Abstract
RbpA is an RNA polymerase (RNAP)-binding protein whose presence increases the tolerance levels of Mycobacteria to the first-line anti-tuberculosis drug rifampicin by an unknown mechanism. Here, we show that the role of Mycobacterium tuberculosis RbpA in resistance is indirect because it does not affect the sensitivity of RNAP to rifampicin while it stimulates transcription controlled by the housekeeping σA-factor. The transcription regulated by the stress-related σF was not affected by RbpA. The binding site of RbpA maps to the RNAP β subunit Sandwich-Barrel Hybrid Motif, which has not previously been described as an activator target and does not overlap the rifampicin binding site. Our data suggest that RbpA modifies the structure of the core RNAP, increases its affinity for σA and facilitates the assembly of the transcriptionally competent promoter complexes. We propose that RbpA is an essential partner which advantages σA competitiveness for core RNAP binding with respect to the alternative σ factors. The RbpA-driven stimulation of the housekeeping gene expression may help Mycobacteria to tolerate high rifampicin levels and to adapt to the stress conditions during infection.
INTRODUCTION
Tuberculosis appeared ∼40 000 years ago, has infected almost one-third of the world population and continues to kill approximately 1.5 million people each year (1,2). Rifampicin, an antibiotic targeting the bacterial RNA polymerase (RNAP), remains the first-line drug used to cure tuberculosis infections. However, the bacteria have developed multiple mechanisms to escape from the effects of antibiotic treatment (3). Most of these mechanisms, such as activation of stress-response pathways and switching to the persistent (dormant) state are linked to the regulation of transcription.
The bacterial RNAP holoenzyme, the central enzyme of transcription, is a complex molecular machine composed of the catalytic core (5 subunits α2ββ'ω) and a promoter-specific σ factor directing promoter recognition. The σ subunit confers to the holoenzyme an ability to recognize the −10 and −35 promoter elements, to melt promoter DNA at the transcription start site and to initiate RNA synthesis. Transcription initiation starts with the reversible promoter binding, leading to the formation of the ‘closed complex’ (RPc). The closed complex isomerizes into the transcriptionally competent ‘open complex’ (RPo), in which the ∼13 bp of promoter DNA around the transcription start site are melted to form a transcription bubble (4,5). As a result of local DNA melting, the antisense DNA strand enters to the RNAP active site and serves as a template for the initiation of RNA synthesis. The isomerization from the RPc to RPo involves several intermediate complexes (RPi) and is modulated by numerous transcriptional activators, repressors and small regulatory molecules (5–7). Transcription of the essential genes during exponential growth is driven by RNAP containing the housekeeping (principal) σ factor (σ70 in Escherichia coli or σA in Mycobacterium tuberculosis). Alternative σ factors activate the transcription of the specialized genes implicated in the stress response, virulence and the switch from exponential to stationary growth phase or to a persistent state (8,9). In E. coli only one σ factor, σS, controls stationary phase gene expression and adaptation to stress. In M. tuberculosis, which must adapt to various stress conditions in the host, this process is more complex and involves several σ factors, including σB and σF (10,11). Competition between σ factors for a limited amount of core RNAP provides the basal regulatory mechanisms for the fine-tuning of bacterial gene expression in response to environmental signals including antibiotic treatment (9,12). Several core RNAP-binding proteins that do not bind DNA (e.g. DksA, CarD) provide additional level of regulation for RNAP activity in growth phase dependent fashion (13,14). The interplay between σ factors and RNAP-binding proteins and the role of these proteins in modulation of bacterial cell sensitivity to the antibiotics is poorly understood.
RbpA is a 14 kDa RNAP-binding dimeric protein, specific to Actinomycetes, that was first identified in Streptomyces coelicolor (15,16). The S. coelicolor RbpA stimulates transcription from ribosomal promoters regulated by the housekeeping σHrdB factor (16). Expression of the rbpA gene in S. coelicolor is induced during the disulfide stress and rifampicin treatment, while the ΔrbpA mutant exhibits ∼15-fold increased sensitivity to rifampicin and displayed growth defects (16). In M. tuberculosis, the rbpA gene is upregulated ∼8-fold during stationary phase (17). The M. smegmatis RbpA protein was suggested to play a role in basal resistance to rifampicin by increasing tolerance to the antibiotic (18,19). The following three alternative mechanisms of RbpA-mediated rifampicin resistance can be deduced from the published studies: (i) ‘direct’ competition between RbpA and rifampicin for the binding site, (ii) an ‘allosteric’ change in the rifampicin-binding site and consequent decrease in the affinity of rifampicin for RNAP, (iii) ‘indirect’ mechanism through the stimulation of genes implicated in the control of cell proliferation or membrane permeability. A recent cross-linking study mapped the binding site of M. smegmatis RbpA close to the rifampicin-binding cluster I of the β subunit and suggested that RbpA induces the dissociation of the antibiotic from RNAP, indicating that the effect is obtained through mechanism 1 or 2 (19).
The M. tuberculosis RbpA has not yet been studied, and its role in transcription regulation and rifampicin resistance remains obscure. In the current study, we explored the mechanism of action of M. tuberculosis RbpA in vitro using reconstituted M. tuberculosis RNAP and two σ factors, the principal σA and the alternative σF, which is involved in immunopathogenesis, host–pathogen interactions and virulence (20,21). Our results support the ‘indirect’ role of RbpA in rifampicin resistance and suggest that RbpA functions as a σ-specific transcriptional activator that regulates the access of the principal σA factor to the core RNAP during the stress response and stationary phase. The binding site of RbpA maps to the region of the RNAP β subunit distant from the previously characterized regulatory sites, revealing a new regulatory mechanism for transcription initiation.
MATERIALS AND METHODS
Proteins and DNA templates
Plasmids pSR52, pJF09, pJF10, pSR01 and pSR05 were used to express the α, β, β′, σA and σF subunits, respectively, and were a generous gift from Dr Sébastien Rodrique (22). The ω subunit and rbpA gene (Rv2050) were amplified from M. tuberculosis H37Rv genomic DNA and cloned into pET21a. The N-terminal Flag and C-terminal HA tags were introduced to the β subunit by PCR and cloned into pET21a. Proteins were all expressed in the E. coli BL21(DE3)pLysS strain and purified by Ni2+-agarose affinity chromatography from either a soluble fraction (α, RbpA, σA) or inclusion bodies (β, β′, σF and ω) as previously described (23). The RNAP core enzyme was reconstituted from the α, β, β′ and ω subunits as previously described (23). Reconstituted RNAP was precipitated with 60% ammonium sulfate, purified by Ni2+-agarose affinity chromatography and concentrated using Ultracel-100 membrane filter unit (Millipore). The E. coli RNAP core enzyme was purchased from Epicentre. The rrnAP3, sigAP and usfXP1 promoter DNA fragments were amplified from M. tuberculosis H37Rv genomic DNA by PCR. The sinP3 promoter was amplified from Bacillus subtilis genomic DNA. All primers used in this study are listed in Supplementary Table S1.
In vitro transcription and EMSA
Transcription was performed in 5 μl transcription buffer (TB, 40 mM HEPES pH 7.9, 50 mM NaCl, 5 mM MgCl2 and 5% glycerol). The RNAP holoenzyme was assembled by mixing 900 nM σ subunit and 300 nM core RNAP in the presence of 5% DMSO and then incubating for 5 min at 37°C. The mixture was then incubated with a promoter fragment (15 nM) at 37°C for 10 min. Transcription was initiated by the addition of 50 μM ATP, GTP and CTP (100 µM GTP was used in the case of rrnAP3) and 3 μCi [α-32P] UTP. RbpA at 1.2 μM or at the concentrations indicated in the figure legends was added to the assembled RNAP holoenzyme and incubated for 5 min before promoter binding. Heparin (10 μg/ml) was added together with NTPs to start the reactions in the single-round transcription assay. For the abortive-initiation assay, a mixture of three NTPs (omitting ATP for rrnAP3 and omitting GTP for usfXP1) was added. The reactions were stopped by adding 1 volume of the stop buffer (2× TBE, 8 M urea). RNA was analyzed on a 24% PAGE-7 M urea denaturing gel. For the EMSA experiments, the promoter fragments end-labeled by fluorescein were mixed with RNAP in TB as described above and were incubated with 10 μg/ml of poly (dA-dT) or 10 μg/ml heparin for 5 min at 37°C and loaded on a 5% native 0.5× TBE–PAGE. Different concentrations of RbpA (as indicated in the figure legends) were pre-incubated with RNAP for 5 min at 37°C before RNAP–promoter complex formation. Gels were scanned with Typhoon 9400 Imager (GE Healthcare) and quantified using ImageQuant software (Molecular Dynamics).
Quantification of the σ-core titration experiments
To determine the maximum level of transcripts synthesized after 5 min of transcription at saturating concentration of the σ subunits (Amax) each data set was fitted to exponential function: 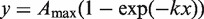 . Afterwards, the RNA amounts were normalized to the Amax value obtained in the presence of RbpA. An average values obtained in two experiments were plotted against the σ concentrations. To determine the concentration of σ subunit at the half-maximal transcriptional activity, K0.5, the data points were fitted to the Hill equation:
. Afterwards, the RNA amounts were normalized to the Amax value obtained in the presence of RbpA. An average values obtained in two experiments were plotted against the σ concentrations. To determine the concentration of σ subunit at the half-maximal transcriptional activity, K0.5, the data points were fitted to the Hill equation:
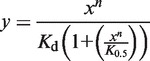 |
where y is the fraction of abortive transcripts and x is the concentration of σ subunit.
Pull-down and western blotting
σA at 2 μM or σF at 4 μM was incubated with 0.6 μM core enzyme (containing the FLAG-tagged β subunit) in the presence or absence of 5 μM RbpA in TB at 37°C for 15 min. Then, 50 μl of the reaction mixture was added to 20 μl anti-Flag M2 Affinity Gel (Sigma-Aldrich) in 500 μl TBS [50 mM Tris (pH 7.5) and 150 mM NaCl] and rotated for 1 h at room temperature. The matrix was then washed three times with TBS. The protein was eluted with 0.1 M glycine–HCl (pH 3.5) as described in the manufacturer’s protocol. For western blotting analysis, the input and elution samples were loaded on a 12% SDS–PAGE gel, run and transferred to a PVDF membrane. Monoclonal anti-FLAG (Sigma-Aldrich, Lot#: F1804), anti-polyhistidine (Sigma-Aldrich, Lot#: H1029) and anti-6xHis(C-term)-HRP (Invitrogen, #R931-25) antibodies were used to detect β, σ and RbpA proteins.
FeBABE conjugation and RNAP cleavage
FeBABE conjugation was performed according to the PIERCE protocol (see ‘Supplementary Data’ for details). The RNAP core (2 µM) or holoenzyme was mixed with a 2-fold molar excess of FeBABE–RbpA and incubated at 30°C for 30 min. Cleavage reactions were initiated by the addition of ascorbic acid, EDTA and H2O2 (5 mM final concentration) and allowed to proceed for 30 s at 30°C. Control reactions using non-conjugated RbpA were treated identically. Reaction mixtures were quenched by the addition of 1 volume of Laemmli loading buffer, separated on 10% SDS–PAGE, blotted and visualized by immunostaining with monoclonal anti-Flag or anti-HA (Sigma-Aldrich, Lot #: H3363) antibodies. Cleavage at cysteine residues was performed with 2-nitro-5-thiocyanobenzoic acid (NTCBA) as previously described (24).
RESULTS
RbpA stimulates σA-dependent transcription initiation
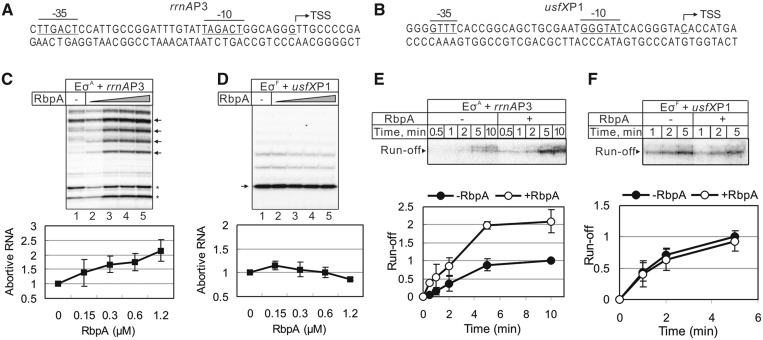
To explore the role of RbpA in transcription, we purified recombinant M. tuberculosis RbpA and reconstituted the M. tuberculosis RNAP core enzyme from the individually expressed subunits: α, β, β′ and ω (Supplementary Figure S1A). The reconstituted M. tuberculosis core RNAP displayed activity similar to that of the commercially available E. coli core RNAP (Supplementary Figure S1B and S1C). Two M. tuberculosis σ factors, the housekeeping σA and the stress response σF, were over-expressed in E. coli, purified and used to initiate transcription from the σA-dependent ribosomal promoter rrnAP3 and the σF-dependent usfXP1 promoter (25,26) (Figure 1A and B and Supplementary Figure S1D and S1E). First, we tested whether RbpA affects the σA-dependent abortive transcription on the rrnAP3 promoter initiated by the addition of [32P]-UTP, CTP and GTP, allowing the synthesis of abortive RNAs up to 9 nt. Transcription was performed in the presence of increasing concentrations of RbpA (Figure 1C, lanes 2–5). The short RNA products (indicated by asterisks) observed even during initiation performed by the core RNAP alone appeared to be due to the initiation at the DNA fragment ends (Supplementary Figure S1D). The addition of RbpA changed the pattern of transcripts and enhanced the overall amount of abortive RNAs ∼2-fold. Approximately 50% level of activation was reached at 300 nM of RbpA (1:1 molar ratio to core RNAP). To determine whether RbpA also stimulates the formation of a productive elongation complex, a single-round run-off transcription in the presence of all four NTPs and the competitor heparin was performed on the rrnAP3 promoter (Figure 1E). The quantification of the experiment showed that the rate of synthesis of the 50 nt run-off RNA product was ∼2-fold higher in the presence of RbpA (apparent rate constants: 0.005 s−1 versus 0.0024 s−1) (Figure 1E), suggesting that RbpA either stimulates promoter complex formation or increases the efficiency of promoter escape (or both). The σF-dependent transcription initiation from the usfXP1 promoter was not affected by RbpA either in the abortive initiation assay in presence of [32P]-UTP, ATP and CTP or in the single-round run-off transcription (Figure 1D and F), which suggests that the activation effect of RbpA is specific to σA or to the promoter.
Figure 1.
Effects of RbpA on σA and σF dependent transcription. (A and B): Sequences of the rrnAP3 and usfXP1 promoters. The −10 and −35 consensus elements are underlined, and transcription start sites (TSS) are marked by arrows. (C and D) Abortive transcription carried out by the σA-containing RNAP (EσA) on the rrnAP3 template and σF-containing RNAP (EσF) on the usfXP1 template. Lanes 2–5: RbpA was added to 0.15, 0.3, 0.6 and 1.2 µM, respectively. Quantification of the bands indicated by arrows is shown on the bottom graphs. The RNA amounts were normalized to the value obtained without RbpA (lane 1). The error bars are the SD of triplicate experiments. (E and F) Kinetics of single-round run-off transcription carried out by σA-containing RNAP on the rrnAP3 and by σF-containing RNAP on the usfXP1 template, respectively. RbpA (1.2 μM) was added to the reaction when indicated. The quantification of the run-off RNA is shown on the bottom graphs. The RNA amounts were normalized to the amount of RNA synthesized without RbpA after 10 min of transcription (rrnAP3) or 5 min of transcription (usfXP1). The error bars are the SD of duplicate experiments.
Transcription initiation from the rRNA promoters is a special case due to the very short half-life of the promoter complexes (27). To determine whether or not the RbpA activity is specific to the rRNA promoter, we performed a single-round run-off transcription assay using the following two other σA-dependent promoters: the M. tuberculosis housekeeping sigAP promoter (28), and the B. subtilis sinP3 promoter, which is known to be transcribed by Mycobacterium RNAP (22). Examination of the sinP3 promoter sequence reveals that it belongs to the ‘extended-10’ class of promoters lacking the −35 consensus element which is compensated by the ‘TG motif’ 1-bp upstream of the −10 element. Similar to the rrnAP3 promoter, transcription on the sigAP and sinP3 promoter templates displayed a 2- to 3-fold increase in the amount of run-off products in the presence of RbpA (Supplementary Figure S2A and S2B). The above result suggests that the activation effect of RbpA is specific to the σA factor but not to the promoter type. However, we noticed that during transcription performed in the presence of RbpA at the sinP3 promoter, the amount of abortive products decreased while for the sigAP promoter, they increased. Therefore, the activation effect of RbpA can be modulated by the promoter DNA sequence. The observed activation effect is consistent with RbpA action on either the open complex formation or on the initiation of RNA synthesis. The function of RbpA is restricted to transcription initiation because no influence of RbpA on the rate of transcription elongation on the promoter-less scaffold DNA template was observed (Supplementary Figure S3).
RbpA stimulates the formation of the stable promoter complexes
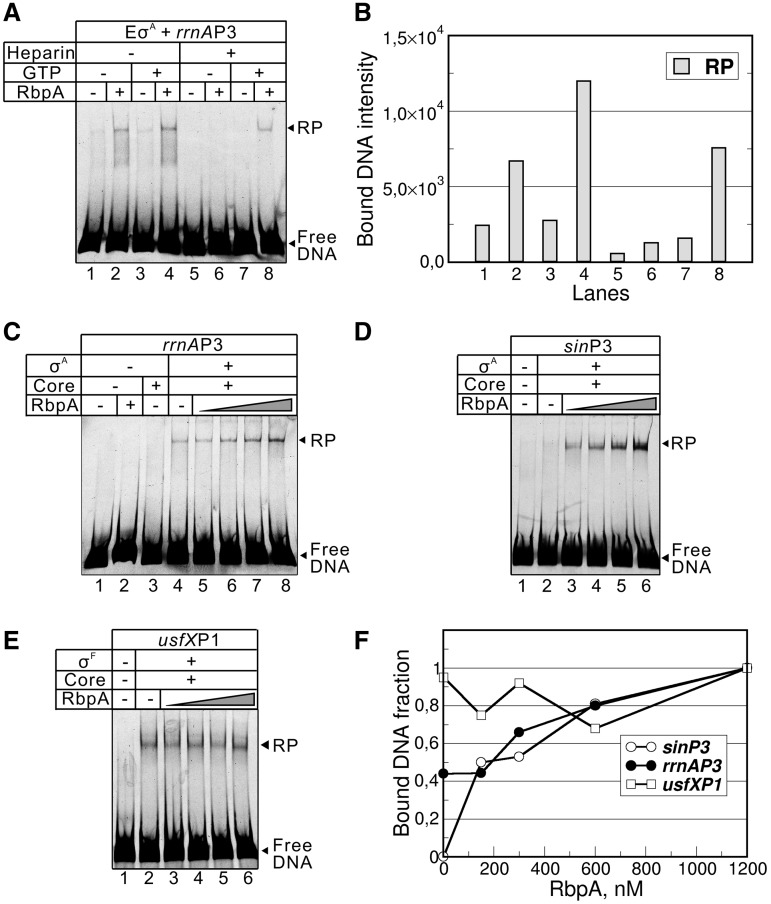
We next explored whether RbpA influences the formation of stable open promoter complexes in the presence of the competitor heparin. Complexes between the σA-containing RNAP holoenzyme and the end-labeled rrnAP3 promoter DNA fragment in the presence or absence of RbpA were monitored with an electrophoretic mobility shift assay (EMSA). Without RbpA and when no competitor was present in the reaction, only a small amount of the complex was formed (Figure 2A and B, lane 1). The addition of RbpA enhances promoter complex formation; however, overall binding remains weak (Figure 2A and B, lanes 1 and 2). The observed weak binding may reflect the low affinity of RNAP for the promoter, the reduced stability of the RNAP–promoter complex or both. Indeed, the addition of heparin, which chases unstable closed complexes, abolishes binding independent of the presence of RbpA (Figure 2A and B, lanes 5 and 6). This observation is consistent with the known reduced stability of open complexes formed at the ribosomal promoters that can be stabilized by a high concentration of priming NTP (27). Indeed, the addition of 1 mM GTP, a nucleotide that is complementary to the +1 position of the rrnAP3 promoter template, enhances complex formation (Figure 2A and B, lane 4). Importantly, the formation of the heparin-resistant complex was observed only if RbpA and GTP were present together (Figure 2A and B lane 8). These experiments suggest that RbpA acts synergistically with GTP in increasing the population of the heparin-resistant complex at the ribosomal promoter. We noticed that if poly(dA–dT) was used as a competitor, stable complex formation can be observed even without RbpA (Figure 2C, lane 4). This difference between heparin and poly(dA–dT) is likely due to different mechanisms of competition. Heparin is known to be an ‘active’ competitor that destabilizes the σ-core RNAP interactions, while poly(dA–dT) is a concurrent competitor that chases free RNAP without actively displacing it from the promoter (29). Neither the core RNAP nor RbpA can form promoter complexes in the conditions used (Figure 2C, lanes 2 and 3). Titration of the RNAP–rrnAP3 complex with increasing amounts of RbpA in the presence of 1 mM GTP and poly(dA–dT) (Figure 2C and F) showed that the amount of the complex increases gradually with increasing RbpA concentration. This result correlates with the effect of RbpA on abortive RNA synthesis (Figure 1) and suggests that the fraction of transcriptionally active promoter complexes is proportional to RbpA binding to the core RNAP.
Figure 2.
RbpA stimulates the formation of the stable RNAP–promoter complexes. (A) EMSA of the σA-containing RNAP (EσA) and the rrnAP3 promoter fragment. RbpA at 1.2 μM, GTP at 1 mM and heparin at 10 µg/ml were added when indicated. The promoter fragments were end-labeled by fluorescein. (B) Quantification of the experiment shown in (A). The bar graph shows the fluorescence intensity of promoter DNA bound to RNAP. (C) EMSA of the complexes of the σA-containing RNAP and the rrnAP3 promoter fragments formed in the absence or in the presence of 0.15, 0.3, 0.6 or 1.2 µM RbpA. (D) EMSA of the complexes of the σA-containing RNAP and the sinP3 promoter fragment formed in the absence or in presence of 75, 150, 300 or 600 nM RbpA. (E) EMSA of the σF-containing RNAP and the usfXP1 promoter fragment in the absence or in the presence of 0.15, 0.3, 0.6 or 1.2 µM of RbpA. The promoter complexes shown in (C–E) were challenged with poly(dA–dT). The RNAP–promoter complex is indicated as ‘RP’, and non-bound DNA is indicated as ‘Free DNA’. (F) Quantification of the experiments shown in (C–E).
The above experiments showed that the σA RNAP holoenzyme has a low affinity for the ribosomal promoter in the absence of RbpA. To verify that RbpA-mediated stimulation of the promoter complex formation is not limited to the ribosomal promoter, we performed EMSA using the end-labeled sinP3 promoter DNA fragment. Complexes of RNAP and promoter DNA were formed in the absence or presence of increasing concentrations of RbpA, chased by poly(dA–dT) and analyzed on a gel (Figure 2D). In contrast to the rrnAP3 promoter, no detectable amount of the RNAP–sinP3 promoter complexes was observed without RbpA (Figure 2D and lane 2). Strong enhancement of RNAP–promoter complex formation was detected when RbpA was added to the reaction (Figure 2D, lanes 3–6), suggesting that RbpA acts on both types of promoters. The lack of detectable promoter complexes on sinP3 in the absence of RbpA is in striking contrast with the relatively high transcriptional activity detected at the same conditions on this promoter (Supplementary Figure S2A). Indeed, the total amount of abortive RNA was equivalent or slightly higher in the absence of RbpA compared to the amounts found in the presence of RbpA. Therefore, the σA-containing RNAP holoenzyme is formed and binds to promoter even without RbpA but it is highly unstable in the non-equilibrium conditions of the EMSA experiment. Also, NTPs present in the transcription experiment may contribute to the stabilization of the promoter complex (27).
To prove that the effect of RbpA is specific to σA-dependent promoters, we performed EMSA using a σF-dependent usfXP promoter DNA fragment. In agreement with the transcription results, the binding of the σF-containing RNAP to the usfXP promoter was not influenced by RbpA (Figure 2D). To test whether the activation effect of RbpA is specific to the M. tuberculosis core RNAP, we used a hybrid RNAP assembled from the E. coli core and M. tuberculosis σA. The hybrid enzyme efficiently formed stable promoter complexes at the rrnAP3 promoter and was fully active in transcription. No effect of RbpA on the activity of the hybrid enzyme was observed (Supplementary Figure S4A and S4B). Therefore, the RbpA activity is specific for the M. tuberculosis core RNAP. Taken together, our results showed that the σA-containing RNAP forms highly unstable promoter complexes on the −10/−35 consensus and the perfect extended −10 consensus promoters. We propose that RbpA modifies RNAP and shifts the equilibrium towards the formation of the stable open promoter complex.
RbpA stabilizes σA- RNAP holoenzyme
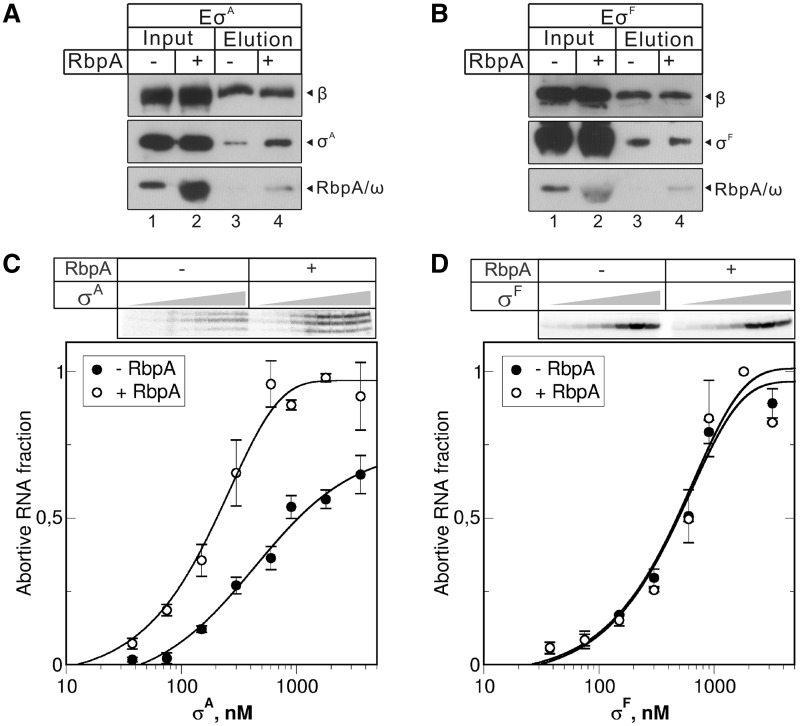
An increase in the amount of the active promoter complex in the presence of RbpA may be a result of the stabilization or reorganization of the σA-core interactions in holoenzyme. To determine whether RbpA influences the holoenzyme formation, we performed pull-down experiments using core RNAP with a FLAG-tag at the N-terminus of the β subunit (Figure 3). The σA or σF subunits were incubated with the FLAG-tagged core enzyme either in the presence or absence of RbpA and then captured by anti-FLAG-conjugated agarose. The total proteins in the reaction sample and agarose-bound proteins were analyzed by western blotting using either anti-FLAG antibodies to detect β subunit (Figure 3, panel β) or anti-6xHIS tag antibodies to detect RbpA, σA, σF and the ω subunits (Figure 3, panels σA and RbpA/ω). The signals for the β subunit with and without RbpA were similar, indicating that the same amounts of core enzyme were captured by the anti-FLAG agarose. Retention of RbpA was visible with both the σA- and σF-containing RNAPs, suggesting that the type of σ factor does not influence RbpA binding to the core RNAP. The retention of σA was higher in the presence of RbpA compared to in its absence (Figure 3A). A control experiment performed with σF showed no influence of RbpA on σF binding (Figure 3B), which confirmed that the function of RbpA is specific to σA.
Figure 3.
RbpA stabilizes σA-core RNAP interactions. (A and B) FLAG-tag pull-down experiments were performed using the RNAP core enzyme containing a FLAG-tagged β subunit. RNAP was incubated with the 6×His-tagged σA (A) or σF (B) in the presence or absence of 6×His-tagged RbpA and captured by anti-FLAG agarose. After washing, the proteins retained by the resin (lanes 3 and 4) and the total proteins in the input (lanes 1,2) were analyzed by western blotting using anti-FLAG (β panel), anti-polyhistidine (σ panel) and anti-C-term 6×His (RbpA/ω panel) antibodies. (C and D) Abortive transcription initiated by core RNAP (300 nM) and increasing concentrations (37.5, 75, 150, 300, 600, 900, 1800 and 3600 nM) of σA (C) and σF (D) at the rrnAP3 and usfXP1 promoters, respectively. RbpA was added to 2.4 µM when indicated. The [32P]-labeled abortive RNA products are shown in the top panels. The graphs show normalized amounts of abortive RNA products synthesized at the indicated concentrations of the σ subunit. Mean values and SD of two independent experiments are shown.
To estimate the relative core-binding activities of σA and σF in the presence and absence of RbpA, we performed titration experiments using transcription assay similar to the one used for analysis of the E.coli σ subunits affinities to core RNAP (30,31). Fixed amount of core RNAP (300 nM) and increasing amounts of σ subunits were used to initiate abortive transcription from the rrnAP3 and usfXP1 promoters (Figure 3C and D). The amounts of abortive transcripts synthesized after 5 min of transcription were normalized to the maximal amount obtained in the presence of RbpA at saturating concentration of σ subunit. The quantification of the titration experiments shows that the half-maximal level of transcription was reached at ∼627 nM of σA (2:1 σA/core ratio) in the absence of RbpA and at ∼208 nM (0.7:1 σA/core ratio) in the presence of RbpA. The σF concentrations required for half-maximal transcriptional activity were similar in the presence or absence of RbpA (∼606 and ∼568 nM correspondingly). Thus, we concluded that the relative affinities of σA and σF to core are quite similar while RbpA increases the σA affinity ∼3-fold but does not affect the σF affinity. Taken together, these results suggest that RbpA stabilizes the σA-core interaction with the holoenzyme and thereby increases the transcriptional activity of RNAP.
RbpA binds to the β subunit SBH motif outside the RNAP main channel
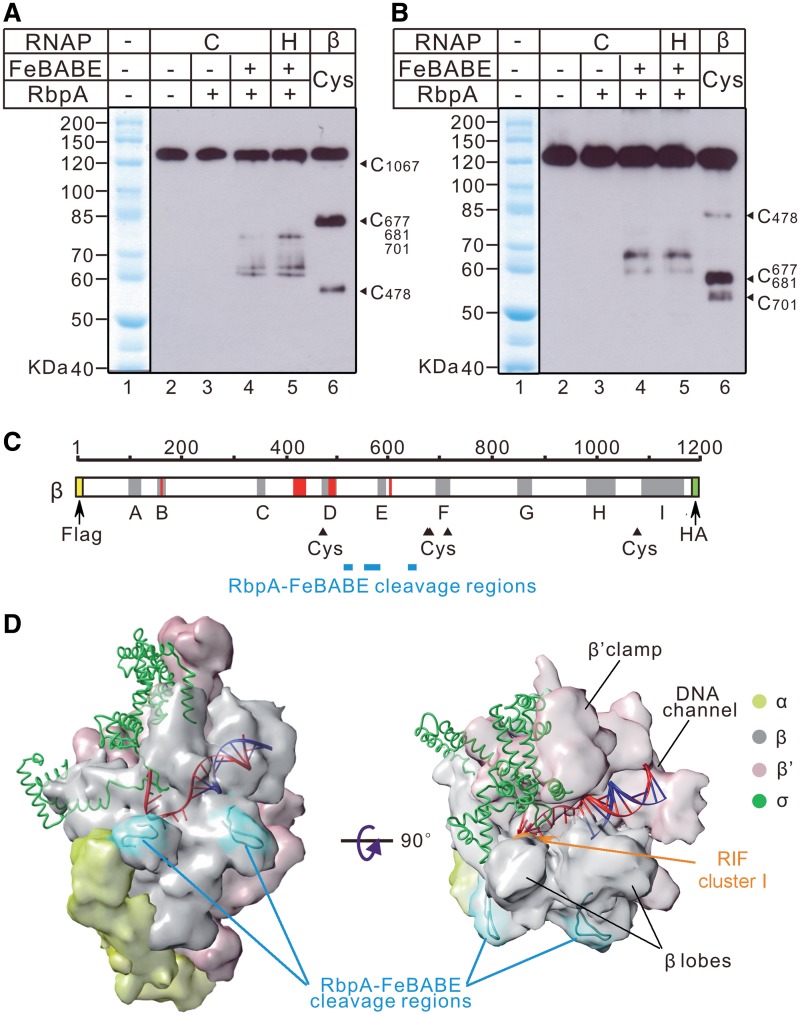
The β subunit of RNAP was identified as a principal binding site for RbpA (18). To map the binding region of RbpA on the β subunit of the RNAP core and holoenzyme, we used the chemical protease Fe(III) (s)-1-(p-bromoacetamidobenzyl) ethylenediamine tetra-acetic acid (FeBABE) covalently tethered to the single Cys56 residue of RbpA. The activity of the FeBABE-tethered RbpA tested in the transcription assay and by EMSA was indistinguishable from the activity of the unmodified protein (Supplementary Figure S5). The FeBABE–RbpA was incubated with either core or holoenzyme, and the cleavage products were resolved on SDS–PAGE and analyzed by western blotting. Staining of the β subunit N-terminus with the FLAG-tag antibodies or the C-terminus with anti-HA antibodies revealed three bands in the cleavage reactions performed with either core or holoenzyme (Figure 4A and B, lanes 4 and 5). In the control lanes with or without unlabeled RbpA, no cleavage was observed (Figure 4A and B, lanes 2 and 3). The FeBABE–RbpA cleavage was specific to the β subunit because no cleavage was detected in the σA subunit, either alone or in the holoenzyme (Supplementary Figure S6). The size of the cleavage products was calculated based on the size of the protein ladder generated with 2-nitro-5-thiocyanobenzoic acid (NTCBA) cleavage at Cys residues of the β subunit (Figure 4, lanes 1 and 6). Based on the calculated size of the cleavage products, we attributed the positions of the cleavage sites to the residues 523 ± 10, 578 ± 26 and 659 ± 8 of the M. tuberculosis β subunit (Figure 4C). Because no difference in cleavage between the RNAP core and holoenzyme was observed, we suggest that the σ subunit is not required for and does not affect RbpA binding to RNAP, in agreement with the idea that only the β subunit forms the RbpA-binding site (18). Using the structure of the Thermus thermophilus RNAP holoenzyme (32,33), we modeled the three cleavage sites that delineate the RbpA-binding region on the surface of RNAP. Strikingly, the FeBABE cleavage sites were clustered at two symmetric protrusions of the β subunit located on the outer side of RNAP (Figure 4D). The revealed configuration of the binding region is consistent with a dimeric structure of RbpA that should carry two FeBABE reactive groups. Because the RbpA-binding region is located outside of the RNAP main channel where the σ subunit is docked, we suggest that RbpA action upon σ binding is allosteric.
Figure 4.
Cleavage of the β subunit of the RNAP core and holoenzyme by FeBABE–RbpA. Western blot analysis of the β subunit fragments stained either at the N-terminus by the anti-FLAG antibodies (A) or at the C-terminus by the anti-HA antibodies (B). FeBABE–RbpA cleavage reactions were performed either with core RNAP (marked ‘C’, lanes 2–4) or holoenzyme (marked ‘H’, lane 5). Control cleavage reactions were performed either without FeBABE and RbpA (lane 2) or with unmodified RbpA (lane 3). The protein ladder was generated by the cleavage of the β subunit at Cys residues using Cys-specific cleavage reagent, TNCBA (lane 6). Numbering along the right edge of the gels shows the positions of the Cys-specific cleavages on the β subunit. A commercial molecular weight marker is shown in lane 1 of each gel. (C) A diagram showing the β subunit with the FLAG-tag shown as an N-terminal yellow box and the HA-tag as a C-terminal green box. The gray shaded areas correspond to the evolutionarily conserved regions A–I. The red shaded areas mark the clusters of rifampicin resistance (3,49). NTCBA-cleaved Cys residues are indicated by black triangles. The positions of the FeBABE–RbpA cleavage sites calculated from the experiments presented in (A and B) are shown as cyan rectangles. (D) A model of the FeBABE–RbpA cleavage sites on the structure of T. thermophilus RNAP holoenzyme in complex with DNA (32,33). DNA is shown as a ladder model with the template strand in red and the non-template strand in blue. The β (gray), β′ (pink), α (yellow) subunits are shown as molecular surface models. The σ subunit (green) is shown as a ribbon model. The RbpA-binding regions are colored in cyan. The surface of the rifampicin-resistance cluster I (RIF cluster I), spanning the M. tuberculosis β residues 425–454, is colored in orange. Molecular graphics images were produced using the UCSF Chimera package (50).
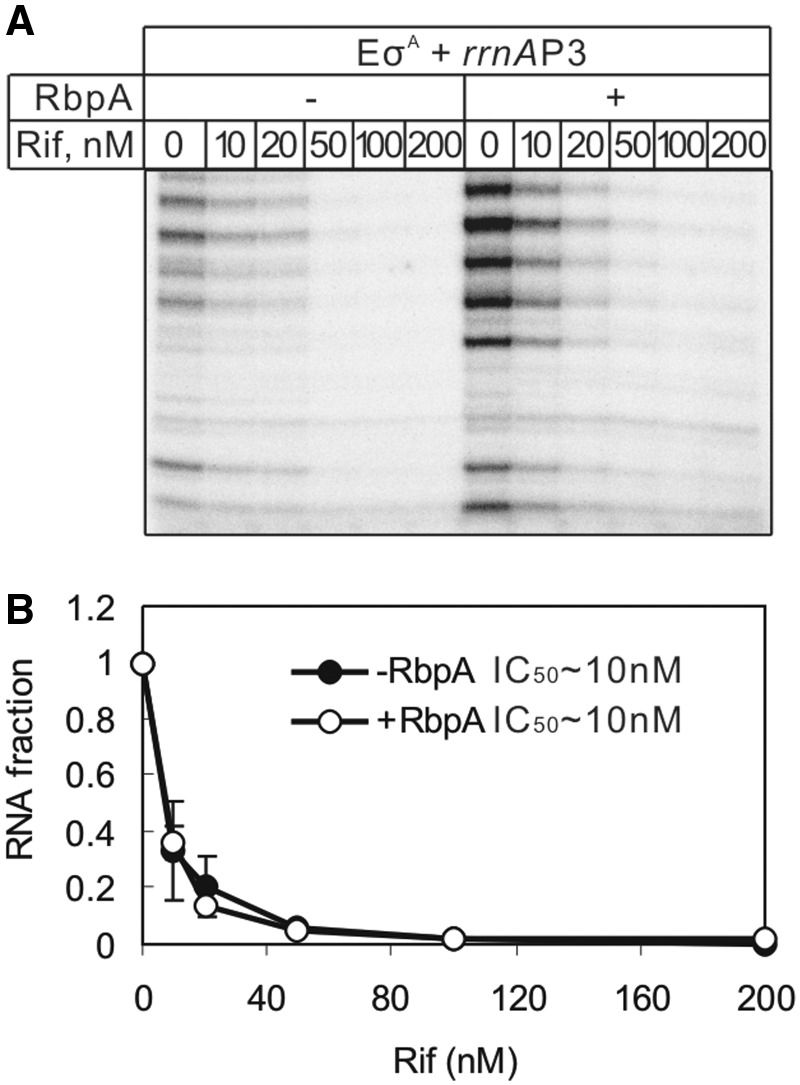
RbpA does not affect the sensitivity of RNAP to rifampicin
RbpA has been shown to confer a basal level of rifampicin resistance in S. coelicolor (16) and to increase the rifampicin tolerance level in M. smegmatis (18,19). Localization of the RbpA-binding site on the RNAP outer surface rules out the direct competition model for RbpA-induced rifampicin resistance. However, an allosteric change in RNAP that affects the rifampicin affinity is still possible. To explore whether RbpA changes the sensitivity of RNAP to rifampicin, an abortive initiation assay at the rrnAP3 promoter was performed in the presence of increasing concentrations of the antibiotic (Figure 5). As expected, the overall yield of transcripts was higher in the presence of RbpA, but transcription was also strongly inhibited by rifampicin. The calculated half inhibitory concentrations (IC50) of rifampicin in the presence and absence of RbpA was ∼10 nM, suggesting that RbpA does not affect the binding of rifampicin to the M. tuberculosis RNAP.
Figure 5.
Influence of RbpA on RNAP inhibition by rifampicin. (A) Abortive transcription initiation from the rrnAP3 promoter by the σA-containing RNAP in the presence of [32P]-UTP, ATP and GTP. Rifampicin (Rif) was added to the reactions at the indicated concentrations. RbpA (1.2 μM) was added to the reactions when indicated. (B) Quantification of the experiment shown in (A). The amounts of abortive RNA synthesized in the presence of different concentrations of rifampicin were normalized to the amount of the RNA synthesized without rifampicin (the first lane) and plotted against rifampicin concentrations. The error bars are the SD of duplicate experiments.
DISCUSSION
Role of RbpA in reprogramming the σA competitiveness in transcription
The principal finding of our study is a novel mechanism of transcription activation employed by the M. tuberculosis RbpA protein in its binding to the β subunit of RNAP. We showed that RbpA stabilizes the RNAP holoenzyme containing the housekeeping σA factor and increases the fraction of the active promoter complexes at σA-dependent promoters. The expression of the rbpA gene is known to be activated during the stationary phase, oxidative stress and rifampicin treatment, though RbpA does not affect transcription driven by the stress response, stationary phase σF subunit. We suggest that RbpA belongs to a new class of transcriptional regulators that increases the competitiveness of the principal σ factor over the alternative σs during the stress response. In turn, the stimulation of σA activity is required for pathogen proliferation during the infection of human macrophages and for the expression of genes involved in virulence (20). The contribution of RbpA in the regulation of gene expression likely differs between Actinomycetes species because the protein is essential for survival of M. tuberculosis (34) but dispensable for survival of S. coelicolor (16).
Switching between different transcriptional programs in bacteria is achieved through the regulation of the access of various σ factors to the RNAP core enzyme (12). The best studied model is the E. coli housekeeping σ70 which has the highest affinity (Kd ≤ 0.5 nM) to the core RNAP compared to alternative σ factors (35,36). Consequently, the most widespread regulatory strategy is the sequestration of either the free σ70 or the σ70-containing holoenzyme by anti-σ factors (9) or 6SRNA in favor of transcription driven by alternative σ factors (7,37). We speculate that the strategy used by M. tuberculosis for the regulation of the transcriptional activity of the principal σ factor may be different from that of E. coli. According to our estimation, M. tuberculosis σA core-binding affinity is lesser or similar to that of the alternative σF subunit. Thus, σA requires RbpA as a helper to reach the maximal activity in transcription. Because M.tuberculosis is a slow growing bacterium that exists in a persistent form for long periods after infection, relatively low affinity of σA to core may be one of the factors required to sustain the slow multiplication rate of the pathogen that was shown to be controlled by σA (20). Paradoxically, the properties of σA resemble the properties of the E. coli σS. Indeed, σS displays low affinity for the core RNAP and requires σS-binding Crl protein that favors assembly of the active holoenzyme (38,39).
RbpA and regulation of the rRNA gene transcription
Bacterial growth rate is proportional to the rate of ribosomal biosynthesis regulated at the transcriptional level by the alarmone ppGpp and priming NTP concentrations (27). RbpA was proposed to be an activator of rRNA transcription in Actinomycetes (16). Based on our results, RbpA also activates other σA-dependent promoters; therefore, its function is not specific to rRNA promoters. However, we hypothesize that RbpA can provide an additional level of control of rRNA transcription in combination with other RNAP-binding proteins and small molecule regulators. Recently, a new regulator of rRNA transcription in Mycobacterium, the CarD protein, was described (13). In a manner strikingly similar to RbpA, the CarD expression is induced by oxidative stress, and the protein also interacts with the RNAP β subunit. However, CarD functions as an antagonist of RbpA by repressing rRNA gene transcription. Future studies should explore how the interplay between these two proteins may be used in the regulation of gene expression.
Regulation of σA docking through the β subunit SBH motif
We show that σA still binds to the core RNAP in the absence of RbpA and can form transcriptionally active promoter complexes that are highly unstable and display reduced transcriptional activity in comparison to when they are in the presence of RbpA. The low affinity of M. tuberculosis σA for the core RNAP is likely due to the non-optimal structure of the σ-core RNAP-binding interface, which involves the RNAP β′ subunit clamp, downstream DNA-binding channel and RNA-binding channel (40–42) (Figure 4D). We suggest that the major function of RbpA is to tune this interface for optimal docking of σA to the core enzyme. In support of this idea, a hybrid enzyme containing M. tuberculosis σA and the E. coli core forms stable promoter complexes at the rRNA promoter and is insensitive to RbpA (Supplementary Figure S4).
We mapped the binding site of RbpA to the outer surface of the RNAP β subunit (between the Cys478 and Cys677) outside of the RNAP active site cleft. Several transcription factors (e.g. Mfd, CarD and Gre) are known to regulate transcription through binding to the β subunit (13,14,43). However, the RbpA-binding site does not overlap any known regulatory site and encompasses the ‘sandwich-barrel hybrid motif’ (SBHM), which has been suggested to be a potential binding site for small regulatory molecules (44). How does binding to the β subunit SBHM affect holoenzyme stability and the open complex formation? The formation of the open complex requires rearrangements in the σ-core interface and the opening or closing of the RNAP ‘jaws’ domains formed by the β subunit lobes and the β′ subunit clamp (4,42,45). The jaws hold promoter DNA in the open complex (4,39). Therefore, any factor affecting these rearrangements can modulate the process of open complex formation. Indeed, mutations and deletions in the β subunit (E. coli R454H, Δ436-445, Δ186-433) have been shown to affect promoter melting and open complex stability, likely by changing the β lobes conformation or mobility (46–48). Based on the RNAP structure, the σ factor interacting with the β lobe (σ region 3.1) and the β′ clamp (σ region 2) links the ‘jaws’ of RNAP and should sense the jaws movements (40). We hypothesize that RbpA can function allosterically by changing the conformation of the β lobes, which in turn, affects σA binding and open complex stability.
RbpA and resistance to rifampicin
Previous studies showed that S. coelicolor RbpA decreases RNAP sensitivity to rifampicin at the rRNA promoter (16). Additionally, RbpA from M. smegmatis was shown to rescue σA-dependent transcription from the rel promoter in the presence of 100 µM rifampicin (18). The cross-linking study localized the binding site of M. smegmatis RbpA to the β subunit R381 (T. thermophilus R345) in the RNAP active-site cleft near the rifampicin binding site (19). An ‘exclusion model’ of RbpA action was proposed, suggesting that RbpA binding leads to the dissociation of rifampicin from RNAP (19). Here, we showed that RbpA binds to a region distant from the rifampicin-binding site and does not affect the IC50 of rifampicin. Therefore, our results are inconsistent with the ‘exclusion model’. Analysis of the T. thermophilus RNAP structure shows that the R381 residue is buried deeply between the β subunit lobe domains and faces the outer surface of RNAP. Therefore, RbpA is unlikely to be cross-linked to R381 from inside the RNAP active-site cleft. However, the cross-link to the β subunit R381 is consistent with our data if one assumes it occurs from the outer enzyme surface.
We showed that RbpA does not prevent rifampicin binding to RNAP, and transcription was fully inhibited by 100 nM rifampicin at the M. tuberculosis rrnAP3 promoter. However, the background level of transcription in the presence of RbpA remains ∼2-fold higher. We suggest that the impact of RbpA on rifampicin resistance is indirect and occurs through the activation of σA-dependent transcription, which may act to stimulate cell proliferation (20) and allows bacteria to survive in the presence of the antibiotic. Further studies should uncover a spectrum of genes regulated by RbpA and to explain how the activation of these genes can affect the sensitivity of Mycobacteria to rifampicin and the survival of the pathogen in the host. Finally, because RbpA activity is essential for Mycobacteria growth, we propose that the RbpA/RNAP-binding interface may be a promising target for the development of highly specific anti-tuberculosis molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–6, Supplementary Materials and Methods and Supplementary References [51,52].
FUNDING
Centre National de la Recherche Scientifique (CNRS); Institut National de la Santé et de la Recherche Médical (INSERM). Y.H. was supported by fellowships from Chinese Academy of Sciences and CNRS. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Sébastien Rodrique (Universite de Sherbrooke, Canada) for providing the expression vectors for M. tuberculosis RNAP subunits.
REFERENCES
- 1. WHO. (2011) Global tuberculosis control. Available online at http://www.who.int/entity/tb/publications/global_report/2011/gtbr11_full.pdf.
- 2.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tupin A, Gualtieri M, Roquet-Banères F, Morichaud Z, Brodolin K, Leonetti J. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int. J. Antimicrob. Agents. 2010;35:519–523. doi: 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MTJ. Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saecker RM, Record MTJ, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase–promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigue S, Provvedi R, Jacques P, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 9.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Karakousis PC, Bishai WR. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J. Bacteriol. 2008;190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMaio J, Zhang Y, Ko C, Young DB, Bishai WR. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl Acad. Sci. USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 2001;42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 16.Newell KV, Thomas DP, Brekasis D, Paget MSB. The RNA polymerase-binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Mol. Microbiol. 2006;60:687–696. doi: 10.1111/j.1365-2958.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- 17.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 18.Dey A, Verma AK, Chatterji D. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology. 2010;156:873–883. doi: 10.1099/mic.0.033670-0. [DOI] [PubMed] [Google Scholar]
- 19.Dey A, Verma AK, Chatterji D. Molecular insights into the mechanism of phenotypic tolerance to rifampicin conferred on mycobacterial RNA polymerase by MsRbpA. Microbiology. 2011;157:2056–2071. doi: 10.1099/mic.0.047480-0. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Howard ST, Lakey DL, Kipnis A, Samten B, Safi H, Gruppo V, Wizel B, Shams H, Basaraba RJ, et al. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol. Microbiol. 2004;51:1551–1562. doi: 10.1111/j.1365-2958.2003.03922.x. [DOI] [PubMed] [Google Scholar]
- 21.Williams EP, Lee J, Bishai WR, Colantuoni C, Karakousis PC. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 2007;189:4234–4242. doi: 10.1128/JB.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques J, Rodrigue S, Brzezinski R, Gaudreau L. A recombinant Mycobacterium tuberculosis in vitro transcription system. FEMS Microbiol. Lett. 2006;255:140–147. doi: 10.1111/j.1574-6968.2005.00071.x. [DOI] [PubMed] [Google Scholar]
- 23.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Exp. Purif. 1993;4:503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson GR, Schaffer MH, Stark GR, Vanaman TC. Specific chemical cleavage in high yield at the amino peptide bonds of cysteine and cystine residues. J. Biol. Chem. 1973;248:6583–6591. [PubMed] [Google Scholar]
- 25.Gonzalez-y-Merchand JA, Colston MJ, Cox RA. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996;142(Pt 3):667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 26.Beaucher J, Rodrigue S, Jacques P, Smith I, Brzezinski R, Gaudreau L. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol. Microbiol. 2002;45:1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaal T, Bartlett MS, Ross W, Turnbough CLJ, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Coates AR. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeffer SR, Stahl SJ, Chamberlin MJ. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. J. Biol. Chem. 1977;252:5403–5407. [PubMed] [Google Scholar]
- 30.Joo DM, Ng N, Calendar R. A sigma32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc. Natl Acad. Sci. USA. 1997;94:4907–4912. doi: 10.1073/pnas.94.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu TK, Kusano S, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase sigmaF holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 33.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 34.Forti F, Mauri V, Deho G, Ghisotti D. Isolation of conditional expression mutants in Mycobacterium tuberculosis by transposon mutagenesis. Tuberculosis. 2011;91:569–578. doi: 10.1016/j.tube.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 37.Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J. Bacteriol. 2004;186:4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 2006;188:7966–7970. doi: 10.1128/JB.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 41.Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber TM, Markov D, Sharp MM, Young BA, Lu CZ, Zhong HJ, Artsimovitch I, Geszvain KM, Arthur TM, Burgess RR, et al. Binding of the initiation factor sigma(70) to core RNA polymerase is a multistep process. Mol. Cell. 2001;8:21–31. doi: 10.1016/s1097-2765(01)00292-1. [DOI] [PubMed] [Google Scholar]
- 43.Park J, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 44.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodolin KL, Studitsky VM, Mirzabekov AD. Conformational changes in E. coli RNA polymerase during promoter recognition. Nucleic Acids Res. 1993;21:5748–5753. doi: 10.1093/nar/21.24.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl Acad. Sci. USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nechaev S, Chlenov M, Severinov K. Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J. Biol. Chem. 2000;275:25516–25522. doi: 10.1074/jbc.M002511200. [DOI] [PubMed] [Google Scholar]
- 48.Brodolin K, Zenkin N, Severinov K. Remodeling of the sigma70 subunit non-template DNA strand contacts during the final step of transcription initiation. J. Mol. Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 49.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 52.Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the -10 promoter element. EMBO J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.