometer facility.
RESULTS
Identification of histone H3K56ac as a TORC1-regulated chromatin modification
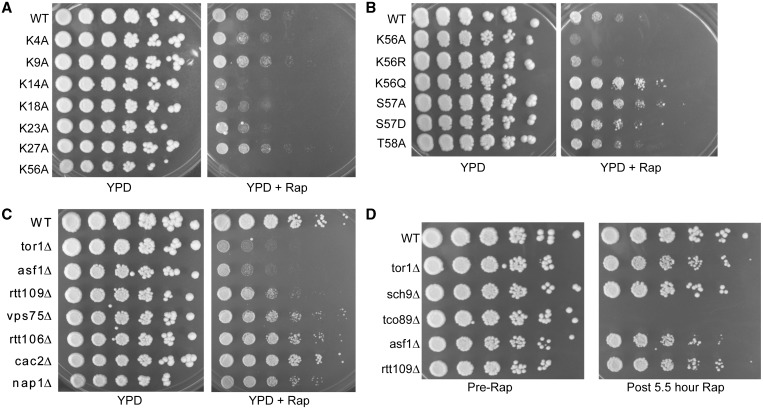
We have completed an initial rapamycin-based chemical genomics screen of a recently described library of histone H3/H4 amino acid mutants by comparing how each histone mutant grows relative to wild-type cells in the presence of a sub-inhibitory (25 nM) concentration of rapamycin. These screens identified lysine to alanine changes at histone H3 residues 14 (H3K14A), 18 (H3K18A), 23 (H3K23A) and 56 (H3K56A) as causing rapamycin sensitivity relative to wild-type cells, suggesting they contribute positively to TORC1 signaling (Figure 1A). These effects are specific as alanine changes at positions 9 (H3K9) and 27 (H3K27), also sites of acetylation, do not affect rapamycin sensitivity. The alanine change at histone H3 lysine 4 (H3K4), a site of both acetylation and methylation, results in a slight slow growth phenotype but upon longer incubation it does not exhibit significant rapamycin sensitivity (Figure 1A and data not shown). Of the mutants analyzed, the histone H3K56A mutation resulted in the most severe rapamycin hypersensitivity (Figure 1A) implicating this residue in TORC1-dependent cell growth. H3K56 is positioned close to where DNA enters and exits the nucleosome (22) and this lysine residue makes key DNA contacts that stabilize histone–DNA interactions and are disrupted by acetylation (20,23). Given the importance of this residue in regulating transcription, DNA repair and replication (21,28), we further analyzed its contribution to TORC1 signaling.
Figure 1.
The H3K56ac pathway is required for TORC1-regulated growth. (A) H3 lysine to alanine mutants exhibit variable sensitivity to rapamycin. Wild-type (WT) and the indicated histone H3 lysine to alanine mutants were grown overnight at 30°C and then cell density for each strain was measured by taking the OD600. Equivalent numbers of cells were serially diluted 5-fold and replica spotted to control YPD or YPD plates containing 25 nM rapamycin and incubated at 30°C for 4 days before photographing. (B) Acetylation of H3K56 is key in TORC1-regulated growth. Experiment was performed as in (A) except photographs were taken at 3 days to highlight the growth difference between the H3K56Q and WT strains. (C) H3K56ac regulators are selectively required for TORC1-dependent cell growth. The experiment was performed as in (A) with the WT and indicated gene deletion mutants. Photographs were taken after 4 days at 30°C. (D) H3K56ac regulators do not undergo cell-cycle arrest after an inhibitory rapamycin treatment. The indicated yeast strains were cultured to log phase and then equal numbers of cells were pelleted, 5-fold serially diluted and spotted to YPD plates. The remaining cultures were treated with 200 nM rapamycin for 5.5 h and then equal numbers of cells were pelleted, washed and 5-fold serially diluted before spotting to YPD plates. Photographs were taken after incubation at 30°C for 3 days.
We next determined whether the H3K56A rapamycin phenotype is due either to loss of charge or acetylation. The H3K56 to arginine (H3K56R) mutant exhibits comparable rapamycin sensitivity as the H3K56A mutant, whereas the H3K56 to glutamine (H3K56Q) change, which mimics acetylation, results in rapamycin resistance relative to control cells (Figure 1B). These results implicate H3K56ac, and not disruption of charge, as the contributor to TORC1-dependent growth. A change of H3 serine 57 to either alanine (H3S57A) or to the phosphomimetic residue aspartic acid (H3S57D) also resulted in rapamycin resistance (Figure 1B), which is likely due to its influence on H3K56ac as H3S57 has been suggested to regulate H3K56ac dynamics (38). Importantly, these rapamycin phenotypes are highly specific for H3K56 and H3S57 since a change of H3 threonine 58 to alanine (H3T58A) exhibits wild-type growth (Figure 1B). We next determined if deleting the genes for the histone chaperone Asf1 or the histone acetyltransferase Rtt109, which are essential for H3K56ac (24–27), also resulted in comparable rapamycin sensitivity. Both asf1Δ and rtt109Δ exhibited profound rapamycin sensitivity whereas other histone chaperone mutants, including vps75Δ, rtt106Δ, cac2Δ and nap1Δ, were either rapamycin insensitive or exhibited only minor sensitivity (Figure 1C). Interestingly, while asf1Δ was as rapamycin sensitive as tor1Δ, rtt109Δ mutants consistently exhibited a slightly milder phenotype (Figure 1C) suggesting Asf1 may provide other functions in addition to H3K56ac regulation that are critical for TORC1-regulated growth. A subset of TORC1 pathway mutants, specifically tco89Δ and cells deleted for EGO complex components, undergo an irreversible G1 cell-cycle arrest upon rapamycin treatment (8). To determine if the rapamycin phenotype of asf1Δ and rtt109Δ is due to a similar cell-cycle defect, we cultured wild-type, asf1Δ, rtt109Δ and multiple TORC1 pathway mutants to log phase before serially diluting equal numbers of cells and spotting onto nutrient rich YPD plates. The remaining cultures were treated with an inhibitory concentration of rapamycin (200 nM) for 5.5 h before washing and spotting onto control plates. Consistent with a previous report, tco89Δ mutants did not reenter the cell cycle and failed to grow (8), whereas both tor1Δ and sch9Δ resumed growth (Figure 1D). Under these conditions both asf1Δ and rtt109Δ fully resumed growth, thus demonstrating their rapamycin sensitivity is not due to a permanent cell-cycle arrest. Overall, these results specifically identify Asf1 and Rtt109 mediated H3K56ac as a chromatin modification pathway essential for TORC1-dependent cell growth.
The TORC1 subunit Tco89 regulates global H3K56ac
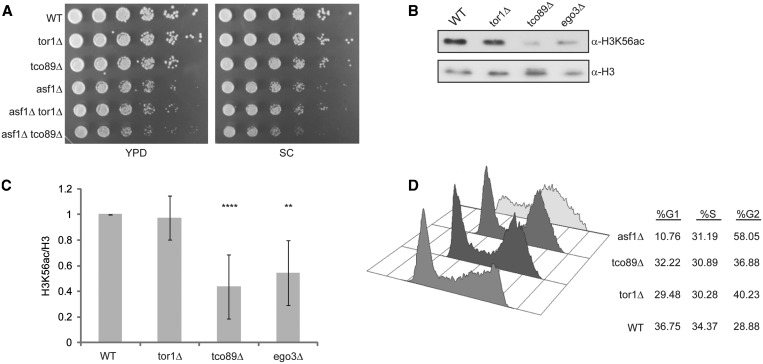
To further characterize the link between TORC1 signaling and the H3K56ac pathway, we combined asf1Δ with either tor1Δ or tco89Δ and analyzed the growth of the individual and double mutants on both nutrient rich (YPD) and nutrient defined (SC) media. The asf1Δ tor1Δ cells grew identically to asf1Δ cells (Figure 2A) demonstrating that the double mutant did not result in a negative (synthetic sick) genetic interaction. In contrast, on YPD plates the asf1Δ tco89Δ resulted in a minor synthetic sick phenotype compared to asf1Δ alone after 2 days of growth, and this synthetic sick phenotype was slightly exacerbated on nutrient defined media (Figure 2A). These data support the negative genetic interactions between asf1Δ and tco89Δ previously identified (39,40), and suggest that the TORC1 and H3K56ac pathways may be functionally linked. We next examined whether perturbation of TORC1 affected global H3K56ac. WCE from log phase wild-type, tor1Δ and tco89Δ cultures were generated and immunoblotted with antibodies against H3K56ac (α−H3K56ac) (Supplementary Figure S1a) or general histone H3 (α−H3) (Figure 2B). We also included ego3Δ in these analyses since the EGO complex is required for TORC1 activation and EGO mutants share many of the same phenotypes as tco89Δ mutants. Intriguingly, while the tor1Δ mutant had no impact on H3K56ac, both tco89Δ and ego3Δ caused significant reductions in global H3K56ac levels (Figure 2B and C). Since EGO activates TORC1 signaling in response to intracellular amino acid availability (8), these results implicate EGO-dependent activation of TORC1 in the regulation of global H3K56ac.
Figure 2.
The TORC1 subunit Tco89 is required for H3K56ac. (A) Asf1 mutations exhibit negative genetic interactions only with Tco89, but not Tor1, mutants. Wild-type and the indicated single and double gene deletion mutants were grown overnight and then equal numbers of cells were 5-fold serially diluted and spotted to YPD or SC media. Plates were incubated at 30°C and photographs taken after 2 days. (B) Disruption of the EGO complex or TORC1 reduces global H3K56ac. Wild-type or the indicated gene deletion mutants were cultured in YPD to log phase, WCE were prepared and 30 µg resolved by 15% SDS–PAGE. Samples were transferred to PVDF membrane and immunoblotted with α-H3K56ac or α-H3 specific antibodies. (C) Quantitation of immunoblots. α−H3K56ac and α−H3 immunoblots were quantified using ImageJ software and expressed as a ratio of H3K56ac over total H3. The average and standard deviation of a minimum of at least three or more independent experiments for each mutant are plotted and statistical significance determined by t-test. **P < 0.05; ****P < 0.005. (D) H3K56ac reduction in tco89Δ is not due to a significant disruption in cell-cycle progression. Wild-type, tor1Δ, tco89Δ and asf1Δ were grown to an OD600 = 0.8 before staining with SYTOX Green and performing flow-cytometry. The histograms for each strain are presented as well as the percentage of cells in G1, S and G2 phase.
To begin delineating the mechanism by which TORC1 regulates H3K56ac, we next determined if the decreased H3K56ac in tco89Δ was due to reduced expression of either Asf1 or Rtt109. We integrated a 6XHA epitope tag at the ASF1 or RTT109 genomic locus in wild-type or tco89Δ cells and analyzed protein expression by immunoblotting. Neither Asf1 nor Rtt109 protein levels were significantly altered in tco89Δ cells compared to wild-type, demonstrating that the decreased H3K56ac is not a result of reduced expression of these H3K56ac regulators (Supplementary Figure S1b and c). Because TORC1 mutants may affect cell-cycle progression (4,41,42), and since H3K56ac levels peak during S phase (29), we next determined if the H3K56ac defect in tco89Δ was caused by cell-cycle defects. Wild-type, tor1Δ, tco89Δ and asf1Δ cells were grown asynchronously to log phase, stained with SYTOX green and analyzed by flow cytometry. Under these conditions, wild-type, tor1Δ cells and tco89Δ exhibit similar numbers of cells in S-phase whereas the tor1Δ and tco89Δ mutants exhibit slight increases in G2-phase cells relative to wild-type (Figure 2D). Under these conditions, asf1Δ results in a reduction in G1 phase cells with a significant increase in the number of cells in G2, a result consistent with a previous study (43). These data suggest that loss of H3K56ac selectively in tco89Δ mutants is not solely due to defects in cell-cycle regulation since H3K56ac is maximal in S-phase (29) and both tor1Δ and tco89Δ show similar cell-cycle profiles. Instead, this effect reflects a mechanistically distinct aspect of TORC1 signaling.
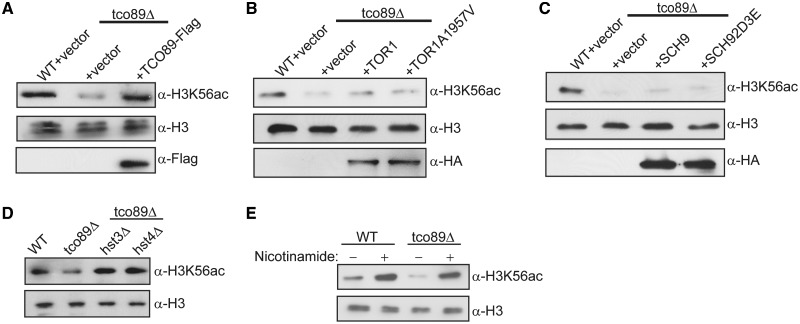
To confirm that the decreased H3K56ac was due solely to loss of Tco89, we transformed tco89Δ cells with either empty vector or vector expressing Tco89 as a C-terminal Flag fusion. H3K56ac was fully rescued upon Tco89-Flag expression (Figure 3A), thus confirming that loss of Tco89 was the sole cause of the H3K56ac defect. Because tco89Δ likely reduces the strength of TORC1 signaling, we attempted to rescue H3K56ac in tco89Δ by overexpressing wild-type Tor1 or a Tor1 mutant (Tor1A1957V) that has increased kinase activity in vitro (44). Surprisingly, neither wild-type nor the hyperactive Tor1 kinase increased H3K56ac in tco89Δ (Figure 3B). Similar overexpression experiments performed in a wild-type background also failed to drive H3K56ac levels higher relative to controls (Supplementary Figure S2) suggesting that TORC1-regulated H3K56ac levels are already maximal in wild-type cells. We do note a minor and reproducible increase in histone H3 levels from wild-type extracts expressing the hyperactive Tor1 allele; however, the significance of this increase is currently unknown. To determine if the effect on H3K56ac in tco89Δ was due to lower levels of Sch9 activation, we overexpressed a wild-type Sch9 kinase or a mutant Sch9 no longer dependent on TORC1-dependent phosphorylation for activation (Sch92D3E) (9). Neither wild-type Sch9 nor the Sch92D3E mutant rescued H3K56ac in tco89Δ cells (Figure 3C) suggesting that this branch of the TORC1-signaling pathway is unlikely responsible for H3K56ac. Histone acetylation is regulated by a balance of both acetyltransferase and deacetylase activity such that lower steady-state H3K56ac could be caused either by defects in acetylation or increased deacetylase activity. To begin to address the mechanism responsible for the decreased H3K56ac, we asked whether deleting the Sirtuin deacetylases Hst3 or Hst4, both of which target H3K56ac (45), restores acetylation in tco89Δ. We engineered tco89Δ hst3Δ and tco89Δ hst4Δ strains and compared the levels of H3K56ac in these double mutants to that in tco89Δ. Deletion of either deacetylase fully rescues H3K56ac (Figure 3D) as does treating tco89Δ cells with the pan-Sirtuin inhibitor nicotinamide (Figure 3E). Taken together, these data suggest that Tco89 provides a unique function within TORC1 essential for H3K56ac regulation that cannot be bypassed by increased Tor1 kinase activity or activation of the downstream effector kinase Sch9. Furthermore, these data suggest that tco89Δ cells are fully competent for generating H3K56ac and that one possible explanation for the lower H3K56ac could be an increase in Hst3 and/or Hst4 deacetylase activity.
Figure 3.
H3K56ac defects in tco89Δ cannot be rescued by increased Tor1 or Sch9 kinase activity but are rescued by loss of the Sirtuin deacetylases Hst3 or Hst4. (A) Reexpression of Tco89 fully rescues global H3K56ac in tco89Δ cells. Wild-type and tco89Δ cells were transformed with either an empty vector or vector expressing Tco89 as a C-terminal mono-Flag fusion and grown in SC-Ura media to select for plasmid maintenance. An amount of 30 µg of WCE were prepared and analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. (B) Increased TORC1 kinase activity does not rescue global H3K56ac in tco89Δ cells. Experiment was performed as in (A) except cells were transformed with control vector or vectors expressing HA-tagged wild-type Tor1 or the mutant Tor1A1957V kinase that has increased kinase activity (44). (C) TORC1 regulation of H3K56ac is independent of the downstream Sch9 kinase. The experiment was performed as in (A) except cells were transformed with control vector or vectors expressing HA-tagged wild-type Sch9 or the Sch92D3E mutant that is active independent of upstream TORC1 activity (9). (D) H3K56ac is rescued in tco89Δ by inactivation of Hst3 or Hst4. Wild-type, tco89Δ, tco89Δ hst3Δ and tco89Δ hst4Δ strains were grown to log phase before preparing WCE . 30 µg of WCE were resolved by SDS–PAGE and analyzed by α-H3K56ac or α-H3 immunoblot. (E) Global inhibition of Sirtuin activity rescues H3K56ac in tco89Δ. Wild-type and tco89Δ cultures were grown to log phase and then mock treated or treated with 25 mM nicotinamide for 1 h before harvesting and preparing WCE. Samples were analyzed as in (D).
TORC1 signaling regulates H3K56ac on the rDNA
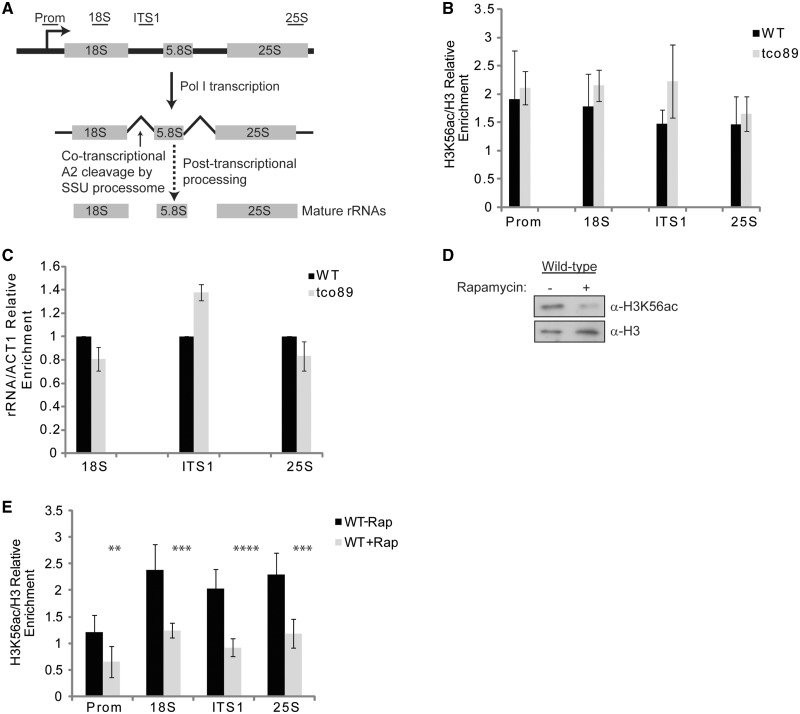
TORC1 signaling directly regulates RNA polymerase I (Pol I) transcription of the rDNA, which is repeated in yeast ∼150–200 times with only half of the repeats being transcriptionally active in growing cells (46,47). To determine if tco89Δ resulted in decreased chromatin-associated H3K56ac on the rDNA, we analyzed H3K56ac levels by chromatin immunoprecipitation coupled with quantitative real-time PCR (ChIP–qPCR) using the PCR primers outlined (Figure 4A). Because we observed a slight exacerbation of the synthetic sick phenotype when asf1Δ tco89Δ cells were grown on nutrient-defined plates (Figure 2A), we grew wild-type and tco89Δ cells in SC media to log phase before performing α-H3K56ac and α−H3 ChIP analysis. Surprisingly, while H3K56ac was readily detectable on the rDNA, tco89Δ did not decrease the level of this histone modification (Figure 4B). To determine if tco89Δ significantly affected rRNA synthesis, we analyzed the levels of 18S and 25S rRNA by qPCR. We also analyzed the levels of ITS1-containing rRNA (Figure 4A) since this region of the rRNA is rapidly cotranscriptionally processed out of the primary transcript and its presence can be used as an indicator of RNA Pol I transcription activity and nascent rRNA synthesis (48). Loss of Tco89 had no effect on 18S, 25S or ITS1-containing rRNAs (Figure 4C), suggesting that the level of TORC1-signaling activity in the tco89Δ mutant was enough to allow for normal rRNA transcription. We also analyzed H3K56ac on two housekeeping genes, ACT1 and TUB1, to determine if tco89Δ affected their acetylation. We found no affect on ACT1 at either the promoter or 3′-end of the gene; however, while we did not detect any effect at the TUB1 promoter, we did see a modest reduction at the 3′ of the gene (Supplementary Figure S3) suggesting the possibility that tco89Δ may have effects on chromatin-associated H3K56ac elsewhere in the genome. To further analyze the impact of TORC1 signaling on H3K56ac, we grew wild-type cells in nutrient rich media (YPD) and mock-treated or treated with an inhibitory concentration of rapamycin (200 nM) for 1 h before repeating the α-H3K56ac and α-H3 immunoblots (Figure 4D) or ChIP experiment (Figure 4E). Under these conditions, H3K56ac was reduced both globally and across the length of the rDNA. These results confirm that TORC1 signaling regulates global H3K56ac and suggest that the H3K56ac maintained on the rDNA in tco89Δ is most likely due to the TORC1-signaling activity still retained in this mutant.
Figure 4.
H3K56ac regulators localize to the rDNA. (A) Simplified schematic of the rDNA locus and the relative locations of primers used in qPCR. Note that the ITS1 specific primers span the A2 site cleaved by the SSU processome during rRNA processing. Detailed description of rRNA processing reactions involved in rRNA maturation are described previously (49). (B) H3K56ac localizes to rDNA but is not decreased in tco89Δ. ChIP using α−H3K56ac or α−H3 antibodies and primers to the promoter, 18S, ITS1 or 25S rDNA sequences. Data are the average and standard deviation of a minimum of two or more independent experiments. (C) tco89Δ does not decrease rRNA levels. Total RNA was prepared from wild-type (WT) and tco89Δ cells grown to log phase and cDNA was synthesized using 1 µg of RNA and random hexamer primers. qPCR was performed with the indicated primer sets, the WT signal was set to 1 and tco89Δ expressed relative to WT The average and standard deviation of two independent experiments are shown. (D) H3K56ac is significantly reduced in WT cells upon rapamycin treatment. WT cells were grown to log phase and either mock treated or treated with an inhibitory (200 nM) concentration of rapamycin for 1 h. WCE were prepared and 30 µg analyzed by SDS–PAGE and α-H3K56ac and α-H3 immunoblotting. (E) Rapamycin mediated inhibition of TORC1 signaling reduces H3K56ac on the rDNA. Experiment was performed as in (D) except samples were processed for α-H3K56ac or α-H3 ChIP. The regions of the rDNA are indicated analyzed are indicated. The data are the average and standard deviation of three or more independent experiments. **P < 0.05; ***P < 0.01; ****P < 0.005.
H3K56ac regulates rDNA transcription and rRNA cotranscriptional processing
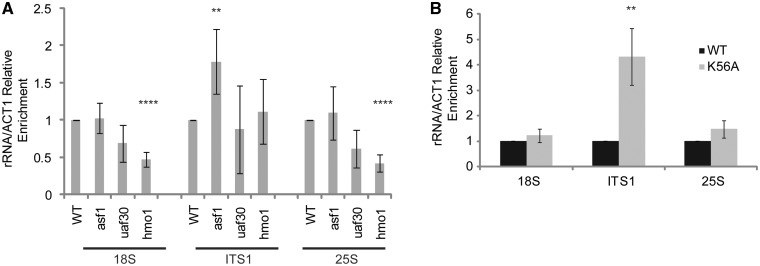
Although tco89Δ alone did not reduce H3K56ac levels on the rDNA, the presence of this histone modification suggested the possibility that H3K56ac may play a regulatory role in rDNA transcription. To test this possibility, we compared the levels of rRNAs in wild-type and asf1Δ. In this analysis, we also included uaf30Δ and hmo1Δ as controls for gene mutants that influence rDNA transcription and/or rRNA levels (50–52). Interestingly, while asf1Δ did not affect 18S or 25S rRNAs under these conditions, we did detect a modest but statistically significant increase (∼1.8-fold) in the non-processed rRNA as indicated by the increased ITS1 signal in asf1Δ relative to wild-type (Figure 5A). In comparison, uaf30Δ, which reduces RNA Pol I levels on the rDNA (52), did not affect 18S, 25S or ITS1 rRNAs whereas the hmo1Δ reduced the levels of both 18S and 25S rRNAs as previously described (50). Although hmo1Δ mutants are known to be deficient in processing the primary rRNA at defined locations (50), we detected no increase in ITS1-containing rRNA in this mutant suggesting that processing at the A2 site in ITS1 was intact (Figure 5A). We next asked whether the increase in ITS1-containing rRNA was due to H3K56ac by analyzing rRNA levels in histone H3 wild-type and H3K56A mutants. Consistent with the results for asf1Δ, the H3K56A mutant exhibited no difference from wild-type in the levels of 18S and 25S rRNA but did have an ∼4-fold increase in ITS1-containing rRNA (Figure 5B). Overall, these results suggest that Asf1 and H3K56ac may regulate RNA Pol I transcription and the processing of the nascent rRNA to promote rRNA biogenesis.
Figure 5.
The H3K56ac pathway regulates rRNA processing. (A) Defects in Asf1 cause accumulation of non-processed rRNA. Wild-type, asf1Δ, uaf30Δ and hmo1Δ were grown to log phase, total RNA was extracted, cDNA was synthesized and qPCR performed with the indicated primer sets. The average and standard deviation of four independent experiments are shown with significance determined by t-test. **P < 0.05; ****P < 0.005. (B) H3K56ac regulates rRNA processing. The histone H3 wild-type and H3K56A mutant were grown to log phase and processed as described in (a) to measure rRNA levels. The average and standard deviation of at least three or more independent experiments is shown and significance was determined by t-test. **P < 0.05.
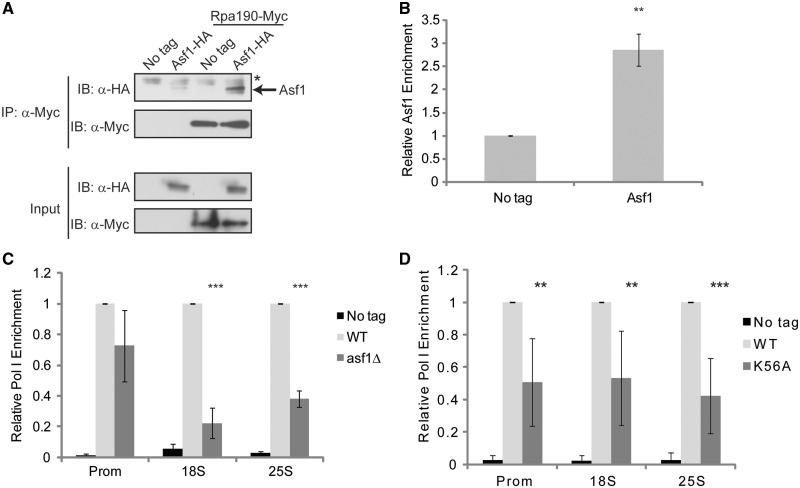
To address if H3K56ac contributes directly to rDNA transcription, we integrated 6XHA and 9XMyc epitope tags at the endogenous ASF1 and RPA190 (catalytic subunit of RNA polymerase I) chromosomal loci and used WCE from these strains to perform coimmunoprecipitation analysis. Immunoprecipitation of Rpa190 readily coprecipitated Asf1 suggesting that Asf1 physically associates with the RNA Pol I complex (Figure 6A). Furthermore, ChIP analysis with α−HA antibodies demonstrated significant Asf1 binding (∼3-fold) to the rDNA promoter suggesting Asf1 is recruited to the rDNA (Figure 6B). We next introduced an asf1Δ into the Rpa190-9XMyc strain and determined what effect this mutation had on RNA Pol I levels at the rDNA. Consistent with the presence of Asf1 and H3K56ac on the rDNA, asf1Δ or the H3K56A mutation led to decreased RNA Pol I engagement throughout the rDNA (Figure 6C and D), which was not due to lower Rpa190 protein levels in these mutants (Supplementary Figure S4a). These results directly implicate the H3K56ac pathway in RNA Pol I transcription and suggest that the increase in nascent, ITS1-containing rRNA is unlikely due to increased rRNA synthesis by RNA Pol I but instead due to decreased cotranscriptional rRNA processing at the A2 cleavage site.
Figure 6.
The H3K56ac pathway regulates rDNA transcription. (A) Asf1 coassociates with RNA Pol I. WCE from the indicated log phase cultures were prepared and α−Myc immunoprecipitations were performed using 750 µg of WCE. Samples were resolved by 10% SDS–PAGE, immunoblotted with α−HA antibody and then membranes were stripped and reprobed with α−Myc antibody. Input samples represent 30 µg of each extract. The asterisk denotes a cross-reactive protein found in all the samples. (B) Asf1 localizes to the rDNA promoter. ChIP experiments using α-HA antibody and either no tag control or Asf1-6XHA expressing cells were performed using the rDNA promoter primer set indicated in Figure 3A. The Asf1-specific signal was expressed as fold increase over the no tag signal which was set to 1. The average and standard deviation of three independent experiments are presented and significance was determined by t-test. **P < 0.05. (C) Asf1 regulates RNA Pol levels on the rDNA. α-Myc ChIP was performed using no tag control, Rpa190-Myc or Rpa190-Myc asf1Δ strains and the level of RNA Pol I determined at the indicated positions on the rDNA. The wild-type Rpa190-Myc signal was set to 1 and the no tag control and asf1Δ signals were expressed relative to this signal. The average and standard deviation of three or more independent experiments are shown with significance determined by t-test. ***P < 0.01. (D) Loss of H3K56ac reduces RNA Pol I recruitment to the rDNA. No tag control, histone H3 wild-type or H3K56A strains expressing Rpa190-9XMyc were used in α−Myc ChIP as described in (C). The histone H3 wild-type strain was set to 1 for each primer set and the no tag control and H3K56A mutant expressed relative to it. The average and standard deviation of four to five independent experiments are shown with significance determined by t-test. **P < 0.05; ***P < 0.01.
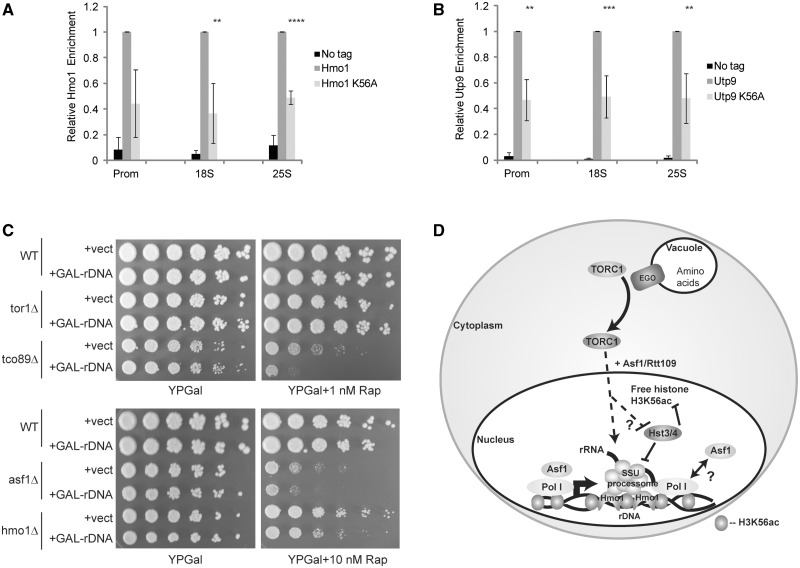
The role of histones in rDNA chromatin structure is somewhat controversial because traditional nucleosomes may not form on the highly transcribed rDNA (53). Instead, nucleosomes may be replaced by the incorporation of the high mobility group protein Hmo1 that forms a specialized rDNA chromatin structure required for high level rDNA transcription and rRNA processing (50,53). However, studies using a yeast strain containing only 42 rDNA copies, all of which are transcriptionally active, clearly detected histones on the rDNA (54), suggesting that histones do contribute to rDNA chromatin. Furthermore, multiple factors involved in chromatin regulation are recruited to transcribed rDNA (54,55) suggesting that modifying chromatin is an important step in rDNA transcription. Since we detected H3K56ac on the rDNA and defects in H3K56ac caused increased levels of non-processed ITS1-containing rRNA but decreased RNA Pol I engagement, we speculated that this histone mark may contribute directly to the formation of the specialized rDNA chromatin structure necessary for RNA Pol I transcription and rRNA processing. To address this issue, we engineered a 6XHA epitope tag at the HMO1 genomic locus in histone H3 wild-type and H3K56A mutant strains, confirmed Hmo1 expression was equivalent between strains (Supplementary Figure S4b) and performed α−HA ChIP. In the H3K56A mutant, we detected a significant reduction of Hmo1 incorporation at the rDNA compared to the wild-type H3 strain (Figure 7A). These data are consistent with the possibility that H3K56ac regulates rDNA chromatin structure. However, the decreased Hmo1 incorporation in H3K56A was not sufficient to result in reduced 18S or 25S rRNA levels as observed in hmo1Δ (Figure 5A and B), nor could it explain the defect in rRNA processing since hmo1Δ mutants did not accumulate ITS1-containing rRNA (Figure 5A). To address these specific points, we next examined whether H3K56ac was necessary for SSU processome rDNA binding since the SSU processome regulates both RNA Pol I transcription and cotranscriptional processing of the nascent rRNA at the A2 cleavage site in ITS1 (56,57). We integrated a 6XHA epitope at the UTP9 (a SSU processome subunit) (56) genomic locus, confirmed the mutation did not alter Utp9 protein levels (Supplementary Figure 4b) and then determined if its recruitment to the rDNA in H3K56A was altered. While we detected strong enrichment of Utp9 throughout the rDNA region in histone H3 wild-type cells, the H3K56A mutant exhibited reduced Utp9 binding along the entire length of the rDNA (Figure 7B), thus demonstrating a crucial role for H3K56ac in SSU processome rDNA binding.
Figure 7.
H3K56ac regulates rDNA chromatin and rDNA transcriptional deregulation sensitizes cells to rapamycin in asf1Δ. (A) Hmo1 rDNA binding is reduced in H3K56A mutants. α-HA ChIP was performed at the rDNA with no tag control, histone H3 wild-type and H3K56A mutants expressing Hmo1-6XHA. The no tag control and H3K56A signals were expressed relative to the histone H3 wild-type signal. The average and standard deviation of three independent experiments are shown with significance determined by t-test. **P < 0.05; ****P < 0.005. (B) H3K56ac is necessary for SSU processome recruitment. Experiment was performed as in (A) except strains used were either a no tag control or expressed the SSU processome subunit Utp9 as a 6XHA fusion in the histone H3 wild-type or H3K56A background. Data are the average and standard deviation of four independent experiments with significance determined by t-test. **P < 0.05; ***P < 0.01. (C) Deregulation of rDNA transcription in tco89Δ, asf1Δ and hmo1Δ mutants sensitizes cells to TORC1 inhibition. Wild-type or the indicated mutants were transformed with control vector or a vector expressing the rDNA from a galactose-inducible promoter. Cultures were grown in SC-Ura media overnight to select for plasmid maintenance and then equal numbers of cells were 5-fold serially diluted and spotted to YPGalactose in the absence or presence of 1 or 10 nM rapamycin. Plates were incubated at 30°C for 3–6 days before photographing. (D) A speculative model for the role of H3K56ac in nutrient regulated TORC1 signaling. TORC1 signaling may regulate histone H3K56ac on a non-chromatin bound pool of histone H3 and/or at a subset of genomic locations in part, but not necessarily exclusively, through suppression of Hst3 or Hst4 activity. At the rDNA specifically, the H3K56ac pathway is required for creating an optimal chromatin environment that allows binding of both Hmo1 and the SSU processome to promote the high level RNA Pol I transcription and rRNA processing necessary for cell growth and proliferation.
These results suggested that the rapamycin sensitivity detected in H3K56ac pathway mutants may be due to a disruption in RNA Pol I transcription and rRNA processing. To determine if increasing rDNA transcription could mitigate some of the rapamycin phenotypes in these mutants, we transformed wild-type, tor1Δ, tco89Δ, asf1Δ and hmo1Δ cells with either a control vector or a vector expressing the rDNA from a galactose-inducible promoter (Gal-rDNA) which is transcribed by RNA Pol II instead of RNA Pol I. In this system, growth of cells on galactose disconnects rDNA transcriptional regulation from its normal upstream signaling pathways (i.e. TOR). We plated cells on YPGalactose plates in the presence or absence of a reduced rapamycin (1 nM for the TORC1 mutants and 10 nM for asf1Δ and hmo1Δ) concentration to allow significant growth of these mutants as they all exhibit enhanced rapamycin sensitivity (Figure 1). Surprisingly, while all samples grew on YPGalactose plates, we found that increasing rDNA transcription in tco89Δ sensitized these cells to exceptionally low rapamycin concentrations (1 nM) compared to vector controls (Figure 7C) suggesting that deregulated rDNA transcription in TORC1 mutants is growth inhibitory. This effect did not occur in tor1Δ which is most likely explained by the presence of the redundant Tor2 kinase. Interestingly, similar experiments with asf1Δ and hmo1Δ also sensitized these mutants to rapamycin, albeit at a 10-fold higher concentration of drug than that used for tco89Δ. Although Hmo1 controls transcription of TORC1-regulated genes, in these experiments the asf1Δ mutant exhibited far greater sensitivity to deregulated rDNA transcription, and to rapamycin in general, than did hmo1Δ. These results suggest increasing expression of rRNA in cells deficient for chromatin pathways contributing to rDNA transcription and rRNA processing can sensitize cells even further to rapamycin-induced TORC1 inhibition. These results implicate the H3K56ac pathway as a regulator of the specialized rDNA chromatin environment necessary for optimal rDNA transcription and rRNA processing in part by controlling both Hmo1 and SSU processome binding. These data also suggest the H3K56ac pathway may function to coordinate TORC1 signaling with correct rRNA expression and processing necessary to promote an optimal cell growth response to nutrient availability.
DISCUSSION
The signaling mechanisms that transmit environmental information to regulate chromatin structure and elicit changes in gene transcription and chromatin states are poorly understood but critical as they may impact gene-environment interactions and epigenetic processes (1). We have utilized the TORC1 pathway to begin identifying mechanisms by which environmental (i.e. nutrient) signals influence chromatin regulation and gene transcription. Our rapamycin screens determined that TORC1-regulated cell growth requires the H3K56ac pathway and its disruption sensitizes cells to rapamycin-induced growth inhibition. Furthermore, genetic disruption of TORC1 (tco89Δ) or nutrient-dependent TORC1 activators (ego3Δ), or pharmacological inhibition of TORC1 in wild-type cells (rapamycin treatment), negatively regulates global H3K56ac thus implicating active TORC1 signaling in the control of this chromatin modification pathway. Tco89 provides an essential function within TORC1 for H3K56ac regulation since neither a hyperactive TORC1 mutant nor a TORC1-independent Sch9 mutant rescued H3K56ac. However, the exact mechanism by which Tco89 regulates H3K56ac remains to be determined. Our results demonstrating that H3K56ac is rescued in tco89Δ when the Sirtuin deacetylases Hst3 or Hst4 are deleted suggest one possible mechanism could be that TORC1 signaling negatively regulates the activity of these enzymes. In the absence of normal TORC1 signaling, as in tco89Δ, these deacetylases may be more active and thus reduce the steady-state levels of H3K56ac (outlined in Figure 7D). This regulation could be direct through TORC1-dependent modification of these enzymes or indirect through TORC1 regulation of metabolism since these deacetylases are regulated by cellular metabolic state (58). Another possibility could be that TORC1 directly phosphorylates H3K56ac regulators to control H3K56ac and Tco89 provides an essential role in substrate recognition. To our knowledge, phosphorylation of yeast Asf1 or Rtt109 has not been demonstrated, although Asf1 in animal cells is phosphorylated by Tousled-like kinases (59) suggesting that H3K56ac may be regulated by upstream kinases in yeast as well. Because Tco89 is a newly identified TORC1 subunit (60), its functions within the complex are largely unknown and so how it functions in TORC1 signaling, and in particular H3K56ac regulation, will need to be determined in future studies. Regardless of the specific mechanisms involved, TORC1 regulated H3K56ac likely impacts many aspects of genome regulation since this histone modification functions in transcription, DNA repair and replication.
Surprisingly, tco89Δ alone failed to decrease H3K56ac on the TORC1-regulated rDNA. This effect is most likely explained by the retention in tco89Δ cells of enough TORC1-signaling activity to maintain rDNA H3K56ac and normal rRNA synthesis. Our results demonstrating that an inhibitory concentration of rapamycin could significantly decrease H3K56ac on the rDNA further supports this idea. The observation of the slight synthetic sick phenotype in the tco89Δ asf1Δ double mutant, also detected by other groups (39,40), further suggests there is likely significant, but not necessarily complete, overlap between TORC1 and H3K56ac in regulating cell growth. This overlap would be consistent with the biochemical data demonstrating a significant reduction, but not complete ablation, of H3K56ac in TORC1 pathway mutants. Our analysis of the housekeeping genes ACT1 and TUB1 suggest that TORC1 disruption may have differential impacts on H3K56ac that are gene specific. Whether H3K56ac is decreased elsewhere in the genome in TORC1 mutants, especially on other TORC1 regulated genes, will need to be determined in future studies. Our results due provide evidence that the H3K56ac pathway contributes directly to rDNA transcription since Asf1 coassociates with RNA Pol I, both Asf1 and H3K56ac localize to the rDNA, and their disruption significantly decreases RNA Pol I levels. These results are in contrast to a previous study that did not detect Asf1 recruitment to the rDNA (61), which may reflect that the rDNA ChIP signal from the epitope-tagged Asf1 IP in this study was normalized to an internal control genomic region but a similar no-tag control was not performed. While we normalized our Asf1 ChIP signal to a control genomic region as well, we also compared Asf1 enrichment to a no tag control strain analyzed identically to confirm specific enrichment. Most importantly, our study provides several lines of independent evidence that the H3K56ac pathway functions directly in RNA Pol I transcription.
Besides affecting RNA Pol I levels on the rDNA, we also provide direct evidence that H3K56ac is critical for rDNA binding by Hmo1 and so it likely participates in forming the specialized rDNA chromatin structure necessary for high level RNA Pol I transcription. The rDNA determinants necessary for mediating Hmo1 incorporation are not defined so our data implicating H3K56ac in this process provides new insight into rDNA transcriptional mechanisms and the chromatin pathways essential for Hmo1 rDNA binding. Exactly how H3K56ac regulates Hmo1 incorporation is still not clear but is the subject of ongoing investigation. One possibility is that histone H3, perhaps within the tetrasome or other sub-nucleosomal particle, blocks access to the DNA and prevents Hmo1 binding. H3K56ac could then facilitate the disruption of histone H3–DNA contacts (22) that would allow greater fluidity for histone repositioning or eviction that permits Hmo1 binding. While we demonstrate H3K56ac is crucial for Hmo1 incorporation and RNA Pol I transcription, we do not detect significant alterations in 18S or 25S rRNA expression in H3K56ac pathway mutants, unlike the decrease in 18S and 25S rRNAs detected in hmo1Δ. These results suggest that the amount of Hmo1 bound in the H3K56A mutant is sufficient to maintain steady-state rRNA levels. This result is not surprising given that we did not detect significant changes in rRNA expression in the uaf30Δ mutant which is known to significantly decrease RNA Pol I rDNA binding (52). Yeast cells can adjust their rates of rRNA synthesis such that cells containing only 42 rDNA repeats express equivalent levels of rRNA as control cells carrying a normal repeat (∼143) number (62). Therefore, the H3K56ac pathway mutants likely maintain normal rRNA expression by either increasing the number of transcriptionally active rDNA repeats or altering the turnover of the rRNAs at a post-transcriptional level.
rRNA cotranscriptional processing by the SSU processome occurs in the ITS1 sequence at the A2 cleavage site and SSU processome defects cause accumulation of non-processed rRNAs (56,57). A sub-complex of SSU processome components, the t-Utps, are also essential for RNA Pol I transcription as depletion of t-Utps reduces the number of RNA Pol I complexes engaged on the rDNA and decreases rRNA synthesis (56). How the SSU processome is recruited to the rDNA has yet to be determined but is known to be independent of RNA Pol I transcription (56). Our data implicate H3K56ac as a critical histone post-translational modification pathway necessary for SSU processome rDNA binding, since a H3K56A mutation results in decreased SSU processome recruitment across the rDNA. These data also explain why H3K56ac pathway mutants have elevated levels of ITS1-containing rRNA since decreased SSU processome binding presumably leads to decreased cotranscriptional rRNA processing within ITS1. However, whether the lower SSU processome association in H3K56A mutants also causes the reduction in rDNA-bound RNA Pol I remains to be determined. A previous study demonstrated that hmo1Δ mutants also accumulate non-processed rRNAs (50) suggesting that the specialized chromatin state existing at the rDNA is essential for correct rRNA processing. Although we only detected increased ITS1-containing rRNAs in H3K56ac pathway mutants and not hmo1Δ, it is important to point out that this previous study examined different regions of the primary rRNA transcript in hmo1Δ cells than we have analyzed.
We also demonstrate that deregulating rDNA transcription in tco89Δ, asf1Δ and hmo1Δ mutants significantly sensitizes cells to growth inhibition by lower rapamycin concentrations. While these results were unexpected they are not unprecedented. Two previous studies demonstrated that uncoupling rDNA transcription from its regulation by nutrient-signaling pathways (i.e. TORC1) causes cells to become significantly rapamycin hypersensitive (63,64). Although the exact mechanisms underlying this effect are unknown, one explanation could be that TORC1 signaling also controls the expression of non-ribosomal factors essential for ribosome biogenesis, such as genes of the Ribi regulon which may also include rRNA processing factors like the SSU processome (65). As such, when pathways essential for SSU processome recruitment are compromised, such as H3K56ac, non-processed rRNAs accumulate. Under these conditions, forcing rDNA transcription using an inducible, RNA Pol II transcribed rDNA may generate high levels of non-processed rRNAs that cause significant ribotoxic stress in the cells, perhaps through the formation of non-functional ribosomal sub-complexes. This explanation could also account for why hmo1Δ were not as sensitized as asf1Δ under these same conditions. Since hmo1Δ did not increase ITS1-containing rRNAs and had significantly lower levels of 18S and 25S rRNAs, the galactose-induced increase in rRNA likely could be compensated by the processing factors already expressed in these cells.
Overall, our studies demonstrate that nutrient signaling through TORC1 regulates global H3K56ac and that this histone modification regulates rDNA transcription and rRNA processing by the SSU processome. To our knowledge this is both the first demonstration that TORC1 signaling regulates the occurrence of a specific histone post-translational modification and that a single site of histone post-translational modification can control rDNA transcription and correct rRNA processing. These results implicate the chromatin environment as a regulator of the post-transcriptional rRNA processing necessary for TORC1-regulated cell growth.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
National Institutes of Health (NIH) [CA133322 to L.M.P., CA140346 to M.F., and R21CA155864 to R.N.L.]; and the American Heart Association [National Scientist Development grant 11SDG5260017 to R.N.L]. Research support is also provided by the Muirhead Chair Endowment of the UTHSC to L.M.P. Funding for open access charge: NIH [R21CA155864].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Robbie Loewith, Masayasu Nomura and Ted Powers for kindly providing the Sch9, galactose-inducible rDNA and Tor1 expression plasmids used in this study. The authors are grateful for the critical reading and helpful suggestions made by Mark Kaplan, Brian Strahl, Jason Workman and Zhaohui Wu.
REFERENCES
- 1.Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 6:791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23:1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dazert E, Hall MN. mTOR signaling in disease. Curr. Opin. Cell Biol. 23:744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 5.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 7.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 9.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 13.Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl Acad. Sci. USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- 16.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey EL, Shamji AF, Bernstein BE, Schreiber SL. Rpd3p relocation mediates a transcriptional response to rapamycin in yeast. Chem. Biol. 2004;11:295–299. doi: 10.1016/j.chembiol.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Rohde JR, Cardenas ME. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl Acad. Sci. USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 23.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl Acad. Sci. USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 26.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 28.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 30.Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 2006;5:2602–2608. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- 31.Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 32.Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 35.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl Acad. Sci. USA. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- 38.Aslam A, Logie C. Histone H3 serine 57 and lysine 56 interplay in transcription elongation and recovery from S-phase stress. PLoS One. 2010;5:e10851. doi: 10.1371/journal.pone.0010851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 40.Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helliwell SB, Howald I, Barbet N, Hall MN. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minard LV, Lin LJ, Schultz MC. SWI/SNF and Asf1 independently promote derepression of the DNA damage response genes under conditions of replication stress. PLoS One. 2011;6:e21633. doi: 10.1371/journal.pone.0021633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 2006;281:31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- 45.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 47.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 48.Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, Aragon L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–222. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 50.Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gadal O, Labarre S, Boschiero C, Thuriaux P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hontz RD, French SL, Oakes ML, Tongaonkar P, Nomura M, Beyer AL, Smith JS. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol. Cell Biol. 2008;28:6709–6719. doi: 10.1128/MCB.00703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hontz RD, Niederer RO, Johnson JM, Smith JS. Genetic identification of factors that modulate ribosomal DNA transcription in Saccharomyces cerevisiae. Genetics. 2009;182:105–119. doi: 10.1534/genetics.108.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 58.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sillje HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 2001;11:1068–1073. doi: 10.1016/s0960-9822(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 60.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 61.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 62.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chedin S, Laferte A, Hoang T, Lafontaine DL, Riva M. and Carles, C. (2007) Is ribosome synthesis controlled by pol I transcription? Cell Cycle. 6:11–15. doi: 10.4161/cc.6.1.3649. [DOI] [PubMed] [Google Scholar]
- 65.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.