Abstract
We report the design of an elastin-mimetic triblock copolymer with the ability to guide endothelial cell adhesion, spreading, and migration while maintaining the elastomeric properties of the protein polymer. The V2 ligand sequence from matricellular protein CCN1 (cysteine-rich 61, CYR61) was multimerized and cloned into elastin polymer LysB10, creating LysB10.V2. Cell adhesion studies demonstrated that a LysB10.V2 surface density of at least 40 pmol/cm2 was required to elicit cell attachment. Peptide blocking studies confirmed V2 specific engagement with integrin receptor αvβ3 (P < 0.05) and we observed the formation of actin stress fiber networks and vinculin clustering, characteristic of focal adhesion assembly. Haptotatic migration assays demonstrated the ability of LysB10.V2 surfaces to stimulate migration of endothelial cells (P < 0.05). Significantly, we illustrated the ability of LysB10.V2 to support a quiescent endothelium. The CCN1 molecule functions to support many key biological processes necessary for tissue repair and thus presents a promising target for bioengineering applications. Collectively, our results demonstrate the potential to harness CCN1 specific function in the design of new scaffold materials for applications in regenerative medicine.
Keywords: Protein polymer, Elastin-mimetic, Extracellular matrix, Matricellular protein, Endothelial cell
1. Introduction
Genetic and recombinant protein engineering has enabled the synthesis of elastin-like protein polymers (ELP) composed of repetitive amino acid sequences and peptide blocks, whose structural complexity impart specific mechanical, chemical, and biological properties. The most significant impact of this strategy is the capacity to introduce precise changes in the amino acid sequence to modulate properties of the entire protein network. Thus, ELPs can serve as scaffolds that are amenable to further functionalization via recombinant techniques. A variety of bioactive peptide sequences have been incorporated into ELPs to mimic functional properties of the extracellular matrix (ECM) and guide cellular behavior. Selected sequences have provided structural, mechanical, biochemical, and degradative properties for a wide range of tissue engineering applications. For example, Girotti and coworkers have integrated elastase sensitive sequences to control proteolytic degradation in response to cellular infiltration [1]. Similarly, sequences sensitive to urokinase plasminogen activator (uPA) and laminin-derived sequences have been genetically engineered into ELPs in order to induce local neuronal remodeling and neurite extension of an ECM-mimetic [2]. Tirrell and colleagues have been able to increase endothelial cell attachment by incorporating the cell-binding domains, RGD and REDV [3,4].
We have recently reported the synthesis of a recombinant elastin-mimetic triblock copolymer, LysB10, designed with relatively large block sequences to promote tunable physical and mechanical properties [5]. This protein polymer contains identical 75 kDa hydrophobic endblocks flanking a central 58 kDa hydrophilic midblock with crosslinkable lysine residues engineered at block interfaces. As reviewed elsewhere, multiblock elastin-mimetic systems reversibly self-assemble above the inverse transition temperature defined by the hydrophobic endblocks [6–8]. Block phase separation yields a stable, solvated network that can be processed into hydrogels, particles, films, and fibers [9–11]. While extensive validation of the mechanical properties of LysB10 has demonstrated its functionality as an effective load-bearing scaffold, it does not display cell binding sequences and, consequently, does not inherently support cell adhesion.
CCN1, or cysteine-rich 61, is a secreted heparin-binding protein of 40 kDa that displays pro-angiogenic activities, including endothelial cell adhesion, migration, proliferation, and tubule formation [12,13]. The protein contains four structural domains with sequence similarities to insulin-like growth factor-binding protein (domain I), von Willebrand factor type C repeat (domain II), thrombospondin type I repeat (domain III), and carboxyl-terminal domains of extracellular matrix proteins such as mucins (domain IV) [14]. Previous studies have identified a 20 amino acid sequence, V2, in the von Willebrand factor type C repeat (domain II) as a site that binds specifically to integrin αvβ3 [15]. The V2 sequence (NCKHQCTCIDGAVGCIPLCP) supports αvβ3 mediated endothelial cell adhesion in a dose-dependent manner, with the aspartate residue playing a key structural role in presenting the peptide to the ligand-binding pocket of the integrin.
We postulated that designing LysB10 to contain αvβ3 integrin-binding sequences would enhance endothelial cell adhesion and migration on an otherwise non-adhesive substrate. To this end, the CCN1-derived V2 cell binding sequence was multimerized to form a contiguous 10-repeat insert for incorporation into the central domain of this triblock copolymer. Genetic engineering techniques enabled facile control over multiblock molecular assembly, as well as the spatial distribution and presentation of this bioactive peptide sequence. We demonstrate that V2 bearing protein polymers afford robust endothelial cell adhesion, migration, and spreading that are specifically dependant upon binding interactions with αvβ3.
2. Materials and methods
2.1. Synthetic gene construction
Synthetic methods used to generate LysB10 have been described elsewhere [5]. Briefly, LysB10 was designed with a central hydrophilic midblock [(VPGAG)2VPGEG-(VPGAG)2]28 flanked by hydrophobic endblocks ([IPAVG]5)33. Crosslinking residues were engineered between the hydrophobic and hydrophilic domains and at the terminal ends of the protein polymer. LysB10.V2 contains identical elastin-mimetic blocks and crosslinking sequences with the addition of a multimerized CCN1-derived V2 sequence inserted into the central midblock (Table 1, Scheme 1).
Table 1.
Amino acid sequences for block domains employed in construction of LysB10 and LysB10.V2.
| Elastin analog | N-terminal Plastic domain (P) | Elastic domain (E) | Bioactive domain (B) | C-terminal Plastic domain (P) |
|---|---|---|---|---|
| LysB10 | VPAVGK [(VPAVG) (IPAVG)4] [(IPAVG)5]33 | (IPAVG) KAAK (VPGAG) (VPGAG)2 VPGEG (VPGAG)2]28 (VPAVG) KAAK (VPGAG) | – | [(VPAVG) (IPAVG)4 [(IPAVG)5]33 IPAVGKAAKA |
| V2 | VPAVGK [(VPAVG) (IPAVG)4] [(IPAVG)5]33 | (IPAVG) KAAK (VPGAG) (VPGAG)2 VPGEG (VPGAG)2]14–B– [(VPGAG)2 VPGEG (VPGAG)2]14 (VPAVG) KAAK (VPGAG) | [VPGVG-GG-NCKHQCTCIDGAVG CIPLCP-GG-PGVG]10 | [(VPAVG)(IPAVG)4 [(IPAVG)5]33 IPAVGKAAKA |
Scheme 1.
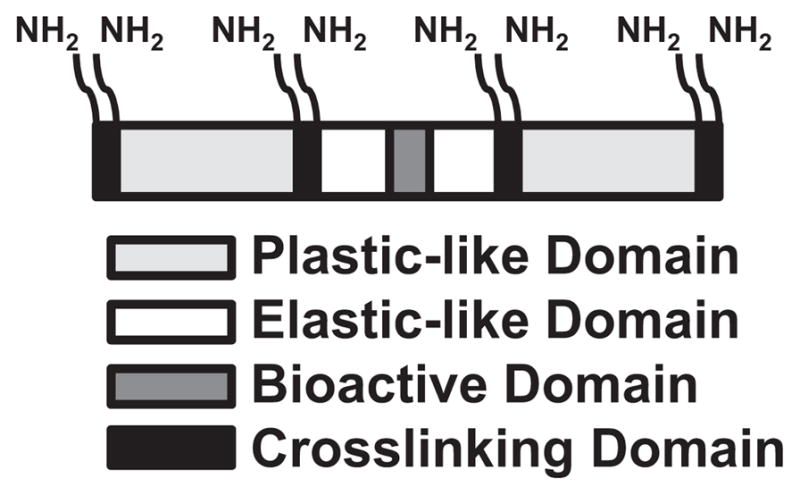
Domain structure of LysB10.V2. Multimerized CCN1-derived V2 sequence (bioactive domain, dark grey) was inserted into the central hydrophilic midblock (elastic-like domain, white) of triblock copolymer LysB10. Lysine-containing cross-linking domains (black) flank each plastic-like (light grey) and elastic-like domain.
The 127 bp V2 monomer sequence was derived from oligonucleotide casettes encoding terminal BamHI/HinDIII restriction sites, as well as internal BbsI/BsmBI restriction sites flanking the monomer sequence. The V2 cassette was inserted into cloning vector pZErO-1 at BamHI/HinDIII for subsequent multimerization reactions. Head-to-tail ligation was performed using BbsI/BsmBI restriction sites, a 10-repeat concatemer of V2 was chosen for insertion into LysB10. To engineer LysB10.V2 in proportion to the parent LysB10 clone, a 14-repeat elastin-mimetic midblock concatemer was generated using a similar strategy.
LysB10.V2 was generated using a modular cloning strategy, where applicable, block sequences from the parent LysB10 construct were utilized All subcloning steps were performed in the pZErO-1 plasmid using LSLB media under Zeocin antibiotic selection. Block ligation and insertion into a lysine adapter sequence was performed as previously reported [5]. All ligation steps were confirmed using DNA sequence analysis. The final construct was inserted into the pQE-80L expression plasmid at BamHI/HindIII and transformed into Escherichia coli expression strain BL21(DE3)
2.2. Expression, purification, and characterization of the recombinant protein triblock copolymer containing V2 peptide sequences
Expression and purification techniques for LysB10 have been detailed elsewhere [5]. Large scale expression of LysB10.V2 was performed in an orbital shaker (225 rpm) at 37 °C in Luria Broth medium supplemented with ampicillin (100 μg/μL). Cells were grown to an optical density of 0.8 before IPTG (1 mM) was added for 4 h to induce protein expression. A hybrid purification protocol was designed for the isolation of LysB10.V2. In brief, the cell lysate was processed through 4 warm/cold centrifugation cycles before purification using TALON (Clontech) cobalt-based immobilized metal affinity chromatography [16]. Eluted LysB10.V2 was dialyzed and lyophilized prior to further characterization.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to characterize the purity of elastin-mimetic proteins. A total of 40 μg LysB10 or LysB10.V2 were run with molecular weight markers (Precision Plus Protein Kaleidoscope, Bio-Rad) on a 7.5% gel and stained with Coomassie stain (Bio-Rad). Amino acid composition analysis of LysB10.V2 was performed by the W.M. Keck Biotechnology Resource Laboratory at Yale University. Lyophilized protein was resuspended in HPLC grade water and filter-sterilized with a 0.22 μm filter. Solutions (1 mg/mL) were submitted for analysis with samples hydrolyzed in 6N HCl and the resulting amino acids examined with a Beckman Model 7300 ion exchange instrument. Improved quantitation of cysteine was obtained via performic acid oxidation. Of note, proline values are artificially elevated after normal acid hydrolysis. Amino acid compositional analysis of LysB10.V2 (observed mol%, expected mol%): Ala (17.4,17); His (0.9,0.6); Thr (0.4,0.2); Glu (0.7,1); Gly (25.7,26); Val (18.3,19.2); Ile (15.3,13); Pro (21.8,18.8); Lys (0.4,0.6); Asn (0.5,0.4); Gln (0.4,0.4); Asp (0.5,0.4); Leu (0.3,0.4); Cys (1.2,1.9).
Experiments were performed using a Setaram Micro DSC III calorimeter (Setaram Inc, France). Lyophilized samples (1 mg/mL) were dissolved at 4 °C in distilled, deionized water and filter-sterilized through a 0.22 μm syringe filter. Thermal transitions were investigated between 4 and 70 °C at a scan rate of 1 °C/min. Reversibility was confirmed by cooling samples back to 4 °C following the initial scan. Data were analyzed using SETSOFT 200 software (Setaram Inc, France).
2.3. Preparation and characterization of protein treated surfaces and crosslinked thin films
Lyophilized protein was dissolved in phosphate buffered saline (PBS) at 4 °C, sterile-filtered through a 0.22 μm syringe filter, and solutions of varying concetration (10, 5, 2.5, 1, 0.5, and 0.1 mg/mL) prepared. A total of 50 μL of each solution was aliquoted into individual wells of a 96-well non tissue-culture treated polystyrene plate and then incubated for 6 h at 4 °C before rinsing each well three times with PBS. Each well was then treated for 1 h with 0.5% heat-inactivated bovine serum albumin (BSA) followed by three rinses with PBS. Reference wells included those incubated overnight at 4 °C with either 0.5% BSA, fibronectin (FN) or vitronectin (VN) (50 μg/mL). Surface adsorption of protein polymers was quantified by the bicinchoninic acid (BCA) method (Pierce, Inc.).Known concentrations of protein solutions were analyzed in parallel to create a calibration curve [3].
LysB10.V2 hydrogels were fabricated by casting 50 μL protein solutions into non tissue culture treated polystyrene wells at 4 °C followed by incubation at 37 °C for 6 h The resulting film was firmly adherent to each well and was subsequently crosslinked with genipin (6 mg/mL) for 24 h after which excess genipin was removed by rinsing three times with PBS over a 12 h period
2.4. Endothelial cell attachment, migration, and morphology
Human umbilical vein endothelial cells (HUVEC) were maintained in endothelial growth medium-2 (EGM-2, 2% serum), passaged every other day, and cells between passage 3 and 10 used for all experiments. Cells were harvested with Cell Dissociation Solution (Sigma) and suspensions prepared at 200,000 cells/mL in basal medium containing 0.2% BSA. For blocking studies, detached HUVECs were treated with LM609 (50 μg/mL) or soluble V2 peptides (V2, scrambled V2, Invitrogen, Inc.; 1 mM) for 30 min at room temperature before plating. A total of 100 μL of cell suspension was plated into each well pretreated with an elastin-mimetic protein polymer. After incubation at 37 °C for 4 h, wells were washed three times with PBS. Cell adhesion was evaluated with the CyQuant Cell Proliferation Assay Kit (Molecular Probes) and results normalized to attachment levels observed on FN-coated wells.
Haptotactic cell migration assays were performed using modified Boyden chambers (Transwell filters, 8 μm pore size) [17]. Chambers for haptotaxis assays were prepared by pre-coating the under surface of the polycarbonate membrane with LysB10 (10 mg/mL) and LysB10.V2 (5 mg/mL), respectively. Reference surfaces were generated with VN or FN (50 μg/mL) using a similar protocol A total of 80,000 cells were placed in the upper chamber of each well and cells allowed to migrate for 6 h at 37 °C. Cells were fixed in 10% formaldehyde and stained in hematoxylin, after which the upper surface was swabbed with a wet cotton swab and rinsed in distilled, deionized water. The average number of cells in six randomly chosen 40× fields was used to quantify the extent of migration in each insert.
Identification of cytoskeletal morphology of cells cultured on elastin-mimetic protein polymers was performed by immunofluorescence confocal microscopy (Zeiss LSM 510 META). A total of 10,000 cells/well were seeded onto protein coated 8-well slides (Nalge Nunc, International) and cultured for 4 h in serum-free medium. Cells were then fixed in 4% paraformaldehyde (10 min), permeabilized with PBS containing 0.5% Triton X-100 (10 min), rinsed once with 100 mM glycine (10 min), and incubated with blocking buffer (PBS+/+, 0.2% Triton X-100, 6% goat serum) for 1 h at room temperature. Cells were incubated with Alexa Fluor 568-conjugated phalloidin for 30 min to stain F-actin and vinculin was visualized by staining with mouse anti-human vinculin IgG1 (10 μg/mL, 1 h), followed by bio-tinylated goat anti-mouse IgG secondary (2.5 μg/mL, 45 min), and streptavidin-AlexaFluor 488 tertiary (2.5 μg/mL, 30 min). Nuclei were counterstained with DAPI Prolong Gold. E-selectin and ICAM-1 monoclonal antibodies (10 μg/mL) were used to evaluate HUVEC activation with reference cells activated by TNFα (100 ng/mL) for 4 h prior to immunostaining. HUVECs cultured on FN-coated polystyrene and maintained at 24 h in serum-free media served as quiescent controls. Pixel intensity was calculated using MATLAB (The MathWorks, Natick, MA) as previously reported [18].
2.5. Statistical analysis
Comparison between groups was analyzed via one-way ANOVA or a paired, two-tailed student’s t-test, with p < 0.05 considered to be significant. Results are presented as mean ± standard deviation. Data represent characteristic results from a particular experimental run, where each group repeated at least in quadruplicate and at least three independent experimental runs conducted.
3. Results
3.1. Synthesis and physical characterization of LysB10.V2 protein copolymer
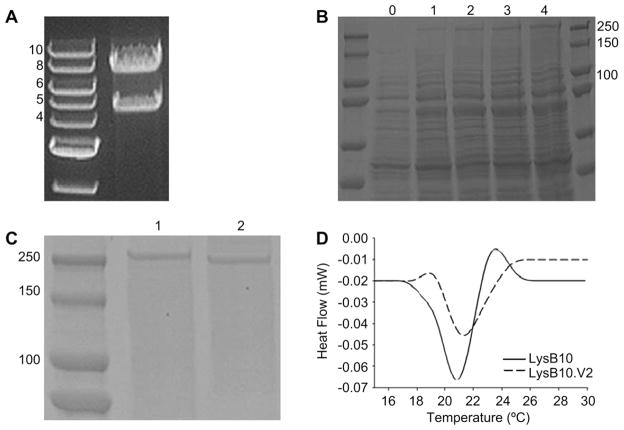
We formulated a cloning strategy to allow seamless insertion of the CCN1-derived V2 multimer sequence into the midblock of the elastin analog LysB10, generating the matricellular-functionalized construct, LysB10.V2. Capitalizing on previously reported modular cloning techniques for the generation of multiblock elastin-mimetic systems, we designed a V2 monomer sequence compatible for head-to-tail multimerization and subsequent ligation into the elastin-mimetic block sequence of LysB10 [5,7]. V2 codon usage was specifically designed for compatibility with E. coli expression systems. A clone encoding 10 repeats of the V2 monomer sequence was chosen for insertion into the LysB10 construct. This 10-mer repeat sequence was estimated to represent ~15% of the ELP molecular weight (Table 1). Final assembly of the LysB10.V2 construct generated an 8.4 kilobase pair insert cloned into the expression vector pQE80-L, which provided an N-terminal His tag for purification (Fig. 1A).
Fig. 1.
Synthesis and physical characterization of LysB10.V2. (A) Analytical restriction digest. Right lane, 1% TAE (Tris-acetate-EDTA) agarose gel displaying LysB10.V2 gene (8.4 kb) and pQE-80L vector (4.8 kb) in the final construct after BamHI/HinDIII digest. Left lane, 1 kb DNA ladder. (B) Protein expression analysis of LysB10.V2 over 4 h time course, protein visualized with Coomassie stain. (C) SDS-PAGE analysis of purified protein, protein visualized with Coomassie stain. Lane 1, LysB10.V2 (233.1 kDa); Lane 2, LysB10 (209 kDa). Molecular weight markers indicate size. (D) Differential scanning microcalorimetry of LysB10.V2 and LysB10.
Small-scale expression cultures of the LysB10.V2 analog were first pursued to verify recombinant protein expression from E coli, and a time course analysis of expression performed to optimize induction conditions. The theoretical molecular weight of LysB10.V2 is 233.1 kDa. SDS-PAGE analysis revealed that the recombinant protein was not expressed before IPTG induction (t = 0 h). However, Coomassie staining displayed increasing protein intensity at ~250 kDa between one to 4 h after IPTG induction (Fig. 1B). As previously reported, molecular weights observed by SDS-PAGE for elastin-mimetic proteins are approximately 20% greater than calculated molecular weights due to their relative hydrophobicity [19,20]. With confirmation of LysB10.V2 protein production over a 4 h time course, large scale expression and purification were performed. Using a combination of hot/cold spin cycles [5] and metal affinity chromatography, recombinant proteins were purified to homogeneity at a final yield of ~35 mg/L (Fig. 1C). Bacterial endotoxin contamination of the resulting protein was evaluated with the Limulus Amebocyte Lysate (LAL) assay. Repeated testing (n = 5) of various purified batches of LysB10 and LysB10.V2 indicated that endotoxin content was below the level of 0.1 EU/mg. Differential scanning microcalorimetry of dilute aqueous solutions of LysB10.V2 (1 mg/mL) confirmed the presence of a single endothermic transition at 21.4 °C consistent with phase separation of the hydrophobic endblocks into beta-turn aggregates (Fig. 1D).
3.2. Endothelial cell attachment and migration on elastin-like copolymers
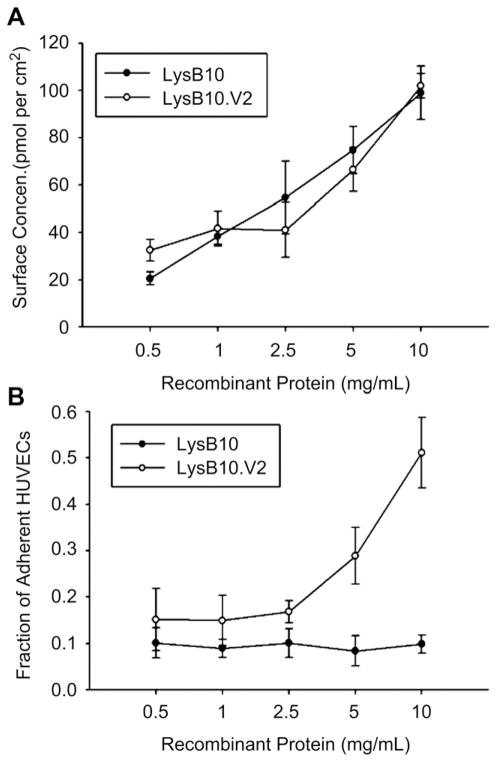
The hydrophobic nature of the elastin-like polypeptides facilitated their passive adsorption onto non-tissue culture-treated polystyrene. Moreover, the ability to precisely characterize the relationship between input ELP concentration and adsorbed ELP surface density assisted studies directed at optimizing cell behavior. To this end, adsorbed surface density increased linearly with the concentration of both LysB10 and LysB10.V2 protein solutions (Fig. 2A). LysB10 was unable to support robust cell attachment, while coating concentrations of LysB10.V2 above 2.5 mg/mL were associated with increasing cell adhesion (Fig. 2B), which suggests a final surface density of at least 40 pmol/cm2 is required to support cell attachment.
Fig. 2.
Endothelial attachment. (A) Adsorbed surface concentrations of LysB10.V2 and LysB10 protein solutions ranging from 0.5 mg/mL to 10 mg/mL. Quantitation was performed with bicinchoninic acid protein assay. (B) HUVEC adhesion to varying amounts of adsorbed proteins. 50 μg/mL FN was adsorbed as a positive control, all data normalized to FN control and reported as fraction of adherent cells compared to that on FN-coated wells. Data represent one of three similar experiments, with each condition run in quadruplicate.
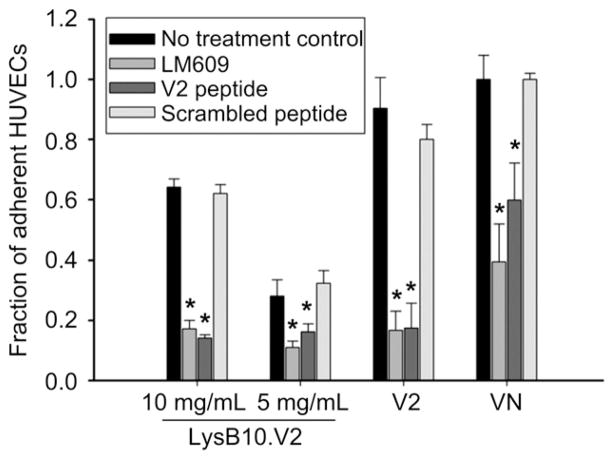
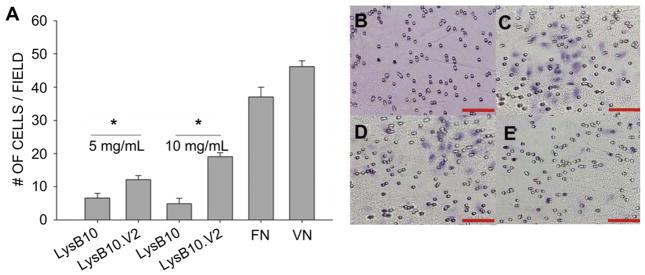
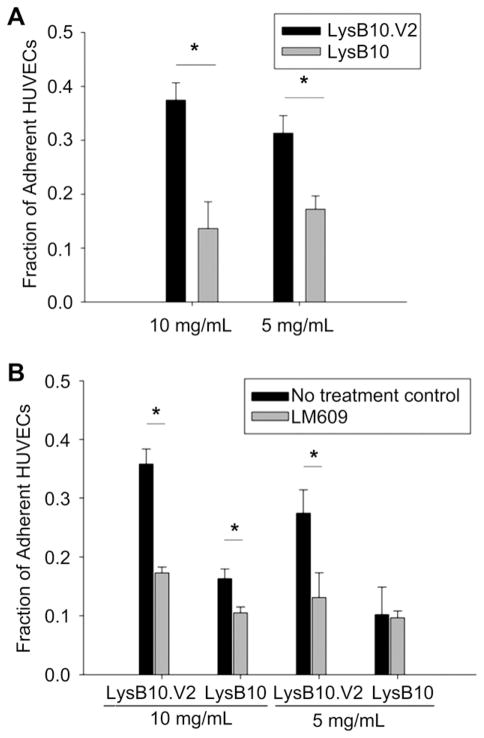
In order to evaluate whether HUVEC adhesion to LysB10.V2 is mediated through integrin specific binding, competitive inhibitors of αvβ3 function were employed. We compared surfaces generated by the passive adsorption of LysB10.V2 with those produced by adsorption of V2 control peptide. Additionally, passive adsorption of VN was also included as a surface that engages multiple integrin receptors, including αvβ3. HUVECs treated with anti-αvβ3 monoclonal antibody (LM609) showed reduced adhesion to all substrates with the greatest effect observed on V2 modified surfaces (Fig. 3). To further confirm V2 peptide engagement of αvβ3, we incubated suspended HUVECs with soluble V2 peptide. Cell adhesion was inhibited on all surfaces, with the greatest reduction on V2 modified surfaces. Scrambled control peptide failed to inhibit HUVEC adhesion. These results indicate that the V2 sequence localized within the elastin-like protein polymer is critical to supporting αvβ3-mediated HUVEC adhesion. Consistent with these results and the established role of αvβ3 in cell migration [21], we examined the ability of LysB10.V2 to induce HUVEC migration. Transwell chambers for migration assays were prepared by pre-coating the under surface of the filter membrane with LysB10.V2 or control material. To initiate migration assays, HUVECs were added to the upper chamber. After 6 h, the cells on the upper surface of the membrane were removed by cotton swabs and the migratory cells on the lower membrane surface were stained and quantified. Significantly, we observed concentration-dependent HUVEC migration to LysB10.V2 (Fig. 4).
Fig. 3.
Integrin specificity. LysB10.V2 protein polymer solutions of 10 mg/mL and 5 mg/mL were passively adsorbed onto polystyrene. The 20-amino acid V2 peptide was adsorbed onto polystyrene at a 1 mM concentration; VN and FN controls were adsorbed at 50 μg/mL. For blocking studies, cells were treated with LM609 (50 μg/mL) antibody, soluble V2 peptide (1 mM), or scrambled V2 peptide (1 mM) for 30 min prior to plating. All data normalized to FN control. Data represent one of three similar experiments, with each condition run in quadruplicate. Competitive inhibitors LM609 and V2 peptide significantly reduced cell attachment to substrates (*P < 0.05 verses no treatment control group).
Fig. 4.
Haptotactic migration assay. 80,000 cells were allowed to migrate across inserts for 6 h at 37 °C. After incubation, cells that remained attached to the upper surfaces of the filters were removed by cotton swabs, and cells that migrated to the lower surfaces of the filters were fixed and stained. The extent of cell migration was determined by calculating the average number of migrated cells in six randomly chosen fields of view per insert. (A) Quantitation of cells counted. Representative images of cells migrated to the lower insert surfaces of hematoxylin stained inserts. Scale bar 500 μm (B) 10 mg/mL LysB10 (C) 50 μg/mL VN (D) 10 mg/mL LysB10.V2 and (E) 5 mg/mL LysB10.V2. *P < 0.05.
3.3. Endothelial cell focal adhesion formation and activation state
Focal adhesion assembly as well as cytoskeletal organization on engineered surfaces was examined. HUVECs demonstrated well-developed actin stress fiber networks and vinculin clustering when plated on LysB10.V2 and FN-coated substrates (Fig. 5). In contrast, when grown on LysB10-coated surface, actin and vinculin were nonspecifically distributed throughout the few adherent cells, which otherwise exhibited rounded morphologies (Fig. 5A,B). These results further confirm that cell adhesion is mediated through integrin-ligand binding, and the LysB10.V2-coated surface is sufficiently robust to induce cytoskeletal organization and focal adhesion formation characteristic of well-spread cells.
Fig. 5.
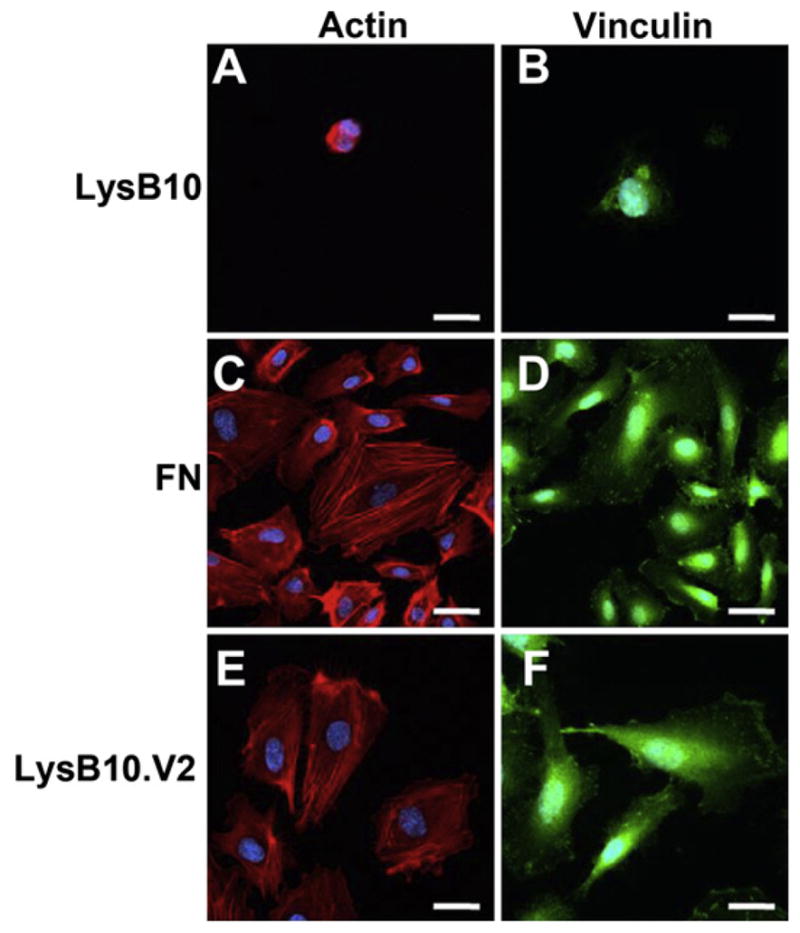
Focal adhesion assembly. 10 mg/mL LysB10 (A & B), LysB10.V2 (E & F), and 50 μg/mL FN solution (C & D) were allowed to adsorb to glass slides and blocked in 0.5% BSA prior to cell seeding. Fluorescently labeled actin is shown in red (A, C, & E), vinculin is displayed in green (B, D, & F). Scale bar 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
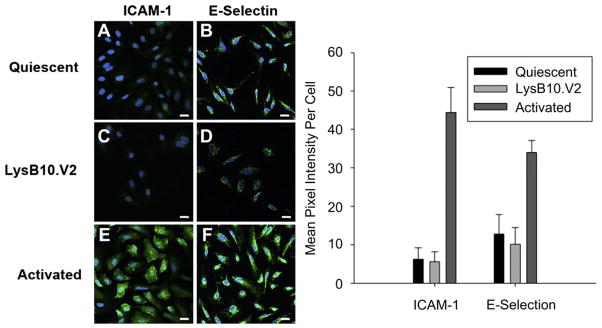
The functional state of an endothelial cell monolayer determines its ability to act as a thromboresistant barrier for blood-contacting applications. Endothelium that express pro-inflammatory markers, such as ICAM-1 and E-selectin, are considered in an activated state and not only attract inflammatory cells, but also promote prothrombotic responses. HUVECs grown on LysB10.V2-coated substrates displayed limited ICAM-1 and E-selectin staining, similar to levels observed in serum-free quiescent control cells (Fig. 6). Thus, endothelial cells are not only able to adhere, spread, and migrate on substrates coated with V2 protein polymer, but also maintain a quiescent phenotype.
Fig. 6.
HUVEC activation state. Cells that were cultured on FN-coated slides and incubated 24 h in serum free media (A & B) maintained a quiescent phenotype. Activation was achieved with the addition of TNFα to the culture medium (E&F). HUVEC activation or quiescence was compared to that on LysB10.V2-coated slides (C & D). Markers of HUVEC activation included ICAM-1 (A,C, and E) and E-selectin (B,D, and F). (G) Quantitation of average green pixel intensity per cell. Scale bar 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Endothelial cell attachment to chemically crosslinked elastin-mimetic hydrogels
In addition to characterizing cell responses to ELPs adsorbed onto polystyrene substrates, alternative ELP formulations were also explored. Triblock copolymers of LysB10.V2 and LysB10 form physical gels at 37 °C due to the association of hydrophobic end-blocks. ELPs can be further stabilized by chemical crosslinking of lysine residues positioned at block interfaces. Aqueous solutions of recombinant ELPs were formulated into soft gels at 37 °C and stabilized by genipin crosslinking. HUVEC attachment was approximately twice as effective on genipin-crosslinked LysB10.V2 as that observed on LysB10 hydrogels (Fig. 7A). Integrin-specific adhesion was observed on V2 hydrogels while a background level of nonspecific adhesion was noted on LysB10 gels (Fig. 7B).
Fig. 7.
(A) HUVEC adhesion on 10 mg/mL and 5 mg/mL genipin-crosslinked LysB10.V2 and LysB10 hydrogels. (B) Integrin specificity was examined with LM609 antibody blocking. Cells were treated with LM609 (50 μg/mL) antibody for 30 min prior to seeding. 50 μg/mL FN adsorbed onto polystyrene served as a positive control, all data was normalized to control. Data represent one of three similar experiments, with each condition run in quadruplicate. *P < 0.05.
4. Discussion
In the present study, we sought to design an elastin-mimetic triblock copolymer with the ability to guide endothelial cell behavior while maintaining the elastomeric properties of the protein polymer. Adhesion-promoting sequences, ligand density and clustering, and ELP morphology were manipulated in order to tailor material properties. To this end, the V2 ligand isolated from the pro-angiogenic, ECM–associated protein CCN1 was cloned into the central, hydrophilic domain of LysB10. To test functional ligand incorporation, we characterized integrin specific endothelial adhesion and migration. We demonstrated density dependent adhesion to LysB10.V2 modified surfaces while blocking studies confirmed specific αvβ3recognition of the V2 ligand within the recombinant protein polymer. Adherent cells displayed a well-developed actin stress fiber network and vinculin clustering characteristic of focal adhesion assembly. Haptotatic motility assays demonstrated concentration-dependent migration of endothelial cells to LysB10.V2 surfaces. Significantly, we illustrate the ability of the ligand functionalized elastin-mimetic protein polymer, LysB10.V2, to support growth of an endothelium displaying a quiescent phenotype.
Integrating practical knowledge derived from cellular biology provides a biologically inspired strategy to advance the design of new scaffold materials for applications in tissue engineering and regenerative medicine. Investigations by our group and others have demonstrated that cell-instructive sequences can enhance the biological properties of a variety of elastin analogs [3,22–28]. We chose to incorporate the V2 ligand sequence from matricellular protein CCN1 to promote endothelialization of our elastin polymer LysB10.Recent studies have revealed CCN molecules as dynamic regulators of tissue development and repair [12]. The CCN family includes six matricellular proteins (CCN1-6) that associate with the ECM as signaling molecules. CCN expression is tightly regulated and designed to translate a specific environmental stimuli through cell adhesion receptors. As such, the CCN1 molecule presents a promising target for bioengineering applications, functioning to support many key biological processes necessary for tissue repair, including cell adhesion, migration, differentiation, angiogenesis, and survival [29]. Significantly, adult tissues display low basal level expression of CCN1 with elevated expression observed at sites of injury and repair [12,15]. Such regulated expression suggests a level of biological specificity that may prove advantageous over more promiscuous cellular binding domains such as RGD. The 20 amino acid V2 peptide sequence isolated from CCN1 functions through integrin αvβ3 to support endothelial adhesion and migration [15]. Thus, affording endothelialization through facile recombinant expression of a discrete domain.
Genetic engineering provides experimental control over the ultimate design and subsequent number of V2 binding sites within the parent protein polymer. As an initial approach, we cloned a multimerized V2 block into LysB10. The multimeric structure of native ECM molecules such as FN, a dimer with dual adhesion sites, and tenascin-C, which presents six repeats of a cell adhesion domain, suggests that ligand clustering, as well as ligand density regulates cell signaling [30]. Several studies have shown that the clustering of ligand-bound integrin receptors is essential in propagating intracellular signaling for proper cell function. For example, αvβ3 integrins can undergo affinity maturation, resulting in the recruitment of αvβ3 to focal adhesions in the cell periphery [31]. Nanoscale arrangement of RGD peptides has revealed that clusters of at least nine peptides per molecule induce cell attachment comparable to native matrix proteins [32]. Cell migration and spreading are also dependent on this clustering mechanism. Thus, to translate the biological benefit of ligand clustering, we empirically isolated a 10-repeat V2 sequence for ligation into the midblock region of the triblock protein polymer. Cell adhesion studies demonstrated that at least 40 pmol/cm2 or ~1014 cell binding domains per cm2 was required to elicit cell attachment. This surface density is similar that reported by Tirrell and colleagues using RGD-modified elastin analogs in which significant HUVEC adhesion required the presentation of ligand densities of 1012–1013/cm2 [3,24,27].
Many tissue engineering applications require the use of gel based scaffolds. Therefore, we explored ligand on LysB10 and LysB10.V2 gels. The lysine residues within the protein polymers were crosslinked with genipin, a cytocompatible chemical cross-linker, in order to further stabilize intermolecular physical cross-links within the elastin-like network [33–36]. While the LysB10 gel presented a relatively non-adherent surface to cells, LysB10.V2 effectively supported cell attachment. Although the exact three-dimensional structure of the V2 active site is poorly understood, mutational analysis has identified an aspartate residue, as well as two flanking cysteine residues that are critical for sequence binding to the αvβ3 receptor [15]. In particular, the two cysteine residues flanking the core binding sequence may form a disulfide bond to present the binding site as a loop [37,38]. If a tertiary looped structure is involved in V2 presentation to the integrin-binding site, admittedly, any conformational strain induced upon the V2 sequence might limit cell adhesion. Nonetheless, despite the potential of covalent crosslinking to alter local protein conformation, LysB10.V2 gels proved to be an effective surface for αvβ3 mediated cell attachment.
5. Conclusions
Integration of biologic and structural functions of the extracellular matrix provides a robust design strategy for generating biomaterials that promote reparative responses. The studies reported herein demonstrate that a biomolecular engineering approach, which introduces cell-instructive peptide motifs within a recombinant elastin-like protein polymer elicits integrin-specific cellular responses. The LysB10 triblock copolymer provides a versatile, non-fouling platform for presenting bioactive ligands mimicking selective functions of the extracellular matrix. The ability to precisely control ligand presentation is an important design parameter, and ultimately directs cell fates such as adhesion, migration, focal adhesion assembly, and spreading. Thus, endothelial cell attachment, migration, morphology, and activation state were evaluated as markers of surface functionality on recombinant protein polymers. Improved biological activity of LysB10.V2 engineered surfaces can be attributed to enhanced binding to αvβ3. The generation of CCN1 mimicking protein polymers presents a rationale approach for promoting endothelialization of tissue-engineered scaffolds and coatings.
References
- 1.Girotti A, Reguera J, Rodriguez-Cabello JC, Arias FJ, Alonso M, Matestera A. Design and bioproduction of a recombinant multi(bio)functional elastin-like protein polymer containing cell adhesion sequences for tissue engineering purposes. J Mater Sci Mater Med. 2004;15(4):479–84. doi: 10.1023/b:jmsm.0000021124.58688.7a. [DOI] [PubMed] [Google Scholar]
- 2.Straley K, Heilshorn SC. Designer protein-based scaffolds for neural tissue engineering. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:2101–2. doi: 10.1109/IEMBS.2009.5334310. [DOI] [PubMed] [Google Scholar]
- 3.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extra-cellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5(2):497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 4.Liu JC, Tirrell DA. Cell response to RGD density in cross-linked artificial extracellular matrix protein films. Biomacromolecules. 2008;9(11):2984–8. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sallach RE, Cui W, Wen J, Martinez A, Conticello VP, Chaikof EL. Elastin-mimetic protein polymers capable of physical and chemical crosslinking. Biomaterials. 2009;30(3):409–22. doi: 10.1016/j.biomaterials.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagapudi K, Brinkman WT, Thomas BS, Park JO, Srinivasarao M, Wright E, et al. Viscoelastic and mechanical behavior of recombinant protein elastomers. Biomaterials. 2005;26(23):4695–706. doi: 10.1016/j.biomaterials.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv Drug Deliv Rev. 2002;54(8):1057–73. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 8.Wright ER, Conticello VP, Apkarian RP. Morphological characterization of elastin-mimetic block copolymers utilizing cryo- and cryoetch-HRSEM. Microsc Microanal. 2003;9(3):171–82. doi: 10.1017/S1431927603030150. [DOI] [PubMed] [Google Scholar]
- 9.Caves JM, Cui W, Wen J, Kumar VA, Haller CA, Chaikof EL. Elastin-like protein matrix reinforced with collagen microfibers for soft tissue repair. Biomaterials. 2011;32(23):5371–9. doi: 10.1016/j.biomaterials.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caves JM, Kumar VA, Martinez AW, Kim J, Ripberger CM, Haller CA, et al. The use of microfiber composites of elastin-like protein matrix reinforced with synthetic collagen in the design of vascular grafts. Biomaterials. 2010;31(27):7175–82. doi: 10.1016/j.biomaterials.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallach RE, Wei M, Biswas N, Conticello VP, Lecommandoux S, Dluhy RA, et al. Micelle density regulated by a reversible switch of protein secondary structure. J Am Chem Soc. 2006;128(36):12014–9. doi: 10.1021/ja0638509. [DOI] [PubMed] [Google Scholar]
- 12.Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41(4):771–83. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau LF, Lam SCT. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res. 1999;248(1):44–57. doi: 10.1006/excr.1999.4456. [DOI] [PubMed] [Google Scholar]
- 14.Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327(2):125–30. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Leu SJ, Todorovic V, Lam SC, Lau LF. Identification of a novel integrin alphavbeta3 binding site in CCN1 (CYR61) critical for pro-angiogenic activities in vascular endothelial cells. J Biol Chem. 2004;279(42):44166–76. doi: 10.1074/jbc.M406813200. [DOI] [PubMed] [Google Scholar]
- 16.McPherson DT, Morrow C, Minehan DS, Wu J, Hunter E, Urry DW. Production and purification of a recombinant elastomeric polypeptide, G-(VPGVG)19-VPGV, from Escherichia coli. Biotechnol Prog. 1992;8(4):347–52. doi: 10.1021/bp00016a012. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy JB, Furcht LT. Laminin and fibronectin promote the haptotactic migration of B16 mouse melanoma cells in vitro. J Cell Biol. 1984;98(4):1474–80. doi: 10.1083/jcb.98.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson JT, Cui W, Kozlovskaya V, Kharlampieva E, Pan D, Qu Z, et al. Cell surface engineering with polyelectrolyte multilayer thin films. J Am Chem Soc. 2011;133(18):7054–64. doi: 10.1021/ja110926s. [DOI] [PubMed] [Google Scholar]
- 19.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Bio-macromolecules. 2002;3(2):357–67. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 20.Trabbic-Carlson K, Setton LA, Chilkoti A. Swelling and mechanical behaviors of chemically cross-linked hydrogels of elastin-like polypeptides. Bio-macromolecules. 2003;4(3):572–80. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 21.Brooks P, Clark R, Cheresh D. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264(5158):569–71. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 22.Heilshorn SC, DiZio KA, Welsh ER, Tirrell DA. Endothelial cell adhesion to the fibronectin CS5 domain in artificial extracellular matrix proteins. Biomaterials. 2003;24(23):4245–52. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 23.Heilshorn SC, Liu JC, Tirrell DA. Cell-binding domain context affects cell behavior on engineered proteins. Biomacromolecules. 2005;6(1):318–23. doi: 10.1021/bm049627q. [DOI] [PubMed] [Google Scholar]
- 24.Kobatake E, Onoda K, Yanagida Y, Aizawa M. Design and gene engineering synthesis of an extremely thermostable protein with biological activity. Bio-macromolecules. 2000;1(3):382–6. doi: 10.1021/bm000284f. [DOI] [PubMed] [Google Scholar]
- 25.Massodi I, Moktan S, Rawat A, GLB, Raucher D. Inhibition of ovarian cancer cell proliferation by a cell cycle inhibitory peptide fused to a thermally responsive polypeptide carrier. Int J Cancer. 2010;126(2):533–44. doi: 10.1002/ijc.24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panitch A, Yamaoka T, Fournier MJ, Mason TL, Tirrell DA. Design and biosynthesis of elastin-like artificial extracellular matrix proteins containing periodically spaced fibronectin CS5 domains. Macromolecules. 1999;32(5):1701–3. [Google Scholar]
- 27.Richman GP, Tirrell DA, Asthagiri AR. Quantitatively distinct requirements for signaling-competent cell spreading on engineered versus natural adhesion ligands. J Control Release. 2005;101(1–3):3–12. doi: 10.1016/j.jconrel.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 28.Straley KS, Heilshorn SC. Design and adsorption of modular engineered proteins to prepare customized, neuron-compatible coatings. Front Neuroeng. 2009:2. doi: 10.3389/neuro.16.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai J, Norng M, Modak C, Reavis KM, Mouazzen W, Pham J. CCN1 induces a reversible epithelial-mesenchymal transition in gastric epithelial cells. Lab Invest. 2010;90(8):1140–51. doi: 10.1038/labinvest.2010.101. [DOI] [PubMed] [Google Scholar]
- 30.von der Mark K, Park J, Bauer S, Schmuki P. Nanoscale engineering of biomimetic surfaces: cues from the extracellular matrix. Cell Tissue Res. 2010;339(1):131–53. doi: 10.1007/s00441-009-0896-5. [DOI] [PubMed] [Google Scholar]
- 31.Kiosses WB, Shattil SJ, Pampori N, Schwartz MA. Rac recruits high-affinity integrin [alpha]v[beta]3 to lamellipodia in endothelial cell migration. Nat Cell Biol. 2001;3(3):316–20. doi: 10.1038/35060120. [DOI] [PubMed] [Google Scholar]
- 32.Maheshwari G, Brown G, Lauffenburger D, Wells A, Griffith L. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(10):1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 33.Yao C-H, Liu B-S, Chang C-J, Hsu S-H, Chen YS. Preparation of networks of gelatin and genipin as degradable biomaterials. Mater Chem Phys. 2004;83(2–3):204–8. [Google Scholar]
- 34.Chang WH, Chang Y, Lai PH, Sung HW. A genipin-crosslinked gelatin membrane as wound-dressing material: in vitro and in vivo studies. J Biomater Sci Polym Ed. 2003;14:481–95. doi: 10.1163/156856203766652084. [DOI] [PubMed] [Google Scholar]
- 35.Sung H-W, Chang W-H, Ma C-Y, Lee M-H. Crosslinking of biological tissues using genipin and/or carbodiimide. J Biomed Mater Res. 2003;64A(3):427–38. doi: 10.1002/jbm.a.10346. [DOI] [PubMed] [Google Scholar]
- 36.Touyama RIK, Takeda Y, Yatsuzuka M, Ikumoto T, Moritome N, Shingu T, et al. Studies on the blue pigments produced from genipin and methylamine. II. On the formation mechanisms of brownish-red intermediates leading to the blue pigment formation. Chem Pharm Bull. 1994;42:1571–8. [Google Scholar]
- 37.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268(27):20205–10. [PubMed] [Google Scholar]
- 38.McLane MA, Vijay-Kumar S, Marcinkiewicz C, Calvete JJ, Niewiarowski S. Importance of the structure of the RGD-containing loop in the disintegrins echistatin and eristostatin for recognition of [alpha]IIb[beta]3 and [alpha]v [beta]3 integrins. FEBS Lett. 1996;391(1–2):139–43. doi: 10.1016/0014-5793(96)00716-8. [DOI] [PubMed] [Google Scholar]