SUMMARY
Pluripotent embryonic stem cells (ESCs) maintain self-renewal and the potential for rapid response to differentiation cues. Both ESC features are subject to epigenetic regulation. Here we show that histone acetyltransferase Mof plays an essential role in the maintenance of ESC self-renewal and pluripotency. ESCs with Mof deletion lose characteristic morphology, alkaline phosphatase (AP) staining and differentiation potential. They also have aberrant expression of core transcription factors Nanog, Oct4 and Sox2. Importantly, the phenotypes of Mof null ESCs can be partially suppressed by Nanog overexpression, supporting that Mof functions as an upstream regulator of Nanog in ESCs. Genome-wide ChIP sequencing and transcriptome analyses further demonstrate that Mof is an integral component of ESC core transcription network and Mof primes genes for diverse developmental programs. Mof is also required for Wdr5 recruitment and H3 K4 methylation at key regulatory loci, highlighting complexity and interconnectivity of various chromatin regulators in ESCs.
INTRODUCTION
Embryonic stem cells (ESCs) are pluripotent cells capable of indefinite self-renewal and differentiation into all cell types. The maintenance of ES pluripotency status requires specific core transcription factors such as Oct4 (also known as Pou5f1), Sox2 and Nanog, which are the corner stones of an intricate and highly interconnected ESC transcription network or core regulatory circuitry (Chen et al., 2008; Macarthur et al., 2009; Orkin et al., 2008). They recruit multiple chromatin regulatory factors or complexes to promote activation of stemness genes while simultaneously allow for repression of differentiation genes (Orkin and Hochedlinger, 2011; Young, 2011). Two antagonistic chromatin methylation activities (i.e. Polycomb repression complex 2 (PRC2) and MLL family complexes) are shown to function coordinately with these core transcription factors in ESCs. The PRC2 complex methylates histone H3 K27 and functions to silence developmentally regulated genes. On the other hand, MLL family histone methyltransferases (HMTs) deposit histone H3 K4 methylation, which keeps lineage specific genes poised for activation as cells enter various differentiation pathways. The significance of H3 K4 and K27 methylation in regulating the ESC transcription program is best exemplified by the presence of ‘bivalent domains’ at many important regulatory regions, defined by high levels of both H3 K4 and K27 tri-methylation. These ‘bivalent domains’ are evolutionarily conserved and its resolution during ESC differentiation serves to commit ESCs into a specific lineage (Azuara et al., 2006; Bernstein et al., 2006; Pan et al., 2007).
In addition to histone methylation, the pluripotency status of ESCs is also regulated by histone acetylation. Addition of histone deacetylase (HDAC) inhibitors prevents ESC differentiation and increases efficiency of iPSC (induced pluripotency stem cells) induction (Feng et al., 2009). Histone acetylation also supports ‘hyper-dynamic’ chromatin conformation (Meshorer, 2007; Niwa, 2007) and hyperactive transcription states (Efroni et al., 2008), two common signatures of pluripotent cells. Upon differentiation, the chromatin structure of ESCs becomes more compact and overall transcription is reduced (Aoto et al., 2006; Park et al., 2004). This process is accompanied by global reduction of pan-acetylation of histone H3 and H4 (Kobayakawa et al., 2007). Consistent with the importance of histone acetylation in ESC function, genetic ablation or knockdown of several histone acetyltransferases (HATs) such as Tip60, p300, Gcn5 led to aberrant expression of lineage specific genes and profound defects in ESC differentiation (Chen et al., 2008; Fazzio et al., 2008; Lin et al., 2007; Zhong and Jin, 2009). Notably, these HATs do not affect expression of core pluripotency factors Oct4, Nanog and Sox2 (Fazzio et al., 2008; Lin et al., 2007; Zhong and Jin, 2009). Instead, they function mostly at downstream differentiation processes.
Histone acetyltransferase Mof (also called MYST1 or KAT8) is a highly conserved MYST family HAT. MOF was originally described as an essential component of the X chromosome dosage compensation complex (DCC) in Drosophila, causing a two-fold increase in expression of X-linked genes in male flies (Conrad and Akhtar, 2011; Gelbart and Kuroda, 2009; Lucchesi et al., 2005). In mammals, MOF is essential for vertebrate development and constitutive ablation of Mof leads to peri-implantation lethality in mouse embryos (Gupta et al., 2008; Thomas et al., 2008). Mof −/− embryos showed massive abnormal chromatin aggregations, suggesting a crucial role for Mof in maintenance of chromatin structures in vivo. Mammalian MOF was initially purified in a WDR5-containing complex (Dou et al., 2005). Later in vitro biochemical studies show that Mof resides in two distinct complexes in mammals: the MOF-MSL complex and the MOF-MSL1v1 complex (Li and Dou, 2010), which are either physically or functionally connected with H3 K4 methyltransferase MLL. In brief, the MOF-MSL1v1 complex physically interacts with the MLL complex through the commonly shared component WDR5 and coordinates with MLL in transcription activation (Dou et al., 2005; Li et al., 2009); On the other hand, the MOF-MSL complex is able to stimulate H3K4me3 through H2BK34ub mediated trans-tail regulation (Wu et al., 2011). Given the close connection of MOF and H3 K4 methylation, the direct interaction between MOF and WDR5 as well as the recent demonstration that WDR5 mediates self-renewal and reprogramming (Ang et al., 2011), we decided to examine whether MOF plays a role in ESC fate determination and whether MOF mediated H4 acetylation contributes to ESC pluripotency.
Using the conditional knockout ES cell lines for Mof, here we show that Mof is essential for ESC self-renewal and pluripotency. Mof deletion leads to loss of ESC self-renewal and defects in embryoid body (EB) formation, which are accompanied by reduced H4 K16 acetylation (K16ac) and global changes in ESC transcriptome. Importantly, unlike other HATs, Mof directly regulates expression of ‘core’ ESC transcription factors Nanog, Oct4 and Sox2 and Mof null phenotypes can be partially rescued by ectopic Nanog expression. Altogether, our data provide strong support for a critical and unique role of Mof in regulating ESC core transcription network.
RESULTS
Mof expression and H4 K16ac are down regulated during ESC differentiation
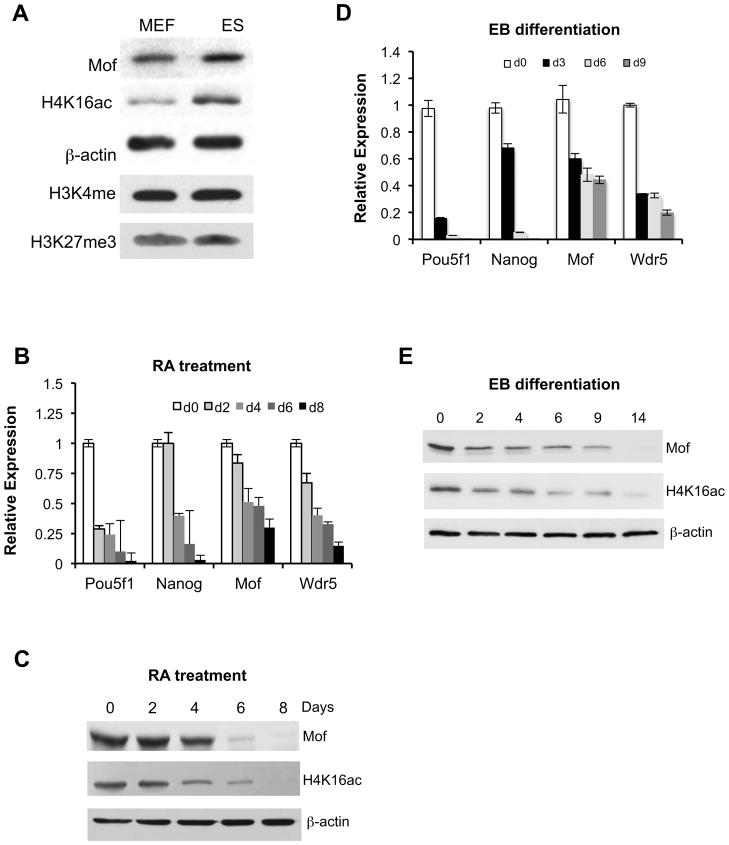
Recent studies show that histone modifications, especially histone H3K4me3 and H3K27me3 play important roles in regulating ESC self-renewal and pluoripotency. Given the interaction of MOF with MLL (Dou et al., 2005) and the correlation of H3K4me3 and H4K16ac at transcriptionally active genes (Ruthenburg et al., 2011), we decided to examine whether Mof and its acetyltransferase activity play a role in murine ESC functions. To this end, we first compared levels of Mof and H4K16ac in ESCs to those of mouse embryonic fibroblasts (MEFs). As shown in Figure 1A, the levels of Mof and H4K16ac were significantly higher in ESCs than those in MEFs. Differences in histone H3 K4me3 and K27me3 in ESCs versus MEFs were moderate in comparison (Figure 1A). Furthermore, when ESCs were subjected to either retinoic acid (RA) induced differentiation (Figure 1B and 1C) or spontaneous differentiation (Figure 1D and 1E), Mof transcript and protein levels were gradually down regulated, which were in parallel with down regulation of ESC pluripotency genes Pou5f1 (Oct4) and Nanog (Figure 1B and 1D). As a control, we also observed a similar down regulation of Wdr5 in differentiating ESCs, consistent with the previous report (Ang et al., 2011).
Figure 1. Mof is down regulated during ESC differentiation.
(A) Immunoblots for proteins from mouse embryonic fibroblasts (MEFs) and embryonic stem cells (ESCs) as indicated on top. Antibodies were indicated on left. (B) Real-time PCR and (C) Immunoblot analyses for RA-induced ESC differentiation. (D) Real-time PCR and (E) Immunoblot analyses for ESC differentiation during EB formation. In (B, D), fold changes of each transcript relative to its expression in day 0 EB formation were presented. For C, E, β-actin was used as the loading control.
Establishing 4-OHT inducible Mof knockout ESC lines
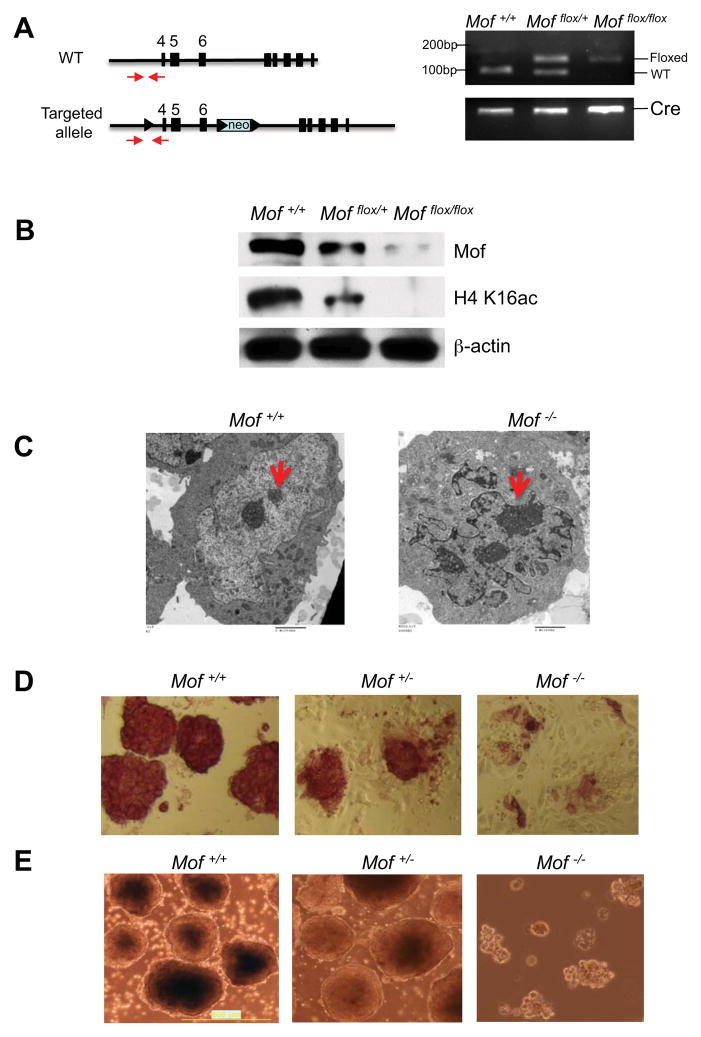
Down regulation of Mof expression during ESC differentiation is intriguing since this process is concomitant with changes in chromatin conformation and dynamism (Gaspar-Maia et al., 2011). To determine the role of Mof in ESCs, we derived inducible Mof knockout ESC lines from the Mof flox/flox, Cre-ER™ mouse model we previously described (Li et al., 2010). In this model, floxed Mof alleles (i.e. exons 4–6) can be deleted upon 4-OHT induced expression of Cre recombinase (Figure 2A). This leads to Mof protein degradation and loss of global H4K16ac (Li et al., 2010). The primary ESC cell lines including Cre-ER™ positive Mof flox/flox, Mof flox/+ and Mof +/+ were obtained from E3.5 dpc embryos after intercrossing of Mof flox/+, Cre-ER™ mice (Figure 2A). Successful generation of Mof +/+, Mof +/− and Mof −/− ESCs was confirmed by genotyping and immunoblots (Figure 2A and 2B). For Mof deletion, these cells were subjected to continuous 4-OHT treatments for four days. As shown in Supplementary Figure 1A, day 4 is the earliest time point that we were able to achieve complete Mof deletion and observe significant reduction in the Mof protein level. We decided to use this time point for all the experiments described in this study. Consistent with the role of Mof and H4K16ac in regulating higher order chromatin structures (Robinson et al., 2008; Shogren-Knaak et al., 2006), Mof deletion led to massive chromatin compaction with significant increase of densely stained heterochromatin in the nucleus by electron microscopy studies (Figure 2C). Mof deletion eventually led to growth arrest and cell death of ESCs (Supplementary Figure 1B). However, the chromatin aggregation shown here (day 4) was not a result of cell death. At this time point, cell cycle index of Mof knockout ESCs was comparable to that of wild type cells (Supplementary Figure 1C–E). The generation of inducible Mof knockout ESC lines allowed us to study the effects of Mof deletion on ESC functions in a defined genetic background.
Figure 2. Mof is essential for ESC self-renewal.
(A) Left, schematic for wild type and Mof knockout alleles. Genotyping primers (red arrows) were indicated. Right, genotyping results for wild type, floxed Mof alleles as well as Cre-ER™ by PCR. (B) Immunoblots for Mof and H4 K16ac in Mof +/+, Mof flox/+ and Mof flox/flox cells after 4-OHT treatment. Immunoblot for β-actin was used as the loading control. (C) Electron microscopy images of wild type (left) and Mof knockout nuclei (right). Densely stained heterochromatin was indicated by arrow. Scale bars, 2 micron. (D) Alkaline phosphatase staining of Mof +/+, Mof +/− and Mof −/− ESCs. (E) Light microscopy images of day 4 EB for Mof +/+, Mof +/− and Mof −/− ESCs. Scale bars, 0.5mm. Also see Supplementary Fig 1.
Mof is required for ESC self-renew and differentiation
Apparent changes in ESC morphology were observed upon Mof deletion. Mof −/− ESCs became flattened and elongated with reduced cell-cell contacts and failed to form compact colonies in culture (Figure 2D, right panel). These morphological changes were not due to defects in ESC proliferation since similar changes, albeit to a less extent, were also observed for Mof +/− ESC (Figure 2D, middle panel), which had no detectable growth difference from the wild type cells (Supplementary Figure 1F and data not shown).
Consistent with morphological changes, Mof −/− ESCs had very weak alkaline phosphatase (AP) staining compared to that of Mof +/+ and Mof +/− ESCs (Figure 2D), suggesting loss of ES self-renewal capability. Moderate decrease of AP staining was also observed for Mof +/− cells (Figure 2D and Supplementary Figure 2A). We further examined the ability of Mof −/− ESCs to aggregate in suspension to form embryoid bodies (EBs). As shown in Figure 2E, Mof −/− ESCs failed to aggregate and most cells remained dispersed in suspension culture. In contrast, both Mof +/+ and Mof +/− ESCs efficiently aggregated and eventually developed into cystic structures (Figure 2E and data not shown). We further examined differentiation of three primitive germ layers in Mof +/− EBs. We found that expression of marker genes for all three germ layers were down regulated in Mof +/− EBs (Supplementary Figure S2B-E). The effects of deleting one Mof allele were moderate, consistent with largely normal phenotypes of Mof +/− mice (data not shown). Since Mof −/− ESCs were not able to form EBs, we decided to delete Mof after ESC aggregation and examine whether Mof played a role at later differentiation steps. We examined expression of hematopoietic genes (i.e. Tal 1, Lmo2 and Runx1), which were highly expressed in late EBs. These genes were significantly compromised in Mof −/− EBs, suggesting impairment of hematopoietic differentiation (Supplementary Figure S3A). Taken together, ESCs with Mof deletion had defects in several characteristic features of stem cells: morphology, AP activity and EB formation/differentiation. These results suggest that Mof is essential for ESC functions.
Mof deletion led to aberrant expression of ESC core transcription factors and differentiation marker genes
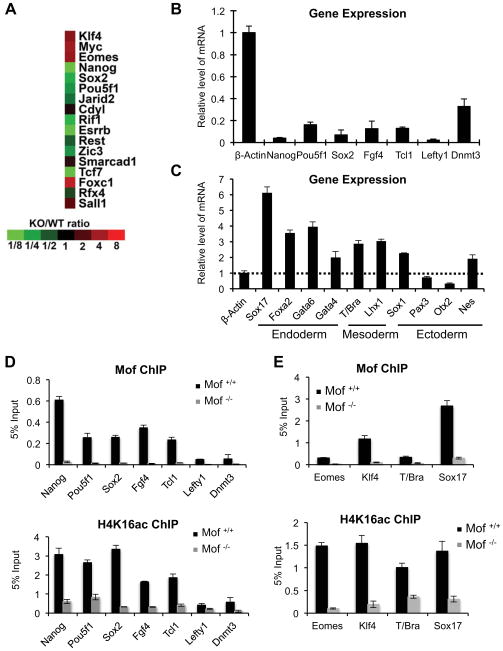
To gain insights into the function of Mof in ESCs and to rule out that loss of self-renewal and pluripotency observed in Mof −/− cells was due to a general defect in cell proliferation and/or increased apoptosis, we performed gene expression analyses for Mof +/+ and Mof −/− ESCs by microarray. We found that Mof deletion had profound impacts on ESC transcriptome. There were 4,475 genes that were differentially expressed by more than 2 fold upon Mof deletion (Table S1 and S2). About equal number of genes were up- (2,081) or down (2,394) regulated in Mof −/− ESCs (Table S1 and S2). Of note, fold changes in gene expression upon Mof deletion were generally small, with mean fold change at ~2.8 (Figure 7C, total) for both up and down regulated genes. Consistent with Mof playing an important role in ESCs, we found changes in expression of Oct4, Nanog and most of their conserved joint targets (Figure 3A and (Loh et al., 2006)). Most of these genes (e.g. Oct4, Nanog, Rif1, Esrrb, Zic3, Tcf7, Jarid2 and Rest) were significantly down regulated with the exception of Klf4 and Myc, whose expressions were increased. Microarray results for key ESC regulators were confirmed by RT-PCR (Figure 3B). In addition to changes in expression of ESC core transcription factors, Mof deletion also led to aberrant expression of differentiation markers for all three primitive germ layers. They included Sox17, Foxa2, Gata6 and Gata4 for primitive endoderm, T/Bra and Lhx1 for primitive mesoderm as well as Sox1, Pax3, Otx2 and Nestin for neuroectoderm (Figure 3C). Most of these differentiation genes were up regulated, supporting that Mof null phenotypes were not simply due to general loss of cell viability.
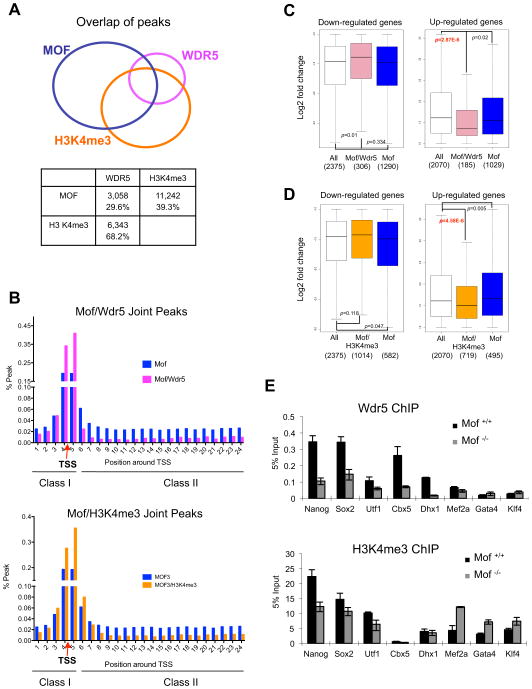
Figure 7. Mof regulates Wdr5 binding at key ESC loci.
(A) Top, Venn diagram for direct physical overlap of binding peaks for Mof (blue), Wdr5 (pink) and H3 K4me3 (orange) (Ang et al., 2011). Total number and percentage of overlapping peaks relative to Wdr5, or H3 K4me3 peaks were summarized in the table below. (B) Distribution of Mof and Mof/Wdr5 joint peaks (top) or Mof/H3K4me3 joint peaks (bottom) as class I or class II peaks. Red arrow, TSS. Y-axis, % peaks relative to total peaks within the defined region. X-axis, each bin represents a 500bp region. (C) The box plots for fold changes in expression of total (white), Mof/Wdr5 (pink) and Mof only (blue) target genes. (D) The box plots for fold changes in expression of total (white), Mof/H3K4me3 (orange) and Mof only (blue) target genes. For (C, D), bottom and top of the boxes correspond to the 25th and 75th percentiles and the internal band is the 50th percentile (median). The plot whiskers extending outside the boxes correspond to the lowest and highest datum within 1.5 interquartile ranges. p-values were calculated using non-paired Wilcoxon tests as indicated. The number of genes in each category was indicated on bottom. Left, down regulated gene set. Right, up regulated gene set. (E) ChIP experiments for Wdr5 (top) and H3 K4me3 (bottom) at selected joint target genes in WT and Mof −/− ESCs. The antibodies used for ChIP were indicated on top. Signals for each experiment were normalized to 5% input. Means and standard deviations (as error bars) from at least three independent experiments were presented. Also see Supplementary Figure 7.
Figure 3. Mof regulates ESC core transcription network.
(A) Heat map of expression of conserved Nanog and Oct4 joint targets (Loh et al., 2006) in WT and Mof −/− ESCs. Fold change of gene expression relative to WT ESCs was indicated at bottom. (B, C) Real-time PCR analyses for pluripotency (B) and differentiation genes (C) in Mof −/− and Mof +/+ ESCs as indicated. All mRNA levels were normalized against β-actin and were presented as relative expression in Mof null versus wild type ESCs. (D) ChIP for pluripotency genes that were down regulated in Mof −/− ESCs. (E) ChIP for differentiation genes that were up regulated in Mof −/− ESCs. For (D–E), primer sets were designed corresponding to Mof binding peaks identified by ChIP-seq (indicated in Supplementary Figure 6). The antibody was indicated on top. Signals for each experiment were normalized to 5% input. For (B–E), Means and standard deviations (as error bars) from at least three independent experiments were presented. Also see Supplementary Fig 6.
To examine whether genes with changed expression were direct Mof targets, we performed ChIP analyses for Mof and H4K16ac on selected gene promoters. As shown in Figure 3D, Mof directly bound to pluripotency genes including Nanog, Pou5f1, Sox2, Fgf4 Lefty1 and Tcl1 (Figure 3D). Down-regulation of these genes in Mof −/− ESCs (Figure 3B) coincided with loss of Mof binding and H4K16ac (Figure 3D). Our result that Mof directly regulates Nanog and Oct4 makes Mof a unique histone acetyltransferase in regulating ESC self-renewal genes. In contrast, all other HATs studied insofar including Tip60, Gcn5 and p300/CBP showed little effects on transcription of Oct4, Nanog and Sox2 after knockout or knockdowns. Instead, they were important for regulating downstream ESC differentiation processes (Fazzio et al., 2008; Lin et al., 2007; Zhong and Jin, 2009). In addition to examining Mof binding at ESC core transcription factor loci, we also checked Mof binding at several genes whose expression was up regulated by Mof deletion. Surprisingly, we found that some of the up regulated genes (i.e. Eomes, Klf4, T/Bra and Sox17) had Mof binding at coding regions and their expression changes were concurrent with loss of Mof and H4K16ac (Figure 3E). Bindings of Mof at these genes and at Sox1, GATA4 and Nestin were also confirmed by ChIP-sequencing (ChIP-seq) analyses (Supplementary Figure 6C, see below). Despite modest fold changes, these results suggest that Mof deletion can lead to both increased and decreased expression of its direct targets.
Genome-wide mapping of Mof binding sites in ESC by ChIP-sequencing
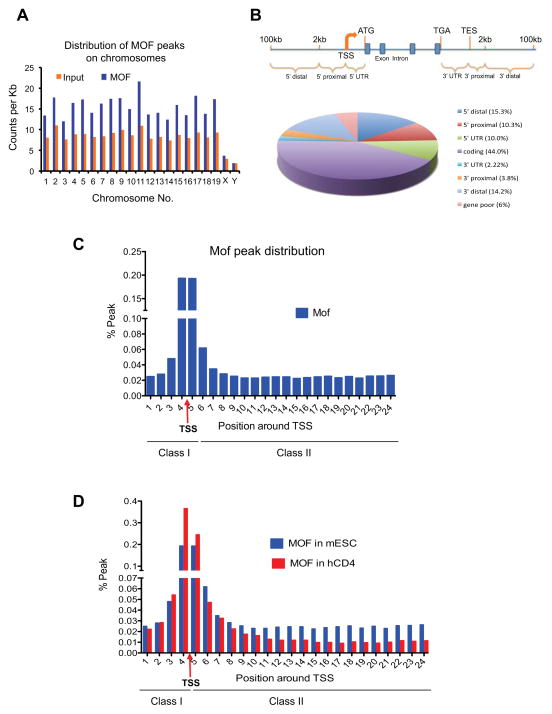
Increased expression of Mof direct targets in Mof −/− ESCs is surprising considering its widely accepted role as a transcription co-activator. In order to assess the function of Mof at global levels, we decided to identify Mof direct targets in murine ESC genome by ChIP-seq. ChIP-seq of input DNA was used as the control and duplicated biological samples were sequenced and analyzed. The ChIP-seq results showed that Mof distributed broadly in ESCs, enriched (over input) on all autosomes in the mammalian genome (Figure 4A). In contrast to highly enriched male X-chromosome binding in Drosophila melanogaster, Mof had minimal binding on sex chromosomes in mammal (Figure 4A). To examine Mof distribution relative to gene structure, we divided the genome into eight categories: 5′ distal (2–100kb upstream of transcription start site (TSS)), 5′ proximal (0–2kb upstream of TSS), 5′UTR (TSS to ATG), coding, 3′UTR (TGA to transcription end site (TES)), 3′ proximal (0–2kb downstream of TES) and 3′ distal (2–100kb downstream of TES) regions respectively. The rest of loci were referred to as gene poor regions. Peak counts after normalizing against category sizes were included in Supplementary Figure 4A. Majority of Mof binding sites (~56%) were mapped to the transcribed region in the genome including ~44% peaks in coding regions, ~10% in 5′UTR and ~2% in 3′ UTR respectively. The distribution of Mof was more pronounced towards 5′ end of genes with 20% peaks at 5′ proximal or 5′UTR as compared to ~6% binding at 3′UTR and 3′ proximal regions (Figure 4B). Furthermore, ~30% of Mof peaks were at either distal or gene-poor regions (Figure 4B). The functional significance of gene distal binding for Mof remained to be explored. Screen shots for Mof peaks at representative genes were included in Supplementary Figure 6 and Supplementary Figure 7.
Figure 4. ChIP-seq analysis of Mof binding sites in ESCs.
(A) Chromosome-distribution of Mof binding peaks in mouse ESCs. Y-axis, count of ChIP-seq reads per kilo base. X-axis, chromosome name. (B) Distribution of Mof binding sites relative to nearest Refseq genes. Top, schematic for eight counting categories. Bottom, pie chart for percentage distribution of Mof peaks in each category. (C) Distribution of Mof peaks in a 12kb region from −2kb to +10kb around TSS (indicated by red arrow). Y-axis, percentage of Mof peaks relative to total Mof peaks within the defined region. X-axis, bin numbers, with each represents a 500bp region. Mof peaks were indicated as class I and class II peaks on bottom. (D) Comparison of Mof distribution within the defined 12kb region in mESCs (blue) and human CD4+ cells (red). TSS and Class I and II sites were indicated on bottom. Also see Supplementary Fig 4
We further analyzed Mof binding peaks within a 12kb region surrounding annotated TSS. To this end, Mof peaks were counted and grouped into 24 bins with 500bp intervals starting from −2kb to +10kb regions. As shown in Figure 4C, Mof peaks centered on TSS and ~40% Mof peaks were within 500bp of TSS. Furthermore, relatively low but persistent Mof binding was found throughout the 10kb region downstream of TSS, which accumulatively accounts for 50% Mof peaks within the defined 12kb region. To gain further insights on Mof binding in ESCs, we divided Mof peaks as two classes: class I includes peaks at −2 to +0.5 kb region (bins 1–5), representing promoter and TSS proximal Mof binding; and class II includes peaks at +0.5 to +10 kb region (bins 6–24), representing Mof binding at downstream coding region (Figure 4C and Supplementary Figure 4B). We then compared our Mof ChIP-seq results with those of primary human resting CD4+ cells (hCD4+) (Wang et al., 2009). Consistent with ESC specific regulation, no MOF binding was found at Nanog, Oct4 or Sox 2 genes in hCD4+ cells (Supplementary Figure 6B, Wang et al., 2009). At global level, Mof binding in hCD4+ cells was significantly enriched at the 5′ end of genes (41.8%), in contrast to 23.2% of MOF binding at coding regions (Supplementary Figure 4C). The difference in Mof peak distribution between mESCs and hCD4+ cells was not due to differences in category breakdown of two genomes, which was about the same (Supplementary Figure 4A and 4C). Consistently, analyses of Mof peaks in the defined 12kb region near TSS showed significant enrichment of Mof class I peaks (71.4%) and less class II peaks (29.6%) in the differentiated hCD4+ cells as compared to mESCs (48.8% class I and 51.2% class II respectively). The basis for different Mof distribution in these two cells remains to be decided. However, broader Mof distribution downstream of TSS is consistent with the ‘hyper-dynamic’ chromatin conformation (Meshorer, 2007; Niwa, 2007) and hyperactive transcription states (Efroni et al., 2008) of ESCs.
Mof has a broad role in regulating ESC transcriptome
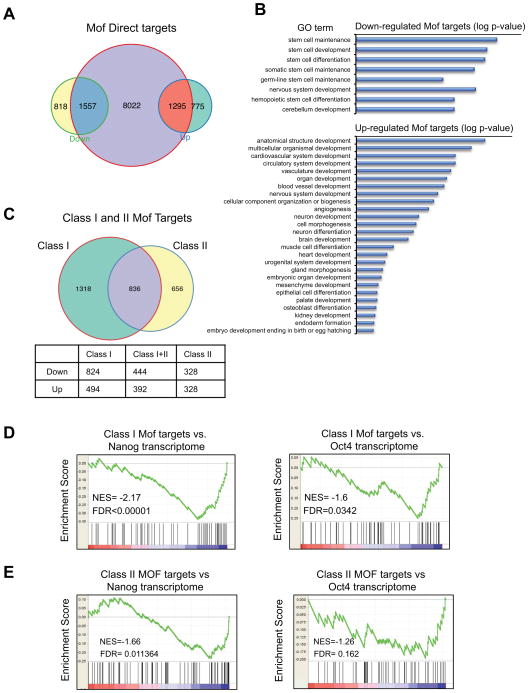
To understand the direct function of Mof in regulating ESC transcriptome, we cross-referenced the ChIP-seq results with gene expression analyses. We found that among genes changed expression in Mof −/− ESCs, 1,557 down- (~65%) and 1,295 up- (~62.5%) regulated genes had Mof binding sites (Figure 5A). Consistent with global changes in Mof transcriptome, changes in expression of Mof direct targets were modest, with mean fold change around 2.5 (Figure 7C). Gene ontology (GO) term enrichment analyses of differentially expressed Mof targets confirmed that Mof, as a general transcription cofactor, is indeed involved in many biological processes such as gene expression, cell cycle regulation, DNA repair and metabolic process (Supplementary Table S3 and S4) (Li et al., 2010). These pathways were largely down regulated upon Mof deletion. When developmental pathways were examined, we found that down-regulated Mof targets were highly enriched for stem cell maintenance, development and differentiation (p < 10−5, Figure 5B). In contrast, up-regulated Mof targets were highly enriched in cellular differentiation and various developmental programs (Figure 5B). The down regulation of stem cell genes and up regulation of multi-lineage differentiation genes in Mof −/− ESCs at global level support our results at selected gene targets. Interestingly, most of the up regulated differentiation genes shown in Figure 3 had Mof binding sites at the downstream coding regions (Supplementary Figure 6).
Figure 5. Mof regulates Nanog specific ESC core transcription network.
(A) Venn diagram for overlap of Mof bound genes (yellow) and genes that were either up regulated (blue) or down regulated (orange) in Mof −/− ESCs. Fisher’s exact test (p< 2.2×10−16) was performed to test for statistical significance of enrichment of up or down regulated genes with direct Mof binding. (B) GO term analyses for Mof down regulated genes (top) and Mof up regulated genes (bottom). Selected developmental pathways were presented and log p-value was used to rank the enrichment. (C) Top, Venn diagram for overlap of Mof targets with class I (green) or class II (yellow) binding sites. Bottom, a table for number of genes that were up or down regulated in each category. (D, E) GSEA of Mof targets with class I binding sites (D) or class II binding sites (E) and Nanog (left) or Oct4 (right) transcriptome (Ang et al., 2011). NES, normalized enrichment score; FDR (p value), false discovery rate. Also see Supplementary Figure 4 and 5.
Given the distinct Mof binding within the 12kb region of TSS, we further characterized Mof target genes based on whether Mof binding is in promoter and TSS proximal region (class I) or at gene bodies (class II) (Figure 5C). We found that significantly more genes with exclusive class I Mof binding sites were down regulated (824 vs. 494) upon Mof deletion (single-sided Fisher’s exact test, p=3.6E-6, Figure 5C). In contrast, significant number of genes with exclusive class II Mof binding sites were up regulated upon Mof deletion (single-sided Fisher’s exact test p=0.0098, Figure 5C). These results imply that distinct Mof binding patterns along target genes may reflect real functional difference for Mof in transcription regulation. Consistent with GO term analyses for Mof transcriptome, class II Mof targets were enriched for genes involved in cell differentiation or tissue/organ development, many of which were up regulated upon Mof deletion (data not shown).
Mof specifically regulates Nanog core transcription network
Given that Mof deletion in mESCs led to loss of self-renewal (Figure 2) and down regulation of stem cell maintenance genes (Figure 5C) including core transcription factors Nanog, Pou5f1 (Oct4) and Sox2 (Figure 3), we hypothesized that Mof may play an important role in ESC core transcription network. To test this, we first compared the Mof transcriptome with those of core transcription factors reported in the literature to see if there were any interconnectivity (Ang et al., 2011; Ivanova et al., 2006; Loh et al., 2006). We performed gene set enrichment analyses (GSEA) for Mof direct targets with those of several core ESC transcription factors including Nanog, Oct4, Esrrb, Tbx3 and Sall4. Interestingly, significant enrichment was only found for Mof and Nanog transcriptome (p<0.00001, Figure 5D), whereas there was no statistically significant enrichment for other ESC core transcription factors (i.e. Oct4, Esrrb, Tbx3 and Sall4) (Figure 5D and Supplementary Figure 5A–C). Furthermore, when we performed separate GSEA for Nanog transcriptome and Mof targets that had class I or class II binding sites, only Mof targets with class I binding sites show significant enrichment of Nanog regulated genes (Figure 5D). No enrichment between Mof targets with class II only binding sites and Nanog transcriptome were found (p=0.011, Figure 5E). Importantly, in corroborate with the observation that class I Mof targets were largely down regulated upon Mof deletion, correlation of Mof and Nanog transcriptome is mostly for the down-regulated gene sets (NES=−2.17, Figure 5D). Similar GSEA for Oct4 transcriptome did not identify significant correlation with either class I or class II Mof targets (Figure 5D and 5E, right panel).
In addition to transcriptome analyses, we also compared Mof binding peaks with those of Nanog (Ang et al., 2011). We found that ~79% Nanog target genes had Mof binding (Fisher’s exact test p< 10−16, Supplementary Figure 4C). Importantly, distribution of average Mof/Nanog joint peaks showed enrichment toward 5′ end of genes compared to Mof alone peaks (Supplementary Figure 4D), in agreement with our finding that genes with class I Mof binding sites are specifically involved in Nanog dependent transcription regulation in ESCs (Figure 5D). Altogether, these results strongly argue that Mof is an integral part of Nanog mediated ESC core transcription network.
Overexpression of Nanog rescues Mof null phenotypes in ESCs
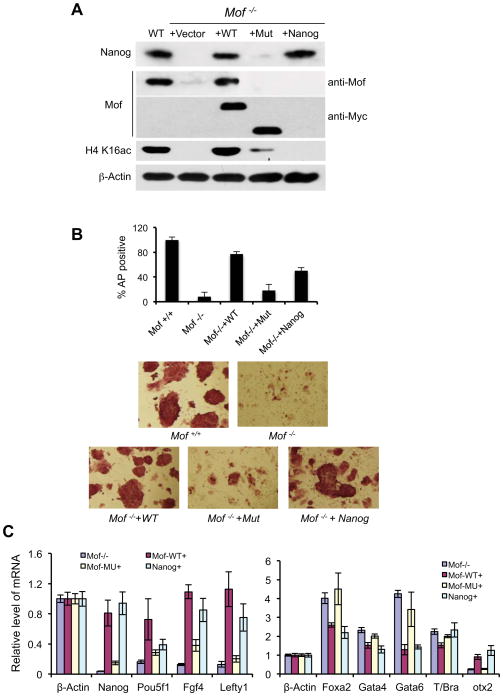
Nanog is a highly divergent homeodomain-containing protein commonly bestowed a central position in the transcriptional network of pluripotency (Chambers et al., 2007; Mitsui et al., 2003; Silva et al., 2009). Given the direct regulation of Nanog expression by Mof (Figure 3B) and specific GSEA enrichment of Mof and Nanog regulated genes (Figure 5D), we decided to test whether overexpression of Nanog rescues self-renewal defects of Mof −/− ESCs. To this end, we stably transfected Mof flox/flox, Cre-ER™ cells with a Nanog expressing vector (Ito et al., 2010). As controls, we also made cell lines that stably express wild type Mof or Mof truncation mutant (i.e. Δ173–257aa) that is enzymatically deficient (Li et al., 2010). As shown in Figure 6A, levels of exogenous Mof, Mof mutant and Nanog proteins were comparable to endogenous protein levels in Mof flox/flox, Cre-ER™ ESCs. Endogenous Mof was then deleted by 4-OHT treatments before the experiment. Consistent with the result that Mof regulated Nanog expression (Figure 3B), Nanog protein level was drastically lower in Mof −/− cells, which could be fully restored by expression of either wild type Mof or exogenous Nanog. In contrast, inactive Mof mutant could not rescue Nanog expression (Figure 6A). When we examined ESC morphology, AP activity as well as expression of key regulators of the rescued cell lines, we found that wild type Mof was able to rescue most Mof null phenotypes (Figure 6B and 6C). The Mof −/− ESCs expressing exogenous Mof had indistinguishable morphology from those of wild type ESCs (Figure 6B) and ~70–80% of colonies were AP staining positive (Figure 6B). This result confirmed that phenotypes observed in Mof −/− ESCs were due to Mof deletion, but not other non-specific secondary mutations. In contrast, the Mof mutant failed to rescue Mof deficient phenotypes. The Mof −/− + Mof mut ESCs had loose cell-cell contacts and poor AP staining, which were similar to Mof −/− ESCs (Figure 6B). This result suggests that Mof acetyltransferase activity was essential for its functions in ESCs. Strikingly, although exogenous Nanog could not rescue loss of viability associated with extended culturing of Mof −/− ESCs (data not shown), it rescued most defects associated with ESC self-renewal. About 50–60% Nanog expressing Mof −/− ES cells formed compact colonies and demonstrated strong AP staining, indicative of restoring ESC features (Figure 6B). The lower numbers of AP positive clones from Nanog rescue cells probably result from variation in exogenous Nanog expression level as well as low Oct4 expression in these cells (see below). In addition to morphological changes, we also examined whether ectopic Nanog expression restored expression of Mof-dependent ESC genes. In most cases, Nanog expression led to changes in gene expression similar to that of wild type or wild type rescue ESCs. As shown in Figure 6C, exogenous Nanog reactivated several genes repressed in Mof −/− ESCs including Fgf4, Lefty1 and Otx2, to the level of wild type or wild type Mof rescued ESCs. The notable exception is Oct4, which remained low in Nanog rescued Mof −/− cells (see discussion). Similarly, Nanog suppressed induction of differentiation regulators such as Foxa2, Gata4 and Gata6 in Mof −/− ESCs (Figure 6C). One thing worth noting is that global H4K16ac remained very low in Nanog rescued ESCs. This suggests that Mof and its acetyltransferase activity are probably required for expression of Nanog, which in turn regulates a cascade of pluripotency and/or differentiation genes. Taken together of the genomic analysis and the Nanog rescue experiment, our results suggest that Nanog is a major target and a functional mediator of Mof in ESCs.
Figure 6. Nanog overexpression rescues Mof null phenotypes in ESCs.
(A) Immunoblots for Nanog, Mof and H4 K16ac in WT or Mof −/− ESCs rescued with control vector, wild type Mof, mutant Mof or Nanog. Antibodies used in the experiments were indicated on right. Both exogenous wild type and mutant Mof were Myc-tagged. (B) AP staining of wild type ESCs and Mof −/− ESCs expressing exogenous wild type Mof, Mof mutant or Nanog as indicated. Top, percentage of AP positive clones of Mof −/− and three rescue ESCs relative to wild type ESCs was presented. Means and standard deviations (as error bars) from two independent experiments were presented. Bottom, images (20×) for each cell lines as indicated on bottom. (C) Real-time PCR analyses for pluripotency (left) and differentiation genes (right) in Mof −/− and three rescue cell lines as indicated. All mRNA levels were normalized against β-actin and were presented as relative fold changes to wild type ESCs. Means and standard deviations (as error bars) from at least three independent experiments were presented. Also see Supplementary Figure 3.
Mof regulates Wdr5 binding at key regulatory regions in ESCs
Several groups recently studied the function of H3K4me3 in ESCs by knocking down key components of the MLL complex Dpy30/Rbbp5 or Wdr5 (Ang et al., 2011; Jiang et al., 2011). Although knocking down these genes affected global H3K4me3, only Wdr5 knock down significantly attenuated expression of self-renewal genes (i.e. Nanog, Oct4 and Sox2) and induction of cell differentiation (Ang et al., 2011). The differences of Wdr5 and Dpy30/Rbbp5 knockdown phenotypes in ESCs raise an interesting question: is the function of Wdr5 in ESCs solely to establish H3K4me3 or does Wdr5 play roles in other yet uncharacterized epigenetic pathways to influence ESC self renewal? Given that Wdr5 is a stable component of the Mof-Msl1v1 complex (Dou et al., 2005; Li et al., 2009), we decided to examine whether Wdr5 plays a role in Mof mediated ESC regulation. We first confirmed that both global H3K4me3 and Wdr5 expression were not affected by Mof deletion in ESCs (Supplementary Figure 7A and 7B). We then compared Mof ChIP-seq data with those of Wdr5 and H3K4me3 in ESCs (Ang et al., 2011). We surveyed the extent that Mof binding peaks fell within 100bp of the peak centers for Wdr5 or H3K4me3 respectively. Strikingly, Mof binding peaks physically overlapped with close to 30% of Wdr5 and 39% of H3 K4me3 peaks across the genome (p<10−16, Pearson’s Chi-squared test, Figure 7A). The close proximity of these binding sites suggested that they co-localized on either the same or adjacent nucleosomes. We further analyzed the distribution of Mof/Wdr5 and Mof/H3K4me3 joint peaks along the defined 12kb region surrounding TSS. As shown in Figure 7B, joint peaks for Mof/Wdr5 and Mof/H3K4me3 were highly enriched around TSS, with 83% (pink, 1293 genes) and 71% (orange, 2,944 genes) peaks in the class I region respectively. In contrast, larger proportion of Mof peaks without Wdr5 or H3K4me3 resides in the class II region (Figure 7B). The promoter enrichment of Mof/Wdr5 was consistent with our previous finding that the Wdr5 containing Mof-Msl1v1 complex functions in transcription initiation (Li et al., 2009).
Consistent with the observation that more genes with class I Mof peaks were down regulated, among ~491 Mof/Wdr5 joint targets that changed expression upon Mof deletion, a significant percentage of genes (306, 62.3%,) were down regulated as compared to ~55% of Mof targets without Wdr5 binding (single sided Fisher’s test p=4.2E-9, gene list see Table S7). Another feature for Mof/Wdr5 joint targets is that their up regulation upon Mof deletion was significantly less than Mof targets without Wdr5 binding (non-paired Wilcoxon test, p=2.87E-6, Figure 7C right panel). A similar distinction of Mof/H3K4me3 joint targets was also found (non-paired Wilcoxon test, p=4.58E-6, Figure 7D right panel). Given the co-localization of Mof/Wdr5 peaks and down regulation of their targets upon Mof deletion, it is likely that regulation of ESC core transcription network by Wdr5 could be partially mediated by Mof. Indeed, ChIP analyses confirmed that binding of Wdr5 and H3K4me3 at selected pluripotent gene targets were Mof dependent. As shown in Figure 7E, Mof was essential for Wdr5 binding at the Nanog promoter (Figure 6B). Mof deletion led to both reduced Wdr5 binding and H3K4me3. Similar regulation was also observed at Sox2 and Utf1 gene promoters (Figure 7E). These results suggest that both Mof and Wdr5 are important for regulating pluripotent genes such as Nanog and Sox2 (Figure 7E). Furthermore, we also identified a couple of cases (i.e. Cbx5 and Dhx1) where Mof deletion led to reduced Wdr5 binding with no change in H3K4me3 in Mof −/− ESCs (Figure 7C), supporting that Wdr5 can play roles independent of H3K4me3 at some gene promoters. All these genes had decreased expression upon Mof deletion. As controls, we performed ChIP assays for Wdr5 and H3K4me3 at several Mof targets without Wdr5 binding. At gene loci such as Mef2a, GATA4 and Klf4, Wdr5 binding was very low and did not change upon Mof deletion. However, we observed slight increase of H3 K4me3 at these loci (Figure 7E), which accompanied increased expression of these genes in Mof −/− ESCs (Table S7). Although interplays between Mof and Wdr5 binding were complex, nonetheless, we were able to establish Mof as an important upstream regulator of Wdr5 at important ESC loci.
Mof regulates H3 K4 methylation at some ‘bivalent domains’ in ESCs
Since we found that H3K4me3 at some gene loci depended on Mof, we decided to further examine whether Mof is involved in setting up and/or regulating the H3K4me3 in ESCs, especially at the functionally important ‘bivalent’ domains (Azuara et al., 2006; Bernstein et al., 2006). To this end, we cross-examined Mof binding peaks with reported ‘bivalent’ regions (Bernstein et al., 2006). Among 8041 peak regions that were marked with both H3K4me3 and H3K27me3, there were 2046 peaks (~26.3%) that physically overlap with Mof binding sites (i.e. centering in the same region). A significant proportion of overlapping peaks (i.e. 564 or 27.6%) was within 2kb of TSS (p< 10−16, full list see Table S7). Among them, 106 genes were down regulated and 41 genes were up regulated in Mof −/− ESCs (Table S7), highlighting the potential regulatory role of Mof at ‘bivalent domains in ESC. ChIP assays for direct Mof binding as well as H3K4me3 and H3K27me3 at selected loci were shown in Supplementary Figure 7D. Several Mof regulated ‘bivalent genes’ (e.g. Olig1 and Fgf15) have been shown to play important roles in ES differentiation (Fischer et al., 2011; Zhou and Anderson, 2002), further supporting the importance of Mof in ESC regulation.
DISCUSSION
Nanog is a key downstream target for histone acetyltransferase Mof in ESCs
Among chromatin regulators, only a handful of chromatin modifiers have been reported to regulate embryonic stem cell self-renewal (Orkin and Hochedlinger, 2011; Young, 2011). Here, we have firmly established that Mof is a critical epigenetic regulator for this important stem cell feature. ESCs with Mof deletion exhibit loss of self-renewal and aberrant expression of both pluripotency genes and differentiation marker genes. Importantly, we have shown that Mof function is largely mediated by ESC core transcription factor Nanog. Using combined gene expression and ChIP-seq analyses for wild type and Mof −/− ESCs, we demonstrate that Mof has profound and direct impact on ESC transcriptome. GSEA for Mof direct targets and ESC core transcription network shows significant and specific enrichment between Mof and Nanog transcriptome (Figure 4D). The enrichment is mostly for the down-regulated gene set, supporting Mof as a transcription co-activator in Nanog pathways. A prominent role of Mof in regulating the ESC transcription network is further supported by the fact that ~80% of Nanog target genes have direct Mof binding sites (Supplementary Figure 5D) and ectopic expression of Nanog can partially suppress loss of self-renewal phenotype in Mof −/− ESCs.
There are several things we would like to point out in the Nanog experiments:
First, although Mof targets overlap significantly with those of Nanog, they do not necessarily bind to the same DNA sequences. In fact, when we performed ‘motif’ search for Mof binding sites, consensus sequences for Nanog binding sites were not identified as top hits (data not shown). It is possible that Nanog may preferably bind to genes that already have Mof bindings. The recent finding that Nanog weakly interacts with Wdr5, a Mof-interacting protein, is consistent with this scenario (Ang et al., 2011). Alternatively, given the wide distribution of Mof peaks in genome, it is possible that Mof and H4K16ac modulate chromatin ‘milieu’ (Orkin and Hochedlinger, 2011), which in turn influences Nanog recruitment. The exact mechanism for the functional interplays between Mof and core transcription factors in ESCs remains to be studied.
Second, Nanog expression suppressed most Mof null phenotypes without restoration of H4K16ac (Figure 6A). This result points out that although Mof is essential for regulating Nanog and/or other ESC core transcription factors, it is probably functionally redundant with other chromatin regulators in regulating downstream targets. Therefore, once Nanog protein level is restored by ectopic expression, Mof is largely dispensable for downstream regulatory events. Precedence has recently been reported for an Eed/Sox2 regulatory loop, in which overexpressing Sox2 can rescue phenotypes of Eed deficient ES cells without restoring H3K27me3 (Ura et al., 2011). This hypothesis is further supported by previous studies that multiple histone acetyltransferases including Tip60, p300 and Gcn5 function downstream of ONS in ESC. These enzymes are able to acetylate histones for transcription activation. Indeed, although genetic ablation or knocking down these enzymes has no effects on expression of ONS themselves, they affect expression of ONS target genes and ESC differentiation processes to various degrees (Fazzio et al., 2008; Lin et al., 2007; Zhong and Jin, 2009). Future characterization of Nanog-dependent gene regulation in Mof −/− ESCs and interplays of Mof with other chromatin regulatory complexes at Nanog target genes will provide insights in this regard.
Third, although Nanog expression rescued most Mof null phenotypes, it failed to restore Oct4 expression in Mof −/− ESCs (Figure 6C). This result suggests that Mof regulation of Oct4 expression is independent of Nanog in ESCs. The failure for Nanog overexpression to restore Oct4 expression may explain partial rescue phenotypes of the Mof −/− ESCs (Figure 6B). Unexpectedly, although most Nanog expressing Mof −/− ESCs have low Oct4 level, they did not differentiate into the trophectoderm lineage. The levels of trophectoderm marker genes such as Hand1 and Cdx2 that were normally activated by Oct4 knockdown (Niwa et al., 2000) remained unchanged in Mof −/− ESCs (Supplementary Figure 3B). This result suggests that Mof is important for activation of at least some trophectoderm markers and for differentiation of trophectoderm lineage. The requirement of Mof during cell differentiation is also supported by the fact that differentiation markers for all three germ layers are modestly but consistently down regulated by loss of a Mof allele (Supplementary Figure 2). Therefore, it is likely that Mof is important for both ESC ‘stemness’ and differentiation. Whether these two processes involve the same or distinct Mof complexes will be subject to future studies.
Distinctive Mof binding in ESCs
Our ChIP-seq analyses reveal that unlike Drosophila, where Mof exhibits a bi-modular Mof binding pattern at TSS and 3′ end of genes (Kind et al., 2008), Mof binding in mammals is enriched at TSS but also distributed evenly in downstream coding regions. The difference may be a reflection of distinct gene structures for mammal and Drosophila. Interestingly, we find that the elevated Mof binding at gene coding regions is a unique feature in ESCs. The coding bound Mof peaks account for ~50% of total Mof peaks within the 12kb defined regions in ESCs while they compose of ~20% in hCD4+ cells (Figure 4D). GO term analyses for genes with coding bound Mof show drastic differences between these two cells. When pathways specific for developmental processes are analyzed, genes with coding bound Mof in ESCs are heavily involved in tissue/organ development and programs for multi-lineage cell differentiation (Supplementary Figure 5A). However, those in hCD4+ cells are only involved in hematopoietic, lymphoid organ development as well as leukocyte differentiation (Supplementary Figure 5A). As control, parallel GO term analyses on genes with TSS Mof binding show no cell specific enrichment in differentiation (Table S5). This result shows that significant and cell specific enrichment of Mof targets is intriguingly linked to the differentiation potential of respective cells. Since most differentiation genes are not expressed in wild type ESCs, Mof binding serves to mark these ‘poised’ genes for later activation.
One remaining question for this ESC specific Mof binding pattern is how it is established and how Mof is recruited to these ‘poised’ loci in ESCs? Since these loci are not actively transcribed, Mof binding at these regions cannot be simply explained as transcription coupled events. One feature of ESC cells is their highly dynamic chromatin states. It would be interesting to test if broad binding of Mof is a result of less compact higher order chromatin structure in ESCs and if Mof is selectively targeted to regions with yet-to-be-characterized epigenetic marks that destine genes for differentiation induced activation. Notably, the epigenetic marks are not necessarily ‘bivalent domains’, which show no significant enrichment at Mof binding sites in ESCs.
Mof mediated transcription regulation in ESCs
One surprising finding of our study is that Mof deletion leads to both increased and decreased expression of its direct targets. Although we cannot rule out that gene up regulation is due to indirect effects, significant number of up regulated genes have Mof binding sties near TSS or in gene bodies (Figure 3C). Notably, the fold changes for both the down- and up-regulation of gene expression upon Mof deletion seem modest, with mean changes ~2.5–2.8 fold. It is likely that Mof functions as a chromatin modulator, regulating chromatin environment and fine-tuning the transcription machinery as they pass the transcribed region. The moderate effects on transcription are consistent with chromosome wide two fold gene activation observed in the Mof-mediated Drosophila dosage compensation process. To understand whether different transcription outcome upon Mof deletion are due to regulation by distinct Mof complexes (i.e. Mof-Msl1v1 and Mof-Msl) (Li et al., 2010), we divide Mof targets based on 1) relative position of Mof binding sites to TSS and 2) whether they have Mof/Wdr5 joint peaks. The results show that genes with Mof binding exclusively at TSS are more likely to be down regulated upon Mof deletion (824 vs. 494, Figure 5C). This bias is also observed for Mof/Wdr5 joint targets (306 vs. 185, Figure 7C). Given that Mof/Wdr5 joint peaks are overwhelmingly located at TSS (Figure 7B), these two results corroborate with each other in supporting a specific function of the Mof-Msl1v1 complex at TSS.
On the contrary, for genes with coding bound Mof, especially those with exclusive class II sites, Mof deletion leads to equal chance of up- or down- regulation (Figure 5C). These Mof targets (without Wdr5 peaks) also seem to be more up regulated upon Mof deletion compared to those with Mof/Wdr5 or Mof/H3K4me3 joint peaks (Figure 7C and data not shown). Altogether, these results argue for a distinct role for coding bound Mof in transcription regulation. Since the Wdr5 independent Mof-Msl complex is important for transcription elongation, it is tempting to suggest that coding bound Mof mostly resides in the Mof-Msl complex. It will be important to further dissect the Mof binding pattern and corresponding transcriptome based on presence of other Mof interacting proteins (i.e. MSL1–3) to prove this point in future. Intriguingly, two recent studies on Drosophila DCC complex (dMof-Msl) show that components of DCC are capable of reducing gene expression in the presence of Mof and its H4K16ac activity (Prestel et al., 2010; Schiemann et al., 2010). It would be interesting to examine whether this is conserved in mammal and whether MSL proteins serve to restrain expression of some Mof-Msl targets in ESCs. In latter case, Mof deletion could lead to dissemble of the Mof-Msl complex and thus relieve the repressive effects of MSL proteins at specific loci.
Mof, H3K4me3 and ESC regulation
Several recent studies explored the function of another transcription activation related chromatin modification, H3K4me3, in ESC regulation. One group showed that knocking down Wdr5, a component of the MLL methyltransferase, significantly attenuated expression of self-renewal genes (i.e. Nanog, Oct4 and Sox2) and resulted in loss of pluripotency and induction of cell differentiation (Ang et al., 2011). However, another group showed that knocking down Dpy30 and Rbbp5 of the same complexes had minimal effects on ESC self-renewal and ONS expression despite reduction of global as well as loci-specific H3K4me3 (Jiang et al., 2011). The differences of Wdr5 and Dpy30/Rbbp5 knockdown phenotypes in ESCs raise an interesting question: does H3K4me3 independent function of Wdr5 contributes to ESC regulation? In light of our results here, one likely explanation for the reported paradoxical observation is that Wdr5 functions as part of the Mof complex to regulate transcription in ESCs. This explains why Wdr5 depletion has broader ESC phenotypes than knocking down Dpy30 or RbBP5. In support, we show that Mof deletion affects Wdr5 recruitment at important gene loci including Nanog and Sox2 and at some loci, changes in Wdr5 binding and gene expression (e.g. Cbx5) are not always accompanied by changes in H3K4me3 (Figure 7C). Future studies on the detailed mechanisms for how Mof and H3K4me3 coordinate to activate ONS genes and how Wdr5 contributes to Mof function in this context will provide insights in this aspect. The ability of Mof to regulate Wdr5 and H3K4me3 at some loci in ESCs has prompted us to examine its role at setting up the ‘bivalent domains’, epigenetic regulatory elements that govern ESC transcription program (Azuara et al., 2006; Bernstein et al., 2006). Indeed, we find that Mof regulates H3K4me3 at some important bivalent domains including those at promoters of Nanog and Sox2 (Figure 7D). Genome wide analyses further support extensive interconnection between Mof and H3K4me3 in ESCs (Figure 7A). Importantly, deletion of Mof in ESCs leads to aberrant expression for genes with nearby ‘bivalent domains’ (Figure 7D and Table S7). The close interactions between Mof and Wdr5/H3K4me3 probably underlie the essential functions of Mof in regulating ONS expression and their regulatory circuitry.
MATERIALS AND METHODS
Generation of embryonic stem (ES) cell lines and ES cell differentiation
Inducible Cre-expressing mouse line CAGG Cre-ER™ was as previously described (Li et al., 2010). The Mof ES cell lines were derived from the inner cell mass of 3.5dpc blastocysts which were obtained from timed mating of Mof flox/flox; CAGG Cre-ER™ mice.
Alkaline Phosphatase Staining of ES cells
The Stemgent™ Alkaline Phosphatase (AP) Staining Kit was used for the detection of the AP activity according to the manufacturer’s instructions. For AP staining, 1000 ESCs for each genotype were plated and cultured with or without 4-OHT for 4 days before the staining.
Immunoblot, quantitative RT-PCR and ChIP analyses
These experiments were performed as previously described (Byun et al., 2009; Dou et al., 2006). Anti-Mof (Santa Cruz), anti-H4K16ac (Millipore), anti-H3K27me3 (Millipore), anti-H3K4me3 (Millipore), anti-Wdr5 (Millipore), anti-mouse or anti-rabbit IgG (Sigma) antibodies were used. All RT and ChIP-PCR primers were listed in Supplemental Experimental Procedure.
Gene expression microarray, GO and GSEA analyses
Microarray analyses for wild type (GSM910916) and Mof null (GSM910917) ESCs were performed on Affymetrix GeneChip Mouse Genome 430 2.0 arrays (Affymetrix). The expression change of a gene was calculated using the geometric mean of all probes aligned on the gene. R package GOstats (Falcon and Gentleman, 2007) and GO.db (http://stuff.mit.edu/afs/athena.mit.edu/software/) were used for GO term association studies. For each gene list, conditional single-sided hypergeometric tests were used to calculate P value of GO term enrichment. GSEA (Isakoff et al., 2005) was performed using JavaGSEA software provided by http://www.broadinstitute.org/gsea/. GSEA of two gene sets representing differentially expressed genes were ranked as a list by fold changes. GSEA was run on this pre-ranked list with number of permutations=1000.
ChIP-seq analyses
ChIP-seq analysis for Mof (GSE37109) was performed at NCI Sequencing Facility. Images acquired were processed through the image extraction pipeline and aligned to mouse NCBI build mm9 using ELAND. Peaks were called using HPeak (Qin et al., 2010), a hidden Markov model-based software program for identifying ChIP-enriched regions. Pearson’s Chi-squared test with Yates’ continuity correction or Fisher exact test was used for calculating P-values when evaluating overlaps between lists of genes.
Supplementary Material
HIGHLIGHTS.
Histone acetyltransferase Mof is essential for ESC self-renewal.
Mof directly regulates Nanog mediated ESC core transcription network.
Overexpression of Nanog partially rescues Mof deletion phenotypes in ESCs.
Mof regulates Wdr5 recruitment and H3 K4 methylation at important loci in ESCs.
Acknowledgments
We thank Dr. Yi Zhang for anti-Nanog antibody as well as the Nanog expression vector. This work is supported by NIGMS (R01GM082856) and American Cancer Society (RSG 117573) grants to YD, NHGRI (R01HG005119) grant to ZQ and NSFC (31171428) grant to XL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. doi: 10.1016/j.ydbio.2006.04.450. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, Bilke S, Haggerty CM, Player A, Wang YH, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci U S A. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Fischer T, Faus-Kessler T, Welzl G, Simeone A, Wurst W, Prakash N. Fgf15-mediated control of neurogenic and proneural gene expression regulates dorsal midbrain neurogenesis. Dev Biol. 2011;350:496–510. doi: 10.1016/j.ydbio.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, Wang X, Biegel JA, Pomeroy SL, Mesirov JP, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Dou Y. New perspectives for the regulation of acetyltransferase MOF. Epigenetics. 2010:5. doi: 10.4161/epi.5.3.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Macarthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol. 2008;73:195–202. doi: 10.1101/sqb.2008.72.001. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Park SH, Kook MC, Kim EY, Park S, Lim JH. Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct Pathol. 2004;28:229–238. doi: 10.1080/01913120490515595. [DOI] [PubMed] [Google Scholar]
- Prestel M, Feller C, Straub T, Mitlohner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Mol Cell. 2010;38:815–826. doi: 10.1016/j.molcel.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Yu J, Shen J, Maher CA, Hu M, Kalyana-Sundaram S, Chinnaiyan AM. HPeak: an HMM-based algorithm for defining read-enriched regions in ChIP-Seq data. BMC Bioinformatics. 2010;11:369. doi: 10.1186/1471-2105-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann AH, Li F, Weake VM, Belikoff EJ, Klemmer KC, Moore SA, Scott MJ. Sex-biased transcription enhancement by a 5′ tethered Gal4-MOF histone acetyltransferase fusion protein in Drosophila. BMC Mol Biol. 2010;11:80. doi: 10.1186/1471-2199-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura H, Murakami K, Akagi T, Kinoshita K, Yamaguchi S, Masui S, Niwa H, Koide H, Yokota T. Eed/Sox2 regulatory loop controls ES cell self-renewal through histone methylation and acetylation. Embo J. 2011;30:2190–2204. doi: 10.1038/emboj.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–44. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Jin Y. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J Biol Chem. 2009;284:9168–9175. doi: 10.1074/jbc.M805562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.