Abstract
In eukaryotic Okazaki fragment processing, the RNA primer is displaced into a single-stranded flap prior to removal. Evidence suggests that some flaps become long before they are cleaved, and that this cleavage involves the sequential action of two nucleases. Strand displacement characteristics of the polymerase show that a short gap precedes the flap during synthesis. Using biochemical techniques, binding and cleavage assays presented here indicate that when the flap is ∼30 nt long the nuclease Dna2 can bind with high affinity to the flap and downstream double strand and begin cleavage. When the polymerase idles or dissociates the Dna2 can reorient for additional contacts with the upstream primer region, allowing the nuclease to remain stably bound as the flap is further shortened. The DNA can then equilibrate to a double flap that can bind Dna2 and flap endonuclease (FEN1) simultaneously. When Dna2 shortens the flap even more, FEN1 can displace the Dna2 and cleave at the flap base to make a nick for ligation.
INTRODUCTION
In both prokaryotic and eukaryotic DNA replication, one strand of DNA, called lagging, is initially made in discontinuous segments termed Okazaki fragments. In humans, each fragment is initiated by an RNA primer 8–10 nt in length and extended 150–200 nt with DNA (1). The fragments need to be joined to make a continuous strand. This involves strand displacement synthesis from one fragment to raise the RNA primer region of the adjacent fragment into a flap. Genetic experiments and reconstitutions in vitro suggest that flap creation and removal occurs via two pathways. One employs the flap endonuclease (FEN1), which removes the flap containing the initiator RNA leaving a nick for ligation. If a flap escapes FEN1 to grow long enough to acquire the eukaryotic DNA-binding protein (RPA), nucleolytic cleavage will be blocked. If so, the flap is processed by the second pathway in which the nuclease/helicase Dna2 displaces the RPA. Dna2 then successively cleaves the flap until it is too short to bind RPA and becomes a substrate for FEN1 (2,3).
Biochemical analyses have demonstrated that both FEN1 (4–13) and Dna2 (14) employ a threading mechanism for binding and cleavage of the flap substrate. Recent results show that both nucleases recognize and bind to the base of the flap and then thread the 5′ end of the flap through the protein (15–17). Dna2 cleaves periodically during the threading (18,19), and FEN1 cleaves specifically at the base (4,20,21).
Previous analyses with purified proteins from Saccharomyces cerevisiae clarified how these proteins act in succession to process Okazaki fragments (8,22–25). On a long flap substrate, S. cerevisiae Dna2 (scDna2) was shown to displace bound scRPA in a reaction that did not require cleavage (26). scFEN1 was shown to displace the scDna2, so that it could have access to its cleavage site at the flap base (17).
In addition to the role it plays in Okazaki fragment processing, Dna2 functions in conjunction with 3′–5′ helicases and RPA to resect the 5′ end of DNA during double-strand break (DSB) repair and telomere maintenance (27). Human and yeast systems employ redundant pathways during these processes, one of which utilizes Dna2 (28–30). Once recruited to the DSB, a 3′–5′ helicase separates the two strands of the DNA creating a pseudo-Y structure consisting of a 3′ single-stranded (ss) DNA tail, a 5′ ssDNA tail and a double-stranded region. These single-stranded tails can then be coated by RPA. Similar to its role in the long flap Okazaki fragment pathway, Dna2 cleaves the 5′-ssDNA. Surprisingly, even though Dna2 possesses a 3′ nuclease activity, RPA prevents cleavage of the 3′-ssDNA (30,31). Dna2 interacts with the RPA1 (70 kDa) subunit of RPA (22). The N terminus of RPA1 interacts with the DNA with specific polarity showing strong binding to 5′ sequences and weak interactions with 3′ sequences (32). This polarity guides the orientation of Dna2 so that it has 5′ cleavage specificity. This ability of Dna2 to process the 5′ flap but not 3′ flap generates a 3′-tail structure necessary to initiate recombinational DNA repair and promote telomere maintenance (30). These activities on the pseudo-Y structure imply that Dna2 recognizes different features of substrate structure than FEN1 for initial binding, threading and cleavage.
Human Dna2 (hDna2) has been reported to have different functional characteristics than scDna2, specifically a much weaker DNA helicase function (33,34). This suggested that the sequential interactions of Dna2 and FEN1 differ in the human and yeast systems. In this report, we set out to explore binding and cleavage characteristics of the human nucleases to clarify their interactions during Okazaki fragment processing.
MATERIALS AND METHODS
Materials
Oligonucleotides were synthesized by Integrated DNA Technologies (IDT). DNA substrates were radiolabeled with [α-32P]dCTP (6000 Ci/mmol) purchased from PerkinElmer Life Sciences. The Klenow fragment of DNA polymerase I purchased from Roche Applied Science was used to 3′ radiolabel oligonucleotides. All other reagents were purchased from the best commercial sources available.
Oligonucleotides
Oligonucleotides are listed in Table 1. Upstream primers and downstream flap primers are listed in the 5′–3′ direction, and templates in the 3′–5′ direction, for annealing alignment. Oligonucleotides were 3′ radiolabeled, annealed and purified for binding and nuclease visualization as previously described (35). Double-flap and gap-flap experimental substrates containing a 3′ downstream radiolabeled primer, template and upstream primer were annealed in a ratio of 1:2:4, respectively. Substrates containing a 3′ radiolabeled downstream 5′-tail and template were annealed in a ratio of 1:2. The location of the radiolabel on the experimental substrate downstream flap primer is noted in each figure with an asterisk.
Table 1.
Oligonucleotide sequences
| Oligo | Length (nt) | Sequence |
|---|---|---|
| Upstream (5′–3′) | ||
| U1 | 26 | CGCCAGGGTTTTCCCAGTCACGACCA |
| U2 | 26 | CGACCGTGCCAGCCTAAATTTCAATT |
| U3 | 20 | CGACCGTGCCAGCCTAAATT |
| U4 | 23 | CGACCGTGCCAGCCTAAATTTCA |
| Downstream (5′–3′) | ||
| D1.2 | 25 | GTCGTTTTACAACGACGTGACTGGG |
| D1.5 | 28 | GCCGTCGTTTTACAACGACGTGACTGGG |
| D1.10 | 33 | CACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.15 | 38 | TAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.20 | 43 | TTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D1.30 | 53 | TTCACGCCTGTTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D2.15 | 41 | ATACGCGCTATAACCCCCGTCCACCCGACGCCACCTCCTGC |
| D2.30 | 56 | TTCTACTTCCAATTGATACGCGCTATAACCCCCGTCCACCCGACGCCACCTCCTGC |
| Template (3′–5′) | ||
| T1 | 49 | GCGGTCCCAAAAGGGTCAGTGCTGGGCAAAATGTTGCTGCACTGACCCG |
| T2 | 51 | GCTGGCACGGTCGGATTTAAAGTTAGGGCAGGTGGGCTGCGGTGGAGGACG |
| T3 | 26 | GGGCAGGTGGGCTGCGGTGGAGGACG |
Enzymes
Recombinant human FEN1 was cloned into the pET-FCH plasmid, over-expressed in Escherichia coli strain BL21 (DE3)/pLysS and purified using a C-terminal histidine tag as previously described (6). Recombinant human Dna2 was over-expressed using pFastBac HTc vector in baculovirus High Five cells and purified using a C-terminal FLAG tag as previously described (33).
Binding assays
FEN1 and Dna2 binding affinities with various experimental substrate configurations were measured using an electrophoretic mobility shift assay (EMSA). All binding assays were performed using a reaction buffer containing 50 mM Tris–HCl (pH 8.0), 2 mM DTT, 0.25 mg/ml bovine serum albumin (BSA), 30 mM NaCl, 20 μM EDTA and 5% glycerol. For non-competition experiments, reactions were initiated by incubating either Dna2 for 10 min or FEN1 for 5 min with the experimental substrate on ice. After the pre-incubation period, the reactions were placed at 37°C for an additional 10 min. For competition reactions, Dna2 was incubated with the experimental substrate for 5 min on ice followed by the addition of FEN1 for 5 more min on ice. Reactions were further incubated at 37°C for an additional 10 min. Reactions were then loaded onto a 6% native polyacrylamide gel in 1× TBE and subjected to electrophoresis at 200 V for 45 min.
Nuclease assays
FEN1 and Dna2 nuclease activities were measured by incubating the enzymes with various experimental substrates at 37°C for 20 min. Reactions were performed in a reaction buffer containing 50 mM Tris–HCl (pH 8.0), 2 mM DTT, 0.25 mg/ml BSA, 30 mM NaCl, 4 mM MgCl2, 2 mM ATP and 5% glycerol. Reactions were terminated by the addition of 20 μl of 2× termination dye and heating to 95°C for 5 min. Termination dye contained 90% formamide (v/v), 10 mM EDTA, as well as xylene cyanole and bromophenol blue markers. Nuclease activity was measured using 15% polyacrylamide, 7 M urea denaturing gels in 1× TBE that were subjected to electrophoresis for 60 min at 80 W.
Experimental result analysis
For both native and denaturing assays, gel drying and exposure were conducted as previously described (35). Autoradiography images were generated using Storm hardware (GE Healthcare) with Molecular Dynamics PhosphorImager technology and quantified utilizing Molecular Dynamics ImageQuant software version 5.2. All reactions were performed at least in triplicate and representative gels are shown. The percent DNA substrate bound by either FEN1 or Dna2 is defined as [bound/(bound + unbound)] × 100. The percent DNA substrate cleaved by either FEN1 or Dna2 is defined as [cleaved/(cleaved + uncleaved)] × 100. Error bars indicate ±standard deviation. The relative DNA substrate cleaved by Dna2 (Figure 3E) is normalized to the amount of 30 nt double-flap DNA substrate cleaved by 0.63 nM Dna2. The relative DNA substrate bound by Dna2 (Figure 5D) is normalized to the amount of DNA substrate bound by 25 nM Dna2 in the absence of FEN1.
Figure 3.
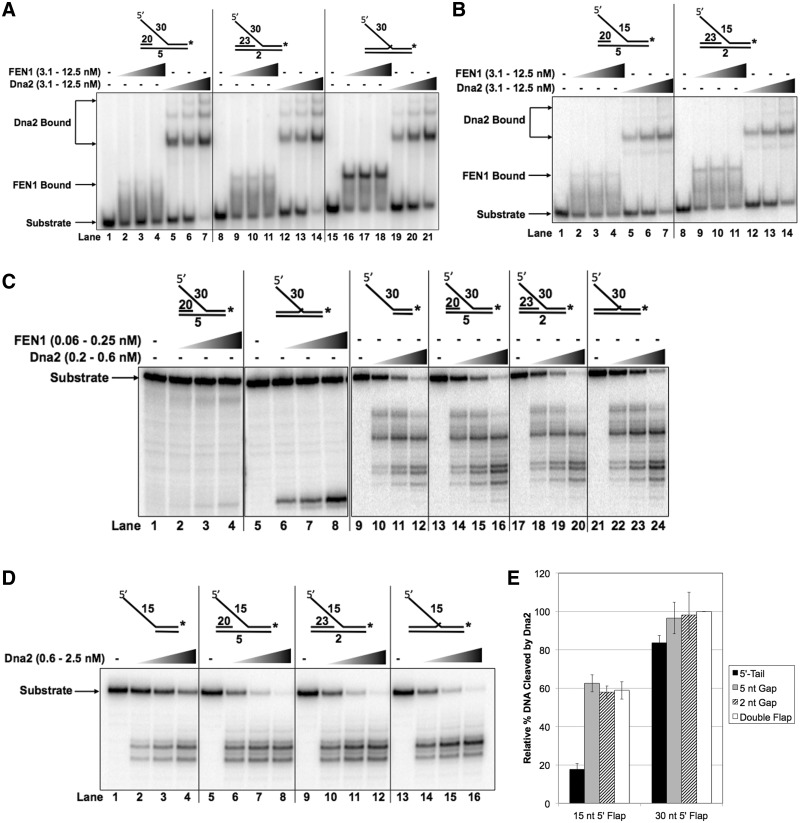
Dna2 binding is improved by the upstream region for short flap structures while stable FEN1 binding requires the full upstream region. (A) shows Dna2 or FEN1 binding to the 5 nt gap-flap (U3:T3:D2.30), 2 nt gap-flap (U4:T3:D2.30) and double-flap (U2:T3:D2.30) substrates with 30 nt 5′ flaps. (B) shows Dna2 or FEN1 binding to the 5 nt gap-flap (U3:T3:D2.15) and 2 nt gap-flap (U4:T3:D2.15) structures with 15 nt 5′ flaps. Binding was measured in (A) and (B) by EMSA using increasing Dna2 and FEN1 concentrations (3.1, 6.25 or 12.5 nM). (C) shows FEN1 cleavage of the 5 nt gap-flap (U3:T3:D2.30) and double-flap (U2:T3:D2.30) structures (lanes 1–8) and Dna2 cleavage of the 5′-tail (T4:D2.30), 5 nt gap-flap (U3:T3:D2.30), 2 nt gap-flap (U4:T3:D2.30) and double-flap (U2:T3:D2.30) structures (lanes 9–24) with 30 nt 5′ flaps. (D) shows Dna2 cleavage of the 5′-tail (T4:D2.15), 5 nt gap-flap (U3:T3:D2.15), 2 nt gap-flap (U4:T3:D2.15) and double-flap (U2:T3:D2.15) structures with 15 nt 5′ flaps. Nuclease activity was measured by denaturing gel electrophoresis. (E) shows the graphical quantitation of the relative cleavage of 0.63 nM Dna2 from (C) and (D) as defined in the ‘Materials and Methods’. In (A) and (B) the position of the substrate alone, Dna2–substrate complex and FEN1–substrate complex are indicated to the left of the figure. In (C) and (D) the position of the radiolabeled oligonucleotide is indicated to the left of the figure. The experimental substrate configurations are shown above figures (A)–(D).
Figure 5.
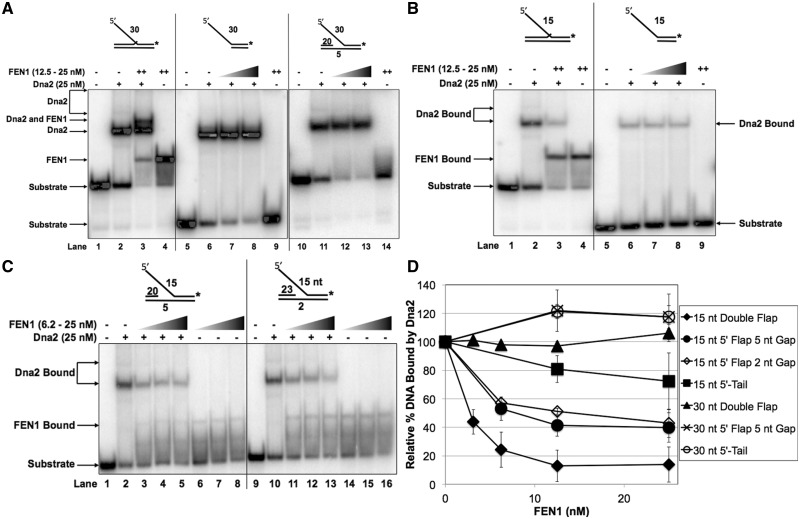
Binding affinity from the upstream DNA as well as a short 5′ flap is necessary for Dna2 displacement by FEN1. Dna2 (25 nM) was pre-incubated with the experimental substrate prior to the addition of increasing concentrations of FEN1 (6.25, 12.5 or 25 nM as indicated in the figure headings). (A) shows Dna2 and FEN1 binding competition for the 30 nt double-flap substrate (U2:T3:D2.30), the 5′-tail substrate (T4:D2.30) and the 5 nt gap-flap (U3:T3:D2.30). (B) shows Dna2 and FEN1 binding competition for the 15 nt double-flap substrate (U2:T3:D2.15) and the 5′-tail (T4:D2.15). (C) shows Dna2 and FEN1 binding competition for the 5 nt gap-flap (U3:T3:D2.15) and 2 nt gap-flap (U4:T3:D2.15) having a 15 nt 5′ flap. (D) shows the graphical quantitation of the relative percent Dna2 bound to the substrate in the absence of FEN1 or with increasing concentrations of FEN1 as defined in the ‘Materials and Methods’ section. In (A)–(C) the position of the Dna2–substrate complex and FEN1–substrate complex are indicated to the left and/or right of the figure. In (A), the FEN1–Dna2–substrate complex is indicated to the left of the figure. ‘++’ represents the maximum concentration of FEN1 used.
RESULTS
Human Dna2 preferentially binds and processes long flap Okazaki fragment substrates
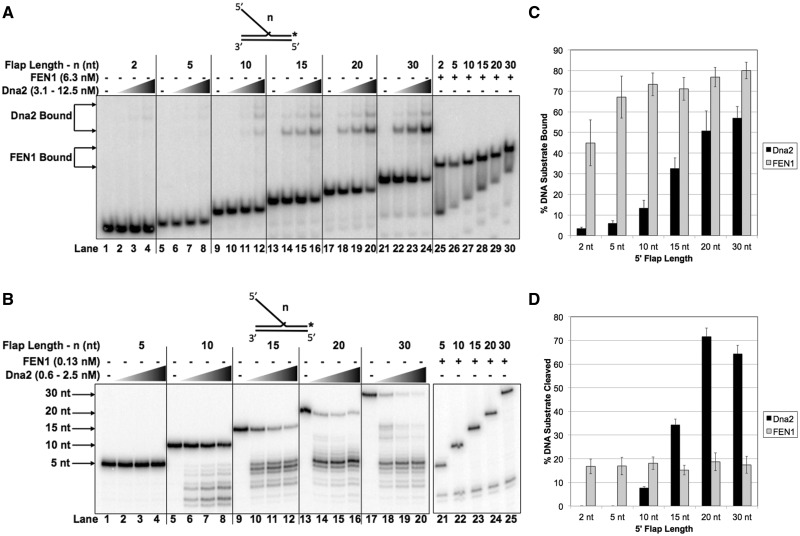
Prior work showed that the cleavage activity of scDna2 on 5′ flap substrates improved as the length of the 5′ flap increased (23). In order to correlate the binding properties of Dna2 with the cleavage function, we first measured the binding characteristics of hDna2 on substrates containing a 1 nt 3′ overhang and varied lengths of 5′ flaps, called a double-flap substrate. Using an EMSA, we compared the binding of hDna2 to substrates with increasing 5′ flap lengths (Figure 1A). These experiments were done in the absence of Mg2+ and ATP to prevent nuclease and helicase activities, respectively. hDna2 showed negligible binding to 2 and 5 nt 5′ flap substrates (lanes 1–8) but improved binding affinity as the flap length increased from 10 to 30 nt (lanes 9–24). In addition to the predominant hDna2 band, additional super-shifted bands, presumed to represent multiple proteins bound likely resulting from protein–protein interactions, were visualized on all of the double-flap substrates in a concentration-dependent fashion. Consistent with our prior results (15), hFEN1 bound substrates containing 5′ flaps from 5 to 30 nt with high affinity (lanes 26–30) but exhibited a slight decrease in binding for the 2 nt flap (lane 25). Quantitation of the percent DNA substrate bound by 12.5 nM hDna2 or 6.3 nM hFEN1 is shown in Figure 1C.
Figure 1.
Dna2 binding correlates to cleavage on Okazaki fragment double-flap structures. Dna2 and FEN1 binding and nuclease activity of double-flap substrates having a 5′ flap of 2 nt (U1:T1:D1.2), 5 nt (U1:T1:D1.5), 10 nt (U1:T1:D1.10), 15 nt (U1:T1:D1.15), 20 nt (U1:T1:D1.20) or 30 nt (U1:T1:D1.30) were measured by EMSA and denaturing gel electrophoresis, respectively, as described in the ‘Material and Methods’ section. (A) shows Dna2 (3.1, 6.25 and 12.5 nM) and FEN1 (6.25 nM) binding to the double-flap substrates. (B) shows Dna2 (0.6, 1.25 and 2.5 nM) and FEN1 (0.13 nM) cleavage of the double-flap substrates. (C) shows the graphical quantitation of 12.5 nM Dna2 and 6.3 nM FEN1 from (A) and (D) shows the quantitation of 0.63 nM Dna2 and 0.13 nM FEN1 from (B). In (A), the position of the Dna2–substrate complex and FEN1–substrate complex are indicated to the left of the figure. In (B), the position of the radiolabeled oligonucleotide is indicated to the left of the figure. The experimental substrate configuration is shown above each figure where ‘n’ represents the length of the 5′ flap.
In contrast with the scDna2 cleavage specificity, it was shown that scFEN1 cleaves Okazaki fragments of varying 5′ flap lengths with similar efficiency (4). We now asked whether the human FEN1 and Dna2 would cleave double-flap substrates with similar characteristics as the yeast homologs. We incubated hFEN1 and hDna2 with the varying 5′ flap Okazaki fragment substrates in the presence of Mg2+ and ATP. Consistent with scDna2, hDna2 nuclease activity was not detected on the 2 nt (data not shown) or 5 nt flaps (Figure 1B, lanes 1–4). hDna2 cleavage progressively increased as the 5′ flap was lengthened from 10 to 20 nt (lanes 5–8, 9–12 and 13–16, respectively). hDna2 cleaved the 20 and 30 nt flaps with similar efficiency (lanes 13–16 and 17–20, respectively). Consistent with the previous reports about scFEN1, hFEN1 cleaved substrates with 5′ flaps of 2, 5, 10, 15, 20 and 30 nt similarly (lanes 21–25). Quantitation of the percent DNA substrate cleaved by 0.63 nM hDna2 or 0.13 nM hFEN1 is shown in Figure 1D.
Dna2 demonstrates reduced binding for short 5′ tail structures relative to double flaps
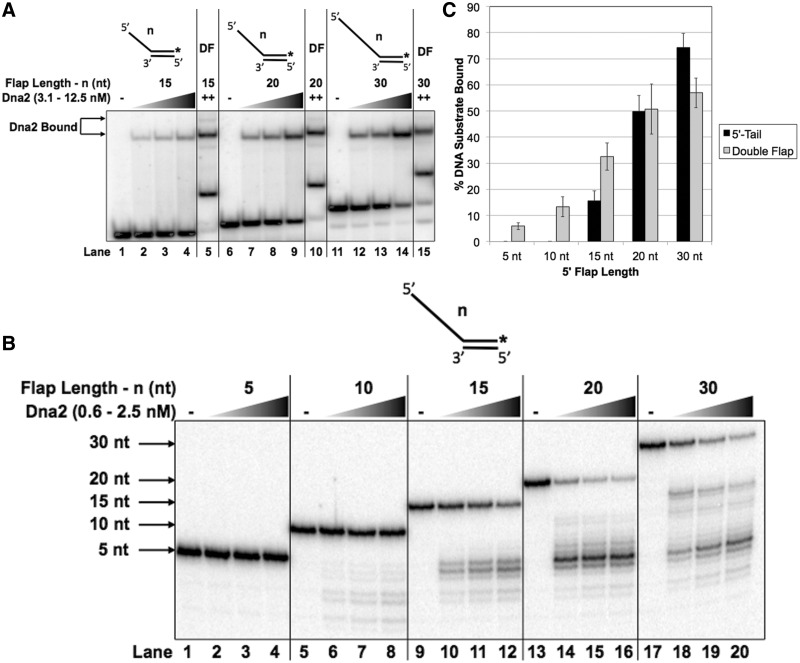
Since hDna2 showed reduced binding affinity to substrates containing intermediate length 5′ flaps (15 nt), we assessed the minimal 5′ flap length requirement for stable hDna2 binding. We measured hDna2 binding to substrates with various 5′ flap lengths in the absence of the upstream double-stranded (ds) DNA, referred to here as a 5′-tail structure (Figure 2A). Significantly, hDna2 demonstrated lower binding affinity for the 15 nt 5′-tail structure relative to the 15 nt double-flap structure (compare lanes 4 and 5). This suggests a role for the upstream DNA region in Dna2 binding stability. Conversely, hDna2 bound the 20 nt 5′-tail structure with similar affinity to the 20 nt double flap (compare lanes 9 and 10) and bound the 30 nt 5′-tail with slightly higher affinity than the 30 nt double-flap substrate (compare lanes 14 and 15). Quantitation of the percent DNA 5′-tail substrate versus the double-flap substrates bound by 12.5 nM hDna2 is shown in Figure 2C. At the maximum concentration used, stable hDna2 binding was visualized neither for the 5 nor 10 nt 5′-tail structures (data not shown). hFEN1 binding was not visualized for any of the 5′-tail structures (Figure 5A, lane 9 and Figure 5B lane 9, and data not shown).
Figure 2.
Dna2 demonstrates reduced binding for intermediate 5′-tail structures relative to double flaps. (A) shows Dna2 binding to the 5′-tail substrates having a 5′ flap of 15 nt (T2:D1.15), 20 nt (T2:D1.20) or 30 nt (T2:D1.30). The double-flap substrates have a 5′ flap of 15 nt (U1:T1:D1.15), 20 nt (U1:T1:D1.20) or 30 nt (U1:T1:D1.30). Binding was measured by EMSA using increasing Dna2 concentrations (3.1, 6.25 or 12.5 nM). (B) shows Dna2 cleavage of 5′-tail substrates having a 5′ flap of 5 nt (T2:D1.5), 10 nt (T2:D1.10), 15 nt (T2:D1.15), 20 nt (T2:D1.20) or 30 nt (T2:D1.30). Cleavage was measured by denaturing gel electrophoresis using increasing concentrations of Dna2 (0.6, 1.25 or 2.5 nM). (C) shows the graphical quantitation of 12.5 nM Dna2 from (A). In (A), the position of the Dna2–substrate complex is indicated to the left of the figure. ‘++’ represents the maximum concentration of Dna2 used. ‘DF’ represents the lane containing the double-flap structure. In (B), the position of the radiolabeled oligonucleotide is indicated to the left of the figure. The experimental 5′-tail substrate configuration is shown above each figure where ‘n’ represents the length of the 5′ flap.
We then asked whether this relative decrease in binding affinity for the short 5′-tail substrate would translate to a comparable decrease in nuclease activity. We measured the hDna2 nuclease activity on the 5′-tail substrate with flaps ranging from 5 to 30 nt (Figure 2B). hDna2 cleavage was not visualized on the 5 nt 5′-tail substrate (lanes 1–4), while cleavage efficiency improved with increasing flap length for the 10, 15, 20 and 30 nt 5′-tail structures (lanes 5–8, 9–12, 13–16 and 17–20, respectively). Notably, while the cleavage efficiency trend was similar, hDna2 cleavage was reduced on short 5′-tail structures relative to short double-flap structures.
On short flap substrates, an upstream region improves Dna2 binding while stable FEN1 binding requires the full double-flap structure
In addition to its nuclease function, Dna2 also possesses 5′–3′ helicase activity, although the reported helicase function of hDna2 is much weaker compared to scDna2 (33,34). Nevertheless, Dna2 may have evolved to recognize substrates with a gap between the upstream synthesis primer and downstream 5′ flap. In order to assess the influence of the 3′ end of the upstream primer on Dna2 binding affinity, we measured hDna2 binding to substrates with gap-flap structures, i.e. having an upstream primer separated by a gap from the flap base (Figure 3A). We were surprised to find that compared to the 30 nt double-flap structure (lanes 19–21), hDna2 demonstrated slightly higher binding affinity for the gap-flap structures containing either a 5 nt (lanes 5–7) or 2 nt single-stranded gap (lanes 12–14). Conversely, hFEN1 exhibited a substantial loss in binding affinity as the upstream primer single-stranded gap was increased from 2 nt (lanes 9–11) to 5 nt (lanes 2–4) relative to the double-flap structure (lanes 16–18). The amount of stably bound hFEN1 was decreased as the single-stranded gap size was increased, as evident from the smearing between the hFEN1–DNA complex band and the free substrate band indicative of FEN1 dissociation during electrophoresis.
To further explore the combined impact of flap length and upstream DNA structure on binding affinities of hFEN1 and hDna2, we measured affinity for 15 nt 5′ gap-flap structures (Figure 3B) containing a 5 nt (lanes 5–7) or 2 nt (lanes 12–14) single-stranded gap. Consistent with results using the double-flap and 5′-tail structures, hDna2-binding affinity for the 15 nt flap was reduced ∼2-fold with respect to the 30 nt gap-flap structures. hFEN1 binding affinity was generally low for the 30 and 15 nt gap-flap structures with 5 and 2 nt gaps. Figure 3A showing the 30 nt gap-flap structures with 5 nt (lanes 2–4) and 2 nt (lanes 9–11) gaps can be compared with Figure 3B showing the 15 nt gap-flap structures with 5 nt (lanes 2–4) and 2 nt (lanes 9–11) gaps. Irrespective of whether the flap length was 15 or 30 nt, binding affinity to the double-flap configuration was much higher. For the 15 nt gap and double-flap substrates, compare Figure 3B (lane 3 and 10) to Figure 1A (lane 28), respectively.
We next assessed the impact of the upstream single-stranded gap on the cleavage activity of the two nucleases. Prior results have shown that the 3′ flap of the upstream primer significantly influences scFEN1 nuclease activity while scDna2 cleavage remains unchanged (4,20,36). hFEN1 cleavage was greatly reduced with the removal of the upstream 3′ flap relative to the double-flap structure (Figure 3C, compare lanes 1–4 with lanes 5–8). hDna2 cleavage efficiency was similar for 30 nt 5′-tail (lanes 9–12), 5 nt gap-flap (lanes 13–16), 2 nt gap-flap (lanes 17–20) and the double-flap (lanes 21–24) structures. On the long flap structures, both hFEN1 and hDna2 cleavage efficiencies were consistent with their yeast homologs.
The minimal binding characteristics changed for hDna2 on substrates with short flaps (Figure 2A). Therefore, we examined the impact of short flaps on hDna2 cleavage. hDna2 cleavage of the 15 nt 5′-tail structure (Figure 3D, lanes 1–4) was significantly less efficient than the cleavage of the 5 nt gap-flap (lanes 5–8), 2 nt gap-flap (lanes 9–12) and double-flap structures (lanes 13–16). Both the binding and cleavage results suggest that hDna2 uses upstream DNA to stabilize binding and improve cleavage on short flap structures. Quantitation of the relative percent of 5′-tail, 5 nt gap-flap, 2 nt gap-flap and double-flap DNA substrates cleaved by 0.63 nM hDna2 normalized to the 30 nt double-flap structure is shown in Figure 3E labeled ‘5′-Tail’, ‘5 nt Gap’, ‘2 nt Gap’ and ‘Double Flap’, respectively.
Displacement of human Dna2 by human FEN1 requires a short 5′ flap
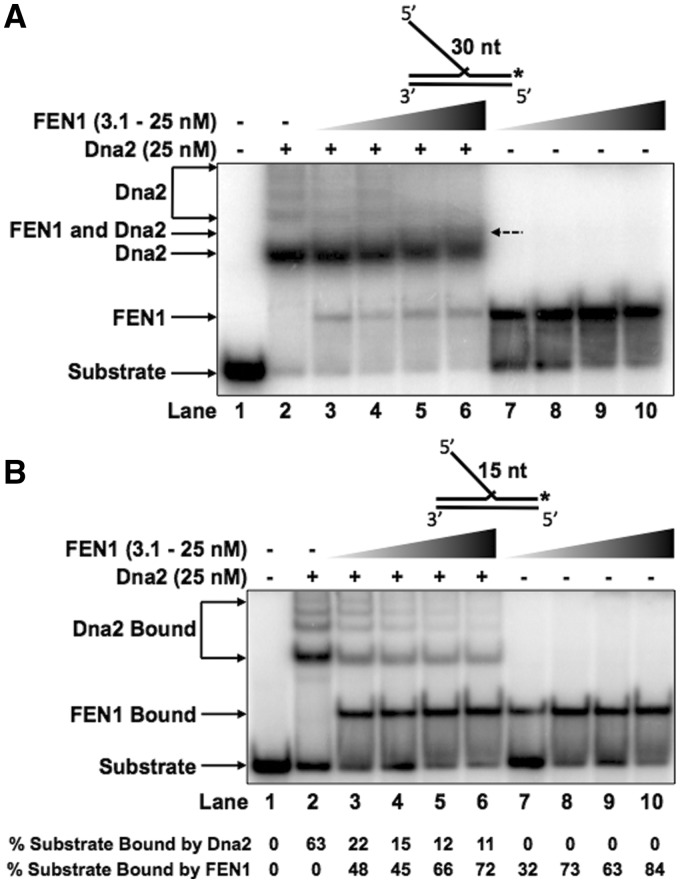
It was previously reported that, after cleaving, scDna2 remains unproductively bound to the base of the flap, requiring displacement by scFEN1 (17,25). Though both FEN1 and Dna2 preferentially bind to the flap base, their modes of binding appear to need different attributes of the 5′ and 3′ flaps, as well as the DNA surrounding the flap base junction. Based on substrate characteristics preferred by the two nucleases, we used DNA-binding competition assays to determine how these proteins acted sequentially on 5′ flap substrates (Figure 4A). To measure hDna2 displacement by hFEN1, we pre-bound hDna2 to a 30 nt double-flap structure prior to the addition of increasing concentrations of competing hFEN1, as described in the ‘Materials and Methods’ section. hDna2 alone bound to the substrate appeared as the predominant hDna2 bound band (Labeled ‘Dna2’) and lesser super-shifted multimeric bands (lane 2). At the lowest concentration of hFEN1 used, the super-shifted hDna2 bands were reduced in intensity, implying multimeric hDna2 displacement from the substrate while the predominant hDna2–DNA complex was not dissociated (lane 3). In addition, a new band appeared corresponding to a hFEN1–DNA complex (Labeled ‘FEN1’). As the hFEN1 concentration was increased (lanes 4–6), a large fraction of the hDna2–DNA alone complex remained visible and a new band appeared retarded more than the predominant hDna2–DNA band but less than the super-shifted hDna2 bands. This new super-shifted band (Labeled ‘FEN1 and Dna2’) was visualized in neither the substrate alone (lane 1), hDna2 alone (lane 2), nor hFEN1 alone titration reactions (lanes 7–10) suggesting that hFEN1 and hDna2 can bind concurrently. Quantitation of the percent DNA 30 nt double-flap substrate bound by hDna2 in the presence of competing hFEN1 normalized to the percent DNA substrate bound by hDna2 alone is shown in Figure 5D indicated as ‘30 nt Double Flap’.
Figure 4.
FEN1 preferentially displaces Dna2 from intermediate length double-flap structures. Dna2 (25 nM) was pre-incubated with the experimental substrate prior to the addition of increasing concentrations of FEN1 (3.1, 6.25, 12.5 or 25 nM in lanes 3–6). Lane 1 shows the substrate alone, lane 2 shows Dna2 bound without FEN1 and lanes 7–10 show FEN1 alone bound to the substrate at the same concentrations as in 3–6. (A) shows Dna2 and FEN1 binding competition for the 30 nt double-flap substrate (U2:T3:D2.30). (B) shows Dna2 and FEN1 binding competition for the 15 nt double-flap substrate (U2:T3:D2.15). In (A) and (B) the position of the substrate alone, Dna2–substrate complex and FEN1–substrate complex are indicated to the left of the figure. In (A), the FEN1–Dna2–substrate complex is indicated to the left of the figure and to the right of lane 6 with a dashed arrow. In (B), the quantitation of the percent substrate bound by hDna2 or hFEN1 is shown below the figure.
The inability of hFEN1 to displace hDna2 was surprising because our previous results with nucleases from yeast showed that scFEN1 efficiently removes scDna2 from double-flap structures (17,25). Because we observed a significant difference between the yeast and human systems, we considered that the variance might have resulted from differences in experimental conditions. In our previous yeast binding and competition assays, Ca2+ was used in place of Mg2+ to inhibit nuclease activity. In our human protein binding and competition assays, we excluded divalent metal ions and added a small amount of EDTA to inhibit nuclease activity. To distinguish between differences in experimental conditions, we repeated our prior yeast competition experiments using the human competition assay conditions. Consistent with prior results, scFEN1 displaced scDna2 in a concentration-dependent fashion in the presence of EDTA and independent of the Ca2+ (Supplementary Figure S1).
Initial results showed that hDna2-binding affinity is highly sensitive to 5′ flap length while hFEN1 binding is relatively insensitive (Figure 1A). We considered that the human system might have adopted a flap length threshold below which hFEN1 could displace pre-bound hDna2. To test this, we repeated the competition assay as in Figure 4A with a 15 nt 5′ flap (Figure 4B). Similar to the 30 nt flap reaction conditions, we allowed hDna2 to pre-bind the 15 nt double-flap substrate prior to the addition of hFEN1. hDna2 alone binding is shown in lane 2. hFEN1 was then titrated into the reaction in competition with hDna2 (lanes 3–6) or alone (lanes 7–10). The relative hDna2 displacement from the 15 nt double flap is shown graphically in Figure 5D as ‘15 nt Double Flap’. In contrast to the 30 nt flap results, hFEN1 demonstrated the ability to displace hDna2 from the 15 nt 5′ flap in a concentration-dependent fashion. Quantitation of the percent substrate bound by hDna2 or hFEN1 is shown below the figure. In addition, the super-shifted hFEN1–hDna2 co-binding complex visualized in the 30 nt 5′ double flap EMSA no longer appeared in the 15 nt 5′ double flap competition experiment.
Dna2 and FEN1 co-binding requires upstream DNA
Prior reports show that optimal FEN1 substrate binding requires an upstream 1 nt 3′ flap (37). To further characterize substrate properties that enable hFEN1 and hDna2 substrate co-binding, we competed hDna2, pre-bound to a 30 nt 5′-tail substrate, with hFEN1 (Figure 5A). When titrated into the reaction (lanes 7 and 8), hFEN1 was unable to compete hDna2 away from the 30 nt 5′-tail. The hDna2/hFEN1 co-binding complex was not visualized on the 5′-tail substrate (compare lane 8 to lane 3). The addition of hFEN1 to the reaction enabled a slight increase in the relative amount of DNA substrate bound by hDna2 alone (graphically represented in Figure 5D; ‘30 nt 5′-Tail’).
In order to determine whether the 1 nt upstream 3′ flap was necessary to generate the co-binding complex mode, we pre-bound hDna2 to 5 nt gap-flap substrate, with a 30 nt 5′ flap, followed by the addition of increasing concentrations of competing FEN1 (Figure 5A). hDna2 alone bound to the gap-flap substrate is shown in lane 11 and hFEN1 alone in lane 14. Similar to the 30 nt 5′-tail competition, when titrated into the reaction, hFEN1 was unable to displace hDna2 from the gap substrate (lanes 12 and 13). Similar to the 30 nt 5′-tail structure, the addition of hFEN1 slightly increased the percent DNA substrate bound by hDna2 (graphically represented in Figure 5D, ‘30 nt 5′ Flap 5 nt Gap’).
Displacement of human Dna2 by FEN1 requires upstream DNA
Considering that hFEN1 was unable to effectively displace hDna2 from the long 5′ flap substrates, we asked whether the hDna2 displacement by hFEN1 on the intermediate double-flap structure resulted from protein–protein interactions or the shift in hDna2-binding affinity based on substrate characteristics. In a model for displacement of hDna2 based on one protein disengaging the other, hFEN1 alters the conformation of hDna2, so that it leaves an intermediate flap 5′ flap structure. In the binding affinity displacement model, the substrate flap base properties favor hFEN1 leading to hDna2 displacement. To test the protein–protein displacement model, we competed pre-bound hDna2 with hFEN1 on the 15 nt 5′-tail structure (Figure 5B). hDna2 alone bound to the 15 nt 5′-tail substrate is shown in lane 6, hFEN1 alone in lane 9 and hFEN1 titrated into pre-bound hDna2 reactions in lanes 7 and 8. While hFEN1 displaced the pre-bound hDna2 from the 15 nt double-flap structure, minimal hDna2 displacement was visualized from the 15 nt 5′-tail structure (compare Lane 3 with Lane 8, respectively, and see Figure 5D; ‘15 nt 5′-Tail’), providing evidence against the protein–protein interaction model of displacement.
To test for displacement based on substrate binding affinity, we utilized the 15 nt gap-flap structures that provided an intermediate hFEN1-binding affinity platform relative to the high affinity double flap and low affinity 5′-tail. We competed hDna2 pre-bound to the 15 nt gap-flap structures with increasing concentrations of hFEN1 (Figure 5C). hDna2 pre-bound to the 5 and 2 nt single-stranded gap structures is shown in lanes 2 and 10, respectively. The titration of hFEN1 alone bound to the 5 and 2 nt single-stranded gap structures is shown in lanes 6–8 and 14–16, respectively. hFEN1 titrated into the reactions containing pre-bound hDna2 to the 5 and 2 nt single-stranded gap-flap structures is shown in lanes 3–5 and 11–13, respectively. hDna2 was displaced similarly by hFEN1 from both gap-flap structures. The comparative displacement of hDna2 from the 5 and 2 nt gap-flap structures relative to the initial amount bound is shown graphically in Figure 5D ‘15 nt 5′ Flap 5 nt Gap’ and ‘15 nt 5′ Flap 2 nt Gap’, respectively. These results suggest that hDna2 displacement by hFEN1 requires upstream DNA to stabilize hFEN1 binding and a shortened flap to reduce hDna2-binding affinity.
DISCUSSION
All homologs of eukaryotic FEN1 display high-affinity binding and cleavage specificity for a double-flap substrate with a 1 nt 3′ flap and a 5′ flap of variable length (15,20,37). During Okazaki fragment maturation, strand displacement synthesis by DNA polymerase δ (pol δ) creates a 5′ flap (38). A number of reports indicate that the steady-state structure during strand displacement synthesis is an annealed 3′ terminus separated by a short gap from the 5′-displaced flap created by steric separation of the template and downstream complementary strand by the DNA polymerase (39,40). The manner in which the double flap becomes available to FEN1 is currently not known. The efficiency and accuracy of short flap processing suggest a simple mechanism in which the polymerase withdraws to expose the upstream synthesis strand and downstream 5′ flap for FEN1 binding and subsequent cleavage (41,42). Prior reports suggest a polymerase idling process during strand displacement, in which the polymerase backs up or dissociates, creating an opportunity for FEN1 cleavage and subsequent ligation at virtually every DNA nucleotide (42). For those flaps that escape FEN1 cleavage and bind RPA and then Dna2, the mechanism must have additional complexity involving a sequential action of Dna2 and then FEN1 (8,43).
To better understand how the two nucleases work together, we characterized the substrate specificities influencing hDna2 binding and cleavage functions and the sequential interaction of hFEN1 and hDna2 on Okazaki fragment intermediates. hDna2 bound and cleaved long 5′ flap structures with similar high affinity and efficiency irrespective of the presence of an upstream segment creating a double-flap structure. Binding affinity and cleavage efficiency on substrates with 5′ flaps decreased progressively with flap length to become negligible at lengths of 4–5 nt. Utilizing substrates with a single-stranded gap between the upstream primer and downstream 5′ flap, termed a gap-flap, we acquired evidence that the upstream DNA improved hDna2 binding affinity and cleavage efficiency on intermediate length 5′ flap structures. The decreased hDna2 binding for the intermediate length 5′-tail substrates compared to the double flap was unexpected. We previously reported that optimal scDna2 substrate recognition involves simultaneous binding of the 5′ flap and downstream dsDNA (36). Either human and yeast Dna2 possess different binding specificities or hDna2 utilizes additional upstream primer-template contacts for intermediate length 5′ flap processing that were undetected previously with the long flap substrates used to characterize scDna2. Evidence for these additional binding contacts suggests that hDna2 reorients to access the upstream DNA after partially processing long flaps.
During DNA replication, pol δ initiates 5′ flap creation through strand displacement synthesis (38,41,44). Foot-printing data show that pol δ protects 5–6 nt of ssDNA ahead of the most recently incorporated nucleotide (45). The combination of the displaced 5′ flap and single-stranded region protected by pol δ comprises a gap-flap structure. Current work and prior reports have shown that hFEN1 neither stably binds nor efficiently cleaves these gap-flap structures requiring that the DNA be remodeled prior to hFEN1 cleavage (4,20,37). Pol δ most likely releases from the strand being synthesized allowing a portion of the 5′ flap to re-anneal to the template filling the gap region. This re-annealing can generate the double flap base structure preferred by hFEN1 for cleavage. During short flap processing, this polymerase dissociation, 5′ flap re-annealing and flap base acquisition by hFEN1 would need to be efficiently coordinated to process the estimated 20–50 million Okazaki fragments made prior to every mammalian cell division (38). The synchronized protein handoff is likely accomplished by the polymerase processivity factor, proliferating cell nuclear antigen (PCNA) (41). PCNA is a homo-trimeric protein known to promote pol δ DNA substrate binding stability and synthesis processivity (46–48). Reports show that pol δ, hFEN1 and DNA ligase I can concurrently bind individual subunits of PCNA improving enzymatic function through increased substrate binding affinity, and presumably coordinating sequential action (49–51).
The steps by which hDna2 hands off the substrate to hFEN1 during long flap processing have not been clear. To improve our understanding, we analyzed hDna2 displacement by hFEN1 from substrates representing expected replication intermediates. Specifically, we measured hDna2 and hFEN1 binding to long 5′ flap substrates with the gap-flap and FEN1-preferred double flap configurations. Surprisingly, in contrast with the yeast system, hFEN1 was unable to displace hDna2 from any of the long flap substrates. Additionally, hFEN1 and hDna2 concurrently bound the long double-flap structure. This co-binding mode was visualized neither with the long gap-flap nor the 5′-tail structures. The inability of hFEN1 to displace hDna2 from long flaps was surprising as we previously reported that scFEN1 displaces scDna2 from the flap base prior to cleaving (17,25). The observed concurrent binding suggests that hFEN1 and hDna2 are able to stably bind different locations of the substrate. hDna2 likely binds the upper portion of the 5′ flap and downstream DNA while hFEN1 binds the upstream dsDNA and the 3′ 1 nt flap (Figure 6A). hFEN1 may also bind the lower portion of the 5′ flap and a portion of the downstream dsDNA.
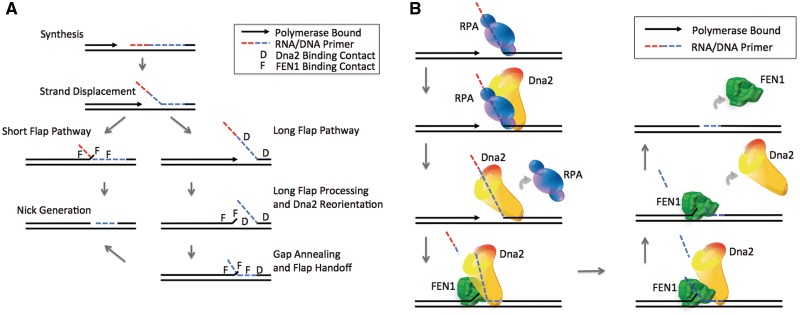
Figure 6.
Models of FEN1 and Dna2 Okazaki fragment substrate recognition and processing. (A) hDna2 and hFEN1 substrate recognition model. Pol δ synthesizes until encountering the downstream Okazaki fragment RNA/DNA primer. Pol δ then physically separates the two strands of the duplex DNA to form a 5′ flap. The active site of the polymerase protects a 5–6 nt single-stranded region on the template creating a structure with a 5′-displaced flap and a single-stranded gap between the upstream and downstream primers, termed a gap-flap. Left arrow: short flap pathway. During short flap processing, the polymerase idles back or dissociates from the synthesis fragment allowing the 5′ flap to re-anneal with the template generating a short double-flap structure. hFEN1 binds the double-flap substrate using three contact locations, indicated by ‘F’, (i) upstream dsDNA, (ii) downstream dsDNA and (iii) the 3′ 1 nt flap. Right arrow: long flap pathway. During long flap processing, flaps escape hFEN1 cleavage. hDna2 binds the long gap-flap using two contact locations, indicated by ‘D’, the downstream dsDNA and the long 5′ flap. Dna2 threads the long flap and begins cleaving. The polymerase idles back or dissociates from the synthesis fragment allowing Dna2 to continue to cleave the long flap. hDna2 binding affinity for the 5′ flap is reduced and it reorients to bind the upstream DNA. This hDna2 reorientation promotes the re-annealing of the 5′ flap with the template, filling the single-stranded gap. The combination of hDna2 upstream DNA binding and the formation of the double flap promote hFEN1 flap base localization. hDna2 releases from the upstream DNA and the 5′ flap allowing hFEN1 binding contacts with the upstream dsDNA and the 1 nt 3′ flap. The 5′ flap transitions to hFEN1 and hFEN1 binds the downstream DNA. hFEN1 is now poised to thread and cleave the substrate and generate a nicked product. (B) Long Flap Pathway Model. As in (A), pol δdisplaces the 5′ flap into a 5–6 nt gap-flap structure. Flaps escaping hFEN1 cleavage become long and are bound by hRPA. hDna2 binds the flap displacing hRPA. hDna2 cleaves the long flap as pol δidles back or dissociates from the synthesis fragment. hDna2 reorients promoting 5′ flap re-annealing with the template, as shown in (A). The double-flap structure formation and hDna2–hFEN1 protein interactions allow hFEN1 to bind the flap base. The reduced hDna2-binding affinity for the shortened 5′ flap permits hFEN1 acquisition of the flap for threading. hDna2 stimulates the cleavage activity of hFEN1 to create a nicked product.
It is noteworthy that the human system has evolved to promote hDna2 binding to long flap structures independent of the flap base configuration even in the presence of hFEN1. Original genetic analyses of the yeast homologs suggested that long flap creation and removal is the predominant pathway of Okazaki fragment processing (8). Later, biochemical analyses demonstrated that most flaps are processed while short and that the long flap pathway is possibly used as a backup (43). While the yeast system may use scRPA in combination with scDna2 to process long flaps, the human system may have evolved to more efficiently use hDna2 to cleave elongating flaps prior to stably binding hRPA. Data supporting this model show that scRPA displacement by scDna2 in vitro is more efficient than hRPA displacement by hDna2 (33,34).
In vivo, hDna2 likely processes these long 5′ flaps, to an intermediate length, at which point its substrate binding affinity is reduced. As noted earlier, the shortened 5′ flap may lead to the reorientation of hDna2 to bind both the shorter 5′ flap and the upstream DNA for both the gap-flap and double-flap structures. Our competition data show that hFEN1 effectively displaces hDna2 from these intermediate length double-flap structures. Considering this displacement and the need for hFEN1 to bind the upstream DNA prior to cleaving, it was counterintuitive that hDna2 bound the upstream DNA of intermediate length flaps. We envision two scenarios in which this additional binding would be advantageous. First, binding the upstream DNA may allow hDna2 to interact with proteins located upstream of the flap base. Second, binding the upstream DNA may prepare hDna2 for cleavage activity needed in Okazaki fragment processing but not DSB repair. Consistent with the first scenario, hDna2 has been shown to directly interact with hFEN1 (52). The additional upstream binding contact may bring hDna2 in contact with hFEN1 across a gap-flap structure. This contact may be part of the mechanism by which hFEN1 displaces hDna2. In line with the second scenario, FEN1 substrate localization and cleavage activity are diminished by certain FEN1 post-translational modifications. Specifically, phosphorylation leads to reduced FEN1 substrate localization (53) and acetylation diminishes substrate affinity and cleavage activity (54). In both binding scenarios, the increased binding affinity imparted by the upstream DNA would stabilize hDna2 substrate binding pending acquisition and activity of hFEN1 with lower binding affinity. The hFEN1–hDna2 protein–protein interaction has been shown to result in stimulation of both nuclease activities (24,52). The ability of hFEN1 to displace hDna2 from intermediate double-flap structures taken together with the improved hDna2 upstream DNA binding for intermediate length supports a mechanism in which the nucleases have evolved to sequentially and cooperatively process elongated 5′ flap structures.
Based on current and prior results, we propose the following model for long flap processing (Figure 6B). During synthesis, pol δ generates a gap-flap structure with a 5–6 nt single-stranded gap between the segment being synthesized and the downstream fragment. Recognizing the 5′ flap and downstream dsDNA, hDna2 binds the flap being extended by pol δ strand displacement. hDna2 cleaves generating an intermediate length flap, disrupting stable RPA binding. hFEN1 is unable to bind or cleave this gap-flap configuration. Upstream DNA must be exposed by displacement or idling of pol δ. When this occurs, hDna2 will reorient to bind the shortened flap and upstream DNA, possibly aiding the re-annealing of the 5′ flap to the template forming a double-flap structure. In line with this idea, prior reports show that hDna2 possesses a DNA strand annealing activity (55) and promotes hFEN1 nick generation in the presence of equilibrating flaps where the long 5′ flap can compete for template annealing with the upstream newly synthesized strand (31,34). The combination of the double-flap configuration and hDna2 movement in the upstream DNA direction would enable hFEN1 to bind hDna2 and provide access to the base of the correctly formed double-flap substrate. hFEN1 bound to the flap base and the protein–protein interaction would enable hDna2 displacement from the substrate and stimulate hFEN1 5′ flap cleavage to efficiently create the nicked structure for ligation.
While the ability of hDna2 to bind upstream DNA during replication may be beneficial, this binding site structure may not be available during DSB repair or telomere maintenance. The loss of the upstream binding contact might lead to the reduction in cleavage activity as visualized for the intermediate 5′-tail structures. The possibility exists that hDna2 is not needed to process 5′-tail substrates in these pathways until they are lengthened. Furthermore, protein cofactors may be required to generate long flaps and recruit hDna2 to these long 5′-tail substrates. Reports supporting this model demonstrate that multi-protein complexes, known to bind DSBs and necessary for telomere maintenance, recruit and stimulate hDna2. These reports also show that (i) the recruitment of RPA, a 3′–5′ helicase (such as BLM helicase), and Dna2 is necessary, (ii) the helicase activity of the 3′–5′ helicase and the 5′ nuclease activity of Dna2 are required and (iii) the helicase activity of Dna2 is dispensable for end resection in vitro (29,30). During these processes, Dna2 is suggested to act on pseudo-Y structures cleaving only the 5′ flap. The abilities of Dna2 to bind and process long 5′ flaps independent of the upstream DNA properties support correct DNA product formation during DSB and telomere maintenance. Our binding and cleavage data taken together with these prior results support the model that protein interactions and flap elongation by a 3′–5′ helicase are likely necessary to activate Dna2 during DSB repair and telomere maintenance.
In summary, we have shown that hDna2 binds flap structures with higher affinity as the flap length increases. In the human system, this increased long flap binding affinity prevents displacement of hDna2 by hFEN1. As hDna2 cleaves the flap to an intermediate length, the nuclease is able to bind upstream DNA improving binding affinity and cleavage efficiency. We suggest a model in which this additional hDna2 binding aids in generating the double-flap structure from the gap-flap created by pol δ during long flap strand displacement synthesis. hDna2 can remain bound to the flap base during acquisition of hFEN1. It is likely that the two proteins transiently bind each other and the substrate and then, as the flap is further shortened, hDna2 dissociates and hFEN1 cleaves at the base. While further analysis is needed to validate the model in a cellular system, the dynamic characteristics of the replication fork do not currently allow for measurement of these properties in vivo. Additionally, lack of a crystal structure for hDna2 limits designing experiments in vivo that would specifically define the sequential action of the proteins on the replication fork. While structural analysis of hDna2 is needed to validate the specific binding contacts and protein–protein interactions in the presence of various physiological DNA substrates, our results in vitro define a model in which hDna2 plays an important role to promote genomic stability during DNA replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
The National Institutes of Health [GM024441 to R.A.B., GM068411 to J.W.G., GM098328-01A1 to L.B., GM078666 to J.L.C.]. Funding for open access charge: The National Institutes of Health [GM024441].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Bambara and Campbell research groups for critical discussion during the course of this work and comments on the manuscript.
REFERENCES
- 1.Arezi B, Kuchta RD. Eukaryotic DNA primase. Trends Biochem. Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 2.Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 2006;106:453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- 3.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 4.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 6.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 7.Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 8.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 11.Ceska TA, Sayers JR, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 12.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 13.Patel N, Atack JM, Finger LD, Exell JC, Thompson P, Tsutakawa S, Tainer JA, Williams DM, Grasby JA. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 2012;40:4507–4519. doi: 10.1093/nar/gks051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao HI, Campbell JL, Bambara RA. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 15.Gloor JW, Balakrishnan L, Bambara RA. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J. Biol. Chem. 2010;285:34922–34931. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart JA, Campbell JL, Bambara RA. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 2006;281:38565–38572. doi: 10.1074/jbc.M606884200. [DOI] [PubMed] [Google Scholar]
- 18.Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 19.Budd ME, Choe W, Campbell JL. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 20.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 21.Murante RS, Huang L, Turchi JJ, Bambara RA. The calf 5′- to 3′-exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J. Biol. Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 22.Bae KH, Kim HS, Bae SH, Kang HY, Brill S, Seo YS. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- 24.Munashingha PR, Lee CH, Kang YH, Shin YK, Nguyen TA, Seo YS. The trans-auto stimulatory activity of Rad27 suppresses dna2 defects in Okazaki fragment processing. J. Biol. Chem. 2012;287:8675–8687. doi: 10.1074/jbc.M111.326470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart JA, Campbell JL, Bambara RA. Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:8283–8291. doi: 10.1074/jbc.M809189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart JA, Miller AS, Campbell JL, Bambara RA. Dynamic removal of replication protein A by Dna2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:31356–31365. doi: 10.1074/jbc.M805965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balakrishnan L, Brandt PD, Lindsey-Boltz LA, Sancar A, Bambara RA. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J. Biol. Chem. 2009;284:15158–15172. doi: 10.1074/jbc.M109.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrington JJ, Lieber MR. DNA structural elements required for FEN-1 binding. J. Biol. Chem. 1995;270:4503–4508. doi: 10.1074/jbc.270.9.4503. [DOI] [PubMed] [Google Scholar]
- 38.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munn MM, Alberts BM. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J. Biol. Chem. 1991;266:20034–20044. [PubMed] [Google Scholar]
- 40.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 41.Maga G, Villani G, Tillement V, Stucki M, Locatelli GA, Frouin I, Spadari S, Hubscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl Acad. Sci. USA. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 45.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J. Biol. Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 46.Mozzherin DJ, Tan CK, Downey KM, Fisher PA. Architecture of the active DNA polymerase delta.proliferating cell nuclear antigen.template-primer complex. J. Biol. Chem. 1999;274:19862–19867. doi: 10.1074/jbc.274.28.19862. [DOI] [PubMed] [Google Scholar]
- 47.Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J. Biol. Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 48.Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 49.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31:1556–1567. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 52.Henry RA, Balakrishnan L, Ying-Lin ST, Campbell JL, Bambara RA. Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J. Biol. Chem. 2010;285:28496–28505. doi: 10.1074/jbc.M110.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hubscher U, Hottiger MO. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 2001;7:1221–1231. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 55.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J. Biol. Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.