Abstract
Tbx4 and Tbx5 are two closely related T-box genes that encode transcription factors expressed in the prospective hindlimb and forelimb territories, respectively, of all jawed vertebrates. Despite their striking limb type-restricted expression pattern, we have shown that these genes do not participate in the acquisition of limb type-specific morphologies. Instead, Tbx4 and Tbx5 play similar roles in the initiation of hindlimb and forelimb outgrowth, respectively. We hypothesized that different combinations of Hox proteins expressed in different rostral and caudal domains of the lateral plate mesoderm, where limb induction occurs, might be involved in regulating the limb type-restricted expression of Tbx4 and Tbx5 and in the later determination of limb type-specific morphologies. Here, we identify the minimal regulatory element sufficient for the earliest forelimb-restricted expression of the mouse Tbx5 gene and show that this sequence is Hox responsive. Our results support a mechanism in which Hox genes act upstream of Tbx5 to control the axial position of forelimb formation.
Keywords: Tbx5, Hox, Limb development
INTRODUCTION
T-box genes encode a family of transcription factors that have been identified in all metazoans and which play diverse roles during embryonic development (Minguillon and Logan, 2003; Naiche et al., 2005; Papaioannou, 2001). From this extensive gene family, the paralogues Tbx4 and Tbx5 have received particular attention due to the striking, mutually exclusive expression domains that they exhibit in various territories of the developing embryo, such as the heart and the limbs (Chapman et al., 1996; Gibson-Brown et al., 1996; Krause et al., 2004). Tbx5 is expressed in the prospective forelimb territory and subsequent forelimb bud, whereas Tbx4 is expressed in the prospective hindlimb territory and hindlimb bud (reviewed by Logan, 2003). Initial misexpression experiments in the chick suggested that these genes play a role in the specification of limb type-specific morphologies (Rodriguez-Esteban et al., 1999; Takeuchi et al., 1999). However, subsequent gene deletion experiments in the mouse confirmed that Tbx5 and Tbx4 play essential roles in the initiation of limb outgrowth (Agarwal et al., 2003; Naiche and Papaioannou, 2003; Rallis et al., 2003). Furthermore, we have shown that neither Tbx5 nor Tbx4 is required for the acquisition of limb type-specific morphologies, but that they play equivalent roles in the initiation of forelimb and hindlimb outgrowth, respectively (Minguillon et al., 2005). We used a combination of transgenic and gene deletion approaches in mice to show that forelimb outgrowth can be induced by Tbx4 in the absence of Tbx5. We concluded that although Tbx5 and Tbx4 normally induce, and are markers of, forelimb and hindlimb outgrowth, respectively, they do not play a role in the specification of limb type-specific morphologies. We hypothesized that the limb type-specific morphologies that ultimately develop are instead dictated by the axial identity of the pre-patterned lateral plate mesoderm (LPM) in which Tbx-mediated limb induction occurs. This is concordant with classical embryological experiments in the chick that show that limb type specification is determined at very early stages of limb development, prior to overt limb outgrowth and the expression of Tbx4 and Tbx5 (Saito et al., 2002; Stephens et al., 1989).
Hox genes encode a family of transcription factors that are found in all eumetazoans and are frequently arranged in chromosomal clusters (reviewed by Duboule, 2007; Lemons and McGinnis, 2006). Hox genes have been implicated in the morphological diversification of many embryonic structures, including the neural tube, somites and gut (Zacchetti et al., 2007) (for a review, see Wellik, 2007). Although the detailed expression of these genes in the LPM has been less well characterised than in other axial tissues, it has been shown that combinations of Hox genes expressed in the LPM correlate well with the type of limb that subsequently develops (Cohn et al., 1997). In a previous study, we hypothesized that limb type identity, and ultimately limb type-specific morphology, are specified by different combinatorial codes of factors in the LPM at different rostrocaudal positions, and that one of these factors could include a particular combination of Hox proteins. We suggested that Tbx5 expression in the LPM is activated as a result of a combinatorial code of ‘rostral’ Hox genes, whereas Tbx4 expression is initiated by a combinatorial ‘caudal’ Hox code. The activation of Tbx5 and Tbx4 at the prospective forelimb and hindlimb level, respectively, is ultimately necessary for the initiation of limb bud outgrowth (Gibson-Brown et al., 1998; Minguillon et al., 2005).
To determine whether Hox genes expressed in the LPM control the limb type-restricted expression of Tbx genes, we have isolated the minimal regulatory element sufficient to drive the earliest forelimb-restricted expression of the mouse Tbx5 gene. We find that a 361 bp sequence located in the second intron is able to recapitulate the earliest forelimb-restricted expression of the mouse Tbx5 gene when linked to a lacZ reporter. This region contains six predicted Hox binding sites that are required for the regulatory properties of this region. We use co-electroporation studies in the chick and site-directed mutagenesis of mouse transgenic constructs to show that, via this regulatory element, Hox proteins regulate the onset of forelimb-restricted expression of Tbx5 in vivo. Furthermore, we show that Hox proteins bind directly to these putative Hox binding sites in vitro. These data provide the first evidence that rostral Hox genes expressed in the LPM directly regulate the forelimb-restricted expression of Tbx5 and thereby control the axial position at which forelimb outgrowth is initiated.
MATERIALS AND METHODS
DNA constructs and analysis of regulatory regions
For reporter gene analyses in both mouse and chick embryos we used the BGZA reporter vector (Summerbell et al., 2000). For chick electroporation experiments, cDNAs coding for murine Hox genes were cloned upstream of the IRES-eGFP element in the pCIG expression vector (Megason and McMahon, 2002). For site-directed mutagenesis, the Stratagene QuikChange XL Site-Directed Mutagenesis Kit was used following the manufacturer’s instructions. Constructs are summarised in supplementary material Fig. S1.
Conserved regulatory regions were scanned using the VISTA genome browser (Bray et al., 2003; Couronne et al., 2003). MatInspector (Genomatix) was used to search for putative binding sites, which were also searched for by eye following published binding sites (Manzanares et al., 2001; Pearson et al., 2005; and references therein).
Generation of transgenic lines
Transgenic mouse embryos were generated by the Procedural Services section, NIMR, by standard pronuclear microinjection techniques.
Embryos
Mouse embryos were staged according to Kaufman (Kaufman, 2001). Noon on the day a vaginal plug was observed was taken to be 0.5 days of development (E0.5). Fertilised chicken eggs (Needle’s Farms, Winter’s Farms) were incubated at 37°C and staged according to Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1992). Mice carrying the lacZ transgene were identified by PCR using primers LacZfwd (5′-GGTCGGCTTACGGCGGTGATTT-3′) and LacZrev (5′-AGCGGCGTCAGCAGTTGTTTTT-3′).
Chick electroporation
In ovo hindbrain electroporation was performed as described (Itasaki et al., 1999). Briefly, the m5-5 promoter fragment (250 ng/μl) was co-injected with Fast Green and the expression vector pCIG (1 μg/μl) into the neural tube of HH10 chicken embryos. This DNA mixture was then electroporated into the right side of the neural tube. Embryos were allowed to develop for a further 22 hours before harvesting. Only those embryos showing robust GFP expression were subsequently processed for β-galactosidase activity.
Whole-mount in situ hybridisation and histochemistry
Whole-mount in situ hybridisations were carried out essentially as described (Riddle et al., 1993). IMAGE clones #40044111 and #3985274 were used as templates to prepare mouse Hoxa4 and Hoxa5 probes, respectively. The full-length cDNAs for mouse Hoxb5, Hoxc4 and Hoxc5 were cloned by RT-PCR into pGEM (Promega) using standard techniques and used as templates to prepare the corresponding probes. The 3′ region of the Tbx5 cDNA was used to create an in situ probe that lacked the conserved T-box domain. A monoclonal antibody against mouse Hoxb4 (a gift from Alex Gould) was used for immunostaining as previously described (Gould et al., 1997).
Electrophoretic mobility shift assay
cDNAs encoding selected Hox genes were cloned into pBluescript pKS (Stratagene) using standard techniques. These constructs were in vitro translated using the Coupled TnT Transcription/Translation System (Promega) in the presence of [35S]methionine (Amersham) following the manufacturer’s instructions. Proteins were visualised by SDS-PAGE followed by autoradiography to check correct translation.
For electrophoretic mobility shift assays (EMSAs), 2 μl lysate containing the desired Hox protein was mixed with 40,000 cpm 32P-labelled double-stranded oligonucleotide in binding buffer [10 mM Tris-HCl pH 7.5, 75 mM NaCl, 1 mM EDTA, 1 mM DTT, 2 μg BSA, 6% glycerol and 1 μg poly(dI-dC)] in a total volume of 20 μl. After 30 minutes incubation in ice, the reactions were separated by 5% PAGE in 0.5×TBE. For supershift, 2 μl anti-flag antibody was incubated with the binding reaction for 30 minutes in ice.
Nuclear extracts were prepared from rostral regions of ten early E9 embryos using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce) following the manufacturer’s instructions (final volume of 50 μl). For EMSA, this nuclear extract was diluted 1:4 in binding buffer and 20 μl incubated with 40,000 cpm 32P-labelled double-stranded oligonucleotide. When used for competition analyses, unlabelled oligonucleotide was added to the binding reactions. After 30 minutes incubation in ice, the reactions were separated by 5% PAGE in 0.5×TBE.
RESULTS
Analysis of the mouse Tbx5 regulatory region
To address whether a particular combination of Hox proteins lies upstream of the forelimb-restricted expression of Tbx5, we first decided to identify the region of the Tbx5 locus in which the regulatory element responsible for driving this rostral LPM expression domain is located. A series of constructs containing fragments of the genomic region of the mouse Tbx5 gene were cloned upstream of a minimal promoter linked to the lacZ reporter gene in the BGZA vector (Summerbell et al., 2000) and assayed by transgenesis for their ability to regulate forelimb-restricted expression (Fig. 1, supplementary material Fig. S1).
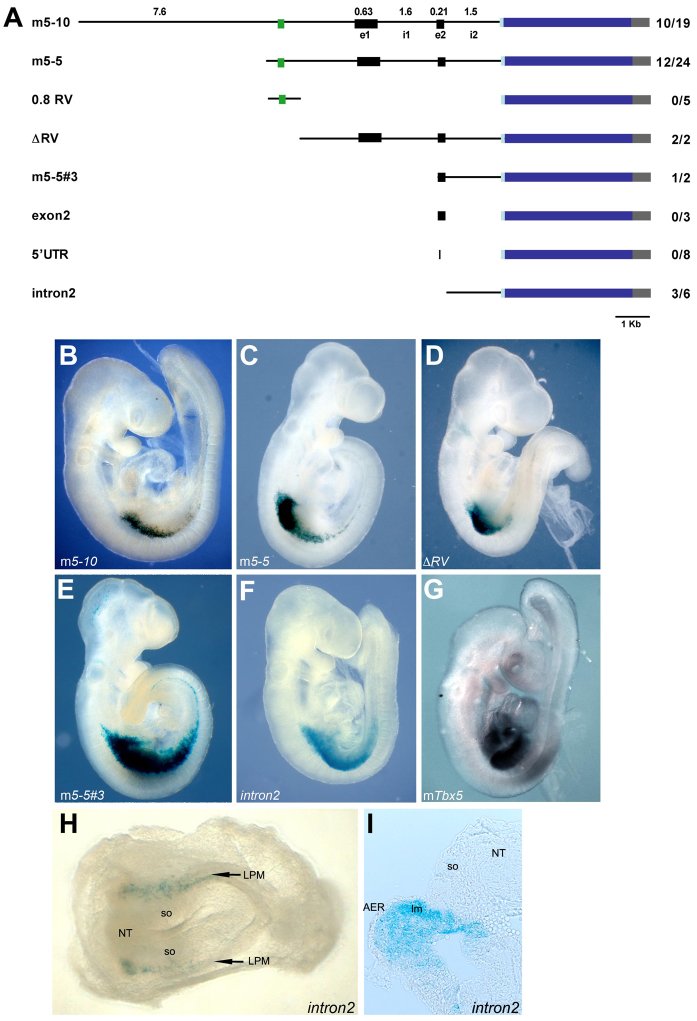
Fig. 1.
Localisation of the forelimb enhancer of the mouse Tbx5 gene. (A) Tbx5 transgenic constructs. Putative regulatory sequences (thin lines) were cloned into the BGZA reporter vector, which contains a chick β-globin minimal promoter (light-blue box), the lacZ gene (dark-blue box) and an SV40 polyadenylation signal (grey box). Black boxes represent exons (e1, e2) and the green box represents a conserved non-coding element (CNE) shared between amniotes. i1, intron 1; i2, intron 2. The numbers denote size in kb. To the right is shown the number of embryos showing forelimb-restricted expression out of the total number of transgenic embryos recovered. (B-F,H,I) Representative β-galactosidase staining for the m5-10 (B), m5-5 (C), ΔRV (D), m5-5#3 (E) and intron2 (F,H,I) constructs. (G) E9.5 mouse embryo showing endogenous Tbx5 expression assayed by whole-mount in situ hybridisation. (I) Transverse section at the forelimb level of an E9.5 intron2 transgenic embryo showing β-galactosidase staining in LPM-derived limb mesenchyme. Lateral views from the right side are shown for E9.5 embryos (B-G), whereas a ventral view with anterior to the left is shown for the E8.5 embryo (H). AER, apical ectodermal ridge; lm, limb mesoderm; LPM, lateral plate mesoderm; NT, neural tube; so, somites.
An 11.5 kb genomic fragment upstream of Tbx5 intron 2 spanning –7634 to +3914 (m5-10; Chr5:120,277,038-120,288,589, Ensembl NCBIM37) was able to drive forelimb-restricted expression of the lacZ reporter (Fig. 1B) similar to the endogenous expression of the Tbx5 gene in the developing forelimb (Fig. 1G). We then generated a series of shorter constructs to better define the location of this forelimb-regulatory region. As assessed by β-galactosidase activity, a fragment containing the most proximal 6.4 kb of this genomic region (Chr5:120,282,130-120,288,589, Ensembl NCBIM37), as represented by the m5-5 construct, was still able to regulate forelimb expression (Fig. 1C).
We then used phylogenetic footprinting to search for conserved non-coding elements within this 6.4 kb region among species with genome sequence available using the VISTA browser (Bray et al., 2003; Couronne et al., 2003). We found a 180 bp DNA fragment (Chr5:120,282,622-120,282,807, Ensembl NCBIM37; green rectangle in Fig. 1A) conserved in both chicken and mammals, which prompted us to test its regulatory abilities in our reporter assay system. We constructed two complementary constructs: an 800 bp construct containing the conserved sequence (0.8RV) and a 5.6 kb construct lacking this conserved region (ΔRV). Despite the conservation of this sequence element, it was not sufficient to drive forelimb expression of the reporter gene (Fig. 1A), nor was it necessary, as the ΔRV construct was still able to drive expression of the lacZ reporter in the forelimb region (Fig. 1D). However, deletion of this conserved sequence resulted in more robust limb staining, suggesting that inhibitory inputs might reside in this element.
We then divided the ΔRV construct into three smaller fragments: one containing the upstream non-coding region (m5-5#1), a second containing Tbx5 exon 1 (5′UTR) and intron 1 (m5-5#2), and a third containing exon 2 (5′UTR+ORF) and intron 2 (m5-5#3). As assayed by β-galactosidase activity, the forelimb-specific regulatory region of Tbx5 resides in m5-5#3 (Fig. 1E). Injection of a construct containing either the 5′UTR of exon 2 (5′UTR) or the whole of exon 2 (exon2) did not produce any forelimb-specific β-galactosidase staining in transgenic embryos (Fig. 1A). Injection of a 1.5 kb construct containing most of Tbx5 intron 2 (intron2; Chr5:120,287,070-120,288,589, Ensembl NCBIM37) revealed that the rostral LPM enhancer resides in intron 2 (Fig. 1F).
We then asked whether the regulatory region contained within intron 2 represents the earliest enhancer required for the onset of Tbx5 expression in the LPM. We performed β-galactosidase staining on early E8.5 mouse transgenic embryos for the Tbx5 intron2 construct. The blue signal in the rostral LPM of these early embryos (arrows in Fig. 1H) demonstrated that this enhancer is sufficient for the onset of Tbx5 expression in the presumptive forelimb area. Sectioning of a stained E9.5 transgenic embryo demonstrated that expression of the Tbx5 intron2 reporter was limited to the forelimb bud mesenchyme (Fig. 1I).
A combination of Hox4 and Hox5 genes is expressed in the rostral LPM of early mouse embryos
Hox genes are expressed in nested domains in various embryonic tissues including the central nervous system, the somites and the gut, where they confer positional (axial) identity to cells in these tissues (Deschamps, 2007; Wellik, 2007; Zacchetti et al., 2007). Hox genes have also been reported to be differentially expressed in the LPM, where they have been implicated in the specification of limb type identity (Cohn et al., 1997). The Hox co-factors Pbx and Meis are also expressed very early in the LPM prior to overt limb bud outgrowth (Capellini et al., 2006; Mercader et al., 2000).
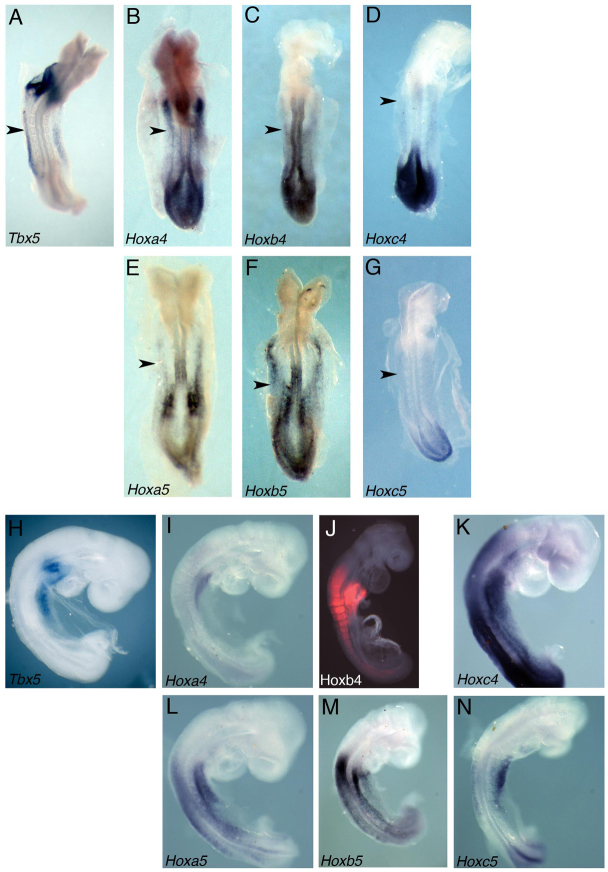
To determine which Hox genes may be regulating the expression of Tbx5 in the presumptive forelimb territory, we first analysed which Hox genes are expressed in the appropriate rostral LPM domain of E8.5 mouse embryos, where Tbx5 expression is first detected. We generated a series of Hox antisense RNA probes to analyse their expression pattern by whole-mount in situ hybridisation (Fig. 2; data not shown). All Hox4 and Hox5 paralogous group (PG) genes, with the exception of Hoxd4 (not shown), are expressed in rostral domains of the LPM of 7- to 9-somite (Fig. 2B-G) and 13- to 14-somite (Fig. 2I-N) mouse embryos, overlapping the first detectable expression of Tbx5 (Fig. 2A,H). This initial domain of expression resolves to a domain lateral to somites 4-10 in a 14-somite embryo (Fig. 2H). Similarly, the domains of PG4 and PG5 Hox genes tend to be initially broad and then become more clearly spatially defined (compare Fig. 2B-G,I-N). In the 14-somite embryo, Hoxa4 expression is detected lateral to somites 3 to 7 (Fig. 2I), whereas Hoxa5 is lateral to somites 4 to 10 (Fig. 2L) and Hoxb4 lateral to somites 3 to 6 (Fig. 2J). Transcripts of Hoxb5 and Hoxc5 can be detected in a 13-somite embryo in the LPM between somites 3 to 8 (Fig. 2M,N). Hoxc4 is expressed in a broader domain of the LPM, between somite 3 and the last formed somite in a 14-somite embryo (Fig. 2K).
Fig. 2.
Hox4 and Hox5 paralogous group (PG) genes are expressed in the presumptive forelimb region of the LPM. (A-G) Tbx5 is expressed in the LPM of the E8 (7- to 9-somite) mouse embryo (A). PG4 and PG5 Hox genes are co-expressed in this domain: Hoxa4 (B), Hoxb4 (C), Hoxc4 (D), Hoxa5 (E), Hoxb5 (F) and Hoxc5 (G). The arrowhead marks the level of somite 3 in each embryo. (H) Fifteen-somite embryo showing endogenous expression of Tbx5. (I-N) Expression of Hoxa4 (I), Hoxb4 (J), Hoxc4 (K) and Hoxa5 (L) at the 14-somite stage and of Hoxb5 (M) and Hoxc5 (N) at the 13-somite stage. Embryos in A-G are dorsal views, anterior to the top, whereas H-N are lateral views of the right side with anterior to the top.
Thus, six Hox genes (Hoxa4, Hoxa5, Hoxb4, Hoxb5, Hoxc4 and Hoxc5; Fig. 2B-G,I-N) are expressed in restricted regions of the rostral LPM that overlap with the area where Tbx5 transcripts are first detected (Fig. 2A,H). These Hox genes are therefore candidates (acting in a combinatorial, semi-redundant manner) to regulate the onset of Tbx5 expression in the presumptive forelimb-forming region.
PG4 and PG5 Hox genes can activate the expression of lacZ in a chick co-electroporation assay
To test whether our candidate Hox genes could activate expression from the Tbx5 locus present in the m5-5 lacZ reporter vector (Fig. 1A), we co-electroporated this construct with an expression vector containing the open reading frames of selected Hox genes upstream of an IRES-GFP cassette in the pCIG vector (Megason and McMahon, 2002) into HH10 chick hindbrains.
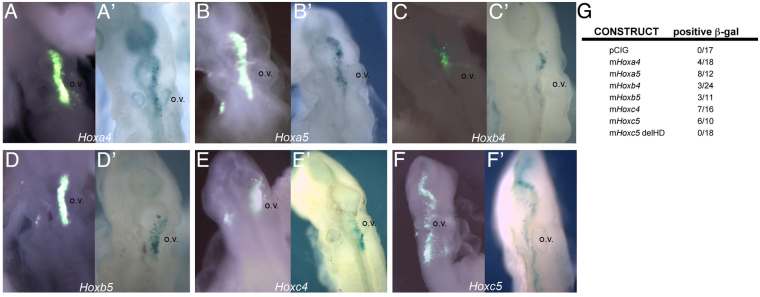
As a negative control, we first co-electroporated the m5-5 reporter construct with the empty expression vector pCIG. After a 22-hour incubation and although the presence of GFP demonstrated the occurrence of electroporated cells, these cells were negative for β-galactosidase activity (Fig. 3G; 0/17), thus validating this assay as a useful tool for assessing the ability of Hox proteins to activate expression of the lacZ reporter. We then co-electroporated the reporter construct with particular Hox-containing pCIG expression vectors. As revealed by β-galactosidase activity, all six Hox candidate genes (i.e. Hoxa4, Hoxa5, Hoxb4, Hoxb5, Hoxc4 and Hoxc5) were able to drive expression of the reporter gene in the chick hindbrain (Fig. 3A-F). Finally, as an additional negative control, and to investigate whether the DNA-binding homeodomain was necessary for the activation of the reporter gene, we engineered a Hoxc5 deletion construct (Hoxc5 delHD) in which a premature STOP codon was produced in the homeobox, similar to that in a previously published Hoxd4 construct (Hasty et al., 1991). Co-electroporation of this construct with the reporter vector and subsequent analysis of β-galactosidase staining did not reveal any lacZ-positive cells, demonstrating that the DNA-binding domain of Hoxc5 is required for this factor to activate this reporter (Fig. 3G; 0/18).
Fig. 3.
Hox genes can activate m5-5-driven lacZ expression in a chick co-electroporation assay. (A-F′) Co-expression of mouse (m) Hoxa4, Hoxa5, Hoxb4, Hoxb5, Hoxc4 and Hoxc5 pCIG expression plasmids (identified by expression of GFP, green in A-F) with the m5-5 BGZA reporter vector in chick hindbrain results in activation of the lacZ reporter gene (A′-F′, blue). (G) The number of β-galactosidase-positive embryos among the total number of GFP-positive embryos as a reflection of lacZ activation. This activation does not occur with the expression plasmid alone (pCIG) and is homeodomain dependent (Hoxc5 delHD). o.v., otic vesicle.
Hox binding sites are required to direct forelimb-restricted expression of Tbx5 in vivo
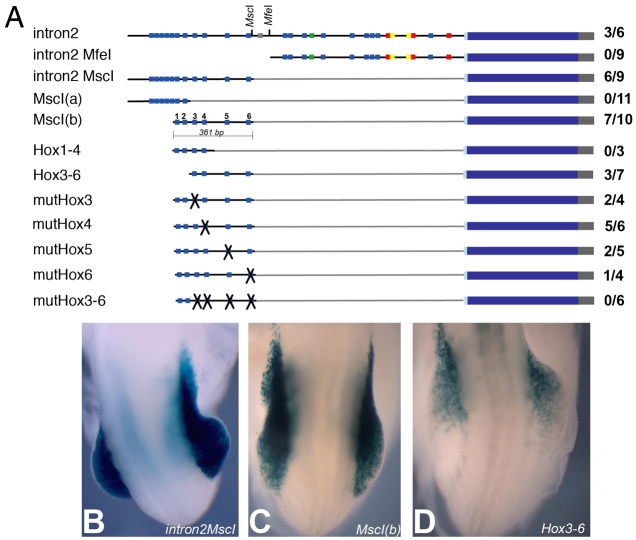
Hox genes encode transcription factors that can regulate target gene expression as monomers or through cooperative interactions on DNA with Pbx and Meis co-factors (reviewed by Pearson et al., 2005). Using MatInspector and visual inspection of published Hox binding sites (Jeong et al., 2006; Manzanares et al., 2001) we found multiple clustered Hox, Hox/Pbx and Pbx/Meis binding sites, and a Hox/Meis site, within the intron 2 sequence. To narrow our search for critical binding sites, we engineered further deletion constructs and cloned them into the BGZA reporter vector to determine their regulatory potential (Fig. 4A, supplementary material Fig. S1). A 865 bp fragment (intron2 MfeI; Chr5:120,287,724-120,288,589, Ensembl NCBIM37; Fig. 4A) containing multiple putative Hox/Pbx/Meis binding sites failed to direct forelimb expression of the lacZ reporter gene in any of the transgenic embryos examined at E9.5. This demonstrated that this fragment is not sufficient for the rostral LPM expression of Tbx5 in vivo. Conversely, a 562 bp fragment containing eleven putative Hox binding sites (intron2 MscI; Chr5:120,287,070-120,287,631, Ensembl NCBIM37; Fig. 4A) was sufficient to drive forelimb expression of our reporter gene (Fig. 4B), demonstrating that the canonical Hox/Pbx/Meis sites in the intron2 MfeI construct are neither sufficient nor necessary to activate expression of Tbx5 in the presumptive forelimb area.
Fig. 4.
Analysis of Hox/Pbx/Meis binding sites in the intron 2 regulatory region of Tbx5. (A) Tbx5 reporter constructs in the BGZA vector used in mouse transgenesis. Blue squares, putative Hox binding sites; green squares, Hox/Meis binding sites; red squares, Pbx/Meis binding sites; yellow squares, Hox/Pbx binding sites; and grey square, a putative Tbx5 binding site. To the right is shown the number of embryos showing forelimb expression out of the total number of transgenic embryos recovered. (B-D) Dorsal views of E9.5 forelimb regions showing representative β-galactosidase staining for the intron2 MscI (B), MscI(b) (C) and Hbs3-6 (Hox3-6) (D) constructs.
To determine which of the eleven predicted Hox binding sites present in the intron2 MscI construct are required for the onset of Tbx5 expression in rostral LPM, we generated deletion constructs and carried out site-directed mutagenesis of specific sites. We first subdivided the intron2 MscI construct into two partially overlapping constructs: MscI(a) and MscI(b) (Fig. 4A; Chr5:120,287,070-120,287,311 and Chr5:120,287,271-120,287,631, respectively; Ensembl NCBIM37). The MscI(a) construct contains the first seven Hox sites and the MscI(b) construct contains the last six; two sites are common between these two constructs. Generation of transgenic embryos showed that the last six putative Hox binding sites are sufficient to drive forelimb expression of the lacZ reporter in the forelimb area (Fig. 4C), but that the first five sites are not required for the regulatory properties of the region. For clarity, we hereafter refer to the six putative Hox binding sites in the MscI(b) construct as Hbs1 (Hox binding site #1) to Hbs6, representing their 5′-3′ order in the construct (see Fig. 4A). We then constructed two additional overlapping constructs: one containing the first four sites (Hbs1-4) and a second containing the last four sites (Hbs3-6). Neither of these was able to recapitulate the endogenous pan-forelimb expression of Tbx5 as judged by β-galactosidase activity. Hbs1-4 failed to drive forelimb expression at all, whereas Hbs3-6 was only able to drive expression in the anterior part of the developing E9.5 forelimb (Fig. 4D).
We next used site-directed mutagenesis to selectively mutate specific Hox sites or a combination of these four sites (Hbs3-6) contained in the MscI(b) construct. Similarly to Hbs3-6, constructs containing mutations of individual Hox binding sites (Hbs3, 4, 5 or 6) in the background of the MscI(b) construct (i.e. containing wild-type Hbs1 and 2) were not able to drive pan-forelimb expression, producing reporter expression similar to that seen with Hbs3-6, suggesting that these sites can operate cumulatively (data not shown). Mutation of all four binding sites in the background of the MscI(b) construct completely abolished the partial anterior forelimb expression of the lacZ reporter (mutHbs3-6; Fig. 4A). From these results, we concluded that all the Hox binding sites (Hbs1-6) are required for the pan-forelimb expression of Tbx5, as different combinations of mutations and/or deletions abolished the early forelimb enhancer function of this construct.
To further demonstrate the requirement of this core region containing Hbs1-6 for forelimb-restricted expression, we produced versions of the original m5-10 and m5-5 constructs (Fig. 1A) with this core sequence deleted. Neither deletion construct was able to drive expression in the forelimbs (supplementary material Fig. S1B,C).
Hox proteins can directly bind to these putative Hox binding sites in vitro
To test whether Hox proteins directly bind to the Hox sites identified in silico and demonstrated to be required for Tbx5 limb expression in vivo (Hbs1-6), we performed EMSAs using labelled double-stranded oligonucleotide probes identical to Hbs2 (not shown) and Hbs5 in the MscI(b) construct (Fig. 5).
Fig. 5.
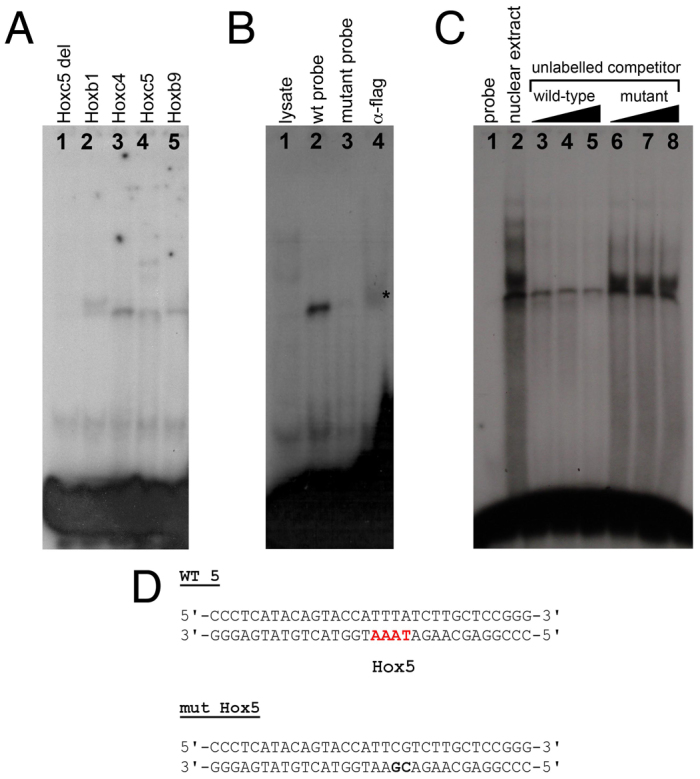
Direct binding of Hox proteins to the in-silico-predicted Hox binding sites. (A) Binding of in vitro translated mouse Hox proteins to the predicted Hox binding site 5 (Hbs5). Hoxc5 delHD provides a control. (B) Binding assay to mutant oligonucleotides and supershift assays for the predicted Hbs5. Lane 1, lysate control; lane 2, flag-tagged mouse Hoxc4 binding to labelled wild-type double-stranded oligonucleotide; lane 3, flag-tagged Hoxc4 fails to bind to labelled mutant double-stranded oligonucleotide; lane 4, supershift of the Hoxc4-Hbs5 complex with anti-flag antibody (asterisk). (C) Nuclear extracts obtained from prospective E9 forelimb regions were subjected to EMSA with labelled double-stranded oligonucleotide containing Hbs5. To test for the specificity of the shifted bands we performed competition analyses with excess unlabelled wild-type or mutant double-stranded oligonucleotide (100× molar excess, lanes 3, 6; 200×, lanes 4, 7; 500×, lanes 5, 8). (D) Double-stranded oligonucleotides used for EMSA contained mutated (mut Hox5, bold text) or wild-type (WT 5) Hbs5 (red).
We first used these oligonucleotides to test the direct binding of specific in vitro translated Hox proteins. As shown in Fig. 5A, murine Hoxb1, Hoxc4, Hoxc5 and Hoxb9 (lanes 2-5) can bind to the oligonucleotide sequence that contains Hbs5. As a control, we used in vitro translated Hoxc5 delHD construct to show that this binding was dependent on the Hoxc5 homeodomain, as this truncated protein did not produce a slower migrating band (Fig. 5A, lane 1). To test the specificity of these DNA-protein complexes, we assayed whether binding could be abolished when labelled double-stranded oligonucleotides with mutations in putative Hox binding sites were included. In addition, we carried out supershift experiments by adding an antibody against a flag epitope present at the C-terminus of our in vitro translated Hoxc4 protein. As shown in Fig. 5B, in vitro translated Hoxc4 is not able to bind to the mutant version of the Hbs5 oligonucleotide (lane 3), and addition of an anti-flag antibody that recognises the epitope in the Hoxc4-flag chimaeric protein supershifts the product of Hoxc4 binding to the wild-type oligonucleotide (lane 4, asterisk), indicating that this binding is specific.
Finally, we investigated whether this specific DNA-protein association occurs in vivo using nuclear extracts from the tissue where this interaction should occur, namely the rostral region of early E9 embryos. We performed EMSAs in which a radiolabelled double-stranded oligonucleotide containing Hbs5 was mixed with nuclear extracts obtained from dissected rostral tissues. As shown in Fig. 5C, we detected bands corresponding to DNA-protein complexes that can be specifically competed by addition of unlabelled wild-type (lanes 3-5) but not mutant (lanes 6-8) double-stranded oligonucleotide.
DISCUSSION
Hox genes regulate forelimb expression of the mouse Tbx5 gene
Our results demonstrate that a 361 bp region located in intron 2 of the mouse Tbx5 gene (+2600 to +2960) is sufficient to initiate the early forelimb-restricted expression of Tbx5. We further show that deletion of this core element from a fragment spanning 10 kb of Tbx5 5′UTR abolishes its ability to drive forelimb expression. Previously, we have shown that this initial pulse of Tbx5 expression is crucial for forelimb initiation and that expression at later stages is dispensable for continued limb outgrowth (Hasson et al., 2007). Using (1) site-directed mutagenesis linked to in vivo reporter analyses in transgenic mice, (2) in vivo chick electroporation and (3) in vitro DNA-protein binding assays, we demonstrate that Hox genes are direct upstream regulators of Tbx5 and that they provide a crucial input that determines the forelimb-restricted expression of the mouse Tbx5 gene, as we previously hypothesized (Gibson-Brown et al., 1998; Minguillon et al., 2005; Ruvinsky and Gibson-Brown, 2000). Following careful analysis of Hox gene expression patterns in the LPM at pre-limb bud stages, we propose that Hox genes belonging to PG4 and PG5, with the exception of Hoxd4, represent good candidates for upstream regulators of Tbx5 expression owing to their expression domains in the rostral LPM that overlap with the region where Tbx5 expression is first detected.
We used our chick transactivation assays and the in vitro binding assays to show that PG4 and PG5 Hox genes can specifically bind to, and activate the expression of, our lacZ reporter construct. As shown in Fig. 3, PG4 and PG5 Hox genes can activate the expression of the reporter when co-electroporated into the developing chick hindbrain. However, murine Hoxb9 was also able to transactivate lacZ expression in this system (data not shown). Similarly, not only PG4 and PG5 Hox proteins but also PG1 and PG9 were able to bind to labelled double-stranded oligonucleotides containing specific binding sites in EMSA experiments (Fig. 5A). This is most likely due to promiscuity in binding of Hox proteins to Hox binding sites. Several Hox binding sites are present in the m5-5 construct used for co-electroporations. Nevertheless, PG4 and PG5 genes are the only Hox genes expressed in the appropriate rostral LPM domain at the relevant time, consistent with them having a role in activating Tbx5 expression in this tissue.
Conservation of the Tbx5 forelimb regulatory element
The forelimb/pectoral fin-specific expression pattern of Tbx5 is conserved in all vertebrate species analysed and one would therefore expect this regulatory element to be conserved. Phylogenetic footprinting has proven very useful for the identification of many essential regulatory regions (reviewed by Boffelli et al., 2004); however, this type of analysis failed to detect any conserved non-coding elements (CNEs) within Tbx5 intron 2. Although we found a CNE among amniotes (i.e. chicken and mammals) in silico, this was distinct from the forelimb-expression regulatory element we have characterised in vivo and in vitro, and this CNE was not able to drive reporter expression when assayed in our system (Fig. 1A, construct 0.8 RV). When we looked for and analysed CNEs more closely within the intron 2 sequence, we found various stretches of DNA that were conserved only between mammals. The most distant phylogenetic conservation detected lay within the opossum genome and the region of conservation contained only one of the six Hox binding sites (Hbs4) that we have shown here to be required for forelimb expression in the mouse.
There are a number of reasons why phylogenetic footprinting analyses failed to identify the forelimb regulatory element identified in vivo. The Hox binding sites we demonstrate here to be necessary to drive forelimb expression are spread over a 361 bp sequence that current predictive programs were unable to identify. Moreover, few direct Hox protein target sequences have been identified (Svingen and Tonissen, 2006) and the specific sequences that these proteins bind to can vary. This study identifies Tbx5 as a direct target of Hox genes. When more direct target sequences of Hox genes are known, it might be possible to align the regions that they bind to and potentially define longer DNA sequence motifs required for Hox responsiveness.
A novel role for Hox genes in limb development
It is well known that Hox genes are required for normal limb development. Loss- and gain-of-function experiments, largely including members of the HoxA and D clusters, have demonstrated the various roles that these genes play in anteroposterior and proximodistal patterning of the developing limb, both upstream and downstream of the sonic hedgehog morphogen (reviewed by Zakany and Duboule, 2007). Genes belonging to the HoxB and C clusters have received less attention because the entire deletion of each of these clusters does not generate any abnormal limb phenotypes (Medina-Martínez et al., 2000; Suemori and Noguchi, 2000). Study of the phenotypes of Hox mutants has been greatly hampered by the high degree of redundancy between Hox genes belonging to the same PG (i.e. Hox genes that occupy the same position in the different clusters). Several single and compound PG Hox mutants have been generated. Mice that are individually null for each of our candidate Hox genes (i.e. Hoxa4, Hoxa5, Hoxb4, Hoxb5, Hoxc4 and Hoxc5) have been generated (Boulet and Capecchi, 1996; Horan et al., 1994; Jeannotte et al., 1993; Ramírez-Solis et al., 1993; Rancourt et al., 1995) and none display any unusual forelimb phenotypes. Compound mutants have been generated for three of the four PG4 genes (Hoxa4, Hoxb4 and Hoxd4) (Horan et al., 1995) as well as for all the PG5 genes (McIntyre et al., 2007). Still, no abnormal forelimb phenotypes were observed. In light of our results, the lack of a limb phenotype in Hoxb1 to Hoxb9 null mice (Medina-Martínez et al., 2000) can simply be explained by redundant function of other PG4 and PG5 Hox genes, such as those from the HoxA and C clusters. Similarly, the wild-type forelimb phenotype of mice carrying a deleted HoxC cluster (Suemori and Noguchi, 2000) can be explained by the redundant action of PG4 and/or PG5 genes in the HoxA and B clusters. Our results predict that only the deletion of at least all Hox genes belonging to PG4 and PG5 would lead to a complete lack of forelimbs, a phenocopy of the forelimbless Tbx5 knockout (Agarwal et al., 2003; Rallis et al., 2003).
Our work and that of others suggests that Hox genes pattern the LPM during early, pre-limb bud stages. Specific combinations of Hox genes expressed in the embryonic LPM correlate well with the type of limb that will subsequently develop (Cohn et al., 1997), and changes in Hox gene expression domains correlate with the absence of forelimbs in snakes (Cohn and Tickle, 1999). We show here that Hox genes belonging to PG4 and PG5 are specifically expressed in rostral LPM and are required to regulate forelimb-restricted expression of the mouse Tbx5 gene. Our data demonstrate that Hox genes act upstream of Tbx5, the earliest factor required for initiation of forelimb outgrowth, and identify Tbx5 as their direct transcriptional target (Fig. 6). These data show Hox gene expression in the LPM as the earliest step in the genetic cascade that leads to the initiation of forelimb outgrowth.
Fig. 6.
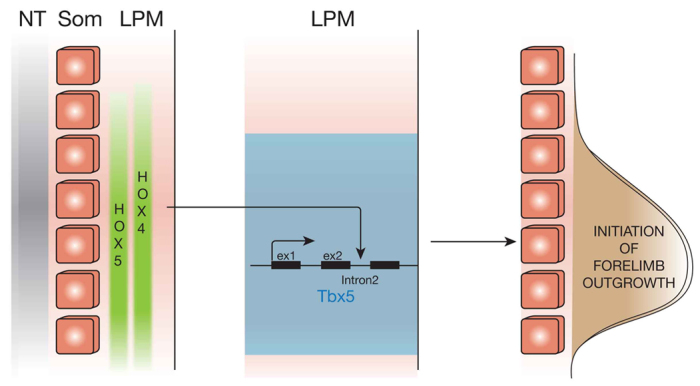
Model for the activation of Tbx5 in the LPM. Hox genes belonging to PG4 and PG5 are expressed in the presumptive forelimb LPM where they bind to the intron 2 early forelimb enhancer of the mouse Tbx5 gene to specifically activate its transcription and allow Tbx5-mediated initiation of forelimb outgrowth. ex1, exon1; ex2, exon2; LPM, lateral plate mesoderm; NT, neural tube; som, somites.
The elements required to drive hindlimb expression of the mouse Tbx4 gene have been described (Menke et al., 2008). Tbx4 is the paralogue of Tbx5 and both genes are thought to play equivalent roles in hindlimb and forelimb initiation, respectively. Two main differences are observed in the regulation of expression of this cognate pair of genes that suggest each has evolved a distinct mode of regulation. First, although a single enhancer is sufficient to drive expression of the Tbx5 gene in the forelimb field, two distinct regulatory regions are required to recapitulate the expression of Tbx4 in the hindlimb. Second, whereas the single Tbx5 forelimb enhancer identified here is present within the gene (+2600 to +2960), the two elements required to drive expression of Tbx4 in the hindlimb are located upstream (–10 kb) and downstream (+78 kb) of the Tbx4 gene. We and others have hypothesized that Hox genes might control the forelimb- and hindlimb-specific expression of Tbx5 and Tbx4, respectively. In this report we demonstrate that PG4 and PG5 Hox genes are indeed required to drive Tbx5-mediated forelimb outgrowth. Menke et al. (Menke et al., 2008) describe Pitx1 as an upstream regulator of the hindlimb expression of Tbx4. However, mutations in conserved Pitx1 binding sites in the hindlimb regulatory region do not abolish lacZ expression in mouse embryo reporter assays, indicating that other factors are required to drive robust Tbx4 expression in this territory (Menke et al., 2008). By analogy to our observations of Hox regulation of Tbx5 in the rostral LPM, it is tempting to speculate that such factors are posterior Hox genes, such as those of PG10 and PG11, that are specifically expressed in caudal regions of the LPM before overt hindlimb outgrowth.
Supplementary Material
Acknowledgments
We thank the staff of the Biological Services and Procedural Services sections, NIMR, for assistance with the animal work; Alex Gould, Patricia Serpente and Fabrice Prin for advice, discussion and exchange of constructs; and Jo Del Buono, Anna Kucharska, Jon Giblin, Xavier Franch-Marro, Lynne Fairclough, Rob Orford and Anita Abu-Daya for valuable technical help.
Footnotes
Funding
This work was supported by an EMBO Long-Term Fellowship and a Medical Research Council Career Development Fellowship to C.M. M.P.O.L. is funded by the Medical Research Council (U117560477). Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.084814/-/DC1
References
- Agarwal P., Wylie J. N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B. G. (2003). Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130, 623–633 [DOI] [PubMed] [Google Scholar]
- Boffelli D., Nobrega M. A., Rubin E. M. (2004). Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 5, 456–465 [DOI] [PubMed] [Google Scholar]
- Boulet A. M., Capecchi M. R. (1996). Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev. Biol. 177, 232–249 [DOI] [PubMed] [Google Scholar]
- Bray N., Dubchak I., Pachter L. (2003). AVID: A global alignment program. Genome Res. 13, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini T. D., Di Giacomo G., Salsi V., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L. (2006). Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133, 2263–2273 [DOI] [PubMed] [Google Scholar]
- Chapman D. L., Garvey N., Hancock S., Alexiou M., Agulnik S. I., Gibson-Brown J. J., Cebra-Thomas J., Bollag R. J., Silver L. M., Papaioannou V. E. (1996). Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn. 206, 379–390 [DOI] [PubMed] [Google Scholar]
- Cohn M. J., Tickle C. (1999). Developmental basis of limblessness and axial patterning in snakes. Nature 399, 474–479 [DOI] [PubMed] [Google Scholar]
- Cohn M. J., Patel K., Krumlauf R., Wilkinson D. G., Clarke J. D. W., Tickle C. (1997). Hox9 genes and vertebrate limb specification. Nature 387, 97–101 [DOI] [PubMed] [Google Scholar]
- Couronne O., Poliakov A., Bray N., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I. (2003). Strategies and tools for whole-genome alignments. Genome Res. 13, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J. (2007). Ancestral and recently recruited global control of the Hox genes in development. Curr. Opin. Genet. Dev. 17, 422–427 [DOI] [PubMed] [Google Scholar]
- Duboule D. (2007). The rise and fall of Hox gene clusters. Development 134, 2549–2560 [DOI] [PubMed] [Google Scholar]
- Gibson-Brown J. J., Agulnik S. I., Chapman D. L., Alexiou M., Garvey N., Silver L. M., Papaioannou V. E. (1996). Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech. Dev. 56, 93–101 [DOI] [PubMed] [Google Scholar]
- Gibson-Brown J. J., Agulnik S. I., Silver L. M., Niswander L., Papaioannou V. E. (1998). Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125, 2499–2509 [DOI] [PubMed] [Google Scholar]
- Gould A., Morrison A., Sproat G., White R. A., Krumlauf R. (1997). Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11, 900–913 [DOI] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H. L. (1992). A series of normal stages in the development of the chick embryo. Dev. Dyn. 195, 231–272 [DOI] [PubMed] [Google Scholar]
- Hasson P., Del Buono J., Logan M. P. (2007). Tbx5 is dispensable for forelimb outgrowth. Development 134, 85–92 [DOI] [PubMed] [Google Scholar]
- Hasty P., Ramírez-Solis R., Krumlauf R., Bradley A. (1991). Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature 350, 243–246 [DOI] [PubMed] [Google Scholar]
- Horan G. S., Wu K., Wolgemuth D. J., Behringer R. R. (1994). Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc. Natl. Acad. Sci. USA 91, 12644–12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan G. S., Ramírez-Solis R., Featherstone M. S., Wolgemuth D. J., Bradley A., Behringer R. R. (1995). Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 9, 1667–1677 [DOI] [PubMed] [Google Scholar]
- Itasaki N., Bel-Vialar S., Krumlauf R. (1999). ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1, E203–E207 [DOI] [PubMed] [Google Scholar]
- Jeannotte L., Lemieux M., Charron J., Poirier F., Robertson E. J. (1993). Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev. 7, 2085–2096 [DOI] [PubMed] [Google Scholar]
- Jeong S., Rokas A., Carroll S. B. (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–1399 [DOI] [PubMed] [Google Scholar]
- Kaufman M. H. (2001). The Atlas of Mouse Development, 2nd edn. Cambridge, UK: Academic Press; [Google Scholar]
- Krause A., Zacharias W., Camarata T., Linkhart B., Law E., Lischke A., Miljan E., Simon H. G. (2004). Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev. Biol. 273, 106–120 [DOI] [PubMed] [Google Scholar]
- Lemons D., McGinnis W. (2006). Genomic evolution of Hox gene clusters. Science 313, 1918–1922 [DOI] [PubMed] [Google Scholar]
- Logan M. (2003). Finger or toe: the molecular basis of limb identity. Development 130, 6401–6410 [DOI] [PubMed] [Google Scholar]
- Manzanares M., Bel-Vialar S., Ariza-McNaughton L., Ferretti E., Marshall H., Maconochie M. M., Blasi F., Krumlauf R. (2001). Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development 128, 3595–3607 [DOI] [PubMed] [Google Scholar]
- McIntyre D. C., Rakshit S., Yallowitz A. R., Loken L., Jeannotte L., Capecchi M. R., Wellik D. M. (2007). Hox patterning of the vertebrate rib cage. Development 134, 2981–2989 [DOI] [PubMed] [Google Scholar]
- Medina-Martínez O., Bradley A., Ramírez-Solis R. (2000). A large targeted deletion of Hoxb1-Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev. Biol. 222, 71–83 [DOI] [PubMed] [Google Scholar]
- Megason S. G., McMahon A. P. (2002). A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129, 2087–2098 [DOI] [PubMed] [Google Scholar]
- Menke D. B., Guenther C., Kingsley D. M. (2008). Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development 135, 2543–2553 [DOI] [PubMed] [Google Scholar]
- Mercader N., Leonardo E., Piedra M. E., Martínez-A C., Ros M. A., Torres M. (2000). Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961–3970 [DOI] [PubMed] [Google Scholar]
- Minguillon C., Logan M. (2003). The comparative genomics of T-box genes. Brief. Funct. Genomics Proteomics 2, 224–233 [DOI] [PubMed] [Google Scholar]
- Minguillon C., Del Buono J., Logan M. P. (2005). Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev. Cell 8, 75–84 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Papaioannou V. E. (2003). Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130, 2681–2693 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Harrelson Z., Kelly R. G., Papaioannou V. E. (2005). T-box genes in vertebrate development. Annu. Rev. Genet. 39, 219–239 [DOI] [PubMed] [Google Scholar]
- Papaioannou V. E. (2001). T-box genes in development: from hydra to humans. Int. Rev. Cytol. 207, 1–70 [DOI] [PubMed] [Google Scholar]
- Pearson J. C., Lemons D., McGinnis W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904 [DOI] [PubMed] [Google Scholar]
- Rallis C., Bruneau B. G., Del Buono J., Seidman C. E., Seidman J. G., Nissim S., Tabin C. J., Logan M. P. (2003). Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130, 2741–2751 [DOI] [PubMed] [Google Scholar]
- Ramírez-Solis R., Zheng H., Whiting J., Krumlauf R., Bradley A. (1993). Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell 73, 279–294 [DOI] [PubMed] [Google Scholar]
- Rancourt D. E., Tsuzuki T., Capecchi M. R. (1995). Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 9, 108–122 [DOI] [PubMed] [Google Scholar]
- Riddle R. D., Johnson R. L., Laufer E., Tabin C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Esteban C., Tsukui T., Yonei S., Magallon J., Tamura K., Izpisua Belmonte J. C. (1999). The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature 398, 814–818 [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Gibson-Brown J. J. (2000). Genetic and developmental bases of serial homology in vertebrate limb evolution. Development 127, 5233–5244 [DOI] [PubMed] [Google Scholar]
- Saito D., Yonei-Tamura S., Kano K., Ide H., Tamura K. (2002). Specification and determination of limb identity: evidence for inhibitory regulation of Tbx gene expression. Development 129, 211–220 [DOI] [PubMed] [Google Scholar]
- Stephens T. D., Beier R. L., Bringhurst D. C., Hiatt S. R., Prestridge M., Pugmire D. E., Willis H. J. (1989). Limbness in the early chick embryo lateral plate. Dev. Biol. 133, 1–7 [DOI] [PubMed] [Google Scholar]
- Suemori H., Noguchi S. (2000). Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev. Biol. 220, 333–342 [DOI] [PubMed] [Google Scholar]
- Summerbell D., Ashby P. R., Coutelle O., Cox D., Yee S., Rigby P. W. (2000). The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development 127, 3745–3757 [DOI] [PubMed] [Google Scholar]
- Svingen T., Tonissen K. F. (2006). Hox transcription factors and their elusive mammalian gene targets. Heredity (Edinb.) 97, 88–96 [DOI] [PubMed] [Google Scholar]
- Takeuchi J. K., Koshiba-Takeuchi K., Matsumoto K., Vogel-Höpker A., Naitoh-Matsuo M., Ogura K., Takahashi N., Yasuda K., Ogura T. (1999). Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature 398, 810–814 [DOI] [PubMed] [Google Scholar]
- Wellik D. M. (2007). Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 236, 2454–2463 [DOI] [PubMed] [Google Scholar]
- Zacchetti G., Duboule D., Zakany J. (2007). Hox gene function in vertebrate gut morphogenesis: the case of the caecum. Development 134, 3967–3973 [DOI] [PubMed] [Google Scholar]
- Zakany J., Duboule D. (2007). The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17, 359–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.