Abstract
OLE (Ornate, Large, Extremophilic) RNAs represent a recently discovered non-coding RNA class found in extremophilic anaerobic bacteria, including certain human pathogens. OLE RNAs exhibit several unusual characteristics that indicate a potentially novel function, including exceptionally high expression and localization to cell membranes via interaction with a protein partner called OLE-associated protein (OAP). In the current study, new genetic and phenotypic characteristics of OLE RNA from Bacillus halodurans C-125 were established. OLE RNA is transcribed at high levels from its own promoter under normal growth conditions and the transcript is exceptionally stable compared to most other RNAs. Expression is increased by ∼7-fold when cells are exposed to near lethal concentrations of short-chain alcohols such as ethanol or methanol. Strains wherein the genes for OLE and/or OAP are deleted are more susceptible to growth inhibition by alcohol and also become more sensitive to cold. Normal growth characteristics can be restored by expressing the genes for OLE and OAP from plasmids or from elsewhere on the chromosome. Our findings confirm a functional link between OLE and OAP and reveal the importance of a large non-coding RNA in the response to alcohol-induced stress.
INTRODUCTION
In addition to the fundamental tasks many RNAs perform in biological information transfer, non-coding RNAs are involved in many diverse biochemical functions in bacteria including RNA processing (1–4), gene regulation (5–8), defense against phage (9) and protein localization (10) among others. The number of discoveries of additional non-coding RNA classes has been increasing as more microbial DNA is sequenced and as bioinformatics search algorithms for structured RNA identification continue to improve. Intriguingly, novel classes of bacterial non-coding RNAs that rank among the largest and most complex known have been reported recently (11–14), suggesting that numerous additional RNAs with distinct biochemical functions important for many species remain to be discovered.
The functions of most newfound classes of large non-coding RNAs remain to be validated. RNAs whose functions were established long ago typically were discovered by investigating the molecular basis for specific cellular processes. For example, processing of precursor tRNAs to yield mature tRNAs necessarily involves RNase activity. Studies revealed the existence of a large non-coding RNA (15) that was determined to function as an RNA-cleaving ribozyme called RNase P (16). In the rare instances, when the specific non-coding RNA was discovered first (e.g. tmRNA and 6S RNA), decades passed between their initial identification and experimental validation of their biological and biochemical functions (17,18). As bioinformatics searches yield new non-coding RNA classes that are not yet linked to biological processes, additional work will be required to gather clues about their biochemical functions. Such efforts are particularly challenging for RNAs that are not involved in processes that are universal in biology or that are essential under typical laboratory growth conditions.
One of the recently discovered classes of large non-coding RNAs is called OLE (Ornate, Large, Extremophilic) RNA (11,19). OLE RNAs are similar in length and structural complexity to several large ribozymes, including Group I and II self-splicing introns and RNase P (13). OLE RNA representatives occur exclusively in extremophilic and/or anaerobic Firmicutes (Gram-positive bacteria) (11), including the causative agents for both botulism (Clostridium botulinum) and tetanus (Clostridium tetani). All organisms that contain OLE RNA also carry a gene for a predicted integral membrane protein of unknown function, called OLE-associated protein (OAP). We recently demonstrated that OLE and OAP form an RNA–protein complex in vitro and in vivo, and this binding is necessary for OLE RNA to localize to the membrane of a cell (19). Membrane localization by OLE RNA is almost unique among known non-coding RNAs. One prominent exception is the signal recognition particle (SRP) RNA from bacteria (10), which binds a soluble protein to form a complex that transiently localizes to the membrane (20,21).
Under normal growth conditions, OLE RNA is one of the most abundant transcripts in Bacillus halodurans C-125, a bacterial alkaliphile and facultative anaerobe (19). The number of copies of OLE RNA is nearly equal to that observed for RNase P or for SRP RNA. Due to its large size, complex structure, unusual phylogenetic distribution, unusual subcellular localization and high expression level, we hypothesized that OLE RNA might perform a biochemical function that has not yet been observed for non-coding RNAs. In an attempt to link OLE RNA to a biological process, we performed a series of bioinformatics analyses and genetic and molecular biology experiments using B. halodurans C-125, which is one of the few genetically tractable organisms that carries OLE RNA and that can grow in the presence of oxygen.
MATERIALS AND METHODS
Bacillus halodurans C-125 was purchased from the ATCC (Catalog #BAA-125). Plasmids pMK4, pHCMC05, pE194 and pBGSC6 were obtained from the Bacillus Genetic Stock Center (The Ohio State University). Plasmid pUC19 was purchased from Invitrogen, and pGFPuv was purchased from Clontech. Unless otherwise specified, B. halodurans was grown in LB (Lysogeny Broth) broth (USB Corporation) that was prepared at 90% volume, autoclaved and adjusted to full volume and pH ∼10.5 with 10% (w/v) filter-sterilized Na2CO3 [1% (w/v) final concentration]. Anaerobic cultures were grown in nutrient broth [NB; 5 g peptone (Difco) and 3 g beef extract (Difco) per liter] supplemented with 1% (w/v) glucose (J.T. Baker), 5 mM KNO3 (Sigma-Aldrich) and 1 μg ml−1 resazurin (Sigma-Aldrich), and adjusted to pH ∼10.5 as described above. Media was solidified with 1.5% (w/v) agar (BD) as needed.Unless otherwise indicated, all cells were grown at 37°C and shaken at ∼200 rpm.
Mutual information (MI; a measure of the dependence of one variable on another, such as covariance of two nucleotides) analysis and other comparative sequence analyses (22,23) were conducted on OLE RNAs from 52 sequenced genomes and 156 unique sequences from environmental DNA samples. Sequences were aligned using CMfinder (http://bio.cs.washington.edu/yzizhen/CMfinder/ 20 April 2012, date last accessed) (24) and MI content was calculated as described previously (5). P-values were calculated by comparing MI scores with those from 100 000 neutral-evolution simulations to determine the probability of a given MI score arising by chance. The OLE RNA secondary structure diagram was generated by first processing the alignment with the RNA structure drawing algorithm R2R (http://breaker.research.yale.edu/R2R/ 20 April 2012, date last accessed) (25) and then manually adjusting the output to improve clarity.
Total cellular RNA was extracted from cells that were pelleted from media and resuspended in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0 at 23°C) containing 3 mg ml−1 lysozyme. The resuspended mixture was incubated either at 23°C or on ice for 5 min. Cells were lysed by freeze–thawing three times, and total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s directions. Residual genomic DNA was removed with RQ1 DNase (Promega) according to the manufacturer’s directions.
To assess possible splicing of OLE RNA, total RNA (1 μg) was reverse-transcribed with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol with 50 ng random hexamer. cDNA was then amplified in 50 μl PCR reactions containing 200 nM primers (each), 200 μM dNTPs (NEB), 2.5 U Taq polymerase (NEB), PCR buffer [1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl (pH 8.3 at 23°C), 0.01% (w/v) gelatin] and 0.5 μl cDNA. Amplification was performed for 30–50 cycles with an extension at 72°C for 2 min, and products were separated and analyzed by agarose (0.8%) gel electrophoresis.
Plasmids for knocking out OLE and OAP were constructed using techniques similar to those reported previously (26). Briefly, the TaqI fragment of pE194 containing its temperature-sensitive oriC was amplified via PCR with primers that introduce BamHI restriction sites. The PCR product was cloned using a TOPO-TA cloning kit (Invitrogen), after which the insert was removed by digestion with BamHI (NEB), gel purified and ligated into pUC19 to generate plasmid pUCE194. Inserts to knockout-specific genes (as described in Figure 4a) were created by overlap-extension PCR and ligated into the EcoRI site of pUCE194. Plasmids were then transformed into B. halodurans C-125 via protoplast transformation as described previously (27) and the cells were recovered on medium containing 3 μg ml−1 chloramphenicol. Subsequent transformations were conducted using improvements in B. halodurans cloning as described elsewhere (28). All constructs were confirmed via sequencing by the W. M. Keck Biotechnology Resource Laboratory at Yale University.
Figure 4.
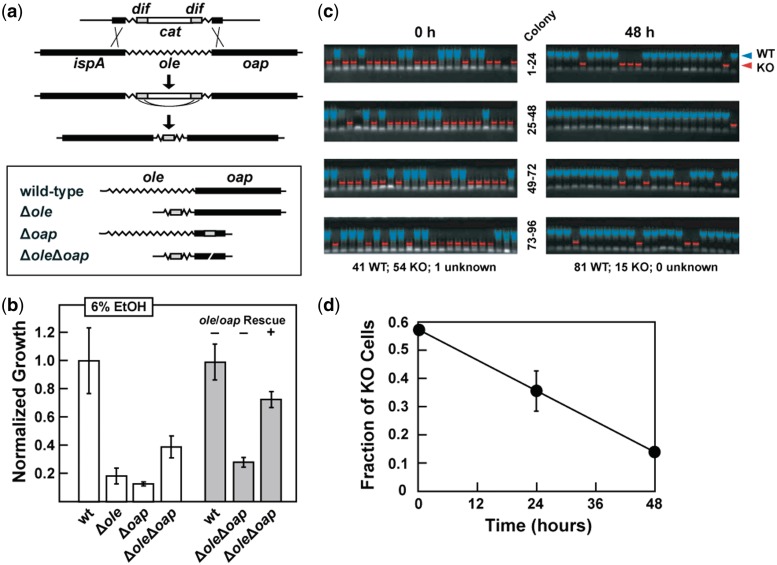
Ethanol tolerance of WT, KO and rescue strains for ole and oap. (a) Strategy for creating various ole and oap deletion strains. Top: after integration of the deletion cassette into the chromosome, spontaneous recombination between flanking dif sites mediated by Xer recombinase (30) deletes the cat gene and leaves only a 30 bp scar. Bottom: schematic representation of the four B. halodurans strains used for phenotype analyses. Since the presence of two dif sites in proximity would cause recombination between them, the ΔoleΔoap double KO was created by including an in-frame internal deletion (white slash) in oap as part of the 3′ homology segment. (b) Growth of the KO and rescue strains compared with WT in medium containing 6% (v/v) ethanol as measured by OD600 values. Cells were inoculated at an OD600 of 0.001 and grown for 48 h at 37°C. Growth is normalized to WT. Data are generated from at least three independent experiments and the error bars represent the standard error of the mean. Rescue strains contain an insertion in lacZ of a chloramphenicol resistance cassette either with (+) or without (−) ole, oap, and their promoter. (c) Agarose gel electrophoresis results of colony PCR reactions from WT and ΔoleΔoap. Bacillus halodurans co-cultures were incubated for 0 or 48 h in LB (pH ∼ 10.5) with 6% (v/v) ethanol. PCR using primers selective for the ole-oap locus yields different products for WT and KO strains as identified by false coloration. (d) Plot of the fraction of KO cells remaining in a competitive co-culture with WT. Values were established by colony PCR as depicted in (c). Error bars are as described for (b) and in some instances are smaller than the point size.
Plasmid integration was achieved using published protocols (26,29) with minor modifications. In brief, overnight cultures grown at 30°C were diluted 1:100 in fresh media with antibiotic (chloramphenicol, 3 μg ml−1) and grown 2 h at 30°C. Cultures were then shifted to 45°C and grown 3 h to dilute plasmid concentration per cell. Serial 10-fold dilutions of each culture were then plated on LB pH ∼10.5 agar with 3 μg ml−1 chloramphenicol and incubated at 45°C overnight to recover colonies that had integrated the plasmid. Double-crossover events were identified via PCR. The chloramphenicol resistance cassette was subsequently excised through Xer recombination (30) as follows. Cells were grown 24 h at 37°C in LB (pH ∼ 10.5; no antibiotic) and plated on LB (pH ∼ 10.5) agar for overnight growth. The resulting colonies were replica-plated onto LB (pH ∼ 10.5) agar with 3 μg ml−1 chloramphenicol and incubated at 37°C overnight. Colonies that failed to grow on the chloramphenicol-containing plates were picked from the initial plate and screened for the desired genetic change by PCR and DNA sequencing.
Competitive co-culture experiments were conducted by inoculating liquid LB medium (pH ∼ 10.5) with equal amounts of exponentially growing wild-type (WT) and knock-out (KO) cells (0.001 initial OD600) and incubating them with shaking for 0, 24 or 48 h in LB (pH ∼ 10.5) containing 6% (v/v) ethanol. Culture samples were serially diluted 10-fold and plated on LB (pH ∼ 10.5) agar, incubated overnight at 37°C, and individual colonies were picked and subjected to PCR using primers specific for the ole-oap locus. PCR products corresponding to amplification of the full-length WT locus or the shortened KO locus were identified by separation using 1% agarose gel electrophoresis and staining with ethidium bromide. Cell growth was estimated by counting the total number of colonies per plate and using the plate’s dilution factor and the PCR genotype proportions to determine the total number of cells of each genotype at each time point.
Transcriptome analyses were conducted in duplicate using wild-type and ΔoleΔoap B. halodurans C-125 strains grown to mid-exponential phase (OD600 0.2–0.3) in LB (pH ∼ 10.5). Triplicate subcultures of 3 ml each were incubated in sealed tubes for 3 h at 37°C with shaking after the addition of ethanol (95% aqueous-distilled, Pharmco-AAPER) to 5% (v/v) concentration. In all instances, ethanol concentrations noted are initial values and may change due to evaporation or bacterial metabolic activities. Cultures were pooled and RNA was extracted as described above, and rRNA was removed by using a RiboMinus Kit for Bacteria (Invitrogen). cDNA library preparation and sequencing was conducted by Otogenetics (Tucker, Georgia, USA). Reads were aligned to the B. halodurans reference genome with SOAP2 (http://soap.genomics.org.cn/ 20 April 2012, date last accessed) (31), and the expression of individual genes was tabulated by custom Perl scripts. Significant differences in gene expression were determined with edgeR (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html 20 April 2012, date last accessed) (32), and relative expression levels were computed by calculating RPKM scores (Reads Per Kilobase per Million reads) (33). Raw and mapped data are available at the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/ 20 April 2012, date last accessed), accession GSE33303.
Constructs for plasmid-based genetic rescues were each constructed using plasmid pHCMC05 carrying the appropriate ole-oap construct controlled by an IPTG-inducible promoter. Plasmids were transformed into the indicated B. halodurans C-125 strain as described earlier. Rescue transformants were grown in media containing 3 μg ml−1 chloramphenicol and induced with 1 mM IPTG for at least 3 h prior to the start of the assays.
Chromosomal rescue constructs were made by inserting the oriC-containing TaqI fragment of pE194 into pUC19 at the HindIII restriction site to create plasmid pUCE195. 5′ and 3′ homology segments of the lacZ gene (BH2723) of B. halodurans and a chloramphenicol resistance cassette from pBGSC6 were amplified by PCR with primers containing the necessary restriction sites for cloning into pUCE195. Integration of the final vector disrupts lacZ and places the chloramphenicol resistance cassette on the chromosome. For rescue, the full ole-oap region, including the promoter (see text), was amplified and inserted upstream of the chloramphenicol resistance cassette. Plasmids were transformed into B. halodurans C-125 and integrated as described for making KOs.
Stress assays were conducted using 3 ml cultures (LB at pH ∼ 10.5) of B. halodurans C-125 cells in exponential growth phase. For ethanol stress, the initial OD600 was 0.001 for 48-h assays, or ∼0.1 for 24-h assays. Ethanol was added to culture tubes to a final concentration of 5% (northern blots) or 6% (growth assays). Tube caps were sealed and the cultures shaken at 37°C for the indicated times. For cold stress, cells were diluted to OD600 0.01 and placed in a 15°C incubator for 7 days.
GFPuv mRNA reporter constructs were prepared by PCR amplification from pGFPuv (Clontech) and fused to the OLE RNA promoter via overlap-extension PCR. The resulting construct was cloned into plasmid pMK4 and transformed into B. halodurans as described above. mRNA levels were assayed via northern blot analysis.
RNA decay rates were established by growing wild-type B. halodurans to mid-exponential phase (OD600 ∼ 0.3) in LB (pH ∼ 10.5). Rifampicin (Sigma-Aldrich) was added to 100 μg ml−1 final concentration, and aliquots of the culture were withdrawn at specific time points. Total RNA was extracted as described above and probed via northern blot analysis.
qRT-PCR was conducted on total RNA samples extracted as described above. Equal amounts of total RNA (0.5–1.0 μg) were reverse-transcribed with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol with 100 ng of random hexamers. The resulting cDNA was diluted by the addition of nine volumes dH2O and subjected to quantitative PCR on an Applied Biosystems 7500 Real-Time PCR system using reactions consisting of 10 μl Power SYBR Green Master Mix (Applied Biosystems), 4 μl water, 1 μl primer mix (10 mM each) and 5 μl template. Expression levels were determined according to published protocols (34).
Northern blot analyses were performed according to published protocols (35). Hybridization was carried out in GE Rapid-Hyb buffer at 44°C according to the manufacturer’s protocol. Signals were normalized to 16S rRNA for quantification unless otherwise specified.
RESULTS AND DISCUSSION
MI analysis reveals additional structural elements in OLE RNAs
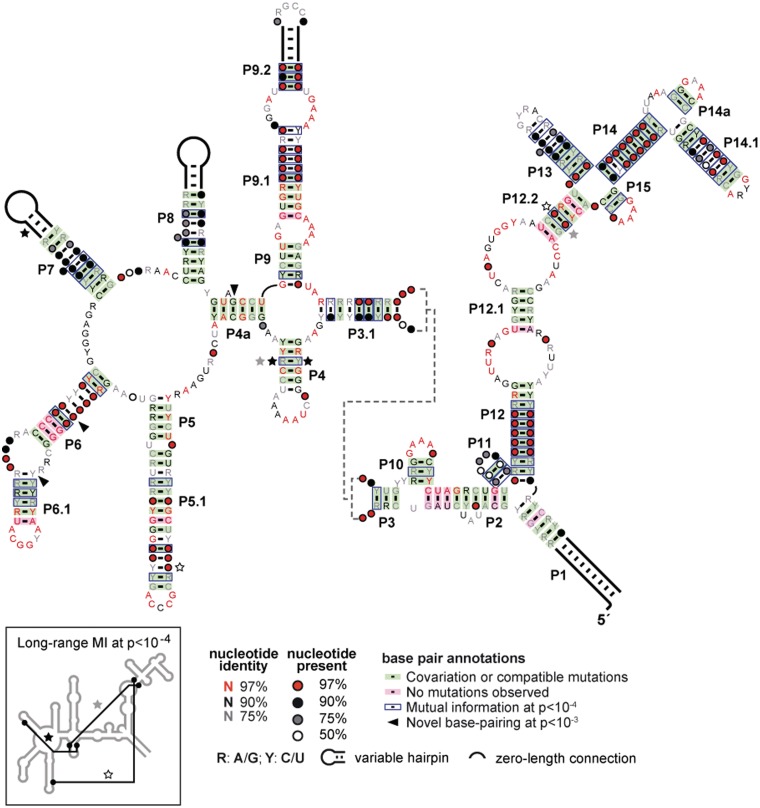
A MI analysis (22,23) was conducted using approximately 200 unique OLE RNA representatives derived from sequenced genomes and from metagenomic data (19). MI is a statistical measure of how predictable the identity of one particular nucleotide is given the identity of another, and this method has been used previously to predict and refine the structures of other conserved RNAs (e.g. 5). MI analysis is particularly well suited to reveal base-paired structures that can co-vary, although other tertiary-structure contacts also can be revealed.
MI scores confirm most of the secondary-structure elements predicted previously (Figure 1). In addition, this analysis revealed the likely existence of three previously unidentified stems: P4a, P14a and P15. P4a is formed in part from one of two short regions of conserved RNA sequence proposed to be important for OLE RNA binding to the protein partner OAP (19). Formation of this newfound P4a stem would make the local structure at this first protein-binding site similar to the structure of the second proposed OAP-binding site (stem P2). It is not known whether protein binding at each site requires stem formation as predicted by our MI analysis, but such a requirement could explain the selective binding of OLE RNA by OAP despite the relatively low information content of the binding site (19).
Figure 1.
Updated consensus sequence and secondary structure model for OLE RNAs based on approximately 200 genomic and metagenomic representatives. Co-variation was calculated as described previously (19), and MI content of all possible nucleotide pairs was computed using published methods (5). P-values represent the probability of the given MI score occurring by chance in simulated neutral-evolution populations. A P-value cutoff of 10−4 is consistent with many base pairs predicted in previous models, and with two additional stems (P14a and P15). Potential tertiary interactions (Inset) are also evident. A P-value cutoff of 10−3 reveals possible base-pairing interactions (including P4a) that can form in at least 80% of the representatives (arrowhead).
Formation of stems P14a and P15 further supports our original suggestion (11) that the nucleotides in this region form a sequence and structural domain with partial 2-fold symmetry. Three of four loop sequences conform to a consensus GNRA tetraloop, and analogous hairpins are known to stabilize RNA structures and can dock to the minor groove of distal helices of specific architectures (36–38). The remaining sequence is typically GAAAN, which closely approximates a GNRA tetraloop. The proximity of these loops to one another could indicate that several distal portions of OLE RNA may form tertiary contacts with this symmetrical domain. This entire region of OLE RNA (P12 through P15) could function as a stabilizing scaffold analogous to Domain I in Group II introns (39). However, there are numerous other feasible reasons for the clustering of these tetraloops, including the possibility that they might form contacts with another RNA molecule.
Our MI analysis did not reveal the presence of potential pseudoknots, which is surprising for an RNA of this size and structural complexity. Perhaps OLE RNAs do not rely on pseudoknots to form their tertiary structure, or perhaps pseudoknots are present but their component nucleotides are subject to other selective pressures that keep their identities too highly conserved for MI analysis to reveal co-variation. However, several distal interactions that indicate non-canonical tertiary contacts (Figure 1 and Inset) were revealed. Additional tertiary contacts are also likely to exist but there were insufficient occurrences of co-variation to declare their existence with confidence. The newfound structural elements further refine our model for OLE RNA and place it firmly among the most complex bacterial RNAs known. Only the two large ribosomal RNAs, Group II self-splicing introns, and the recently identified GOLLD RNA are known to conserve greater lengths and structural complexity (13).
Analysis of OLE RNA transcripts from B. halodurans
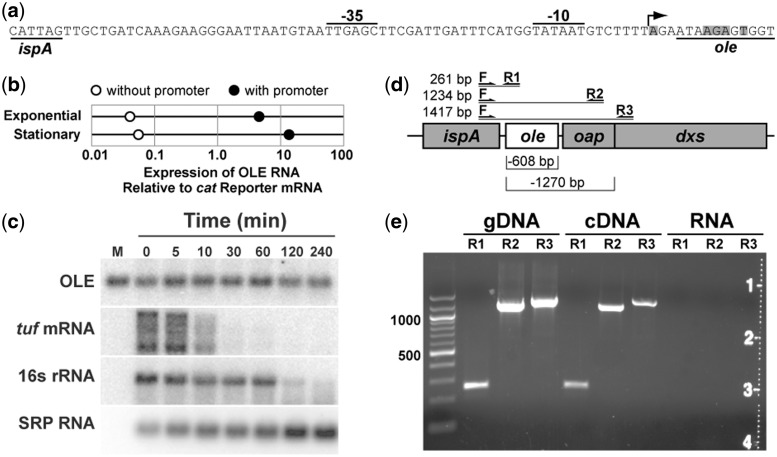
In B. halodurans, OLE RNA is a highly abundant transcript whose expression level is within a few folds of that for the RNase P and SRP RNAs (19). At least some OLE RNA is co-transcribed with neighboring genes (11), but a putative promoter located immediately upstream of the RNA gene also has been identified (40) (Figure 2a). To determine if this proximal promoter is used, we cloned the OLE RNA gene (hereafter called ole) either with or without the putative promoter into a plasmid and transformed the constructs into a B. halodurans strain with an ole deletion (Δole, see description below).
Figure 2.
Expression and stability of OLE RNA. (a) Sequence of the predicted promoter for OLE RNA synthesis. The predicted transcription start site is indicated by an arrow. Shaded nucleotides identify ends of natural OLE RNAs as previously determined by using 5′ RACE (11). (b) Relative expression of OLE RNA from a plasmid with and without the promoter region. Expression levels were determined in B. halodurans lacking its genomic OLE RNA gene under exponential growth or stationary phase as indicated. Analyses were conducted by qRT-PCR and values were normalized to expression of the mRNA for the antibiotic resistance gene on the plasmid. (c) RNA decay analysis in B. halodurans cells treated with 100 μg ml−1 rifampicin. Each lane contains an equal amount of total RNA and was examined by northern blot analysis using radiolabeled probes for the transcripts indicated. M, in vitro transcribed OLE RNA marker; tuf, transcription elongation factor Tu; SRP RNA, SRP RNA component. (d) Locations of PCR primer-binding sites used to search for spliced transcripts. Lines indicate predicted sizes for full-length transcripts, while brackets (below) indicate potential deletions in spliced products. (e) RT-PCR products of OAP RNA transcripts. Primer pairs as described in (d) were used to amplify (30 thermocycles) genomic DNA, cDNA or total RNA (no cDNA synthesis).
Total RNA was extracted from cells isolated either during exponential growth or stationary phases. The RNA was subjected to quantitative reverse transcription and polymerase chain reaction amplification (qRT-PCR), and the levels of OLE RNA transcription were determined and normalized relative to the levels of mRNA expression of the chloramphenicol acetyltransferase (cat) antibiotic resistance gene also carried by the plasmids. Inclusion of the promoter increases OLE expression by >100-fold (Figure 2b), demonstrating that the promoter is indeed active under normal growth conditions.
To assess the stability of OLE RNA, we treated exponentially growing B. halodurans C-125 with rifampicin to halt RNA synthesis and extracted total RNA at various times. A northern blot analysis was conducted to detect several abundant transcripts, including OLE (Figure 2c). In contrast to 16S rRNA and the mRNA for the tuf gene (encoding elongation factor Tu, which is expressed at approximately the same level as OLE RNA) (19), OLE RNA exhibits almost no degradation even 4 h after the addition of rifampicin. However, each lane of the gel was loaded with an equivalent amount of RNA, and SRP RNA (an exceptionally stable RNA) appears to be increasing in abundance because it is becoming a greater proportion of the total RNA remaining. Therefore, although OLE RNA appears to maintain a constant level on the gel, it is likely to be degrading slowly over the course of this assay.
When normalized to the amount of SRP RNA, the half-life of OLE RNA is estimated to be ∼3 h. Given that bacterial doubling time under these conditions is ∼45 min (data not shown), it appears that the decay of OLE RNA does not play an appreciable role in cells under these growth conditions. We also evaluated the stability of OLE RNA in B. halodurans cells grown at neutral pH or in cells lacking the OAP protein, but no variation in stability was observed (data not shown). Our findings indicate that OLE RNA could be considered as a ‘stable’ RNA (41), and might be degraded only in times of stress or damage.
Northern blot analyses reveal that there is only a single product band for OLE RNA (Supplementary Figure S1). Furthermore, previous 5′ RACE data (11) revealed that sites of OLE RNA processing are located only near the start of the P1 stem (Figure 2a). These sites are very close to the predicted transcription start site for the proximal promoter, indicating that the vast majority of independent OLE RNA transcripts and any OLE RNAs processed from larger RNA precursors likely have similar 5′ termini.
Self-splicing ribozymes are similar in size and complexity to OLE RNAs (13), which raises the possibility that OLE RNAs could represent a new type of self-splicing RNA. However, the northern blot data described above suggest that the RNA does not undergo splicing because there is no evidence of smaller processed products. To further evaluate this hypothesis, we performed RT-PCR on RNA extracts from B. halodurans growing under various conditions. No spliced transcripts were ever detected (Figure 2d and e), even after 50 cycles of PCR amplification (data not shown). Additionally, although we have not rigorously ruled out an RNA self-cleavage function, the size and complexity of OLE RNAs relative to known self-cleaving ribozymes (42) implies that if self-cleavage activity exists, this is unlikely to be its sole function.
OLE RNA expression is induced by ethanol
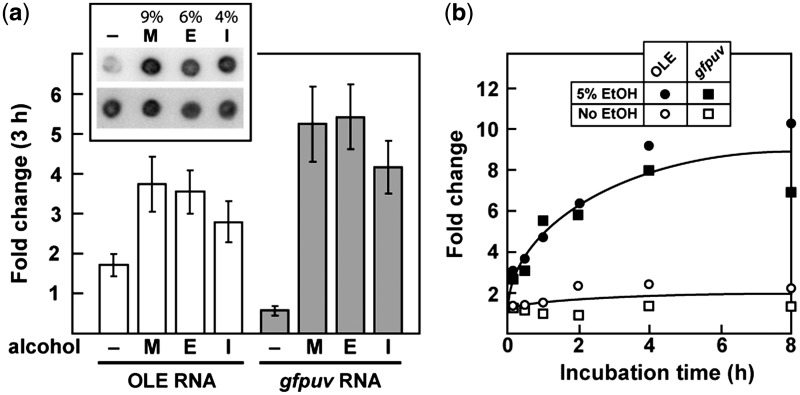
Cell culture conditions were sought under which OLE RNA expression is altered relative to its established baseline. Due to the strong association of OLE and OAP genes with extremophilic organisms (11), we hypothesized that the RNA might play a role in protecting cells against stresses present in extreme environments. Bacillis. halodurans C-125 cells were subjected to a variety of stresses (Supplementary Table S1) and OLE expression changes were monitored via northern blot analyses. Conditions that produced a 2-fold or larger change in OLE RNA levels were reassessed with a reporter construct (gfpuv mRNA fused to the promoter for OLE RNA) (Figure 2a) to confirm changes in transcript levels. The only conditions that consistently altered OLE RNA expression were exposure to short-chain alcohols (Figure 3a). Treatment with heat (55°C, the limit of B. halodurans growth) or shift to anaerobic conditions also appeared to induce OLE (Supplementary Table S1 and Supplementary Figure S2), but these results were not consistently reproducible with the reporter construct.
Figure 3.
OLE RNA expression is induced by short-chain alcohols. (a) Relative expression levels of OLE and a reporter gene (gfpuv mRNA fused to the OLE RNA promoter) after 3 h with either no stress (–) or near-lethal levels of methanol [M, 9% (v/v)], ethanol [E, 6% (v/v)], or isopropanol [I, 4% (v/v)]. Cells were incubated for 3 h with a short-chain alcohol or no added alcohol as indicated prior to RNA extraction. RNA levels were determined via Northern blot analysis (see inset for a representative blot; all dots were formed by loading an equal amount of total RNA; top row was probed for OLE RNA and bottom row was probed for 16S RNA) and normalized to the value measured at time zero for the given alcohol and reporter. Error bars represent the standard error of the mean of three independent experiments. (b) Representative time course of OLE RNA induction by ethanol (see Supplementary Figure S5 for a representative northern blot). Cells were treated either without or with 5% (v/v) ethanol for the indicated time. Transcript levels were determined as described in (a) and the lines approximate the data trends obtained either in the absence or presence of ethanol.
All three short-chain alcohols examined (methanol, ethanol and isopropanol) induce OLE RNA production. Likewise, these alcohols induce expression of the gfpuv reporter gene. Each alcohol was tested at a concentration that is near equivalent in toxicity compared with the other alcohols, and the expression levels are similar in all three analyses. This result suggests that the increase in OLE expression is not due to any specific molecular recognition event, but rather to a common effect caused by these compounds.
Given the high-basal expression of OLE RNA (19), its biochemical function is probably also beneficial under normal growth conditions but becomes more beneficial when cells are stressed with alcohols. Alcohols cause many stresses on cells, but one of the strongest effects is to make cell membranes more permeable to ions and small molecules (43,44). Attempts to duplicate this membrane permeability stress with detergents or ionophoric antibiotics failed (Supplementary Table S1), which suggests that there may be a different aspect of alcohol stress that is responsible for triggering increased OLE production.
Since ethanol is presumably the short-chain alcohol that is most common in the natural environment, we performed a more detailed analysis of the response of B. halodurans to sublethal ethanol concentration (Figure 3b). Addition of ethanol causes an almost immediate increase in both OLE and reporter transcripts. Ethanol-mediated induction also occurs at neutral pH (∼7.0) (Supplementary Figure S2), indicating that this response is independent of the pH of the medium.
Strains deficient in OLE and/or OAP exhibit increased sensitivity to ethanol
A series of KO strains were generated in B. halodurans C-125 cells to assess the effects of the loss of OLE and OAP. These KO strains carry internal deletions of ole, oap or both, and were constructed in a manner that minimizes the possibility of unanticipated polar effects on neighboring genes (Figure 4a). The KO strains were subjected to a variety of phenotype assays (Supplementary Table S2), wherein the vast majority of conditions generally show no difference from wild type. Only under conditions of alcohol stress (Figure 4b) or cold stress (Supplementary Figure S3) are KO cells significantly affected. Notably, the growth defect under alcohol stress corresponds well with the observation that OLE RNA production is increased under these same conditions. This finding supports the hypothesis that OLE RNA is important for resisting alcohol-induced stress. Cold stress causes growth reduction for the KO cells, but does not induce OLE RNA expression (Supplementary Table S1). Even though OLE RNAs are present in many different types of extremophiles, they have not been found in psychrophiles (cold-loving organisms). Therefore, the relevance of OLE RNA to cold stress adaptation is less clear.
Ethanol causes the largest growth defect among the short-chain alcohols tested (Figure 4b; Supplementary Figure S4). For each alcohol type, all three KO strains (Δole, Δoap and ΔoleΔoap) are affected similarly, suggesting that OLE and OAP are part of the same pathway and affect the same biological function. To more precisely quantify growth differences, we established competitive co-cultures with WT and ΔoleΔoap KO cells in the presence of 6% ethanol and removed samples at 24 and 48 h to establish the relative fraction of each cell type by plating, picking colonies and classifying their type by PCR (Figure 4c). The fraction of KO cells relative to WT cells decreases by >3.6-fold (Figure 4d), which is consistent with our observations of reduced growth of KO cells compared with WT when they are cultured independently (Figure 4b). We also performed quantitative PCR of co-culture genomic DNA to ensure that each colony reflected cell growth and not other effects (e.g. cell aggregation), with the same results (data not shown).
To determine if the absence of OLE or OAP is responsible for the observed growth defects caused by exposure to ethanol or to cold, we created a series of B. halodurans rescue strains to supply the missing gene(s) by either plasmid or by chromosomal integration at a site distal from the original locus. Expression of plasmid-borne ole and oap, controlled by an IPTG-inducible promoter, resulted in partial rescue of growth when cells were grown in 6% ethanol (Supplementary Figure S3a). Likewise, the rescue strain carrying ole and oap on the chromosome at a distal locus (lacZ, BH2723) under control of the native promoter partially restores resistance to ethanol (Figure 4b) and yields complete restoration of growth under cold stress conditions (Supplementary Figure S3b). These results indicate that the observed phenotypes are due to OLE and OAP rather than to unanticipated effects on neighboring genes.
In an attempt to link other pathways to the ethanol sensitivity phenotype, RNA-seq (45) was performed on both WT and ΔoleΔoap cells before and after growth for 3 h with 5% ethanol. This concentration was chosen because it strongly induces OLE RNA production in WT cells (Figure 3b) but KO cells experience only minor growth defects (data not shown), implying they are largely able to compensate for lack of OLE RNA at this concentration. Transcript analyses reveal essentially no difference between WT and KO cells before ethanol stress (Figure 5a and data not shown), consistent with the lack of a phenotype under these conditions. After ethanol stress, the expression levels of only a small number of genes are significantly different between the strains (Figure 5b and Supplementary Table S3). However, we did not identify any obvious connections between their functions that would guide the design of additional experiments to directly reveal the biochemical function of OLE RNA. Furthermore, although OLE RNA is observed to increase substantially upon ethanol exposure, the mRNA for OAP increases only modestly (Supplementary Table S4). This suggests that if OAP levels increase, regulation may occur at the translation level and not at the level of mRNA abundance.
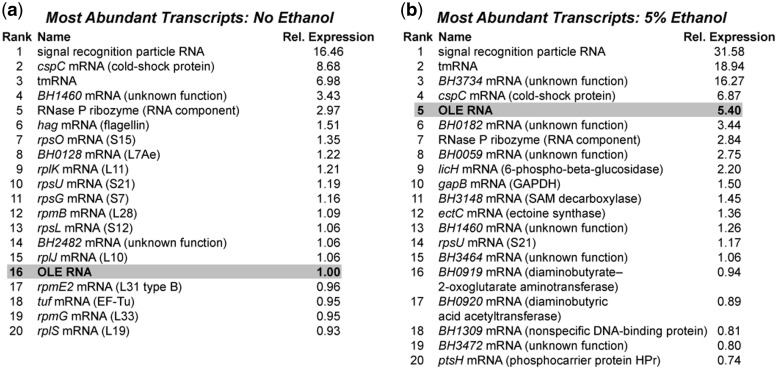
Figure 5.
Whole-transcriptome sequencing (RNA-seq) results for WT B. halodurans cells before (a) and after (b) 3 h exposure to 5% ethanol. The top 20 RNA transcripts in each condition are listed, as determined by RPKM score (reads per kilobase per million reads) (33), excluding rRNAs and tRNAs. All expression levels are relative (Rel. Expression) to OLE RNA in the absence of ethanol, and each list represents the average of RNA-seq data from two independent experiments.
Since the genes with significantly different expression levels between strains represent only 1.5% of the approximately 1600 genes that change expression during the experiment (data not shown), the absence of OLE and OAP appears to have only a minor effect on the transcriptional response of cells to ethanol stress. It is curious that the deletion of such a highly expressed RNA would have so little effect on the transcriptome, especially since under ethanol stress OLE RNA is the fifth most expressed transcript (excluding rRNAs and tRNAs) (Figure 5b). Despite the fact that the point of maximum OLE RNA induction occurs at around 3 h post-exposure to ethanol (Figure 3b), the cell’s transcriptional response to the absence of OLE RNA may occur at a different time. Alternatively, compensation may occur mostly at the post-transcriptional level, or perhaps the functions of OLE and OAP are at the end of a biochemical pathway that has little effect on other pathways.
CONCLUSIONS
Both the induction of OLE RNA in response to short-chain alcohols and the alcohol-sensitive phenotype of ole and oap deletion strains demonstrate that at least one function of OLE and OAP is to protect cells from alcohol-induced stress. Alcohols cause a wide variety of stresses on cells, but one major effect is permeabilization of membranes to ions and small molecules (43,44). In response to ethanol-mediated membrane stress, bacteria make changes in the protein and lipid composition of membranes (46). Since OLE and OAP localize to the cell membrane (19), this complex also may be important for fortifying the membrane against leakage. The large size and complexity of OLE RNA, which is similar to that of RNase P and self-splicing introns, implies that the OLE–OAP complex might function as a ribonucleoprotein particle with enzymatic activity. Other possibilities (e.g. gene regulation, signal transduction, etc.) are still possible, and therefore further experiments will be required to determine the biochemical function of the RNA and its protein partner.
It is unknown how accurately the conditions of our cell culture media approximate the natural environment of OLE-containing organisms. Little data exist on the presence of alcohols in the microenvironments of most organisms that carry OLE RNA (e.g. thermal vents, soda lakes and vertebrate gut), but, in many cases, it would be expected to be low. It is possible that local microenvironments could allow alcohols to build up to appreciable concentrations, or that natural stresses are simply less severe and cause a smaller but still significant fitness benefit for organisms with OLE. Even small fitness benefits can lead to gene fixation, and the extreme ethanol concentrations used in our assays may simply push the cells to the point where the fitness differences become large. Indeed, the 5–6% ethanol concentrations used in this study are at the very limit for viability of B. halodurans.
An interesting question is why cells would use a large and complex non-coding RNA to protect against alcohol stress instead of exclusively exploiting protein factors. Other widely conserved large non-coding RNAs in bacteria appear to be ancient RNAs that carry out fundamental biochemical tasks (e.g. RNAs in translation or RNAs processing RNA) that either are evolutionarily difficult to replace or are particularly well suited for RNA, such as specifically recognizing other nucleic acids. Given these precedents, it seems possible that the biochemical function of OLE may be ancient and fundamental, or involve other nucleic acids, or both.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5 and Supplementary Reference [47].
FUNDING
National Institutes of Health [GM 022778]; Howard Hughes Medical Institute. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Breaker laboratory and to Dr Nick Ornston for helpful discussions.
REFERENCES
- 1.Ellis JC, Brown JW. The RNase P family. RNA Biol. 2009;6:362–369. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- 2.Edgell DR, Chalamcharla VR, Belfort M. Learning to live together: Mutualism between self-splicing introns and their hosts. BMC Biol. 2011;9:22. doi: 10.1186/1741-7007-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De la Peña M, Garcia-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010;16:1943–1950. doi: 10.1261/rna.2130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 7.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genetics. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- 11.Puerta-Fernandez E, Barrick JE, Roth A, Breaker RR. Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl Acad. Sci. USA. 2006;103:19490–19495. doi: 10.1073/pnas.0607493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Tyson GW, Delong EF. Metatranscriptomics reveals unique microbial small RNAs in the oceans water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462:656–659. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stark BC, Kole R, Bowman EJ, Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl Acad. Sci. USA. 1977;75:3719–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 17.Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 18.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 19.Block KF, Puerta-Fernandez E, Wallace JG, Breaker RR. Association of OLE RNA with bacterial membranes via an RNA-protein interaction. Mol. Microbiol. 2011;79:21–34. doi: 10.1111/j.1365-2958.2010.07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol. Chem. 2009;390:775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- 21.Ataide SF, Schmitz N, Shen K, Ke A, Shan S, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu DKY, Kolodziejczak T. Inferring consensus structure from nucleic acid sequences. CABIOS. 1991;7:347–352. doi: 10.1093/bioinformatics/7.3.347. [DOI] [PubMed] [Google Scholar]
- 23.Lindgreen S, Gardner PP, Krogh A. Measuring covariation in RNA alignments: Physical realism improves information measures. Bioinformatics. 2006;22:2988–2995. doi: 10.1093/bioinformatics/btl514. [DOI] [PubMed] [Google Scholar]
- 24.Yao Z, Weinberg Z, Ruzzo WL. CMfinder: a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg Z, Breaker RR. R2R – software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crampton M, Berger E, Reid S, Louw M. The development of a flagellin surface display expression system in a moderate thermophile, Bacillus halodurans Alk36. Appl. Microbiol. Biotechnol. 2007;75:599–607. doi: 10.1007/s00253-007-0869-0. [DOI] [PubMed] [Google Scholar]
- 27.Kudo T, Hino M, Kitada M, Horikoshi K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J. Bacteriol. 1990;172:7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace JG, Breaker RR. Improved genetic transformation methods for the model alkaliphile Bacillus halodurans C-125. Lett. Appl. Microbiol. 2011;52:430–432. doi: 10.1111/j.1472-765X.2011.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas I, Gruss A, Ehrlich SD, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloor AE, Cranenburgh RM. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 2006;72:2520–2525. doi: 10.1128/AEM.72.4.2520-2525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Yu C, Li Y, Lam T, Yiu S, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 34.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb1508s73. Chapter 15, Unit 15.8. [DOI] [PubMed] [Google Scholar]
- 35.Brown T, Mackey K, Du T. Analysis of RNA by northern and slot blot hybridization. Curr. Protoc. Mol. Biol. 2004 doi: 10.1002/0471142727.mb0409s67. Chapter 4, Unit 4.9. [DOI] [PubMed] [Google Scholar]
- 36.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore PB. Structural motifs in RNA. Annu. Rev. Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 38.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA. 2010;16:57–69. doi: 10.1261/rna.1844010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko J, Altman S. OLE RNA, an RNA motif that is highly conserved in several extremophilic bacteria, is a substrate for and can be regulated by RNase P RNA. Proc. Natl Acad. Sci. USA. 2007;104:7815–7820. doi: 10.1073/pnas.0701715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutscher MP. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 42.Ferré-D'Amaré AR, Scott WG. Small self-cleaving ribozymes. Cold Spring Harb.Perspect. Biol. 2010;2:a003574. doi: 10.1101/cshperspect.a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingram LO. Ethanol tolerance in bacteria. Crit. Rev. Biotechnol. 1990;9:305–319. doi: 10.3109/07388558909036741. [DOI] [PubMed] [Google Scholar]
- 44.McKarns SC, Hansch C, Caldwell WS, Morgan WT, Moore SK, Doolittle DJ. Correlation between hydrophobicity of short-chain aliphatic alcohols and their ability to alter plasma membrane integrity. Fund. Appl. Toxicol. 1997;36:62–70. doi: 10.1006/faat.1996.2252. [DOI] [PubMed] [Google Scholar]
- 45.Nagalakshmi U, Waern K, Snyder M. RNA-seq: a method for comprehensive transcriptome analysis. Cur. Protoc. Mol. Biol. 2010;4(Suppl. 89):11.1–11.13. doi: 10.1002/0471142727.mb0411s89. [DOI] [PubMed] [Google Scholar]
- 46.Seydlova F, Halada P, Fišer R, Toman O, Ulrych A, Svobodová J. DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress. J. Appl. Microbiol. 2012;112:765–774. doi: 10.1111/j.1365-2672.2012.05238.x. [DOI] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.