Abstract
Aims
We investigated the sequential gene expression in the gingiva during the induction and resolution of experimental gingivitis.
Methods
Twenty periodontally and systemically healthy non-smoking volunteers participated in a 3-week experimental gingivitis protocol, followed by debridement and 2-week regular plaque control. We recorded clinical indices and harvested gingival tissue samples from 4 interproximal palatal sites in half of the participants at baseline, Day 7, 14 and 21 (‘induction phase’), and at day 21, 25, 30 and 35 in the other half (‘resolution phase’). RNA was extracted, amplified, reversed transcribed, amplified, labeled and hybridized with Affymetrix Human Genome U133Plus2.0 microarrays. Paired t-tests compared gene expression changes between consecutive time points. Gene ontology analyses summarized the expression patterns into biologically relevant categories.
Results
The median gingival index was 0 at baseline, 2 at Day 21 and 1 at Day 35. Differential gene regulation peaked during the third week of induction and the first four days of resolution. Leukocyte transmigration, cell adhesion and antigen processing/presentation were the top differentially regulated pathways.
Conclusions
Transcriptomic studies enhance our understanding of the pathobiology of the reversible inflammatory gingival lesion and provide a detailed account of the dynamic tissue responses during induction and resolution of experimental gingivitis.
Keywords: Experimental gingivitis, microarray, gene expression, tissue response
Introduction
Since its inception approximately 50 years ago (Löe et al., 1965), experimental gingivitis has been used extensively as a clinical research tool in the study of the pathobiology of the reversible gingival lesion that was shown to develop in response to the accumulation of dental plaque on the adjacent tooth surfaces. Over the years, a substantial body of data has accumulated on the microbiologic features of the early gingival lesion (Moore et al., 1982, Moore et al., 1984), including the influence of gingival inflammation on plaque accumulation (Loesche and Syed, 1978, Daly and Highfield, 1996, Hillam and Hull, 1977) as well as the differences in microbial profiles between individuals with high and low propensity for gingival inflammation (Lie et al., 1995) or between subjects with different levels of susceptibility to periodontitis (Abbas et al., 1986). Other studies have used histology and/or immunohistochemistry to characterize the cellular components of the gingival lesion (Payne et al., 1975, Seymour et al., 1983, Kinane et al., 1991, Fransson et al., 1999) or to identify proteins that are secreted into the gingival crevicular fluid (GCF) during the development and the resolution of gingival inflammation (Lamster et al., 1985, Heasman et al., 1993, Deinzer et al., 2007, Offenbacher et al., 2009a, Grant et al., 2010). Lastly, the experimental gingivitis model has been widely used to study the effects of pharmacological agents that inhibit plaque formation and/or modulate gingival inflammation in humans (Wennström, 1988, Jenkins et al., 1993, Ramberg et al., 1995, Quirynen et al., 2001, Sekino et al., 2003, Van Strydonck et al., 2005).
In the past few years, our group has used high throughput microarray technology in the study of the pathobiology of periodontal diseases and was the first to characterize the whole genome gingival tissue transcriptomes in different forms of periodontitis and in states of periodontal health and disease (Papapanou et al., 2004, Demmer et al., 2008), as well as to examine the relationship between subgingival microbial colonization profiles and gene expression signatures in the adjacent tissues (Papapanou et al., 2009). In this paper, we extend our previous work and analyze gingival transcriptional profiles concurrent with the induction and resolution of gingival inflammation during the course of experimental gingivitis. We hypothesized that these profiles would be consistent with known elements of the pathobiology of gingivitis, but would also point to the involvement of novel, yet unrecognized molecules and processes. Thus, the aim of the present study was to systematically investigate the sequential gene expression in the gingival tissues that parallels (i) the gradual conversion from a state of pristine periodontal health to a state of established gingivitis, and (ii) the resolution of gingival inflammation during re-institution of periodontal health.
Material and Methods
The design of the study was approved by the Regional Ethical Review Board, Göteborg, Sweden (#005-09). Informed consent was obtained from all study participants prior to enrollment.
Subject Sample
A total of twenty, systemically healthy volunteers were recruited among the undergraduate students attending the Faculty of Odontology, Sahlgrenska Academy, Göteborg University, Sweden. All participants were free of interproximal attachment loss and had no probing pocket depths of > 4 mm. Buccal recessions of obvious traumatic etiology at single teeth did not automatically disqualify a volunteer from participation. The participants were non-smokers, were not current users of antibiotics, contraceptives, or immunosuppressive drugs, and were not pregnant or lactating. They were divided into two groups, a ‘gingivitis induction’ and a ‘gingivitis resolution’ group comprising 10 individuals each, equally many female and male. With respect to other demographic characteristics, 19 individuals were Caucasian while a single female participant in the resolution group was Asian. The mean age was 24.7 years in the induction group (median 21 years, range 20–31) and 24.4 years in the resolution group (median 24, range 21–29).
Experimental gingivitis protocol
During a 3-week preparatory period prior to the experimental gingivitis phase, all volunteers were instructed in proper oral hygiene measures (tooth brushing and interproximal cleaning using dental floss) and were subjected to between two and three sessions of professional tooth cleaning using a rubber cup and polishing paste until they showed no or only minimal signs of gingival inflammation (average full-mouth Gingival Index (Löe, 1967) <0.2). Maxillary impressions were obtained and acrylic stents that covered the palatal gingival tooth surfaces of all maxillary teeth were fabricated. After establishment of absence of gingival inflammation, experimental gingivitis was induced over a three-week period at the maxillary palatal surfaces. During that time, the participants were asked to abstain from brushing of the palatal surfaces of the maxillary arch and from any means of interproximal cleaning. To prevent accidental removal of plaque from the experimental sites, the individually fabricated stents were always put in place during the regular brushing of the maxillary buccal surfaces and the mandibular teeth. After completion of the 3-week gingivitis induction phase, all participants received thorough oral prophylaxis by the same dental hygienist, including full-mouth debridement and polishing. Oral hygiene measures including tooth brushing and dental flossing at least twice daily were reinstituted in the entire dentition. The “gingivitis resolution” phase was completed two weeks after re-institution of regular oral hygiene.
Clinical assessments
Gingival Index (GI) (Löe, 1967) assessments were carried out bilaterally at the mesio-palatal and disto-palatal aspects of each interdental papilla between the first and second maxillary premolars and between the first maxillary premolar and canine, using a periodontal probe, by a single calibrated examiner (author PR).
Bacterial plaque samples and processing
Immediately after the clinical assessments, a sterile paper point was inserted at the mesio-palatal and disto-palatal aspect of each of the above interdental papillae, left in place for 30 seconds, and transported in sterile Tris-EDTA buffer. The plaque samples were analyzed individually using the checkerboard DNA-DNA hybridization method (Socransky et al., 1994, Papapanou et al., 2001) as earlier described with respect to the following 18 bacterial species: Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Prevotella nigrescens, Prevotella intermedia, Parvimonas micra, Fusobacterium nucleatum, Campylobacter rectus, Capnocytophaga ochracea, Streptococcus sanguis, Streptococcus mutans, Streptococcus intermedius, Streptococcus oralis, Actinomyces naeslundii, Veillonella parvula, Selenomonas noxia, Eikenella corrodens and Aggregatibacter actinomycetemcomitans.
Collection of gingival tissue samples
As mentioned above, to minimize the number of sequentially obtained tissue samples per participant to a maximum of four, we studied the induction of gingivitis separately from the resolution of gingival inflammation in two distinct groups of patients. After local infiltration anesthesia with 2% lidocaine-HCl 2% with 1:100.000 epinephrine, gingival tissue samples amounting to approximately 8 mm3 and comprising both the sulcular epithelium and the underlying connective tissue were obtained from the palatal aspects of four interproximal papillae in the following sequence: the papilla between the upper canine and the first premolar, followed by its contra-lateral site, followed by an interproximal papilla between the two upper premolars, followed by its contra-lateral site. These biopsies were obtained at the following time points: In the induction group, at baseline (Day 0), and at the completion of the first week (Day 7), second week (Day 14) and third week (Day 21) of experimental gingivitis. In the resolution group, biopsies were obtained at the completion of three weeks of experimental gingivitis (Day 21), and at four (Day 25), nine (Day 30) and fourteen days (Day 35) after the provision of full-mouth prophylaxis and re-institution of oral hygiene procedures.
Gingival tissue processing
Immediately after harvesting, each tissue sample was rinsed with sterile saline and placed in an individually labeled Eppendorf tube with an RNA stabilizing agent (RNAlater, Ambion, Inc., Austin, TX). The biopsies were held at 4°C overnight in RNAlater, the liquid was subsequently decanted and the tube was snap-frozen in liquid nitrogen. The samples were held at −70°C until being shipped to the laboratory at the Division of Periodontics, Columbia University in a single batch, in dry ice. The transportation time did not exceed 24 hours and all tissue samples were frozen upon arrival.
Isolation of total RNA, reverse and in-vitro transcription, labeling and hybridization
We largely followed the protocol recently described in detail by Kebschull and Papapanou (2010). In brief, the tissue specimens were homogenized in TRIzol (Invitrogen Life Technologies, Carlsbad, CA, US) and total RNA was isolated and purified using RNeasy cleanup columns (Qiagen, Valencia, CA, US). RNA quantity and quality was evaluated in all cases spectrophotometrically using a NanoDrop 1000 device (Thermo Scientific, DE, USA). In preparatory experiments, sample quality was assessed using an Agilent 2100 Bioanalyzer and RNA Nano Chips (Agilent Technologies, CA, US), consistently demonstrating RNA integrity numbers ≥ 9. One hundred ng of total RNA was reverse- and in vitro transcribed, labeled and fragmented using the 3’IVT kit (Affymetrix, Santa Clara, CA, US), and 15 µg of the labeled RNA was hybridized with a Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, US) which carry > 55,000 probe sets mapping to approximately 38,500 well characterized genes.
Data analysis
Gene expression data were analyzed as previously described (Demmer et al., 2010). In brief, Affymetrix array data were first normalized and summarized using the log scale robust multi-array analysis (RMA; Irizarry et al., 2003) with default settings. For each probe set, a fold change was computed by dividing the mRNA expression value at each time point by the expression value of the immediately preceding time point, or to baseline, i.e., Day 0 in the ‘induction’ group and Day 21 in the ‘resolution’ group. P-values from the aforementioned analyses were input into gene ontology analysis using the Pathway Express software (Draghici et al., 2007, Khatri et al., 2007) to identify biologically-relevant groups of genes that showed changes in expression over time. Gene symbols and descriptions were downloaded from: http://www.bioinformatics.ubc.ca/microannots/.
Results
Clinical Findings
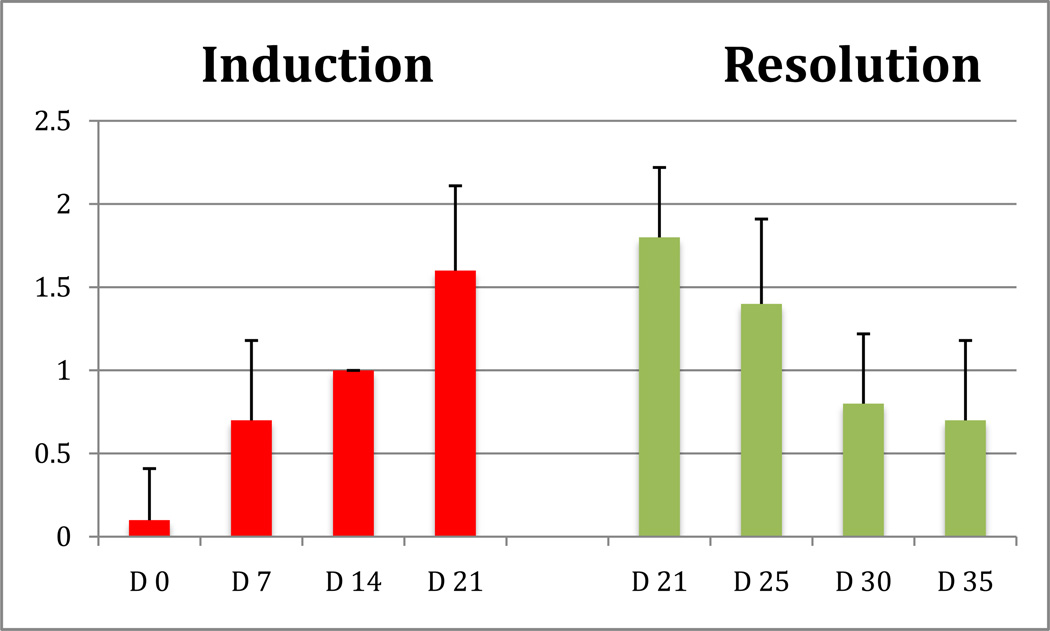
Figure 1 illustrates the development and resolution of experimental gingivitis reflected through the GI scores at the experimental sites. The mean GI was 0.1 at Day 0 (median 0), and increased to 0.7 at Day 7 (median 1), to 1.0 at Day 14 (median 1) and peaked at 1.6 (median 2, range 1–2) at Day 21. In the resolution phase, the average GI was 1.8 (median 2, range 1–2), with 2 of the 10 participants showing a GI of 1 after three weeks of experimental gingivitis induction). The average GI was reduced to 1.4 at Day 25 (median 1), to 0.8 at Day 30 (median 1) and at 0.7 at Day 35 (median 1). Only 3 participants had a GI score of 0 at the end of the resolution phase, with the remaining 7 showing a GI of 1.
Fig 1.
Gingival index at the experimental sites during the induction and resolution of experimental gingivitis. Bars represent means and standard deviations.
Microbiological Findings
Supplemental Fig. 1 describes the bacterial colonization profiles at the gingival crevices adjacent to the harvested gingival tissue samples during the induction and resolution phases. Levels of the “red complex” species P. gingivalis, T. forsythia and T. denticola were much lower than those of all other investigated bacteria throughout both phases. A conspicuous increase in certain “orange complex” bacteria including P. nigrescens and P. intermedia, and to a lesser extent P. micra, was observed during gingivitis induction, and the levels of these species declined during the resolution phase. Likewise streptococcal spp. and levels of Actinomyces naeslundii, which dominated the microbial profiles, showed a similar pattern of increase and decline during the induction and resolution phases, respectively.
Transcriptomic Responses
Dynamics of the sequential gene activation
To initiate the analysis of the sequential activation of genes over time, we first explored the number of probe sets that were statistically significantly (p<0.05) differentially regulated between any two consecutive time points. In the induction phase, 5,278 probes were differentially expressed (p<0.05) during the first week (i.e., Day 7 vs. baseline); 3,660 probes in the second week (Day 14 vs. Day 7); and 6,765 probes during the third week (D21 vs. D14). A comparison between Day 21 and baseline yielded a total of 3,170 differentially regulated probes. In the resolution phase, 7,250 probes were significantly differentially regulated during the first four days after prophylaxis and re-institution of oral hygiene (i.e., Day 25 vs. Day 21); 5,085 probes in the next five days (Day 30 vs. Day 25) and 2,698 probes during the final five days (D35 vs. D30). A comparison between Day 35 and Day 21 yielded 7,763 differentially regulated probes.
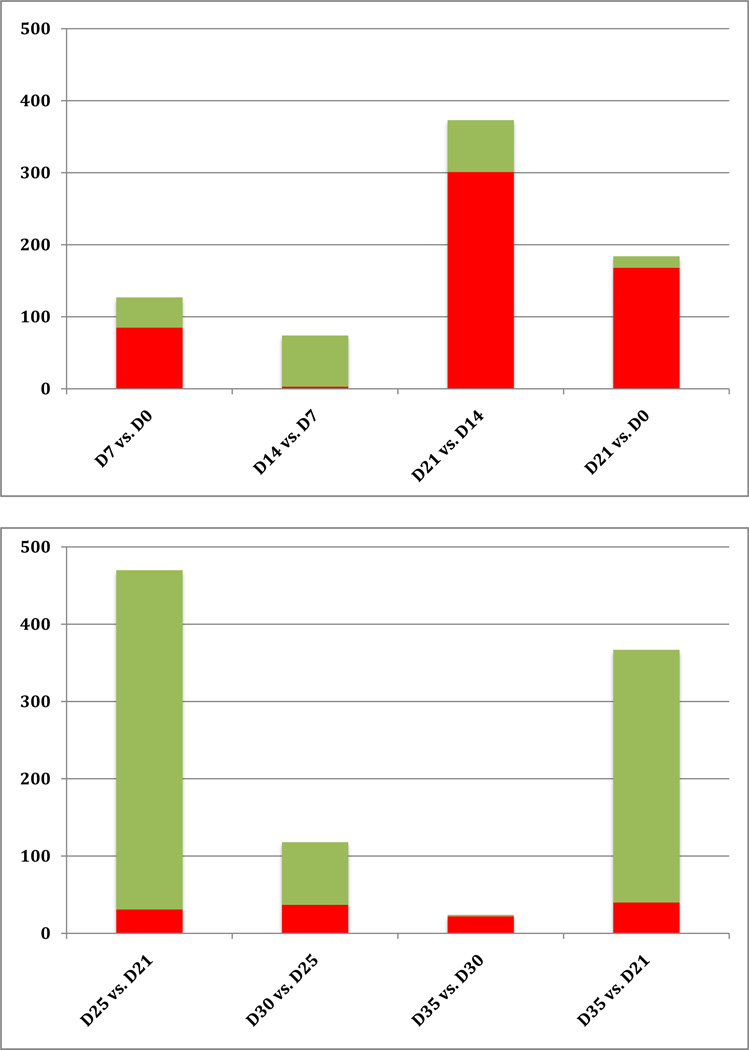
Subsequently, we examined the number of probes with an absolute fold change of >1.5, i.e., probes that were either upregulated or downregulated by at least 50%, between two consecutive time points. In the Induction phase, a total of 127 probe sets were differentially regulated between baseline and Day 7 by more than 1.5-fold (Fig. 2). Of these, 85 were upregulated and 42 were downregulated. During the second week of gingivitis induction, only three probe sets were upregulated while 71 were downregulated. Differential gene expression was maximized during the third week of induction (between Day 21 and Day 14), with a total of 373 probe sets being differentially regulated >1.5 fold, 81% of which (301 probes) were upregulated and 19% (72 probes) downregulated. A comparison between the time point of maximal inflammation (Day 21) and baseline yielded a total of only 184 differentially regulated probe sets, i.e., less than half of the number found to be differentially regulated during the third week of induction alone.
Fig 2.
Number of differentially expressed probe sets with an absolute fold change of >1.5 during the Induction (top) and Resolution phases (bottom). Red bars indicate up-regulation, green bars down-regulation.
The bottom panel of Fig 2 provides the corresponding description of sequential gene activation during the Resolution phase. It is evident that most of the activity in the gingival tissues in terms of differential gene expression occurred within the first four days post-prophylaxis and re-institution of oral hygiene. Out of a total of 470 probe sets that were statistically significantly differentially expressed with an absolute fold change of 1.5, 93% (439 probes) were downregulated and only 7% (31 probes) were upregulated. In comparison, far fewer probe sets were differentially expressed during the subsequent time intervals: 118 between Day 30 and Day 25 (69% down regulated), and only 24 between Day 35 and Day 30. A comparison between the first and last points of the resolution phase showed a total of 367 differentially expressed probe sets, in their vast majority (89%) downregulated.
Tables 1 and 2 list the top 20 probes that mapped to annotated genes and were found to be differentially expressed during the induction and resolution phases, respectively. In these tables, the depicted fold changes in expression were based on data from two consecutive time points and were calculated as the ratio of expression at the latter time point over that of the former time point (i.e., Day 7 / Day 0, Day 14 / Day 7, and Day 21 / 14 in the induction phase, Table 1a–c; and Day 25 / Day 21, Day 30 / Day 25 and Day 35 / Day 30, in the resolution phase Table 2a–c). In both Tables, probes are sorted according to descending absolute fold change, i.e., according to decreasing magnitude of differential regulation irrespective of direction (up- or downregulation). Complete lists of all differentially regulated probes between any two consecutive time points along with the corresponding fold changes and p-values are provided in the Online Supplementary Tables 1–6.
Table 1.
| a. Top differentially expressed probes mapping to annotated genes, during the first week of gingivitis induction, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | CRISP3 | cysteine-rich secretory protein 3 | 5.54 | 0.00261 |
| 2 | MS4A1 | membrane-spanning 4-domains, subfamily A, member 1 |
3.24 | 0.015318 |
| 3 | ATP6V0A4 | ATPase, H+ transporting, lysosomal V0 subunit a4 |
2.64 | 0.00379 |
| 4 | CD177 | CD177 molecule | 2.54 | 0.035596 |
| 5 | CLCA4 | chloride channel, calcium activated, family member 4 |
2.52 | 0.032565 |
| 6 | GYS2 | glycogen synthase 2 (liver) | 2.38 | 0.003151 |
| 7 | TMPRSS2|AK | transmembrane protease, serine 2 | 2.28 | 0.009112 |
| 8 | TMPRSS2|AK | transmembrane protease, serine 2 | 2.13 | 0.027892 |
| 9 | ERO1L | ERO1-like (S. cerevisiae) | 2.05 | 0.014052 |
| 10 | IGFBP6 | insulin-like growth factor 2 mRNA binding protein 3 |
0.49 | 0.010173 |
| 11 | ERO1L | ERO1-like (S. cerevisiae) | 1.98 | 0.017879 |
| 12 | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 |
1.97 | 0.000065 |
| 13 | CDKN3 | cyclin-dependent kinase inhibitor 3 (CDK2- associated dual specificity phosphatase) |
0.51 | 0.016633 |
| 14 | SILV | silver homolog (mouse) | 0.52 | 0.022768 |
| 15 | GPR37 | G protein-coupled receptor 37 (endothelin receptor type B-like) |
0.52 | 0.012444 |
| 16 | PPP1R3C | protein phosphatase 1, regulatory (inhibitor) subunit 3C |
1.92 | 0.013479 |
| 17 | C15orf48 | chromosome 15 open reading frame 48 | 1.91 | 0.003409 |
| 18 | CDKN3 | cyclin-dependent kinase inhibitor 3 (CDK2- associated dual specificity phosphatase) |
0.52 | 0.017647 |
| 19 | FUT3|FUT5| | fucosyltransferase 3 (galactoside 3(4)-L- fucosyltransferase,Lewis blood group)|fucosyltransferase 5 |
1.88 | 0.010279 |
| 20 | PHLDA1 | pleckstrin homology-like domain, family A, member 1 |
1.86 | 0.027904 |
| b. Top differentially expressed probes mapping to annotated genes, during the second week of gingivitis induction, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | CRISP3 | cysteine-rich secretory protein 3 | 0.33 | 0.046981 |
| 2 | POU2AF1 | POU domain, class 2, associating factor 1 | 0.49 | 0.043393 |
| 3 | FAM46C | family with sequence similarity 46, member C | 0.45 | 0.039575 |
| 4 | RSAD2 | radical S-adenosyl methionine domain containing 2 |
0.57 | 0.025385 |
| 5 | TMPRSS2|AK | transmembrane protease, serine 2 | 0.57 | 0.024545 |
| 6 | DSC1 | desmocollin 1 | 1.62 | 0.035139 |
| 7 | PAPSS2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
0.63 | 0.016613 |
| 8 | KIAA0746 | KIAA0746 protein | 0.63 | 0.039536 |
| 9 | USP1 | ubiquitin specific peptidase 1 | 1.37 | 0.027314 |
| 10 | IL6R | interleukin 6 receptor | 1.34 | 0.004456 |
| 11 | CPM | carboxypeptidase M | 1.34 | 0.026291 |
| 12 | UQCRC2 | ubiquinol-cytochrome c reductase core protein II |
1.28 | 0.011865 |
| 13 | QSER1 | glutamine and serine rich 1 | 1.24 | 0.012726 |
| 14 | PYGL | phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) |
1.24 | 0.013103 |
| 15 | RBM25 | RNA binding motif protein 25 | 1.21 | 0.030094 |
| 16 | KLF13 | Kruppel-like factor 13 | 1.20 | 0.023861 |
| 17 | ZFP91 | zinc finger protein 91 homolog (mouse) | 1.16 | 0.035614 |
| 18 | C16orf74 | chromosome 16 open reading frame 74 | 1.15 | 0.033430 |
| 19 | NEK1 | NIMA (never in mitosis gene a)-related kinase 1 |
0.88 | 0.033341 |
| 20 | CUTL1 | cut-like 1, CCAAT displacement protein (Drosophila) |
1.12 | 0.039922 |
| c. Top differentially expressed probes mapping to annotated genes, during the third week of gingivitis induction, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.24 | 0.021738 |
| 2 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.24 | 0.025006 |
| 3 | CXCL13 | chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) |
4.01 | 0.007572 |
| 4 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.24 | 0.021137 |
| 5 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.25 | 0.022495 |
| 6 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.25 | 0.019494 |
| 7 | HBB | hemoglobin, beta | 0.30 | 0.036112 |
| 8 | PDZRN4 | PDZ domain containing RING finger 4 | 3.13 | 0.013944 |
| 9 | CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) |
3.12 | 0.005698 |
| 10 | HBB | hemoglobin, beta | 0.32 | 0.039498 |
| 11 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 0.33 | 0.025507 |
| 12 | RGS4 | regulator of G-protein signalling 4 | 2.99 | 0.013331 |
| 13 | UBD | ubiquitin D | 2.81 | 0.004953 |
| 14 | HBB | hemoglobin, beta | 0.35 | 0.041222 |
| 15 | CCL19 | chemokine (C-C motif) ligand 19 | 2.76 | 0.006895 |
| 16 | SAA1 | serum amyloid A1 | 2.67 | 0.007016 |
| 17 | MGC23985 | similar to AVLV472 | 0.38 | 0.014519 |
| 18 | DCT | dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) |
0.40 | 0.019126 |
| 19 | ARG1 | arginase, liver | 0.40 | 0.003743 |
| 20 | RP1-14N1.3 | filaggrin 2 | 0.41 | 0.000253 |
Table 2.
| a. Top differentially expressed probes mapping to annotated genes, during the first four days of gingivitis resolution, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | ODAM | odontogenic, ameloblast asssociated | 0.12 | 0.00164 |
| 2 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 6.36 | 0.00429 |
| 3 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 6.03 | 0.003739 |
| 4 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 5.81 | 0.003716 |
| 5 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 5.74 | 0.003961 |
| 6 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 5.66 | 0.003054 |
| 7 | C4orf26 | chromosome 4 open reading frame 26 | 0.18 | 0.00814 |
| 8 | HBA1|HBA2 | hemoglobin, alpha 1|hemoglobin, alpha 2 | 4.92 | 0.004938 |
| 9 | HBB | hemoglobin, beta | 4.83 | 0.006822 |
| 10 | HBB | hemoglobin, beta | 4.47 | 0.005677 |
| 11 | HBB | hemoglobin, beta | 4.02 | 0.006566 |
| 12 | SFRP4 | secreted frizzled-related protein 4 | 0.25 | 0.013203 |
| 13 | RGS4 | regulator of G-protein signalling 4 | 0.26 | 0.000218 |
| 14 | MS4A1 | membrane-spanning 4-domains, subfamily A, member 1 |
0.28 | 0.002695 |
| 15 | PPBP | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
3.43 | 0.001211 |
| 16 | CXCL1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
0.30 | 0.001902 |
| 17 | PDZRN4 | PDZ domain containing RING finger 4 | 0.32 | 0.001407 |
| 18 | TNFRSF17 | tumor necrosis factor receptor superfamily, member 17 |
0.32 | 0.001979 |
| 19 | MMP13 | matrix metallopeptidase 13 (collagenase 3) | 0.33 | 0.029987 |
| 20 | OGN | osteoglycin (osteoinductive factor, mimecan) | 0.33 | 0.011285 |
| b. Top differentially expressed probes mapping to annotated genes, during the fifth and tenth day of gingivitis resolution, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | C4orf26 | chromosome 4 open reading frame 26 | 5.82 | 0.046614 |
| 2 | ODAM | odontogenic, ameloblast asssociated | 3.14 | 0.016792 |
| 3 | CXCL11 | chemokine (C-X-C motif) ligand 11 | 2.95 | 0.019572 |
| 4 | CXCL1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
0.38 | 0.030986 |
| 5 | RP1-14N1.3 | filaggrin 2 | 0.47 | 0.02587 |
| 6 | HAL | histidine ammonia-lyase | 2.07 | 0.002678 |
| 7 | HTR3A | 5-hydroxytryptamine (serotonin) receptor 3A |
2.05 | 0.002189 |
| 8 | ANXA9 | annexin A9 | 0.49 | 0.000984 |
| 9 | UGT1A10|UG | UDP glucuronosyltransferase 1 family, polypeptide A10|UDP glucuronosyltransferase 1 family, polypept |
0.49 | 0.019797 |
| 10 | MMP13 | matrix metallopeptidase 13 (collagenase 3) |
0.49 | 0.023574 |
| 11 | S100P | S100 calcium binding protein P | 0.50 | 0.028185 |
| 12 | AADAC | arylacetamide deacetylase (esterase) | 0.50 | 0.029201 |
| 13 | XDH | xanthine dehydrogenase | 1.94 | 0.018272 |
| 14 | TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma- glutamyltransferase) |
1.90 | 0.036086 |
| 15 | ISL1 | ISL1 transcription factor, LIM/homeodomain, (islet-1) |
1.87 | 0.000125 |
| 16 | CXCL9 | chemokine (C-X-C motif) ligand 9 | 0.54 | 0.014364 |
| 17 | POF1B|AK12 | premature ovarian failure, 1B | 1.84 | 0.003542 |
| 18 | LAMC2 | laminin, gamma 2 | 0.54 | 0.033858 |
| 19 | NRCAM | neuronal cell adhesion molecule | 1.82 | 0.011176 |
| 20 | placenta-specific 8 | placenta-specific 8 | 1.70 | 0.024099 |
| c. Top differentially expressed probes mapping to annotated genes, during the last five days of gingivitis resolution, sorted according to descending absolute fold change. | ||||
|---|---|---|---|---|
| Rank | Gene | Description | Fold change |
p-value |
| 1 | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog |
3.19 | 0.02017 |
| 2 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B |
3.08 | 0.040542 |
| 3 | EGR1 | early growth response 1 | 2.83 | 0.005859 |
| 4 | EGR1 | early growth response 1 | 2.71 | 0.004425 |
| 5 | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
2.24 | 0.004138 |
| 6 | ATF3 | activating transcription factor 3 | 2.09 | 0.026379 |
| 7 | RFX2 | regulatory factor X, 2 (influences HLA class II expression) |
1.98 | 0.037945 |
| 8 | HBD | hemoglobin, delta | 1.94 | 0.013261 |
| 9 | HBG1|HBG2 | hemoglobin, gamma A|hemoglobin, gamma> G |
1.90 | 0.036859 |
| 10 | RGS1 | regulator of G-protein signalling 1 | 1.81 | 0.034072 |
| 11 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
1.79 | 0.002562 |
| 12 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
1.75 | 0.004211 |
| 13 | RNASE7 | ribonuclease, RNase A family, 7 | 1.73 | 0.045338 |
| 14 | DUSP1 | dual specificity phosphatase 1 | 1.69 | 0.00139 |
| 15 | SAMD4A | sterile alpha motif domain containing 4A | 0.60 | 0.00811 |
| 16 | CDSN | corneodesmosin | 1.64 | 0.010418 |
| 17 | EREG | epiregulin | 1.64 | 0.002051 |
| 18 | CD55|AX772 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
1.58 | 0.013212 |
| 19 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 |
1.58 | 0.002423 |
| 20 | MNDA | myeloid cell nuclear differentiation antigen | 1.56 | 0.03622 |
Gene ontology analyses
Using the Pathway Express software, we summarized the acquired expression profiles into biological processes. Tables 3 and 4 list the top five differentially regulated pathways between each pair of consecutive time points in the gingivitis induction and resolution phases, respectively. The Tables also list the total number of genes included in each pathway; the percentage of genes in the particular pathway that were statistically differentially regulated (p<0.05); the impact factor of each individual pathway, which is a probabilistic term that takes under consideration both the proportion of the differentially regulated genes in the pathway and the perturbation of each gene; and finally the p-value for the differential regulation of the particular pathway. The top two gene ontology groups in both the first and the second week of gingivitis induction were leukocyte trans-endothelial migration and cell adhesion, while antigen processing and presentation was the top regulated pathway in the third week. Antigen processing and presentation and leukocyte transendothelial migration were the top differentially regulated pathways immediately after debridement and re-institution of oral hygiene, followed by cell adhesion molecules. Leukocyte transendothelial migration was still strongly regulated during the next five days of gingivitis resolution, but all other differentially regulated ontology groups had substantially lower impact factors.
Table 3.
Gene Ontology analysis: Gingivitis induction phase.
| Time Point | Pathway | # Genes in Pathway; % Regulated |
Impact Factor | p-value |
|---|---|---|---|---|
|
D7/D0 |
Leukocyte transendothelial migration | 119; 19% | 172.1 | 3.10E-73 |
| Cell adhesion molecules | 116; 21% | 116.3 | 3.44E-49 | |
| Adherens junction | 76; 32% | 47.1 | 1.73E-19 | |
| Huntington's disease | 171; 39% | 27.1 | 4.57E-11 | |
| Ribosome | 80; 42% | 25.1 | 3.36E-10 | |
| D14/D7 | Leukocyte transendothelial migration | 114; 15% | 325.4 | 1.52E-139 |
| Cell adhesion molecules | 127; 17% | 177.9 | 9.12E-76 | |
| Phosphatidylinositol signaling system | 76; 14% | 15.6 | 2.72E-06 | |
| Tight junction | 131; 23% | 8.6 | 0.00169301 | |
| MAPK signaling pathway | 266; 20% | 8.1 | 0.00260222 | |
| D21/D14 | Antigen processing and presentation | 79; 37% | 102.3 | 3.75E-43 |
| Cell adhesion molecules | 127; 46% | 77.4 | 1.85E-32 | |
| Leukocyte transendothelial migration | 114; 42% | 51.5 | 2.29E-21 | |
| Adherens junction | 76; 29% | 19.2 | 9.44E-08 | |
| Allograft rejection | 33; 55% | 15.1 | 4.33E-06 |
Table 4.
Gene Ontology analysis: Gingivitis resolution phase.
| Time Point | Pathway | # Genes in Pathway; % Regulated |
Impact Factor | p-value |
|---|---|---|---|---|
| D25/D21 | Antigen processing and presentation | 89; 33% | 106.2 | 7.76E-45 |
| Leukocyte transendothelial migration | 119; 35% | 53.2 | 4.17E-22 | |
| Cell adhesion molecules | 134; 41% | 42.7 | 1.25E-17 | |
| Adherens junction | 78; 29% | 25.7 | 1.77E-10 | |
| Phosphatidylinositol signaling system | 76; 41% | 13.9 | 1.42E-05 | |
| D30/D25 | Leukocyte transendothelial migration | 119; 24% | 69.9 | 3.08E-29 |
| Adherens junction | 78; 38% | 29.3 | 5.82E-12 | |
| Cell adhesion molecules | 134; 22% | 29.1 | 6.98E-12 | |
| Antigen processing and presentation | 89; 27% | 26.6 | 7.94E-11 | |
| Ubiquitin mediated proteolysis | 138; 40% | 23.1 | 2.30E-09 | |
| D35/D30 | Adherens junction | 78; 14% | 19.4 | 7.57E-08 |
| Circadian rhythm | 13; 31% | 16.6 | 1.04E-06 | |
| MAPK signaling pathway | 272; 16% | 8.8 | 0.001438 | |
| Phosphatidylinositol signaling system | 76; 14% | 6.8 | 0.008687 | |
| Focal adhesion | 203; 15% | 6.8 | 0.008833 |
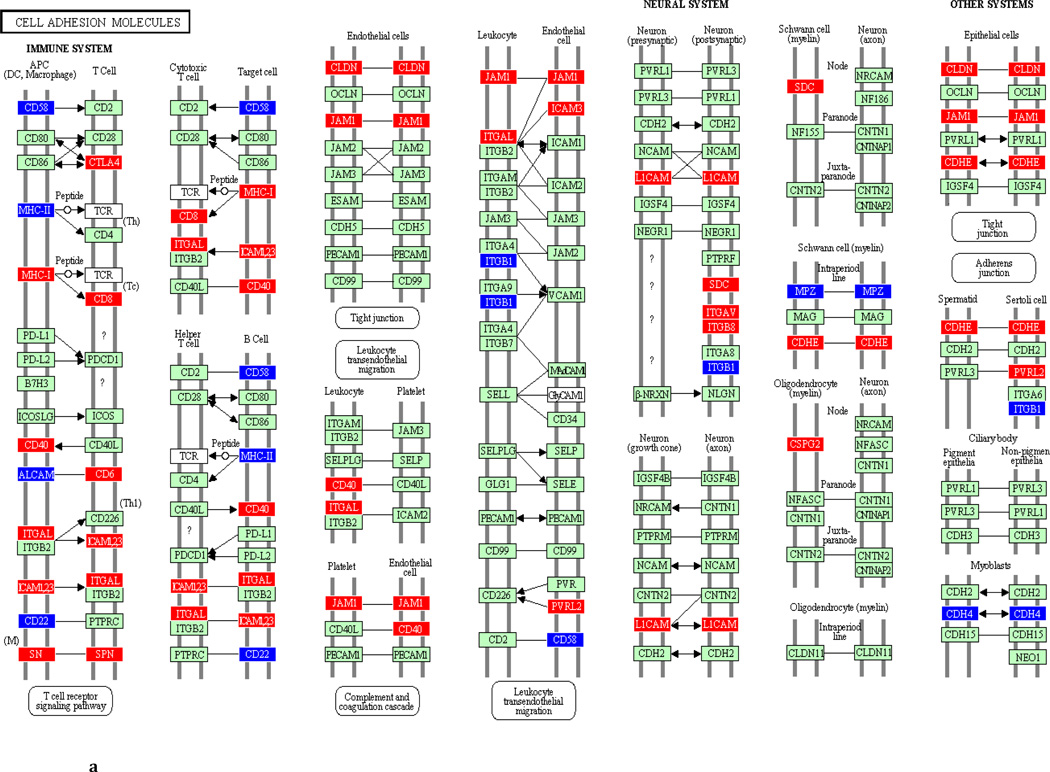
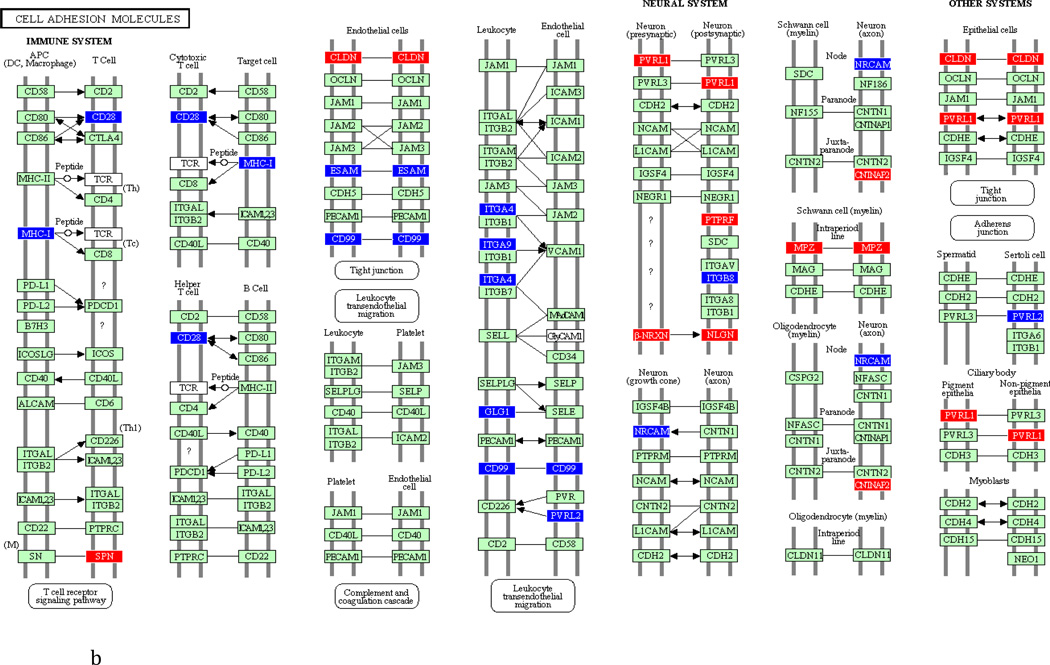
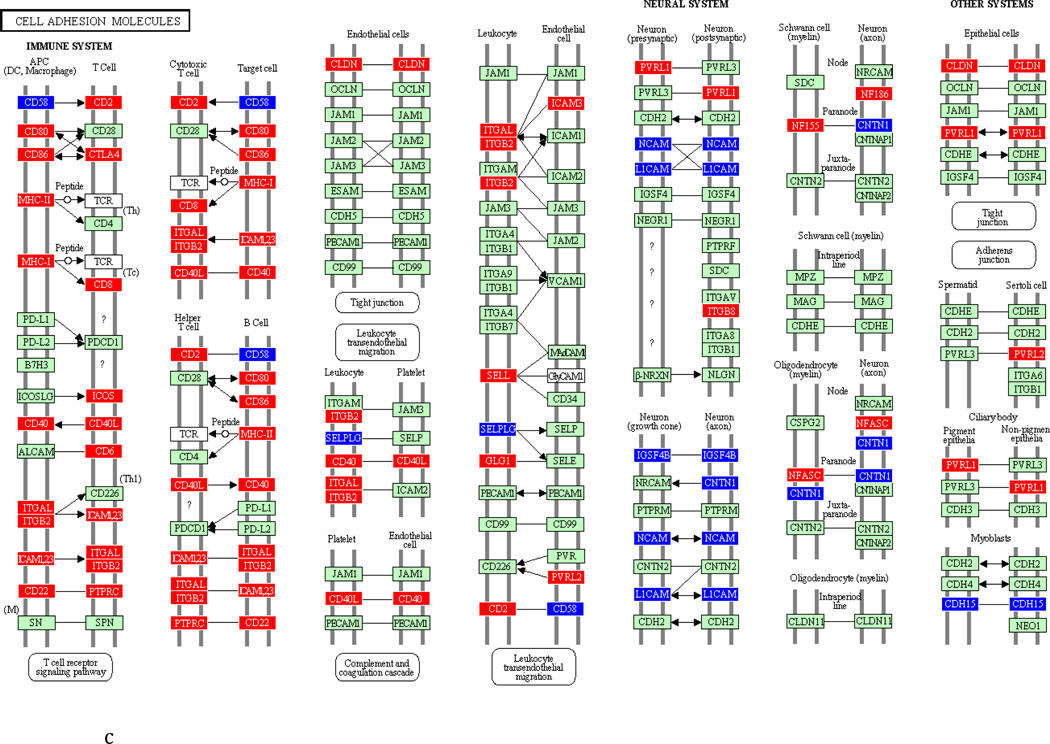
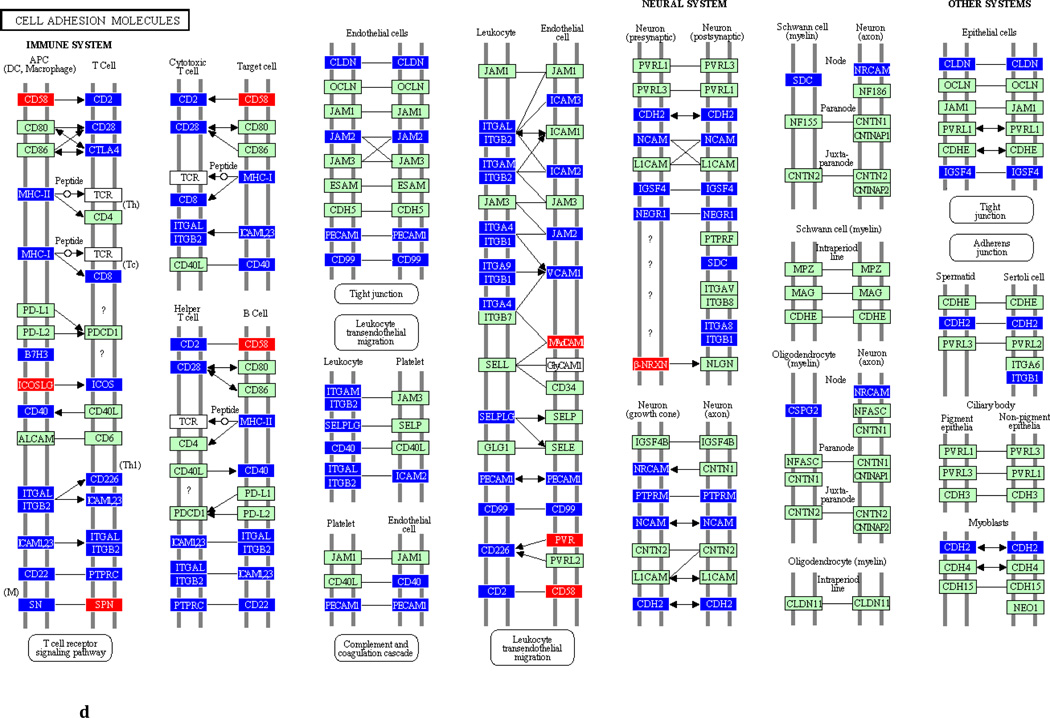
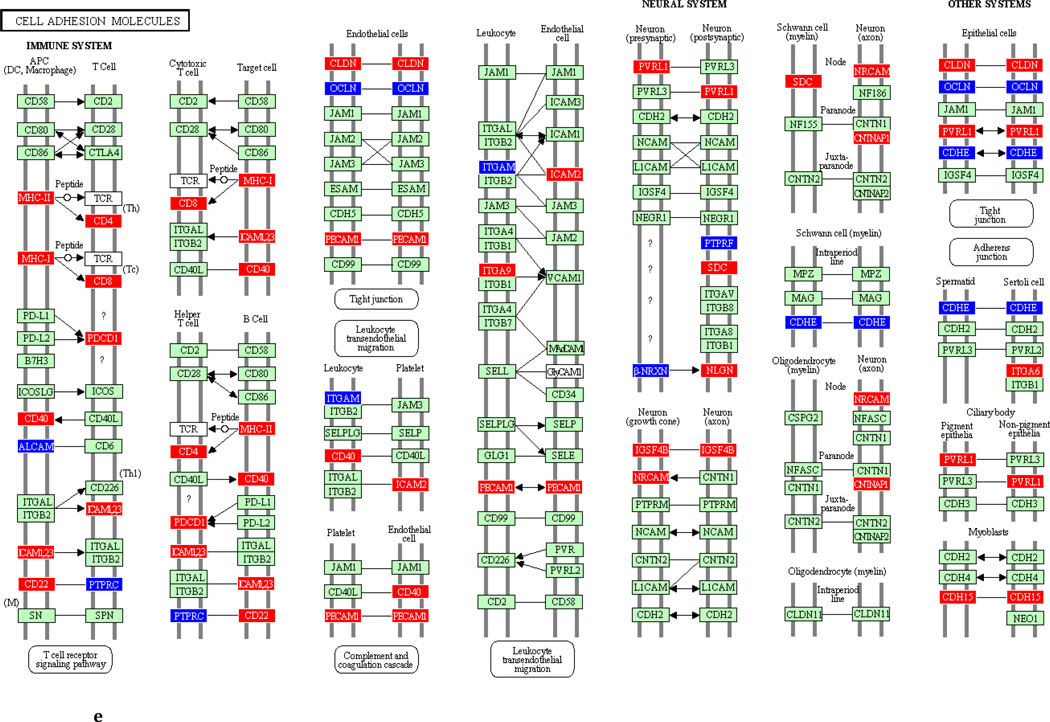
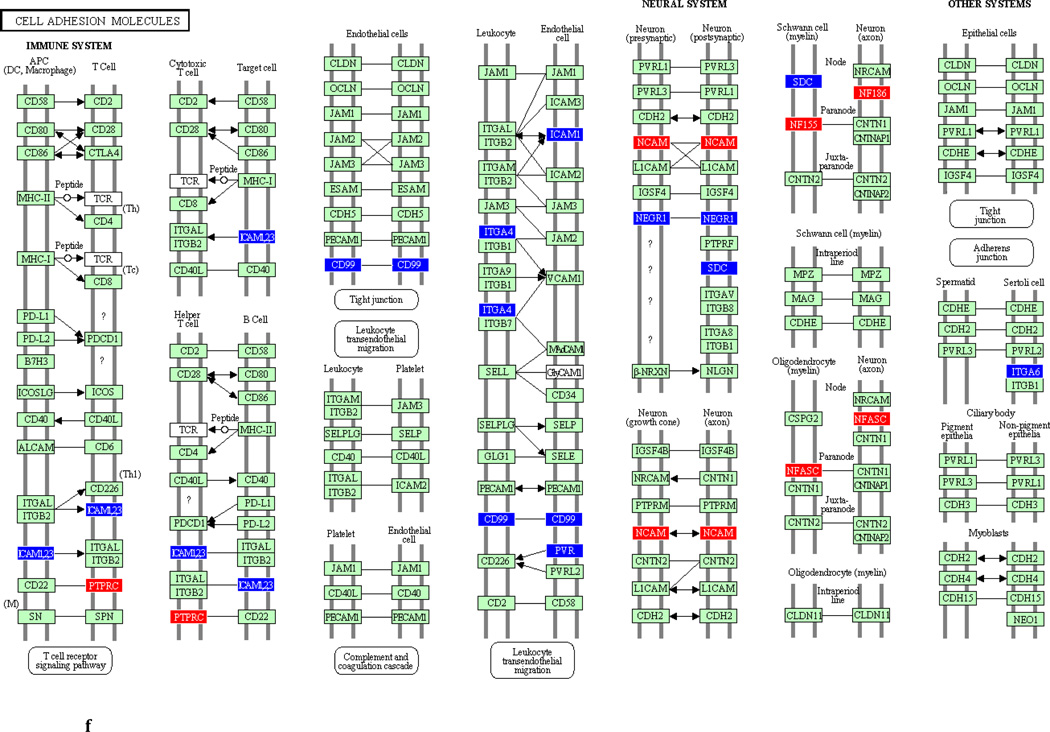
To underscore the distinction between differential regulation on the pathway level and that on the individual gene level, we illustrate in Figure 3 the within-pathway expression dynamics in a single ontology group (Cell Adhesion Molecules) over time. The pathway was more strongly regulated during the induction phase (impact factor range 116.3 and 77.4) than in the resolution phase (impact factor range 42.7–1.7). Individual genes that were upregulated (red color), downregulated (blue color) or unchanged (gene color) are depicted. It is apparent that the direction of differential regulation within this pathway shifted significantly over time: For example, after a relative uneventful second week of gingivitis induction (panel b) there was an obvious up-regulation in multiple genes involved in antigen presentation and T-cell and B-cell signaling (c). In contrast, the first days of gingivitis resolution were characterized by extensive down-regulation of multiple genes in this pathway.
Fig 3.
Graphic illustration of the ‘Cell Adhesion Molecules’ pathway during the first (a), second (b) and third (c) week of gingivitis induction, as well as during the first four (d), subsequent five (e) and final five (f) days of gingivitis resolution. Genes depicted in red are upregulated in the latter versus the former time point, genes in blue are downregulated, and genes in green are unchanged at the p<0.05 significance level.
Discussion
In this study, we used the experimental gingivitis model and whole genome micrroarray technology to study the gingival tissue transcriptomic profiles during the induction and resolution of plaque-induced inflammation in a prospective longitudinal manner. To date, there is only a single report available in the literature that has adopted a similar approach to the study of the pathobiology of the reversible gingival lesion: Recently, Offenbacher et al. (2009b) used an identical microarray platform and presented transcriptomic data from 14 participants in an experimental gingivitis study. Given the uniqueness of the published and the current data set, we briefly summarize some key points in the design of the two studies that are important in the comparative assessment and interpretation of their findings.
First, although experimental gingival inflammation in the Offenbacher et al. report was induced over a four week period, as compared to three weeks in the classic Löe et al., (1965) protocol as well as in the present study, the level of clinical inflammation reached at the peak of gingivitis induction at 28 days was less pronounced than the one observed in our study a week earlier, i.e., at 21 days. Specifically, GI in the Offenbacher et al. study increased from an average of 0.78 at baseline to 1.34 at 28 days, returning to 0.83 at the end of the resolution phase one week later. As shown by our data, our participants displayed almost absolute periodontal health at baseline, reflected by an average GI at the experimental sites of 0.1, reached a mean GI of 1.6 and 1.8 at 21 days in the induction and resolution groups, respectively, and returned to an average GI of 0.7 two weeks after prophylaxis. Although these are obviously crude comparisons, based on averages of categorical indices, it is notable that 6 of the 20 participants in our study did not develop gingival inflammation beyond a GI score of 1 at day 21, consistent with the earlier documented heterogeneity in the clinical inflammatory response during experimental gingivitis (Tatakis and Trombelli, 2004, Trombelli et al., 2008), possibly reflecting lack of full compliance as well. Second, a more important difference between the two studies from a design perspective is the number of gingival tissue samples harvested from each participant and the time interval between the consecutive biopsies. In the Offenbacher et al. study, gingival tissue samples were obtained from all subjects on three occasions (baseline, Day 28 and Day 35). In the present study, we examined gingival tissue transcriptomes at four time points one week apart in the induction phase, and at four time points five days apart in the resolution phase. To minimize the number of soft tissue samples that were obtained from each participant, we inevitably had to study the induction and resolution of gingivitis in two different groups of volunteers, comprising 10 individuals each. The significance of the availability of tissue from multiple time points within each phase is underscored by the data presented in Fig. 2: Thus, the number of up- or downregulated probes by at least 1.5 fold between day 21 and baseline was 184, yet twice as many (373 probes) were differentially regulated within the third week of induction alone. Likewise, 470 probes were differentially regulated within the first four days post-intervention whereas only 367 genes appeared to be differentially regulated between the end of the resolution period and the peak of gingivitis (Day 35 vs. Day 21). Thus, our data suggest that the differential regulation of genes in the tissues over the course of gingivitis is not an additive, cumulative process that closely parallels the development of clinical inflammation but varies significantly among different time points within the five week experimental protocol. Thus, the assessment of gene expression at multiple time points within the induction and resolution phase rather than a three time point, ‘snapshot’ description over the entire experimental period (Offenbacher et al. 2009b), appears to better reflect the kinetics of sequential gene expression, although this approach necessitated involvement of separate groups of individuals in the two phases.
The number of differentially expressed probes by >1.5 fold (Fig. 2), as well the number of probes that were significantly (p<0.05) regulated between any two time points irrespective of fold change, suggest that the two most ‘eventful’ time periods with respect to transcriptomic activation during the course of experimental gingivitis are the third week of gingivitis induction and the first four days of gingivitis resolution. These observations are in agreement with our current understanding of the biological events occurring at the plaque biofilm/gingival tissue interface. Indeed, it makes biological sense that a certain level of maturation of the dental plaque is required to elicit the apparent robust mobilization of the adaptive immune response that occurred during the final week of gingivitis induction. Likewise, the abrupt dispersion of the established biofilm achieved through prophylaxis, likely in combination with an instrumentation-induced mechanical stimulation of the tissues, triggered an immediate and rather profound transcriptomic response.
A closer look at the top differentially regulated genes in the first week of gingivitis induction (Table 1a) identified cysteine-rich secretory protein 3 (CRISP3), an innate host defense gene coding for a protein that is present in peroxidase-negative granules of neutrophils and in exocrine secretions (Udby et al., 2002) to be upregulated by 5.54-fold. Other strongly upregulated genes included MS4A1 (CD20, membrane-spanning 4-domains, subfamily A, member 1), a gene that encodes a B-lymphocyte surface molecule involved in the development and differentiation of B-cells into plasma cells (Petrie and Deans, 2002) upregulated by 3.24-fold; and CD177, a neutrophil-specific, heterophilic binding partner of the platelet endothelial cell adhesion molecule 1 (PECAM-1) (Sachs et al., 2007), upregulated by 2.54-fold. Interestingly, CRIPS3 was also the top differentially expressed gene during the second week of gingivitis (Table 1b), but this time downregulated by approximately 3-fold. In week three (Table 1c), multiple hemoglobin alpha 1 and alpha 2 probe sets were significantly downregulated, while the top upregulated gene (by 4-fold) was CXCL13, a CXC chemokine that promotes the migration of B lymphocytes (Stachowiak et al., 2006), followed by CXCL6, a granulocyte chemoattractant protein recently shown by our group to also be significantly upregulated in periodontitis lesions (Kebschull et al., 2009). CCL19, a CC motif chemokine involved in lymphocyte and dendritic cell trafficking (Leick et al., 2010) was also upregulated by approximately 3-fold. Multiple probes associated with Natural Killer (NK) cell function were also differentially regulated during the third week of induction, including Killer Cell Lectin-Like receptors (KCLLR) B1, C1 and K1 (with fold changes of 1.72, 1.60 and 1.59, respectively) as well as granzyme A, B and K (with fold changes of 1.70, 1.69 and 1.62, respectively). These findings are in accordance with earlier histologic observations (Wynne et al., 1986) demonstrating a gradual increase in the number of NK cells during the course of experimental gingivitis. They are of particular interest since NK cells represent a link between a bacterially-induced immune response and an auto-immune component that has been suggested to play a role in the pathobiology of periodontitis (Yamazaki et al., 2001).
Conversely, several hemoglobin-encoding genes were significantly upregulated during the first four days after prophylaxis and re-institution of oral hygiene (Table 2a), as was pro-platelet basic protein (PPBP), a CXC chemokine family member that is part of the secretory antimicrobial arsenal of the human monocytes (Schaffner et al., 2004). In contrast, CXCL1 was found to be downregulated by approximately 3-fold, as was TNFRSF17 (tumor necrosis factor receptor superfamily, member 17), a receptor preferentially expressed by mature B-lymphocytes, which when bound to its ligand TNFSF13B it mediates NF-kappaB and MAPK8/JNK activation (Hatzoglou et al., 2000). Additional genes that were found to be downregulated by approximately 3-fold during the first days of gingivitis resolution included matrix metallopeptidase 13 (MMP-13), and osteoglycin (OGN), a proteoglycan with osteoinductive capabilities (Kukita et al., 1990). Interestingly, CXCL1 and MMP13 were further downregulated during the next five-day period (Table 2b). The last 5 days of the gingivitis resolution (Table 3b) were characterized by induction of several genes involved in differentiation, including FOS, FOSB, EGR1, PTGS2 and ATF3. The first two belong to the four-member FOS gene family that encodes proteins regulating proliferation, differentiation and transformation (Durchdewald et al., 2009). Early growth response 1 (EGR1) is a nuclear protein that acts as a transcriptional regulator with a role in differentiation and mitogenesis (Braddock, 2001). Prostaglandin-endoperoxide synthase 2 (PTGS2), also known as cyclooxygenase 2, is a key enzyme in prostanoid biosynthesis, and activating transcription factor 3 (ATF3) is a mammalian activation transcription factor (Thompson et al., 2009). Collectively, these top upregulated proteins in the last phase of the resolution period may reflect the ongoing healing processes in the gingival tissues.
Gene ontology analyses identified consortia of genes that broadly orchestrate the soft tissue responses. As shown in Table 3, ‘leukocyte transendothelial migration’ and ‘cell adhesion’, the two pathways with most significant regulation in both the first and the second week of induction, were more strongly regulated during the second than the first week. ‘Antigen processing and presentation’ was the strongest regulated gene ontology group during the final week of induction, indicating a robust mobilization of the adaptive immune response. The ‘antigen processing and presentation’ pathway was in fact stronger regulated during the first four days of gingivitis resolution than in the final week of induction (impact factor 106.2), possibly due to the inoculation of the host with bacteria and their products in conjunction with mechanical prophylaxis. These transcriptomic findings are largely in agreement with earlier histologic observations of the initial and early gingival lesions, first described in detail by Page and Schroeder (1976) primarily based on animal experiments, but also with subsequent human histo-morphometric studies (Seymour et al., 1983, Brecx et al., 1987, Moughal et al., 1992). Nevertheless, it must be recognized that considerable heterogeneity in the histological features of experimental gingivitis lesions has been reported in the literature. For example, data by Kinane et al. (1991) on human gingival biopsies obtained at baseline and after 7, 14 and 21 days of experimental gingivitis demonstrated that infiltration by PMN cells, T-cells, and HLA-DR+ antigen presenting cells, as well as expression of adhesion molecules ELAM-1 and ICAM-1, all peaked at Day 7 and gradually subsided through Day 21. In contrast, the earlier work of Seymour et al. (1983) demonstrated that approximately 70% of the cellular infiltrate throughout the course of experimental gingivitis consisted of T-lymphocytes, and that this proportion remained fairly constant over time despite an increase in infiltrate size. Our gene ontology data do not corroborate the finding by Offenbacher et al. (2009b) of a substantial transient activation of genes involved in neural processes during experimental gingivitis, but differences in the time points of tissue harvesting may partly account for this discrepancy. An attempt to carry out a direct comparison of the probe sets that were statistically (p<0.05) differentially regulated at opposite directions (“up/down”, or “down/up” genes) during induction (Day 28 vs. baseline) and resolution (Day 35 vs. Day 28) in the Offenbacher et al. (2009b) dataset, to those with similar differential regulation at the best corresponding time points in our data set (Day 21 vs. baseline, and Day 30 vs. Day 21, respectively) identified a limited number of transiently regulated genes that were common in both datasets: Genes upregulated in induction and downregulated in resolution included CCL5 (RANTES), a CC cytokine that is chemoattractant for blood monocytes, memory T-helper cells and eosinophiles (Levy, 2009); PYHIN1 (pyrin and HIN domain family, member 1), a primarily nuclear protein involved in transcriptional regulation of genes affecting cell cycle control, differentiation and apoptosis (Ding et al., 2006); Granzyme A (GZMA), a cytotoxic T-cell and natural killer cell-specific serine esterase (Grossman et al., 2004); CD96, a membrane protein involved in antigen presentation and in adhesive interactions between activated T- and NK- cells (Fuchs et al., 2004); Adducing 3 (ADD3), a protein involved in the assembly of spectrin-actin networks and cell to cell contact in epithelial tissues (Kaiser et al., 1989); and Toll-like receptor 7, one of the intra-cellular, nucleic-acid sensing TLRs (Krieg and Vollmer, 2007), whose differential expression during gingivitis likely reflects host cell activation in response to internalized bacteria. Probes common to both data sets that were downregulated during induction and upregulated during resolution included TMEM16A (anoctamin 1), involved in epithelial volume-regulated chloride channels with potential function in proliferation and apoptosis (Almaca et al., 2009); and genes coding for the matrix proteins lamin A/C (LMNA) (Wagner and Krohne, 2007) and CSPG4 (chondroitin sulphate proteoglycan 4; Lorber, 2006).
We acknowledge some important limitations of the current work. First, the transcriptomic data have been derived from a relative small sample of young volunteers, and it is unlikely that they capture the full extent of the variability in gene expression profiles in experimental gingivitis across individuals or age groups. In addition, the limited sample size did not allow for full adjustments for multiple comparisons in the identification of significantly regulated probes, similarly to the published report (Offenbacher et al., 2009b). Second, it must be recognized that longitudinal changes in gingival inflammation and consequently in the gingival transcriptomic profiles cannot be exclusively attributed to plaque accumulation or biofilm dispersion, but are also influenced by additional exposures such as hormonal fluxes in females and dietary effects. Ideally, these could have been accounted for by studying over time gingival units not subjected to experimental gingivitis, but a study design requiring serial harvesting of additional tissue samples was not feasible. Third, due to the exploratory and descriptive nature of this work, we have not yet carried out independent verification of specific genes by a second, mRNA-based method, such as real time RT-PCR. Lastly, verification steps at the protein level need to be performed, either on tissue extracts, or on gingival GCF samples. We have indeed obtained GCF samples over time from the crevices adjacent to the harvested tissue papillae, and are carrying out high throughput proteomic analyses to examine the extent to which gingival tissue mRNA sequences translate into GCF proteins. In the future, we envision that the effects of adjunctive pharmacological therapies on the gingival tissue transcriptome during the induction and resolution of gingivitis will be possible to evaluate against the present data.
Supplementary Material
Periodontal microbiota during the induction (left panel) and resolution (right panel) of experimental gingivitis. Bars represent mean counts × 104. Note the difference in the y-axis scale among the different microbial species.
Clinical Relevance.
Rationale
The study of tissue responses in experimental gingivitis have largely focused on the identification of cell populations using immunohistochemistry, or on the assessment of the levels of selected proteins in the gingival crevicular fluid. Data on the sequential activation of genes in the host tissues during the induction and resolution of gingival inflammation are sparse.
Findings
Our data indicate that the differential expression of genes in the gingival tissues reaches its peak during the third week of experimental gingivitis and during the first four days of re-institution of oral hygiene. Our work identifies networks of genes that orchestrate these tissue responses.
Implications
Our work furthers our understanding of the gingival responses to plaque accumulation and plaque control, and can serve as a basis for a comparison of the effects of adjunctive pharmacological agents in future studies.
ACKNOWLEDGEMENT
The authors are indebted to the participants for their cooperation, and to the dental hygienist Ms. Jessica Skoogh-Ericsson for her valuable contribution to the study.
Sources of Funding Statement: This study was supported by a grant from Colgate-Palmolive, NJ, USA. Dr. Jönsson was supported by stipends from the Swedish Research Council, the American Dental Society of Sweden and the Foundation Blanceflor Boncompagni-Ludovisi, née Bildt. Dr. Demmer was supported by NIH DE K99 018739. Dr. Kebschull was supported by the German Research Foundation (KFO208 TP6 & TP9).
Footnotes
Conflict of Interest: The authors declare that they have no conflicts of interest.
REFERENCES
- Abbas F, van der Velden U, Moorer WR, Everts V, Vroom TM, Scholte G. Experimental gingivitis in relation to susceptibility to periodontal disease. II. Phase-contrast microbiological features and some host-response observations. J Clin Periodontol. 1986;13:551–557. doi: 10.1111/j.1600-051x.1986.tb00846.x. [DOI] [PubMed] [Google Scholar]
- Almaca J, Tian Y, Aldehni F, Ousingsawat J, Kongsuphol P, Rock JR, Harfe BD, Schreiber R, Kunzelmann K. TMEM16 proteins produce volume-regulated chloride currents that are reduced in mice lacking TMEM16A. J Biol Chem. 2009;284:28571–28578. doi: 10.1074/jbc.M109.010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M. The transcription factor Egr-1: a potential drug in wound healing and tissue repair. Ann Med. 2001;33:313–318. doi: 10.3109/07853890109002083. [DOI] [PubMed] [Google Scholar]
- Brecx MC, Gautschi M, Gehr P, Lang NP. Variability of histologic criteria in clinically healthy human gingiva. J Periodontal Res. 1987;22:468–472. doi: 10.1111/j.1600-0765.1987.tb02057.x. [DOI] [PubMed] [Google Scholar]
- Daly CG, Highfield JE. Effect of localized experimental gingivitis on early supragingival plaque accumulation. J Clin Periodontol. 1996;23:160–164. doi: 10.1111/j.1600-051x.1996.tb02071.x. [DOI] [PubMed] [Google Scholar]
- Deinzer R, Weik U, Kolb-Bachofen V, Herforth A. Comparison of experimental gingivitis with persistent gingivitis: differences in clinical parameters and cytokine concentrations. J Periodontal Res. 2007;42:318–324. doi: 10.1111/j.1600-0765.2006.00951.x. [DOI] [PubMed] [Google Scholar]
- Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, Pavlidis P, Papapanou PN. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmer RT, Pavlidis P, Papapanou PN. Bioinformatics techniques in microarray research: applied microarray data analysis using R and SAS software. Methods Mol Biol. 2010;666:395–417. doi: 10.1007/978-1-60761-820-1_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Lee JF, Lu H, Lee MH, Yan DH. Interferon-inducible protein IFIXalpha1 functions as a negative regulator of HDM2. Mol Cell Biol. 2006;26:1979–1996. doi: 10.1128/MCB.26.5.1979-1996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007 doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchdewald M, Angel P, Hess J. The transcription factor Fos: a Janus-type regulator in health and disease. Histol Histopathol. 2009;24:1451–1461. doi: 10.14670/HH-24.1451. [DOI] [PubMed] [Google Scholar]
- Fransson C, Mooney J, Kinane DF, Berglundh T. Differences in the inflammatory response in young and old human subjects during the course of experimental gingivitis. J Clin Periodontol. 1999;26:453–460. doi: 10.1034/j.1600-051x.1999.260707.x. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- Grant MM, Creese AJ, Barr G, Ling MR, Scott AE, Matthews JB, Griffiths HR, Cooper HJ, Chapple IL. Proteomic analysis of a noninvasive human model of acute inflammation and its resolution: the twenty-one day gingivitis model. J Proteome Res. 2010;9:4732–4744. doi: 10.1021/pr100446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman WJ, Verbsky JW, Tollefsen BL, Kemper C, Atkinson JP, Ley TJ. Differential expression of granzymes A and B in human cytotoxic lymphocyte subsets and T regulatory cells. Blood. 2004;104:2840–2848. doi: 10.1182/blood-2004-03-0859. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Roussel J, Bourgeade MF, Rogier E, Madry C, Inoue J, Devergne O, Tsapis A. TNF receptor family member BCMA (B cell maturation) associates with TNF receptor-associated factor (TRAF) 1, TRAF2, and TRAF3 and activates NF-kappa B, elk-1, c-Jun N-terminal kinase, and p38 mitogen-activated protein kinase. J Immunol. 2000;165:1322–1330. doi: 10.4049/jimmunol.165.3.1322. [DOI] [PubMed] [Google Scholar]
- Heasman PA, Collins JG, Offenbacher S. Changes in crevicular fluid levels of interleukin-1 beta, leukotriene B4, prostaglandin E2, thromboxane B2 and tumour necrosis factor alpha in experimental gingivitis in humans. J Periodont Res. 1993;28:241–247. doi: 10.1111/j.1600-0765.1993.tb02090.x. [DOI] [PubMed] [Google Scholar]
- Hillam DG, Hull PS. The influence of experimental gingivitis on plaque formation. J Clin Periodontol. 1977;4:56–61. doi: 10.1111/j.1600-051x.1977.tb01883.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jenkins S, Addy M, Newcombe R. Evaluation of a mouthrinse containing chlorhexidine and fluoride as an adjunct to oral hygiene. J Clin Periodontol. 1993;20:20–25. doi: 10.1111/j.1600-051x.1993.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Kaiser HW, O'Keefe E, Bennett V. Adducin: Ca++-dependent association with sites of cell-cell contact. J Cell Biol. 1989;109:557–569. doi: 10.1083/jcb.109.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull M, Demmer R, Behle JH, Pollreisz A, Heidemann J, Belusko PB, Celenti R, Pavlidis P, Papapanou PN. Granulocyte chemotactic protein 2 (GCP-2/CXCL6) complements interleukin-8 in periodontal disease. J Periodontal Res. 2009;44:465–471. doi: 10.1111/j.1600-0765.2008.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull M, Papapanou PN. The use of gene arrays in deciphering the pathobiology of periodontal diseases. Methods Mol Biol. 2010;666:385–393. doi: 10.1007/978-1-60761-820-1_24. [DOI] [PubMed] [Google Scholar]
- Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane DF, Adonogianaki E, Moughal N, Winstanley FP, Mooney J, Thornhill M. Immunocytochemical characterization of cellular infiltrate, related endothelial changes and determination of GCF acute-phase proteins during human experimental gingivitis. J Periodontal Res. 1991;26:286–288. doi: 10.1111/j.1600-0765.1991.tb01660.x. [DOI] [PubMed] [Google Scholar]
- Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Kukita A, Bonewald L, Rosen D, Seyedin S, Mundy GR, Roodman GD. Osteoinductive factor inhibits formation of human osteoclast-like cells. Proc Natl Acad Sci U S A. 1990;87:3023–3026. doi: 10.1073/pnas.87.8.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamster IB, Hartley LJ, Vogel RI. Development of a biochemical profile for gingival crevicular fluid. Methodological considerations and evaluation of collagen-degrading and ground substance-degrading enzyme activity during experimental gingivitis. J Periodontol. 1985;56:13–21. doi: 10.1902/jop.1985.56.11s.13. [DOI] [PubMed] [Google Scholar]
- Leick M, Catusse J, Follo M, Nibbs RJ, Hartmann TN, Veelken H, Burger M. CCL19 is a specific ligand of the constitutively recycling atypical human chemokine receptor CRAM-B. Immunology. 2010;129:536–546. doi: 10.1111/j.1365-2567.2009.03209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JA. The unexpected pleiotropic activities of RANTES. J Immunol. 2009;182:3945–3946. doi: 10.4049/jimmunol.0990015. [DOI] [PubMed] [Google Scholar]
- Lie MA, Danser MM, van der Weijden GA, Timmerman MF, de Graaff J, van der Velden U. Oral microbiota in subjects with a weak or strong response in experimental gingivitis. J Clin Periodontol. 1995;22:642–647. doi: 10.1111/j.1600-051x.1995.tb00818.x. [DOI] [PubMed] [Google Scholar]
- Löe H. The Gingival Index, the Plaque Index and the Retention Index system. J Periodontol. 1967;38:610–616. doi: 10.1902/jop.1967.38.6.610. [DOI] [PubMed] [Google Scholar]
- Löe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177–187. doi: 10.1902/jop.1965.36.3.177. [DOI] [PubMed] [Google Scholar]
- Loesche WJ, Syed SA. Bacteriology of human experimental gingivitis: effect of plaque and gingivitis score. Infect Immun. 1978;21:830–839. doi: 10.1128/iai.21.3.830-839.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorber B. Role of NG2 in development and regeneration. J Neurosci. 2006;26:7127–7128. doi: 10.1523/JNEUROSCI.2104-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Cato EP, Burmeister JA, Palcanis KG, Ranney RR. Bacteriology of experimental gingivitis in children. Infect Immun. 1984;46:1–6. doi: 10.1128/iai.46.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Good IJ, Burmeister JA, Palcanis KG, Ranney RR. Bacteriology of experimental gingivitis in young adult humans. Infect Immun. 1982;38:651–667. doi: 10.1128/iai.38.2.651-667.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughal NA, Adonogianaki E, Thornhill MH, Kinane DF. Endothelial cell leukocyte adhesion molecule-1 (ELAM-1) and intercellular adhesion molecule-1 (ICAM-1) expression in gingival tissue during health and experimentally-induced gingivitis. J Periodont Res. 1992;27:623–630. doi: 10.1111/j.1600-0765.1992.tb01746.x. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Barros S, Mendoza L, Mauriello S, Preisser J, Moss K, de Jager M, Aspiras M. Changes in gingival crevicular fluid inflammatory mediator levels during the induction and resolution of experimental gingivitis in humans. J Clin Periodontol. 2009a;37:324–333. doi: 10.1111/j.1600-051X.2010.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenbacher S, Barros SP, Paquette DW, Winston JL, Biesbrock AR, Thomason RG, Gibb RD, Fulmer AW, Tiesman JP, Juhlin KD, Wang SL, Reichling TD, Chen KS, Ho B. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009b;80:1963–1982. doi: 10.1902/jop.2009.080645. [DOI] [PubMed] [Google Scholar]
- Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab Invest. 1976;34:235–249. [PubMed] [Google Scholar]
- Papapanou PN, Abron A, Verbitsky M, Picolos D, Yang J, Qin J, Fine JB, Pavlidis P. Gene expression signatures in chronic and aggressive periodontitis: a pilot study. Eur J Oral Sci. 2004;112:216–223. doi: 10.1111/j.1600-0722.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- Papapanou PN, Behle JH, Kebschull M, Celenti R, Wolf DL, Handfield M, Pavlidis P, Demmer RT. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 2009;9:221. doi: 10.1186/1471-2180-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapanou PN, Neiderud AM, Sandros J, Dahlén G. Checkerboard assessments of serum antibodies to oral microbiota as surrogate markers of clinical periodontal status. J Clin Periodontol. 2001;28:103–106. doi: 10.1034/j.1600-051x.2001.280116.x. [DOI] [PubMed] [Google Scholar]
- Payne WA, Page RC, Ogilvie AL, Hall WB. Histopathologic features of the initial and early stages of experimental gingivitis in man. J Periodontal Res. 1975;10:51–64. doi: 10.1111/j.1600-0765.1975.tb00008.x. [DOI] [PubMed] [Google Scholar]
- Petrie RJ, Deans JP. Colocalization of the B cell receptor and CD20 followed by activation-dependent dissociation in distinct lipid rafts. J Immunol. 2002;169:2886–2891. doi: 10.4049/jimmunol.169.6.2886. [DOI] [PubMed] [Google Scholar]
- Quirynen M, Avontroodt P, Peeters W, Pauwels M, Coucke W, van Steenberghe D. Effect of different chlorhexidine formulations in mouthrinses on de novo plaque formation. J Clin Periodontol. 2001;28:1127–1136. doi: 10.1034/j.1600-051x.2001.281207.x. [DOI] [PubMed] [Google Scholar]
- Ramberg P, Furuichi Y, Sherl D, Volpe AR, Nabi N, Gaffar A, Lindhe J. The effect of triclosan on developing gingivitis. J Clin Periodontol. 1995;22:442–448. doi: 10.1111/j.1600-051x.1995.tb00175.x. [DOI] [PubMed] [Google Scholar]
- Sachs UJ, Andrei-Selmer CL, Maniar A, Weiss T, Paddock C, Orlova VV, Choi EY, Newman PJ, Preissner KT, Chavakis T, Santoso S. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31) J Biol Chem. 2007;282:23603–23612. doi: 10.1074/jbc.M701120200. [DOI] [PubMed] [Google Scholar]
- Schaffner A, King CC, Schaer D, Guiney DG. Induction and antimicrobial activity of platelet basic protein derivatives in human monocytes. J Leukoc Biol. 2004;76:1010–1018. doi: 10.1189/jlb.0404261. [DOI] [PubMed] [Google Scholar]
- Sekino S, Ramberg P, Uzel NG, Socransky S, Lindhe J. Effect of various chlorhexidine regimens on salivary bacteria and de novo plaque formation. J Clin Periodontol. 2003;30:919–925. doi: 10.1034/j.1600-051x.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- Seymour GJ, Powell RN, Cole KL, Aitken JF, Brooks D, Beckman I, Zola H, Bradley J, Burns GF. Experimental gingivitis in humans. A histochemical and immunological characterization of the lymphoid cell subpopulations. J Periodontal Res. 1983;18:375–385. doi: 10.1111/j.1600-0765.1983.tb00373.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- Stachowiak AN, Wang Y, Huang YC, Irvine DJ. Homeostatic lymphoid chemokines synergize with adhesion ligands to trigger T and B lymphocyte chemokinesis. J Immunol. 2006;177:2340–2348. doi: 10.4049/jimmunol.177.4.2340. [DOI] [PubMed] [Google Scholar]
- Tatakis DN, Trombelli L. Modulation of clinical expression of plaque-induced gingivitisIBackground review and rationale. J Clin Periodontol. 2004;31:229–238. doi: 10.1111/j.1600-051x.2004.00477.x. [DOI] [PubMed] [Google Scholar]
- Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med. 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombelli L, Farina R, Minenna L, Carrieri A, Scapoli C, Tatakis DN. Experimental gingivitis: reproducibility of plaque accumulation and gingival inflammation parameters in selected populations during a repeat trial. J Clin Periodontol. 2008;35:955–960. doi: 10.1111/j.1600-051X.2008.01315.x. [DOI] [PubMed] [Google Scholar]
- Udby L, Calafat J, Sorensen OE, Borregaard N, Kjeldsen L. Identification of human cysteine-rich secretory protein 3 (CRISP-3) as a matrix protein in a subset of peroxidase-negative granules of neutrophils and in the granules of eosinophils. J Leukoc Biol. 2002;72:462–469. [PubMed] [Google Scholar]
- Van Strydonck DA, Timmerman MF, van der Velden U, van der Weijden GA. Plaque inhibition of two commercially available chlorhexidine mouthrinses. J Clin Periodontol. 2005;32:305–309. doi: 10.1111/j.1600-051X.2005.00681.x. [DOI] [PubMed] [Google Scholar]
- Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- Wennström JL. Mouthrinses in"experimental gingivitis" studies. J Clin Periodontol. 1988;15:511–516. doi: 10.1111/j.1600-051x.1988.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Wynne SE, Walsh LJ, Seymour GJ, Powell RN. In situ demonstration of natural killer (NK) cells in human gingival tissue. J Periodontol. 1986;57:699–702. doi: 10.1902/jop.1986.57.11.699. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Tabeta K, Nakajima T, Ohsawa Y, Ueki K, Itoh H, Yoshie H. Interleukin-10 gene promoter polymorphism in Japanese patients with adult and early-onset periodontitis. J Clin Periodontol. 2001;28:828–832. doi: 10.1034/j.1600-051x.2001.028009828.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Periodontal microbiota during the induction (left panel) and resolution (right panel) of experimental gingivitis. Bars represent mean counts × 104. Note the difference in the y-axis scale among the different microbial species.