Abstract
Telomeres are transcribed into non-coding TElomeric Repeat containing RNAs (TERRA). We have employed a transcriptionally inducible telomere to investigate how telomere transcription affects telomere function in Saccharomyces cerevisiae. We report that telomere shortening resulting from high levels of telomere transcription stems from a DNA replication-dependent loss of telomere tracts, which can occur independent of both telomerase inhibition and homologous recombination. We show that in order for telomere loss to occur, transcription must pass through the telomere tract itself producing a TERRA molecule. We demonstrate that increased telomere transcription of a single telomere leads to a premature cellular senescence in the absence of a telomere maintenance mechanism (telomerase and homology directed repair). Similar rapid senescence and telomere shortening are also seen in sir2Δ cells with compromised telomere maintenance, where TERRA levels are increased at natural telomeres. These data suggest that telomere transcription must be tightly controlled to prevent telomere loss and early onset senescence.
INTRODUCTION
Telomeres protect chromosome ends from being recognized as DNA damage and thus promote general genomic stability (1). When telomere function is compromised, either through telomere shortening or the loss of protective binding (capping) proteins, telomeres become subject to nucleolytic degradation and subsequently activate a DNA damage response (DDR) that may eventually lead to cellular senescence (1). Telomeres shorten upon each cell division in part due to the end-replication problem associated with semi-conservative DNA replication (2). The enzyme telomerase is able to re-elongate chromosome ends and thereby prevent telomere shortening. In some rare instances, homology-directed repair is used to maintain telomere length in the absence of telomerase, likely through a break-induced replication (BIR) mechanism (3). In the majority of human somatic cells, telomeres do however shorten due to insufficient telomerase expression. Most cancer cells, on the other hand, up-regulate telomerase activity to prevent cellular senescence due to telomere loss. Therefore, in the presence of functional cell-cycle checkpoints, telomere shortening can be thought of as a tumor suppressor mechanism, limiting the replicative capacity of potentially tumorigenic cells by promoting cellular senescence. In the budding yeast, Saccharomyces cerevisiae, telomerase is constitutively expressed and telomere length is maintained during cell-cycle progression. Deletion of any of the subunits of the telomerase holoenzyme results in telomere loss and the eventual onset of cellular senescence (4).
Yeast telomeres are silenced through Sir2/3/4-dependent heterochromatin formation via the deacetylation of subtelomeric histones, a conserved phenomenon referred to as the telomere position effect (TPE) (5–7). Despite their heterochromatic state, telomeres are transcribed by RNA polymerase II (RNAPII) resulting in the generation of telomeric repeat containing RNA (TERRA) (8,9). The transcription of telomeres into TERRA is a conserved process, as TERRA has been reported to exist in eukaryotic species ranging from yeast to man (10–14). TERRA transcription initiation occurs in the subtelomeric region, likely at all telomeres, and terminates within the telomeric tract, creating TERRA molecules that are very heterogeneous in length. Whether the heterogeneity stems from the 5′ or 3′ end is not clear, as there is evidence in support of both hypotheses (15,16). These data may also suggest that different telomeres terminate and/or initiate transcription in different manners. In S. cerevisiae, TERRA is negatively regulated by the 5′ to 3′ exonuclease, Rat1 (12). In rat1-1 mutants TERRA levels accumulate, at least in part, due to a decreased rate of RNA turnover (12,17). TERRA regulation is also highly dependent on the type of telomere that it stems from (17). Yeast telomeres are generally sub-divided into two classes: those that harbor a subtelomeric repetitive Y′ element together with an X element (Y′ telomeres) and those that only have an X element and do not harbor the Y′ element (X-only telomeres). TERRA is negatively regulated at Y′ telomeres primarily by the Rap1-binding proteins Rif1 and Rif2, with the Sir2/3/4 histone deacetylase complex playing only a minor repressive role (17). In contrast, at X-only telomeres both the Sir2/3/4 complex as well as the Rif1 and Rif2 proteins are important in promoting TERRA repression (17). Human TERRA levels are also regulated by the heterochromatic state of the telomeres as both inhibition of histone deacetylases via trichostatin A treatment, as well as inactivation of the DNA methyltransferases 1 and 3, result in the up-regulation of TERRA levels (16,18).
Although the regulation of telomeric transcription is becoming better understood, its function remains enigmatic. By placing a galactose inducible upstream activation sequence (UAS) directly in front of the telomeric repeats, a previous study has demonstrated that forced transcription through the telomere results in telomere shortening and loss of subtelomeric silencing specifically at the inducible telomere while other telomeres remain unaffected (19). Moreover, in rat1-1 cells where TERRA accumulates, all telomeres are shorter. Together, these results were interpreted to indicate that TERRA may be inhibiting telomerase and therefore responsible for the telomere shortening (12). Consistent with this interpretation, TERRA potently inhibits telomerase activity in a direct telomerase extension assay in vitro, and was found to co-precipitate with human telomerase (20). It has also been speculated that TERRA has an important role in establishing telomeric heterochromatin as siRNA directed against human TERRA results in the loss of heterochromatin marks like H3 K9me3 (21).
Here we have used a previously described inducible telomere transcription system (19) to understand the effects of deregulated telomere transcription on telomere length regulation in vivo. Our results demonstrate that strong telomere transcription does not exclusively result in telomere shortening through the inhibition of telomerase but rather leads to a replication-dependent loss of telomeric sequence. Furthermore, we show that in the absence of telomere maintenance, forced transcription of a single telomere is sufficient to cause premature cellular senescence. Interestingly, sir2Δ mutants also senesce prematurely when both telomerase and homology directed repair pathways are impaired, likely due to the increased rates of transcription through the telomeres resulting in accelerated telomere shortening. Together, these results suggest that telomere transcription, and hence TERRA production, must be tightly regulated in order to prevent unscheduled telomere loss events that induce cellular senescence in the absence of telomere maintenance. The Sir2/3/4-dependent establishment of silent chromatin in the subtelomeric region may play a critical role in this respect.
MATERIALS AND METHODS
Yeast strains, plasmids and media
Yeast strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. Standard growth conditions and strain manipulation procedures have been described previously (22).
RNA extraction, northern and dot blot
Yeast RNA extraction and northern blotting was performed as described previously (12). For dot blot experiments 0.5 μg and 1 μg RNA was diluted in 300 μl ddH2O, spotted onto a nylon membrane using a Bio-Rad dot blot apparatus and UV-cross-linked. Membrane pre-hybridization was performed in hybridization solution (50% formamide, 5× SSC, 5× Denhardt’s, 5 mM EDTA, 1% PIPES, 0.4 mg/ml filtered yeast RNA, 1% SDS) for 1 h at 47.5°C. DIG-labeled (Roche DIG oligonucleotide 3′-End labeling KIT, 2nd Generation) TERRA-/ARRET-or SRC1-probes (see Supplementary Table S3) were heated for 5 min at 95°C and inoculated overnight at 47.5°C in hybridization solution. For the URA3 probe 61.5°C was used. The blot was washed 2× 5 min in 2× SSC/0.1% SDS and 2× 20 min in 0.5× SSC/0.1% SDS at 47.5°C and rinsed in DIG–wash buffer (0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween-20, pH 7.5) at room temperature. After blocking (Roche blocking reagent in 1× maleic-acid buffer (0.10 M maleic acid, 0.15 M NaCl, pH 7.5) for 30 min at room temperature, the membrane was inoculated 1 h with AP-coupled alpha-DIG Fab (Roche; diluted 1:5000 in blocking solution), washed 4× 15 min in DIG–wash buffer and inoculated 1× 5 min in DIG–detection buffer (100 mM Tris–HCl, 100 mM NaCl, pH 9.5). The signal was detected by CDP-Star (Roche) and the use of LAS (Fujifilm).
Senescence curves
Spore colonies of dissected diploids were re-suspended in water and diluted in 5 ml YPD/YPGal to a final concentration of OD600=0.01. After 24 h of growth at 30°C absorption at 600 nm was measured. Cultures were re-diluted to an OD600=0.01 in 5 ml YPD/YPGal and inoculated for further 24 h at 30°C. Each day cell samples have been stored for genomic DNA extraction. Population doublings (PD) were calculated as log2 (OD60024h/0.01). All PD values refer to PD after the spore colony had grown to saturation on the dissection plate (about 25 generations). Graphs were created with the Prism5 (GraphPad) software package.
Genomic DNA extraction
Yeast genomic DNA was isolated following the Puregen Yeast/Bact. KIT B protocol (Qiagen).
Telomere PCR
One hundred nanograms of genomic DNA was diluted in 1× NEB buffer 4. The sample was denatured for 10 min at 96°C and rapidly cooled to 4°C. Tailing mix (4 U/µl terminal transferase (NEB), 1× NEB buffer 4, 1 mM dCTPs) was added to a final concentration of 10%. Cycling profile for C-tailing reaction: 37°C—30 min; 65°C—10 min; 96°C—5 min. After cooling to 65°C, 3× the volume preheated PCR-mix (1 µM oligo-dG reverse primer, 1 µM telomere-specific forward primer (see Supplementary Table S3), 0.267 mM dNTPs, 0.083 U/µl Phusion polymerase (NEB) in PCR buffer: 89.11 mM Tris–HCl (pH 8.8), 21.28 mM (NH4)2SO4, 6.65% glycerol, 0.0133% Tween-20) was added to the C-tailed-DNA. Cycling profile for PCR reaction: 95°C—3 min; 45 cycles: 95°C—30 sec, Tannealing—15 sec, 72°C—20 sec; 68°C—5 min; hold on 4°C. Samples were separated on a 1.8% agarose gel for 25–40 min at 100 V. Signals were detected by LAS (Fujifilm). Telomere lengths were analyzed in Multi-Gauge (Fujifilm) and graphically displayed using Prism5 (GraphPad).
Cell-cycle arrest and S-phase progression
Exponentially growing PTest2Δ cells in YPRaf medium were washed twice in H2O and re-suspended in YPRaf medium containing 158 μg/ml α-factor (GenScript). After 2.75 h a shmoo efficiency of ≥90% confirmed G1 arrest (sample 1: G1-arrested/uninduced). To induce transcription of the modified telomere 7L galactose was added to a final concentration of 2%. After 1 h of induction (sample 2: G1-arrested/1 h induced) cells were washed twice in H2O. Half of the cells were inoculated for additional 2.5 h in YPGal + α-factor (sample 3: G1-arrested/3.5 h induced) or YPGal + 20 μg/ml nocodazole (AppliChem; sample 4: G2-released/3.5 h induced). A shmoo/budding efficiency of >90% confirmed the maintenance of G1 arrest and arrest in metaphase, respectively. Induced transcription of the modified telomere 7L was confirmed by dot blotting.
RESULTS
Telomere repeat transcription results in cis telomere shortening
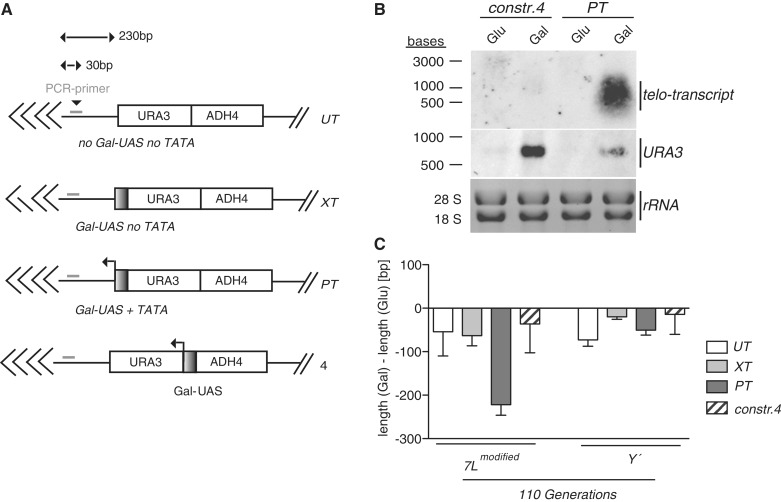
We modified telomere 7L in the BY4741 genetic background so that it harbored either the URA3 gene cassette (UT), a URA3 gene cassette followed by a telomere proximal Gal UAS where the TATA box had been mutated to prevent transcription into the telomere (XT) or a URA3 cassette with a downstream Gal UAS that transcribes into the telomere (PT), as previously described (19) (Figure 1A). Consistent with a previous report (19) we found that only telomere 7L in the PT strain was able to transcribe into the telomeric tract when induced with galactose (Supplementary Figure S1A). Moreover, forced transcription of telomere 7L in the PT strain specifically resulted in a shortening of the modified telomere 7L (Supplementary Figure S1B) as previously described (19). We were able to monitor the telomere shortening in galactose (induced) versus glucose (repressed) via telomere PCR (23) with a PCR primer that specifically bound to the modified telomere 7L (Figure 1A) and found that the shortening was consistent with that previously described by Southern blotting (19), thus qualifying telomere PCR as a legitimate method to follow telomere length in these strains. We detected a slight telomere shortening at XT and UT telomeres after approximately 110 generations in galactose compared to glucose (Supplementary Figure S1B) indicating that galactose alone does affect telomere length to an extent. Y' telomeres were not affected in the PT strain (Supplementary Figure S1B), beyond the effects of galactose, indicating that transcription through the telomere causes cis shortening.
Figure 1.
Telomere transcription causes telomere shortening in cis. (A) The constructs used to modify telomere 7L (see text). The values in bp refer to the distances between the PCR primer/Gal UAS and the start of the telomeric tract. (B) Northern blotting analysis revealed that while PT transcribes into the telomere in the presence of galactose, construct 4 transcription stops short of the telomere due to the presence of the natural URA3 terminator, therefore the production of telomeric transcripts only occurs in the PT strain. (C) Quantification of telomere length for construct 4 modified telomeres, comparing glucose vs. galactose after approximately 110 generations (n = 5). Values from UT, XT and PT have been included from Supplemental Figure 1B as comparison. For all above experiments error bars represent ± SEM.
We assessed whether transcription-induced telomere loss was dependent on transcription through the telomeric tract or whether high levels of transcription in the subtelomeric region (to within 50 bp of the telomeric repeat) may also lead to shortening. We constructed a plasmid (construct 4) where the Gal UAS lies upstream of the URA3 gene (Figure 1A, bottom) and integrated it at telomere 7L. In this context, galactose induction results in high transcription of URA3 (Figure 1B) however due to the presence of the natural URA3 terminator, transcription does not proceed into the telomeric tract as it does with the PT construct (Figure 1B) but rather terminates just upstream of the telomeric repeat sequences. As expected, galactose induction of the PT construct results in slight URA3 expression as a result of telomeric de-silencing (19) (Figure 1B). Galactose induction of a construct 4 containing strain for 110 generations (long-term induction) caused a shortening of telomere 7L similar to that observed in the UT/XT control strains (Figure 1C: UT, XT, PT values are used as comparison from Supplementary Figure S1B). Moreover, induction with galactose had the same effect on construct 4 modified telomere 7L as it did on control Y′ telomeres indicating that the slight shortening observed was non-specific and rather due to the effects of growth in galactose (Figure 1C). Together this suggests that high rates of transcription near the telomere are not sufficient to cause telomere shortening, but rather transcription must proceed through the telomeric tract and produce a telomere repeat containing TERRA-like molecule to negatively affect telomere length. We have also established that we are able to recapitulate the results previously published (19), in that telomere transcription results in cis telomere shortening, in an independent yeast genetic background.
Transcription-induced telomere shortening is additive with telomerase loss and leads to premature senescence
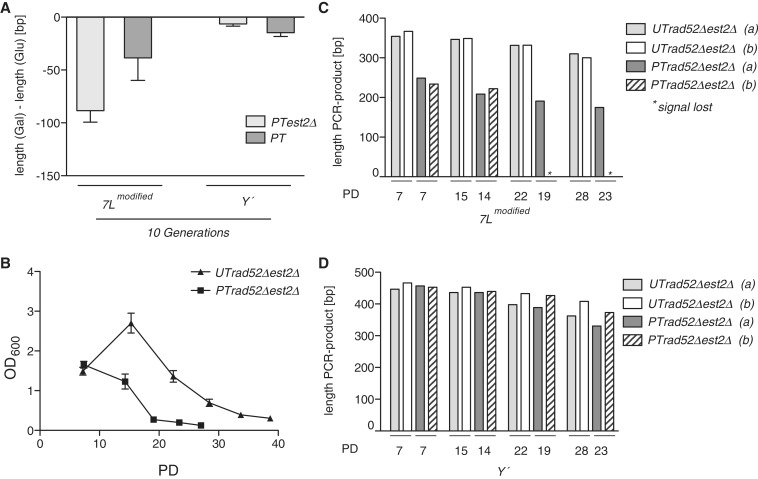
We created telomerase minus (est2Δ) cells that contained the PT construct at telomere 7L by dissecting heterozygous diploids onto YPD (glucose) media. After approximately 25 generations on YPD agar the cells were grown in either glucose or galactose liquid media for approximately 10 further generations (short-term, due to the lack of telomerase), and the telomere loss was compared to isogenic telomerase positive PT cells grown under the same conditions. Surprisingly, the PT est2Δ telomere 7L was still shorter in galactose than in glucose (Figure 2A, column 1) demonstrating that the induced transcription of telomere 7L resulted in telomere shortening in addition to the loss of telomerase. Furthermore, we found that the galactose-induced telomere shortening at the modified telomere 7L in the PT est2Δ strain was more pronounced than that of the PT strain expressing telomerase (Figure 2A compare columns 1 and 2). Together this indicates that transcription-induced telomere shortening is additive with the shortening seen in telomerase mutants and furthermore reveals that telomerase compensates, to an extent, for the loss of telomeric sequence at the transcribing telomere 7L. Importantly, there was no major difference at control Y′ telomeres between cultures grown in glucose or galactose in the same strains after approximately 10 cell divisions (short-term) (Figure 2A) ruling out the possibility that these effects are due to differences between glucose and galactose (∼10 bp difference).
Figure 2.
Transcription-induced shortening is additive with telomerase loss. (A) PT est2Δ cells grown in galactose had shorter telomeres than the same cells grown in glucose as well as PT EST2 cells grown in galactose for approximately 10 PD. Control Y′ telomeres from the same cells did not vary in length after 10 generations in glucose and galactose (n: PT est2Δ = 8, PT = 6). (B) Transcription of modified telomere 7L in PT cells (n = 9) results in an increased rate of senescence as compared to non-transcribing UT (n = 9) cells in an est2Δ rad52Δ genetic background. (C) Telomere PCR performed on modified telomere 7L during the senescence assay shown in (B) which revealed that this telomere was shorter in PT cells as compared to UT cells (two representative cultures (a and b) from each genotype—UT/PT est2Δ rad52Δ). (D) There was no apparent difference in terms of length at Y′ telomeres when comparing UT and PT strains. PD = population doublings following the formation of a colony after tetrad dissection.
In yeast, it has been demonstrated that a single short telomere is sufficient to trigger cellular senescence (24). Our above results (Figure 2A) predict that induced transcription of the modified telomere 7L will lead to a premature senescence phenotype. We sporulated EST2/est2Δ RAD52/rad52Δ heterozygous diploids that had been modified at telomere 7L with either UT (non-transcribing control telomere 7L) or PT (transcribing telomere 7L), in order to obtain UT est2Δ rad52Δ and PT est2Δ rad52Δ strains. RAD52 was deleted in order to rule out effects of HR that may mask senescence to different extents in the PT and UT strains, respectively. We subjected each haploid strain to a liquid senescence assay in galactose, whereby cells were diluted daily to a low density and allowed to re-grow for ∼24 h. Cell density, which was plotted as a function of PD diminished due to the absence of telomerase with ongoing cell divisions as the cells entered telomere-mediated senescence. Importantly, we noticed that cell density as determined via OD600 measurement accurately depicted cell survival in the BY4741 background, as the survival curves were indistinguishable from cell counting with a haemocytometer (unpublished observation). Therefore, we have represented our survival curves with OD600 values representing cell density on the Y-axis. Strikingly, the PT est2Δ rad52Δ strain senesced much faster than the UT est2Δ rad52Δ strain, with the only difference being that the former transcribes its modified telomere 7L (Figure 2B). Consistent with our previous observations, the PT telomere 7L, as measured by telomere PCR, was much shorter than the UT telomere 7L throughout the entire experiment (Figure 2C). Interestingly, there was already a significant difference (∼100 bp) in telomere length after the first seven PD indicating that telomere transcription in the absence of HR and telomerase can lead to rapid telomere loss events. The length of the Y′ telomeres was unchanged between the mutants harboring the UT and PT constructs (Figure 2D). Together these data suggest that controlled telomere transcription is important to maintain telomere length in the absence of telomerase and thereby prevents premature senescence.
Transcription-mediated shortening is DNA replication-dependent
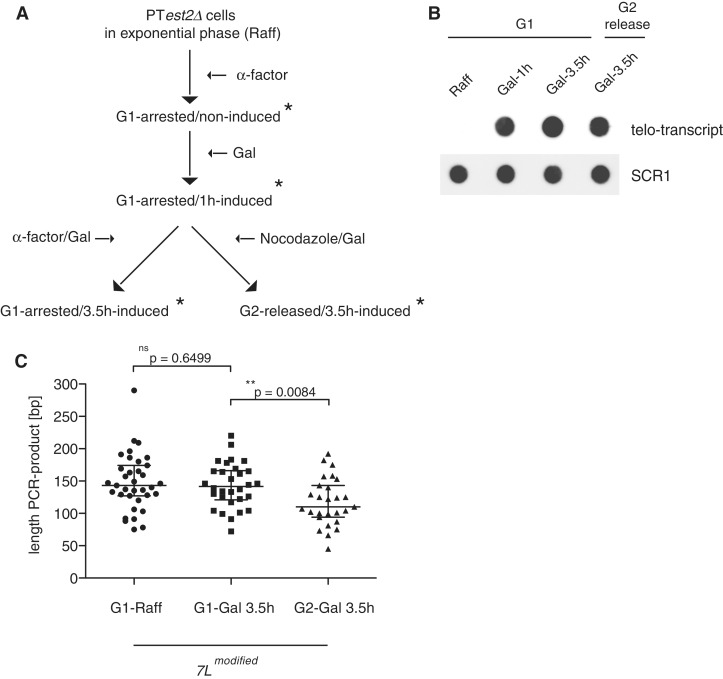
Since transcription-induced telomere loss can occur independent of both telomerase inhibition (Figure 2A and B), as well as recombination-mediated loss (Figure 2B) we explored whether the cell-cycle stage may play a role in transcription-induced telomere shortening. We induced telomere transcription at the modified telomere 7L in PT est2Δ cells in both α-factor (G1) arrested cells (≥90% shmooing) as well as cells that were released through one S-phase, and subsequently arrested in metaphase with nocodazole (>90% large budded cells). Samples for RNA and DNA extraction were taken at the indicated time points (asterisks) (Figure 3A). TERRA-like transcripts were not detected in non-induced G1 cells, but were strongly transcribed following galactose addition in both G1 arrested and released cells (Figure 3B). Due to the fact that small changes in telomere length in a single cell cycle would be difficult to detect via Southern blotting or telomere PCR we opted to PCR amplify telomeres and subsequently sequence cloned telomeres to obtain nucleotide resolution of telomere length. Interestingly, upon release into nocodazole the median telomere length of transcribing telomere 7L decreased in length by ∼25 bp compared to both the induced and non-induced cells that were held in G1 for the same amount of time (Figure 3C). This telomere loss in a single S phase is greater than what has been documented to occur as a result of the end-replication problem in yeast (2–4 bp/generation) (25). Consistently, we were unable to detect such a drastic shortening at a non-induced control telomere (1L) (Supplementary Figure S2A). Together these data indicate that deregulated telomere transcription in G1 arrested cells does not shorten telomeres, however, they are rapidly shortened upon passage through S phase.
Figure 3.
DNA replication is responsible for transcription-induced shortening. (A) Schematic of experimental design to assay for telomere length changes upon induction of telomere transcription in PT est2Δ synchronized cells. RNA and DNA samples were taken at time points indicated with asterisks. (B) Two micrograms RNA [from (A)] was spotted onto a nylon membrane and hybridized with labeled probes to recognize both TERRA and SCR1 as a loading control. (C) DNA was extracted and subjected to telomere PCRs specific for the induced telomere 7L, telomeres were cloned and sequenced and lengths were determined and plotted. A two-tailed unpaired Student’s t-test revealed a significant difference between telomere lengths of the transcribing G1 7 L telomeres (n = 30) and transcribing G2 telomeres (n = 27) (P = 0.0084). No difference between lengths in non-transcribing (n = 35) and transcribing G1 arrested cells (P=0.6499) was detected. Median telomere length is indicated with a line across the data points.
It has previously been demonstrated that unscheduled encounters between RNAPII and the DNA replication machinery can result in stalled replication forks and DNA damage (26). Indeed we speculate that stalled forks within the telomeric tract caused by TERRA transcription may eventually lead to telomere loss. In order to test whether replication fork stalling could account for telomere loss we measured bulk telomere length in telomerase mutants following hydroxyurea (HU) treatment, which leads to replication fork stalling due to decreased nucleotide pools. We grew identical est2Δ isolates (derived via tetrad dissection from heterozygous diploids) either in the presence or absence of sub-lethal concentrations (100 mM) of hydroxyurea for approximately 35 generations and measured length of the natural telomeres 7L and 6R (Supplementary Figure S2B). Similar to induced transcription of the modified telomere 7L, HU treatment leads to telomere shortening that is additive with that caused by the deletion of telomerase (Supplementary Figure S2B). Taken together these experiments suggest that TERRA transcription results in the most dramatic telomere shortening, when high rates of transcription at the telomere coincide with ongoing DNA replication, which could potentially result in replication fork stalling or collapse.
Sir2, a TERRA repressor, delays senescence
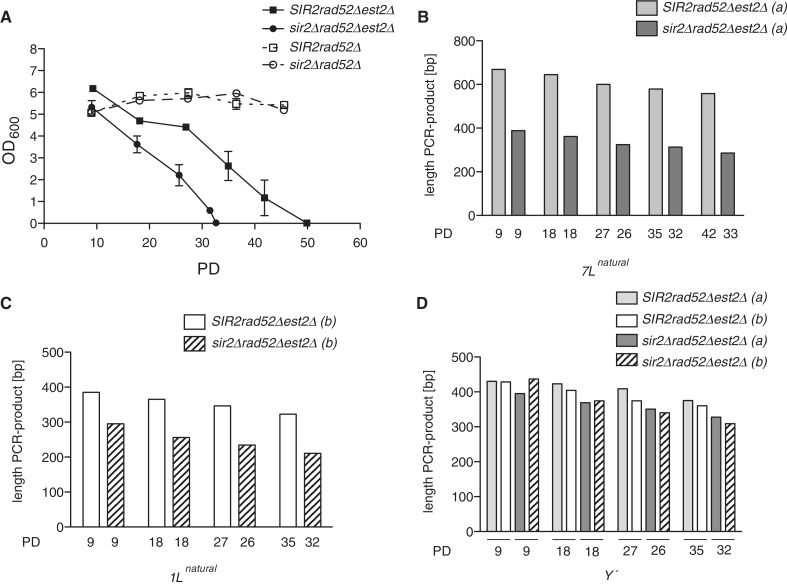
Recently, it has been shown that the Sir2/3/4 histone deacetylase complex is an important negative regulator of telomere transcription and hence TERRA production, especially at X-only telomeres (17). In order to test whether sir mutations also lead to premature senescence, we obtained est2Δ rad52Δ and est2Δ rad52Δ sirΔ haploid yeast strains via tetrad dissection of heterozygous diploids and subjected them to a senescence assay as described above. In agreement with the results obtained in the senescence experiment using the modified telomere 7L, we noticed that est2Δ rad52Δ sirΔ mutants senesced faster than est2Δ rad52Δ mutants (Figure 4A, Supplementary Figure S3A and data not shown). This effect was specifically due to the loss of telomerase and not to the loss of RAD52 as there was no detectable growth defect when comparing rad52Δ cells with rad52Δ sir2Δ mutants (Figure 4A). It has previously been demonstrated that sir2Δ mutants do not show an increased rate of senescence (27), but rather display an apparent delayed rate of senescence, which was explained by premature survivor formation (28). This apparent contradiction can be clarified by the fact that our survival assays were performed in a rad52Δ background, which differs from the above-mentioned studies. Indeed, when we performed a survival curve in the presence of RAD52, we also observed an increased rate of survivor formation (Supplementary Figure S3B), as previously described (28), suggesting that loss of SIR2 and increased TERRA transcription may promote telomere recombination to an extent (see discussion below).
Figure 4.
Telomere transcription leads to early senescence. (A) sir2Δ rad52Δ est2Δ (n = 5) cells lose viability more quickly than est2Δ rad52Δ cells (n = 4) when subjected to a senescence assay, whereas there is no difference in growth profiles when comparing rad52Δ (n = 6) cells with rad52Δ sir2Δ (n = 6), error bars represent ± SEM. PD values refer to PD after the spore colony had grown to saturation on the dissection plate (approximately 25 generations). (B) Telomere lengths were analyzed via telomere PCR at the indicated number of PD for the natural X-only telomere 7L (one representative culture from the curve in (A) (spore a) for the indicated genotypes). (C) Telomere length for natural telomere 1L was analyzed for a second independent culture from the curve in (A) (spore b) by telomere PCR. (D) Y′ telomeres were only slightly shorter upon loss of SIR2 (Y′ telomeres were analyzed for both spores (a) and (b) from panels B and C).
Moreover, in est2Δ rad52Δ sir2Δ cells, natural telomeres 7L and 1L (not modified) were shorter throughout the experiment compared to est2Δ rad52Δ cells (Figure 4B and C) and displayed the same rapid telomere loss that we noticed with the transcribing PT modified telomere (Figure 2C). Y′ telomeres, on the other hand were only slightly shorter in the sir2Δ background (Figure 4D). These results are consistent with the fact that X-only telomeres (including natural 7L and 1L) have highly increased TERRA levels in sir2Δ mutants, whereas TERRA levels from Y′ telomeres are only slightly elevated (17). Together these data lead to the notion that controlled telomere transcription through SIR mediated repression is important to maintain telomere length in the absence of telomerase and thereby prevents premature senescence.
DISCUSSION
Here we have shown that telomere transcription and the subsequent production of telomeric transcripts leads to telomere shortening and, in the absence of telomere maintenance, an increased rate of cellular senescence. Furthermore, we have demonstrated that, in vivo, transcription-induced telomere shortening (TTS) can occur independent of any TERRA-mediated inhibition of telomerase that may take place (Figure 2A). Moreover, TTS does not occur as a result of recombination-dependent loss as telomere shortening was just as prevalent when RAD52 was deleted rendering the cells recombination deficient (Figure 2C). Induced telomere transcription only leads to shortening in cells that have gone through S phase, indicating that the combination of DNA replication and telomere transcription converge to result in a telomere shortening event (Figure 3). Either increased TERRA production or the increased rate of telomere transcription has physiological consequences especially with respect to the rate of cellular senescence. We demonstrate here that over-production of TERRA at a single telomere is sufficient to result in premature senescence (Figure 2B). Consistently, sir2/3/4Δ mutants that have TERRA up-regulated at all X-only telomeres tested (17) also senesce prematurely (Figure 4, Supplementary Figure S3A and data not shown). It is important to note however that the GAL UAS-modified telomere 7L as well as the sir2/3/4Δ mutants produce TERRA far in excess to what can be detected at natural telomeres, and thus it is not yet clear to what extent naturally produced TERRA may influence senescence. It will be essential to find a means to down-regulate TERRA in order to address this important issue.
It has previously been shown that galactose-driven transcription of a single telomere leads to shortening of the induced telomere while other telomeres remain unaffected (19). Upon the discovery that telomeres produce TERRA, an exciting interpretation of this result was that TERRA competitively inhibits telomerase through base-pairing with telomerase’s RNA moiety, a possibility that was demonstrated to occur in an in vitro telomerase assay (20). If this were true in vivo, then the over-expression of TERRA at a single telomere should shorten the telomere to the same extent as the deletion of telomerase (i.e. be non-additive). This however is not the case and telomere shortening is additive when telomere transcription is induced in a telomerase mutant (Figure 2A). Therefore, deregulated transcription of telomeres does not result in telomere shortening exclusively through the inhibition of telomerase. Moreover, telomerase can compensate to an extent for the TTS. It will be interesting to determine whether or not high TERRA levels affect local telomerase activity in addition to its TTS effect.
Inducing telomere transcription in cells deleted for RAD52 still resulted in substantial TTS in cis (Figure 2C) indicating that recombination-dependent events are not accountable for the TTS effect. Previous studies have shown that unscheduled encounters between DNA replication forks and the RNAPII transcription machinery can lead do replication fork pausing and collapse (26,29–31) that potentially cause double strand breaks if not properly repaired. These observations led us to test whether transcription-induced telomere shortening is dependent on a particular cell-cycle stage. Indeed, in synchronized cells we found that the induction of TERRA only had an effect in terms of telomere loss in cells that had gone through S phase in comparison with cells that were held for the same amount of time in G1 (Figure 3C). This effect was specific to the induced transcribing telomere, as negative changes in telomere length were undetectable at telomere 1L (Supplementary Figure S2A). To verify that replication stress could lead to telomere shortening we employed HU treatment, which causes replication fork stalling (32) and upon prolonged exposure may also lead to double strand breaks (33). HU was able to recapitulate telomere shortening in addition to that caused by the absence of telomerase, much like TTS. Although TERRA transcription does occur in spite of the silenced nature of telomeres, we believe that transcriptional silencing at telomeres may have evolved, at least in part, in order to minimize encounters between transcription and ongoing DNA replication through the telomere. Interestingly, TERRA is down-regulated during S phase in human cells, likely to prevent encounters with the replication machinery (15).
Since continued telomere shortening leads to cellular senescence and TTS occurs independently of telomerase inhibition one prediction would be that high TERRA transcription in the absence of telomerase accelerates the onset of senescence. We made use of our transcriptionally inducible telomere to test this notion directly and found that transcription of a single telomere was sufficient to cause early senescence when compared to a strain that harbored a comparable telomere 7L modification, but was non-transcribing (Figure 2B). In agreement with TTS being the causative factor, we found that the modified telomere 7L was significantly shorter in the transcribing as compared to the non-transcribing strain (Figure 2C). Furthermore, the transcribing telomere underwent a dramatic shortening within the first seven PD of the senescence assay indicating that TTS results in rapid telomere shortening events. Such an event would, in theory, be consistent with replication fork stalling and subsequent breakage. The observed premature senescence is in agreement with the fact that a single shortened telomere is sufficient to cause rapid senescence (24). In this regard TERRA transcription must be tightly negatively regulated to ensure that TTS does not occur in telomerase negative cells. On the other hand, one could imagine a scenario whereby TERRA transcription could be used as a tumor suppressor-like response. Thereby dysfunctional telomeres (a source of genome instability) promote TERRA transcription and drive the cells into senescence rather than allowing the potential dangers associated with genomic instability to ensue. It will be of great interest to better understand how TERRA is regulated at both ‘normal’ and dysfunctional or senescent telomeres. Interestingly, Sir2 fails to properly localize to telomeres in old yeast cells as compared to young yeast cells (34), suggesting that TERRA levels may be increased in older cells. This would predict that replicatively aged telomerase negative cells, (like most human somatic cells) should senesce fast if they would start to divide, due to their increased rates of telomere transcription. Interestingly, SIRT1−/– mice have shortened telomeres, increased telomere dysfunction and more chromosomes with multiple telomeric signals (MTS) (35). MTS have been proposed to arise as a result of replication fork stalling and breakage at the telomere (36) suggesting that like sir2Δ yeast, SIRT1−/– mice may be experiencing TTS as a result of replication fork stalling in S phase.
We have shown that sir2/3/4Δ mutants, which have increased TERRA levels (17), display accelerated senescence. Consistently, recent reports have demonstrated that rif1Δ and rif2Δ mutants, which have more TERRA (17), also senesce extremely rapidly (37,38). Although sir2Δ est2Δ rad52Δ cells senesce prematurely due to TTS, it is interesting to note that sir2Δ single mutants do not have short telomeres, suggesting that either telomerase or homology-directed repair (or both) can compensate for the TTS effect. Consistent with the notion of recombination-mediated compensation, sir2Δ est2Δ mutants senesce slower than est2Δ mutants when RAD52 is present (Supplementary Figure S3B).
The RNAPII complex is a large protein complex that melts the DNA duplex and the RNA produced has the potential to base pair behind the polymerase to the ssDNA of the template strand, thereby creating an RNA–DNA hybrid or R-loop (26,39). Interestingly, R-loops are preferentially formed in regions of the genome where the non-coding strand is GC rich (39,40), making the telomeres and TERRA ideal candidates to form R-loops. In support of this, it has been demonstrated that human telomeric sequences lead to in vitro transcription blockage only when the G-rich RNA is transcribed (41). Moreover, R-loops have been demonstrated to be hyper-recombinogenic and a source of DNA replication fork stalling and double-strand breaks (42–44). If TERRA would form such a structure at telomeres, it could lead to replication fork instability and eventual telomere loss as a result of a double strand break. This suggests that TERRA-R-loops may be the source of TTS via induction of replicative stress. Consistent with this notion, we have shown that in order for telomeric transcription to result in telomere shortening, transcription must pass through the repetitive telomeric DNA (Figure 1C). High rates of transcription in the modified ‘subtelomeric’ region directly adjacent to the telomere fail to result in telomere shortening indicating that a repeat containing RNA (TERRA) molecule must be produced. At the telomere the TERRA template strand (the CA-rich strand) also provides the template for leading strand replication meaning that both transcription and replication proceed toward the chromosome end. It has been shown that R-loop-dependent replication fork pausing occurs independent of the direction of transcription (31,45). Interestingly, the over-expression of RNase H which degrades the RNA moiety of an RNA/DNA-hybrid, decreases TERRA levels and restores telomere length in rat1-1 mutants (12). It was proposed that Rat1 promotes telomere lengthening through a telomerase-dependent pathway, based on senescence and telomere length assays performed in rat1-1 est2Δ double mutants (12). Similar to the situation with sir2Δ cells, RAD52-dependent homologous recombination may be compensating for TTS in rat1-1 cells. It will be important to repeat such experiments with rat1-1 in a rad52Δ background setting.
A model outlining our results has been depicted in Figure 5. In summary we show that optimally, TERRA transcription should be kept to a minimum. It is not yet clear whether low (wild-type) levels of TERRA might be important to promote telomere capping as has been proposed in human cells (46). The repression of TERRA transcription likely facilitates DNA replication through the telomere and minimizes loss events at the chromosome ends (Figure 3). In certain situations (sir2Δ mutants, old cells) TERRA transcription increases and potentially through an RNA-DNA hybrid, results in DNA replication fork stalling and collapse (Figure 5). Another non-mutually exclusive explanation for TTS could be that telomere transcription allows nuclease-mediated telomere resection. This would also be consistent with an S phase-dependent shortening, as some nucleases such as Sae2 are only active in the presence of CDK1 activity (47).
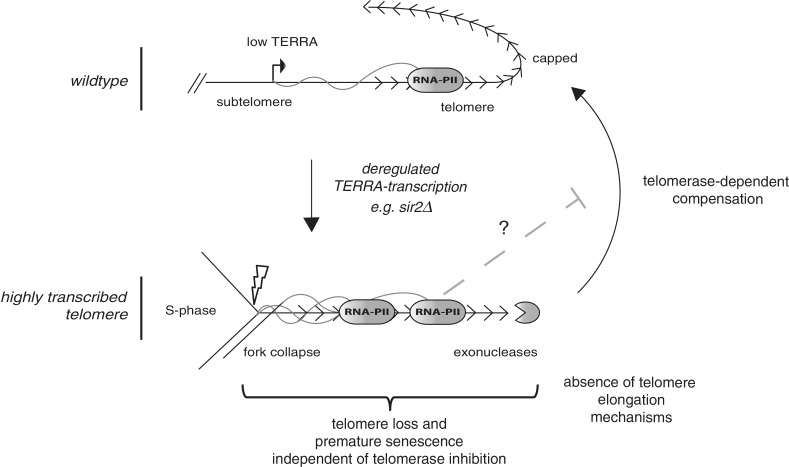
Figure 5.
Transcription-induced telomere shortening. Wild type telomeres transcribe TERRA at very low levels as the TERRA promoter (arrow) is negatively regulated by the Sir2/3/4 complex as well as the Rif1/Rif2 proteins, which allows for a fully functional and capped telomere (top diagram, arrow in subtelomere indicates approximate TERRA transcription start site). When TERRA transcription increases, either by galactose induction in the PT strain or in a sir2Δ, both the RNAPII occupancy at the telomere as well as the number of TERRA molecules increase. Telomere loss could result from a TERRA intermediate that leads to a replication fork stall and eventually a double strand break (lightning bolt). Increased transcription may also render telomeres more accessible to exonucleases, which would result in shortening. Telomerase is able to act at the transcribing telomere to compensate for the loss, although potentially at reduced capacity due to partial TERRA/transcription inhibition (dashed line). In the absence of telomerase critically short telomeres accumulate leading to an earlier activation of checkpoint-induced cellular senescence.
Interestingly, ICF (Immunodeficiency, Centromeric region instability, Facial anomalies) patient cells have increased TERRA levels and very short telomeres with frequent telomere free ends (48). Our results would suggest that the short telomeres in ICF cells may be a direct result of TTS. Indeed in ICF patients telomeres were shorter in both telomerase positive and negative cells when compared to parental controls (48). Finally, the over-expression of telomerase (hTERT) in ICF patient cells was able to re-elongate telomeres despite high TERRA levels (48), suggesting that in this scenario TERRA is not inhibiting telomerase activity in vivo. In support of this, a recent report has shown that 24 h induction of telomere transcription in HeLa cells does not lead to telomere shortening (49). It will be interesting to test the effects of long-term induction on telomere length in human cells as well as the effects of induced telomere transcription in a situation where telomerase has been inhibited. In conclusion, we believe that TERRA transcription must be kept tightly regulated in order to prevent accelerated senescence, especially in cells lacking telomerase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–3.
FUNDING
Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg, Netzwerk Alterns-Forschung; Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS) (to L.K.). Funding for open access charge: Netzwerk Alterns-Forschung, which is funded from the Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDMENTS
The authors thank Virginia Zakian for reagents. They also thank Sarah Luke-Glaser and Michael Chang for critical reading of the manuscript.
REFERENCES
- 1.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 3.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 4.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 5.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 6.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 8.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet. Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 10.Greenwood J, Cooper JP. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012;40:2956–2963. doi: 10.1093/nar/gkr1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 12.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 14.Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011;12:587–593. doi: 10.1038/embor.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 19.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl Acad. Sci. USA. 1994;91:12061–12065. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. London: Academic Press; 1991. [Google Scholar]
- 23.Forstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3' ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, Teixeira MT. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poveda AM, Le Clech M, Pasero P. Transcription and replication: Breaking the rules of the road causes genomic instability. Transcription. 2010;1:99–102. doi: 10.4161/trns.1.2.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak ML, Chavez A, Dang W, Berger SL, Ashok A, Guo X, Johnson FB. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2010;29:158–170. doi: 10.1038/emboj.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowell JE, Roughton AI, Lundblad V, Pillus L. Telomerase-independent proliferation is influenced by cell type in Saccharomyces cerevisiae. Genetics. 2003;164:909–921. doi: 10.1093/genetics/164.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–76. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in saccharomyces cerevisiae. Mol. Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 33.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang M, Dittmar JC, Rothstein R. Long telomeres are preferentially extended during recombination-mediated telomere maintenance. Nat. Struct. Mol. Biol. 2011;18:451–456. doi: 10.1038/nsmb.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H-Y, Lawless C, Addinall SG, Oexle S, Taschuk M, Wipat A, Wilkinson DJ, Lydall D. Genome-wide analysis to identify pathways affecting telomere-initiated senescence in budding yeast. G3. 2011;3:197–208. doi: 10.1534/g3.111.000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 40.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl Acad. Sci. USA. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 45.Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 49.Arora R, Brun CM, Azzalin CM. Transcription regulates telomere dynamics in human cancer cells. RNA. 2012;18:684–693. doi: 10.1261/rna.029587.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.