Abstract
BACKGROUND & AIMS
Cholangiocarcinoma is a heterogeneous disease with a poor outcome that accounts for 5%–10% of primary liver cancers. We characterized its genomic and genetic features and associated these with patient responses to therapy.
METHODS
We profiled the transcriptomes from 104 surgically resected cholangiocarcinoma samples collected from patients in Australia, Europe, and the United States; epithelial and stromal compartments from 23 tumors were laser capture microdissected. We analyzed mutations in KRAS, epidermal growth factor receptor (EGFR), and BRAF in samples from 69 tumors. Changes in gene expression were validated by immunoblotting and immunohistochemistry; integrative genomics combined data from the patients with data from 7 human cholangiocarcinoma cell lines, which were then exposed to trastuzumab and lapatinib.
RESULTS
Patients were classified into 2 subclasses, based on 5-year survival rate (72% vs 30%; χ2 = 11.61; P < .0007), time to recurrence (13.7 vs 22.7 months; P < .001), and the absence or presence of KRAS mutations (24.6%), respectively. Class comparison identified 4 survival subgroups (SGI–IV; χ2 = 8.34; P < .03); SGIII was characterized by genes associated with proteasomal activity and the worst prognosis. The tumor epithelium was defined by deregulation of the HER2 network and frequent overexpression of EGFR, the hepatocyte growth factor receptor (MET), pRPS6, and Ki67, whereas stroma was enriched in inflammatory cytokines. Lapatinib, an inhibitor of HER2 and EGFR, was more effective in inhibiting growth of cholangiocarcinoma cell lines than trastuzumab.
CONCLUSIONS
We provide insight into the pathogenesis of cholangiocarcinoma and identify previously unrecognized subclasses of patients, based on KRAS mutations and increased levels of EGFR and HER2 signaling, who might benefit from dual-target tyrosine kinase inhibitors. The group of patients with the worst prognosis was characterized by transcriptional enrichment of genes that regulate proteasome activity, indicating new therapeutic targets.
Keywords: Genetic Analysis, Gene Expression, CCA, Hepatic
Cholangiocarcinoma (CCA) is the second most common primary hepatic malignancy after hepatocellular carcinoma (HCC). Although overall cancer mortality has declined in the United States, the incidence of pancreatic cancer, melanoma, and liver cancer is increasing, with hepatic tumors presenting with the highest frequency.1 CCA arise in epithelium lining intrahepatic or extrahepatic biliary ducts. Intrahepatic CCA, classified as a peripheral tumor of interlobular bile ducts, accounts for less than 10% of annual CCA cases.2,3 Tumors designated hilar are generally considered extrahepatic and originate from the main hepatic ducts or at the bifurcation of the common hepatic duct. The main difference between peripheral and hilar tumors is in the clinical presentation and gross appearance. If a tumor can be surgically removed, patients may receive postoperative adjuvant chemotherapy to improve chances of cure.4 However, the majority of patients have inoperable intrahepatic CCA and are treated with palliative therapy.4
Although chemotherapy improves the quality of life in patients with inoperable CCA, it does not result in cure.5 Thus, surgery remains the only treatment option with curative intent. The receptor tyrosine kinase (RTK) inhibitors sorafenib6 and erlotinib,7,8 used as first-line therapy for patients with advanced HCC, lung adenocarcinoma, and colorectal cancer, have had limited success in CCA.6–8 This lack of clinical efficacy in management of CCA may in part be the result of inadequate molecular and pathobiological understanding of the disease.
Transcriptomics have been successfully used both for predicting outcome and identifying genetically homogeneous subclasses of patients with diverse malignancies (eg, HCC, lung adenocarcinoma, and breast cancer)9–11 and significantly contributed to improved clinical management. The rapid advancement of cancer genomics and increasing ease of applying these techniques at the bedside suggest growing use in screening, diagnosis, and development of therapeutics for treatment of patients with CCA.12,13
Here, we performed comprehensive genomic profiling accompanied by mutational and immunohistochemical analyses of resected tumors. A subgroup of patients with poor overall survival and early recurrence was characterized by the presence of KRAS mutations and multiple aberrantly regulated oncogenic pathways, including activation of HER2 and epidermal growth factor receptor (EGFR) signaling, as compared with patients with a good clinical outcome. Importantly, treatment of CCA cell lines with activated EGFR and HER2 with tyrosine kinase inhibitors (TKIs) trastuzumab and lapatinib suggested therapeutic potential for lapatinib, a dual-target TKI, in the subclass of patients with activation of HER2 and EGFR signaling.
Materials and Methods
Detailed information is provided in Supplementary Materials and Methods.
Patients and Samples
The data set included 104 surgically resected CCAs obtained from patients diagnosed in 1991–2008 at the Mayo Clinic (Rochester, MN), University of Leuven (Leuven, Belgium), and University of Queensland (Brisbane, Australia). The last update of the patient cohort was in January 2011. The matched surrounding livers were available for 59 patients with CCA. It is not known whether all resections were performed as curative or in some cases as palliative treatment, thus limiting the extrapolation of the data to the nonsurgical candidates. Normal intrahepatic bile ducts (n = 6) resected at the Surgical Branch, National Institutes of Health, were used as reference tissues in the analysis. All samples were obtained with approval by the institutional review board of the National Institutes of Health and collaborating institutions on the condition that patients were anonymized.
Results
Transcriptomic Profiling Identifies 2 Distinct CCA Subclasses With Different Clinical Outcomes
The molecular profiles of the resected tumors were readily distinguishable from a group of matched noncancerous surrounding livers (Supplementary Figure 1A). This classification was confirmed by Bayesian compound covariate prediction modeling with 97% accuracy (95% confidence interval [CI], 0.93–0.99; P < .0001) (Supplementary Figure 1B). Within the cohort, the resected tumors of hilar (36/104) and peripheral type (68/104) were not distinguishable by anatomic location based on overall survival (Supplementary Figure 1C). In fact, the global posttest following the supervised class comparison showed no significant molecular distinction (P > .05) between hilar-type and peripheral-type tumors. However, regardless of tumor location, perineural (PNI) (80/104) and lymphatic (LI) (60/104) invasion were independent prognostic factors for the poor survival groups (Supplementary Figure 1D and E) with 5-year survival rates of 22% (P < .0007) and 13% (P < .0002), respectively.
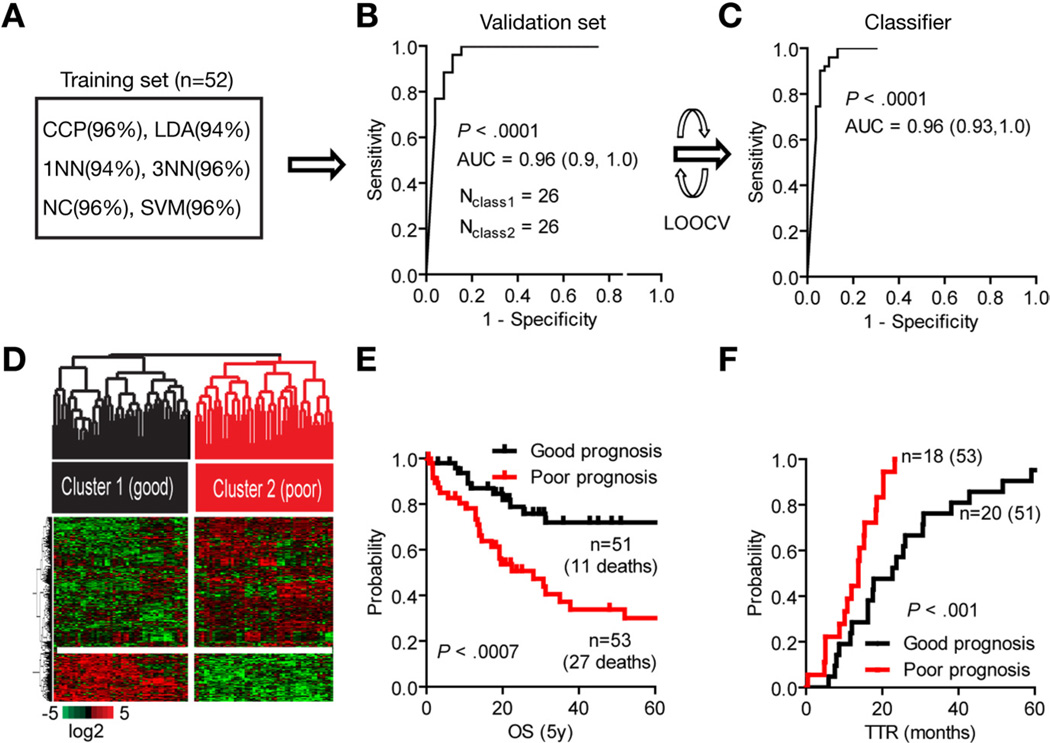
The patient cohort was then randomly divided into 2 equal-size data sets. A total of 1121 significantly expressed genes were identified based on the selection criteria, which included at least 2-fold differences in expression ratios relative to normal intrahepatic bile ducts in at least 80% of samples. The correct classification within the training set (n = 52) was estimated by class comparison applying 6 statistical methods with an accuracy ranging from 94% to 96% (Figure 1A). Bayesian compound covariate prediction modeling confirmed the classification in the validation set (n = 52) with 96% accuracy (95% CI, 0.9–1.0; P < .0001) (Figure 1B). To improve the accuracy of the gene signature, we minimized the misclassification rate and number of significantly differentially expressed genes (P < .001) in the classifier to 238 genes by leave-one-out cross-validation (Supplementary Table 1). Class comparison confirmed the classification (0.96; 95% CI, 0.93–1.0; P < .0001) as shown by the area under the receiver and operator curve (Figure 1C). Hierarchical cluster analysis separated the tumors into 2 distinct subclasses strongly associated with survival (Figure 1D and E). The 5-year survival rate in cluster 1 (n = 51) was 72% compared with 30% in cluster 2 (n = 53; χ2 = 11.61; P < .0007) with a hazard ratio of 0.33 (95% CI, 0.17–0.62). Also, patients with a poor clinical outcome (cluster 2) were characterized by early recurrence (13.7 ± 6.3 vs 22.7 ± 18.1 months; P < .001) (Figure 1F). Having identified 2 prognostic subclasses of CCA, we then examined the association of each cluster with clinical and pathological features (Tables 1 and 2). Consistent with published data, perineural and lymphatic invasion were independent markers of poor prognosis in our cohort (Supplementary Figure 1D and E). We further used these markers as variables in the class prediction modeling and showed their ability to correctly predict our classification (Supplementary Figure 2). Seventy-five percent (27/36) of hilar-type tumors were presented in cluster 2, whereas 62% (42/68) of peripheral-type tumors were found in cluster 1. Despite differences in relative representation of biliary cancer subtypes between the 2 prognostic subclasses, molecular profiles of hilartype and peripheral-type tumors within each cluster were homogeneous, suggesting that similar molecular pathogenesis rather than anatomic location defines the overall prognosis. Patients who received palliative treatment before surgery were distributed evenly between 2 prognostic subclasses (Tables 1 and 2). A univariate analysis did not reveal any statistical difference as a result of treatment, suggesting that it did not influence the classification. Multivariate analysis of the clinical variables showed a significant association of survival with ductal dysplasia, necrosis, stromal infiltration, perineural and lymphatic invasion, and recurrence (Tables 1 and 2).
Figure 1.
Development of a CCA gene classifier. (A) Multiple class comparison models were used to test the robustness of the classification in the training set (n = 52). (B) Sensitivity of the gene signature to correctly predict the classification of patients within the validation set (n = 52). The specificity is represented by the area under the receiver and operator curve (AUC, 95% CI) using Bayesian compound covariate predictor modeling. (C) Development of the gene classifier. To build the classifier, a class random variance model was used, identifying 238 genes significant at α ≤ .001 using Bayesian compound covariate modeling and leave-one-out cross-validation (LOOCV). (D) Hierarchical clustering of the 238-gene classifier separates patients into 2 subclasses according to their clinical outcome. (E) Analysis of survival (OS) and (F) time to recurrence (TTR). Kaplan–Meier and log-rank statistics were used to determine levels of significance.
Table 1.
Clinical and Pathologic Variables
| Clinical outcome | |||
|---|---|---|---|
| Univariatea | Good | Poor | P value |
| Age (y), mean ± SD | 64 ± 10 | 64 ± 13 | |
| Sex (male/female) | 18/33 | 30/23 | .03 |
| Tumor (>50 mm) | 31/50 | 19/50 | .02 |
| Size (mm), mean ± SEM | 67.1 ± 4.3 | 51.3 ± 4.3 | .01b |
| Range (mm) | 5–120 | 10–125 | |
| Anatomic location (%) | |||
| Peripheral | 42 (61.8) | 26 (38.2) | |
| Hilar | 9 (25) | 27 (75) | |
| Histologic differentiation | |||
| Well | 4/25 | 6/28 | |
| Moderately | 17/25 | 15/28 | |
| Poorly | 4/25 | 7/28 | |
| Pathology | |||
| Perineural invasion | 14/47 | 28/45 | .001 |
| Lymphatic invasion | 14/37 | 25/33 | .001 |
| Portal tract invasion | 18/38 | 21/41 | |
| Ductal dysplasia | 13/21 | 21/28 | |
| Necrosis | 13/20 | 14/19 | |
| Stromal infiltration | 18/21 | 20/22 | |
| Lymph nodes | 6/16 | 10/16 | |
| Distal metastasis | 9/28 | 10/25 | |
| Deceased | 19/51 | 29/53 | |
| Palliative treatment | 12/51 | 13/53 | |
| Radiotherapy | 5/12 | 2/13 | |
| Chemotherapy | 5/12 | 7/13 | |
| Genetics | |||
| KRAS mutations (11) | 0 | 17 (24.6%) | <.0001 |
| BRAF (V600E) | 0 | 1 (1.5%) | |
| EGFR mutations (28) | 0 | 0 | |
Fisher exact test.
Unpaired t test with Welch’s correction (2 tailed). Only P values <.05 are presented.
Table 2.
Clinical and Pathologic Variables
| Multivariatea (good vs poor clinical outcome) | Events | Correlation r | P value |
|---|---|---|---|
| Size vs survival | 99 | −0.23 (−0.41, −0.03) | .02 |
| Portal tract vs survival | 77 | −0.33 (−0.51, −0.11) | .004 |
| Ductal dysplasia vs portal tract | 44 | 0.51 (0.25, 0.70) | .0004 |
| Necrosis vs portal tract | 38 | 0.56 (0.29, 0.74) | .003 |
| Necrosis vs ductal dysplasia | 38 | 0.48 (0.20, 0.70) | .002 |
| Stromal infiltration vs ductal dysplasia | 40 | 0.32 (0.01, 0.57) | .04 |
| Perineural invasion vs recurrence | 92 | 0.22 (0.02, 0.41) | .03 |
| Perineural invasion vs size | 89 | −0.37 (−0.54, −0.18) | .0003 |
| Perineural invasion vs portal tract | 21 | 0.34 (0.13, 0.54) | .003 |
| Perineural invasion vs ductal dysplasia | 47 | 0.49 (0.24, 0.68) | .0005 |
| Lymphatic vs survival | 70 | 0.31 (0.08, 0.51) | .01 |
| Lymphatic vs perineural invasion | 61 | 0.42 (0.19, 0.61) | .0008 |
| KRAS mutations vs perineural invasion | 57 | 0.32 (0.06, 0.54) | .02 |
NOTE. Univariate analysis of survival and recurrence was tested by log-rank statistics.
The correlation r is a partial correlation coefficient. Only statistically significant pairwise correlations are presented.
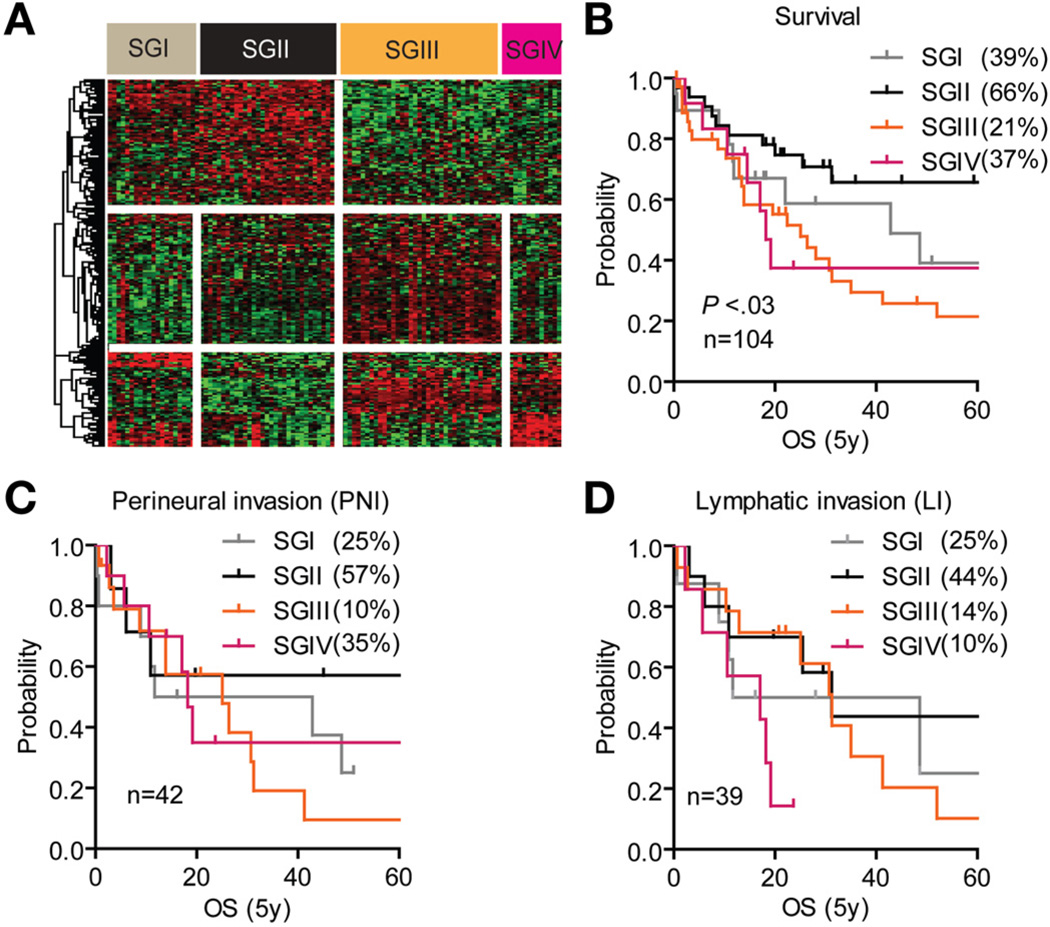
A supervised class comparison of the prognostic subclasses revealed that each subclass could be further subdivided into additional subgroups (SGs) with significant 5-year survival (SGI–IV; χ2 = 8.34; P < .03) (Figure 2A and B). Patients in SGIII showed a dismal clinical outcome, with lymphatic and perineural invasion predicting survival rates of 14% and 10%, respectively, compared with 44% and 57% in SGII (Figure 2C and D). A total of 127 genes at P < .001 distinguished subgroups SGI and SGII, whereas 85 genes differentiated SGIII and SGIV. Genes differentially expressed between SGI and SGII were mainly involved in immune response (Supplementary Figure 3A and C), whereas overrepresentation of genes involved in regulating proteasomal activity distinguished SGIII from SGIV (Supplementary Figure 3B and C). Among these were polo-like kinase 1 (PLK1) and aurora kinase A (AURKA), both currently used as anticancer drug targets. Thus, proteasome and anti-inflammatory inhibitors may represent a novel therapeutic option for a defined subgroup (ie, SGIII) of patients with CCA characterized by severe lymphatic and perineural invasion and shortest survival (Figure 2B–D).
Figure 2.
Supervised class comparison identifies 4 subgroups. (A) Supervised hierarchical cluster analysis of 212 genes identified as differentially regulated between SGI and SGII (127 genes) and between SGIII and SGIV (85 genes). Class comparison using random variance modeling (10,000 permutations; P < .001) was used to identify the significant genes between the subgroups. (B–D) Analysis of (B) overall survival, (C) perineural invasion, and (D) lymphatic invasion as independent prognostic predictors within SGI–IV. Kaplan–Meier and log-rank statistics were used to determine levels of significance.
Network Analysis of 2 Prognostic Subclasses
The network connectivity between the prognostic subclasses was analyzed using the geometric means. Major networks controlled by key molecules, such as tumor necrosis factor (TNF), transforming growth factor (TGF-β), and mitogen-activated protein kinase-1/2 (ERK1/2), were found to be deregulated in cluster 2. We annotated the predominant gene families between each subclass using the Molecular Signatures Database,14 which revealed an aberrant regulation of transcription factors (BCL11B, CDX2, DTX2, HOXB7, KLF5, MYB, NFE2L3, PATZ1, SIX4, SOX21, TFCP2L1, VDR, ZDHHC1), cell differentiation genes (CDCP1, CEACAM5, CEACAM6, FGFR2, ITGA2, PLAUR, TNFRSF12A), and protein kinases (AURKA, BUB1, FGFR2, NUAK2, PTK6, STYK1, TTK, WNK2, WNK4).
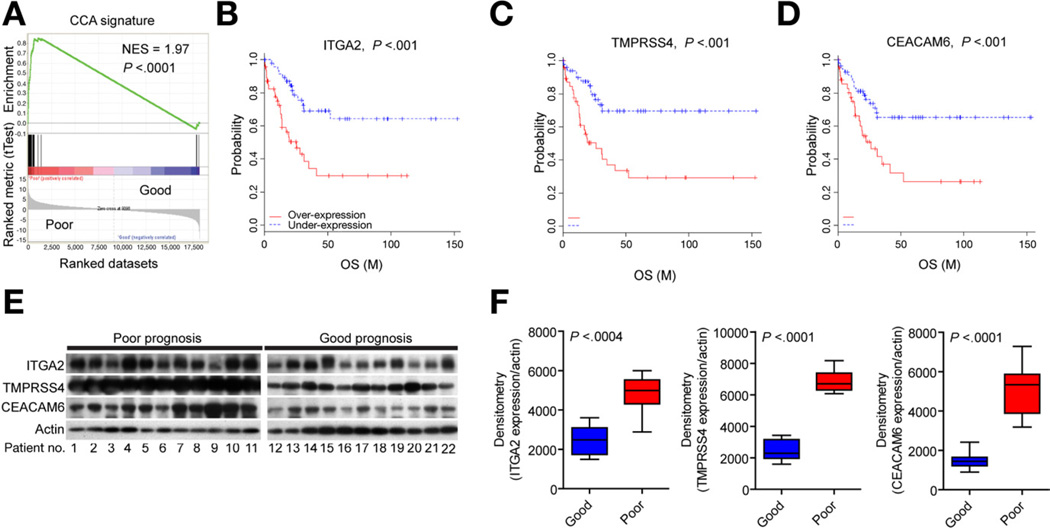
A Cox proportional hazards model was applied in a supervised analysis of genes strongly and independently associated with survival, testing the robustness of the 238-gene classifier. Using Wald statistics, the classifier was reduced to 36 genes (10,000 permutations; P < .01) with at least 2-fold difference in expression ratio (Supplementary Table 2). These genes were found to be enriched in key networks controlled by VEGF/ERRB, CTNNB1/MYC, and TNF. Further, the 36 genes were strongly associated with poor survival (normalized enrichment score [NES] = 1.97; P < .0001), as shown by gene set enrichment analysis (GSEA)14,15 (Figure 3A). To validate the relationship between gene expression and survival, protein levels of 3 of the survival genes (ITGA2, TMPRSS4, CEACAM6) (Figure 3B–D) were examined by Western blotting, confirming a significant overexpression of each marker in CCA with poor prognosis (Figure 3E and F).
Figure 3.
Prognostic survival genes. (A) GSEA of the classifier. Analysis of the survival genes revealed an ES of the rank-ordered genes, which showed a significant positive correlation with the poor survival subclass. The ES was normalized for the gene set (NES). (B–D) Genes significantly associated with the disease outcome were identified by means of a Cox proportional hazards model and Wald statistics. Thirty-six genes independently showed a prognostic ability at P < .01. (E and F) Representative Western blots of 3 survival genes, ITGA2, TMPRSS4, and CEACAM6, in resected tumor material from good and poor prognostic groups, respectively. Western blotting optical densities were normalized to β-actin values and expressed in arbitrary units. Bars represent mean ± 95% CI for each prognostic subtype (n = 11).
Analysis of the Microenvironment in CCA
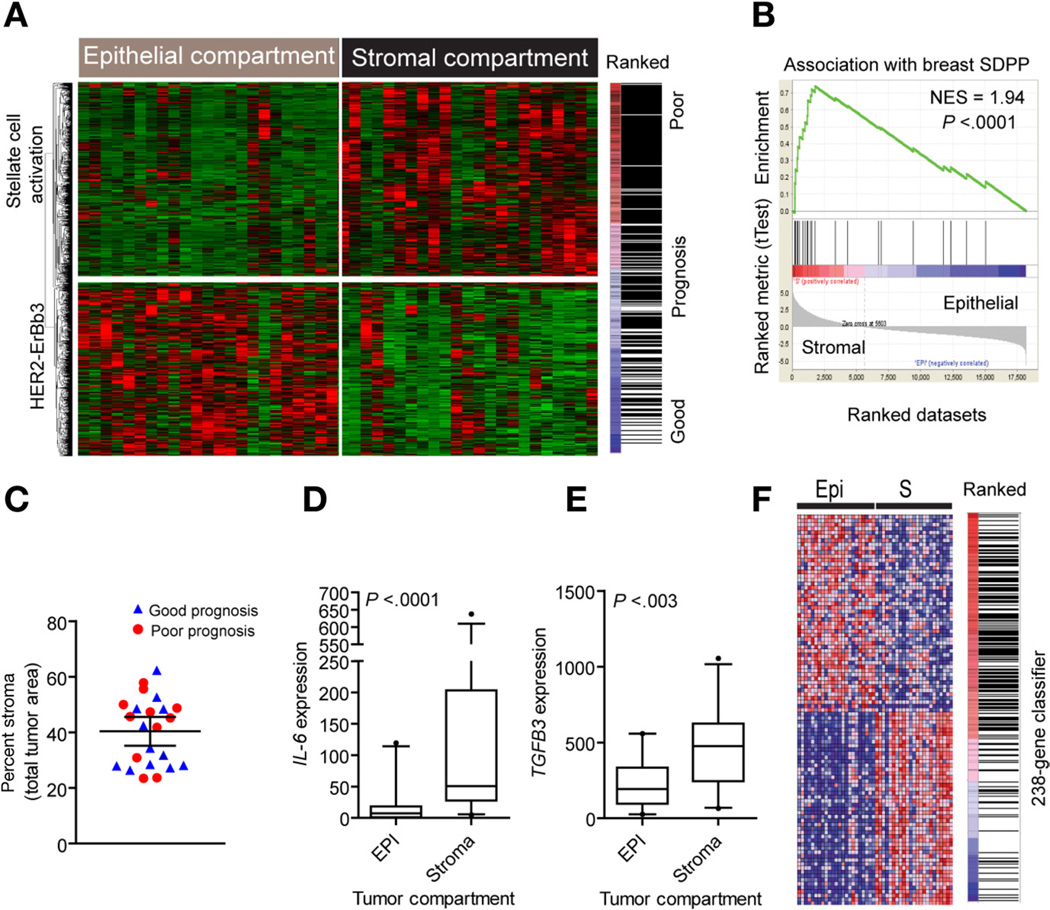
We laser capture microdissected epithelium and stroma from 23 CCAs and analyzed the transcriptome in each cell compartment. Hierarchal clustering revealed 2 groups separating epithelial and stromal tissues (Figure 4A) with a specificity of 83% as assessed by random Forest class prediction. A total of 1442 differentially expressed genes were identified by paired bootstrap t tests (5000 repetitions; P < .001). The stromal signature was significantly associated (enrichment score [ES] = 0.52) with poor prognosis, consistent with contribution of the microenvironment to tumor progression. This was supported by GSEA using a 26-gene breast cancer stromal-derived prognostic predictor (SDPP)16 that showed a significant enrichment (ES = 0.74) and association with the stromal compartment in CCA (Figure 4B). The clinical predictor included cytokines (CXCL14, AMD, SPP1, OGN), cell differentiation markers (CD8A, CD48, CD52, CD247), and transcription factors (HOXA10, RUNX3, VGLL1). To exclude that tumor cellularity could have affected the prognostic classification of CCA, we determined the percent occupied by tumor stroma and compared it with our classification. Although the percentage of tumor stroma was variable between the dissected tumors, it was independent of our prognostic classification and comprised 38.2% ± 12.2% versus 42.8% ± 11.8% (P = .37) in CCA with good and poor prognosis, respectively (Figure 4C). This result was reproduced in an independent set of tumors stained with CK19/4′,6-diamidino-2-phenylindole (DAPI) for a better separation between the epithelial and stromal compartments (n = 24; 39.2% ± 9.5% vs 34.4% ± 15.2% in tumors with good and poor prognosis; P = .36).
Figure 4.
Analysis of the tumor microenvironment. (A) Unsupervised hierarchical clustering of epithelial and stromal cell compartments. Laser capture microdissection was used to isolate distinct cell populations from 23 tumors and identify gene expression differences. A total of 1442 genes were identified as differentially expressed between tumor epithelium and stroma by means of a paired bootstrap t test (P < .001). The stromal gene signature is enriched and associated with the group of patients with overall poor clinical outcome. (B) GSEA using a curated stromal gene set associated with overall poor prognosis in breast cancer. The 26-gene stromal-derived prognostic predictor (SDPP) was significantly enriched and positively associated with the stromal cell compartment in CCA. (C) Quantification of the epithelial-to-stromal composition given as a percent stromal area. The prognostic classification is given as good (blue) and poor (red) survival classes. (D and E) Analysis of (D) IL-6 and (E) TGFB3 gene expression in the tumor epithelial and stromal compartments (n = 23), respectively. Statistical significance was determined by Mann–Whitney test (2 tailed). The box plots show the mean, and whiskers are given for the 5th to 95th percentiles. (F) Association of the 238-gene classifier with the epithelial cell compartment. Hierarchical clustering of the top 50 ranked genes shows the enrichment and positive association with the tumor epithelium.
To identify specific network connectivity for the microenvironment, we applied a quantitative trait analysis using Pearson correlation (10,000 permutations; P < .001). Significant gene ontology classes that distinguished epithelial and stromal compartments were enriched for TGF-β (THY1, CD209, TIMP2, and TIMP3) (observed/expected = 4.05) and TNF receptor superfamilies (observed/expected = 3.47) (Supplementary Table 3). In stroma, IL6 and TGFB3 expression were increased (Figure 4D and E), including a preferential dysregulation of chemokine receptors and ligands (CXCR4, CCR7, CCL2, CCL5, CCL19, CCL21), cytokine receptors (IL3RA, 7R, 10RA, 18RAP), and interleukins (IL6, IL16, IL33). The most significant network (P < .0001) was related to hepatic stellate cell activation (CTGF, PDGFRA, PDGFRB, VEGFC, VCAM1, IGFBP4, IGFBP5, LY96, MMP2). Significantly, the prognostic 238-gene classifier was enriched (ES = 0.33) and associated with the epithelial compartment, indicating that the classifier was predominantly epithelial derived (Figure 4F). Accordingly, the HER2 network (Supplementary Figure 4) and HER2 signaling (CCNE2, ITGB3, PARD6B, SOS2, PI3KCB) were overrepresented (P < .0001) in tumor epithelium (Figure 4A).
KRAS Mutations Are Associated With Poor Prognosis in CCA
To extend the molecular characterization, all CCAs (n = 69) with available genomic DNA were analyzed for 11 somatic mutations in KRAS, 1 in BRAF, and 28 in EGFR. KRAS mutations were identified in 17 of 69 patients, whereas only one tested positive for BRAFV600E. All patients (0/69) in our cohort were wild type for EGFR. No mutations were found in the matched noncancerous control samples from subjects with mutated KRAS, suggesting that mutations were acquired during transformation.
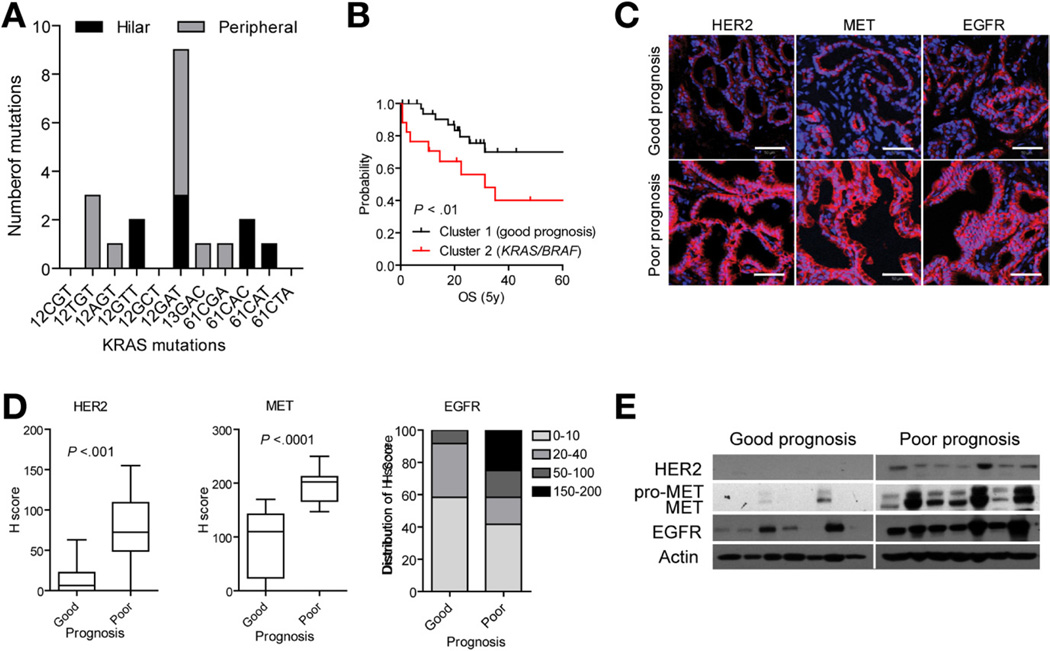
Mutations in KRAS were previously detected in 21%–100% of CCAs17–19 as compared with 24.6% (17/69) in our cohort (Figure 5A). When classified by tumor site, 53.3% (8/15) of hilar versus 16.7% (9/54) of peripheral-type CCAs had mutations in KRAS. Tumors were analyzed for genetic alterations in KRAS codon 12 (CGT, TGT, AGT, GTT, GCT, GAT), 13 (GAC), and 61 (CGA, CAC, CAT, CTA). Only one tumor was identified with more than one mutation in KRAS12TGT/12GAT/13GAC. The most frequent alteration was in codon 12 (14/19) with predominantly mutated sites in 12GAT (8/19) and 12TGT (3/19). Although we were unable to establish KRAS as an independent prognostic factor within our cohort, integrating the KRAS mutational status and the 238-gene classifier grouped all patients with mutated KRAS/BRAF in cluster 2 (patients with poor prognosis) (P < .01) (Figure 5B).
Figure 5.
Characterization of the CCA classification. (A) Frequency of the KRAS mutations in codon 12, 13, and 61 detected by real-time quantitative polymerase chain reaction. The number of mutations is grouped according to hilar (black) and peripheral (gray) tumor subtype. (B) Survival analysis. The mutational status of KRAS/BRAF was significantly associated with poor prognosis as represented by Kaplan–Meier plots and log-rank statistics. (C) Immunohistochemical analysis of HER2, MET, and EGFR protein. Representative images are shown. Scale bar = 50 µm. (D) Semiquantitative assessment of immunohistochemical staining by H-score for HER2, MET, and EGFR. The box plots show the mean H-scores in good and poor prognosis tumor groups (n = 12 each), and whiskers are given for 5th to 95th percentiles. (E) Western blot analysis of HER2, MET, and EGFR expression in 7 patients from each of the prognostic subclasses. Actin was used as loading control.
KRAS regulates cell growth downstream of major RTKs, including EGFR, HER2, and MET. To determine the relationship between RTK expression levels and survival, we performed immunohistochemical analysis in 12 randomly selected tumors from each of 2 prognostic subclasses. To quantify the staining intensity, we used H-scoring, which takes into consideration both the intensity and number of positive cells within the epithelial compartment. Tumors from the poor prognostic subclass were frequently scored moderately to strongly positive for membranous HER2 (P < .001) (Figure 5C and D), corroborating the observed deregulation of HER2-ERBB3 signaling in tumor epithelium (Supplementary Figure 4). In tumors from patients with a good prognosis, HER2 was either not detected (4/12) or weakly expressed in the cytoplasm (8/12), indicating receptor inaction. In comparison, overexpression of EGFR was a common feature of CCA. Although difference in mean H-score for EGFR between survival groups did not reach statistical significance, values were higher overall in tumors from patients with poor survival, suggesting cross-activation within the ErbB receptor family in advanced malignancies (Figure 5C and D). Also, tumors from patients with a poor prognosis exhibited a strong and predominantly membranous staining for MET in the tumor epithelium (10/12) as compared with patients with a good prognosis (Figure 5C and D and Supplementary Figure 5A), indicating possible coactivation of multiple RTKs, which may account for resistance to TKI monotherapy. Western blot analysis of EGFR, HER2, and MET using an independent set of CCA samples confirmed a strong up-regulation of RTKs in patients with a poor outcome (Figure 5E). Selected tumors (n = 48) were additionally analyzed for EGFR, HER2, and MET copy number variations. Although none of the examined genes were amplified (not shown), the detected increases in expression may be explained either by synergistic receptor heterodimerization (HER2-EGFR, HER2- ErbB3, or HER2-MET)20 or receptor stabilization.21
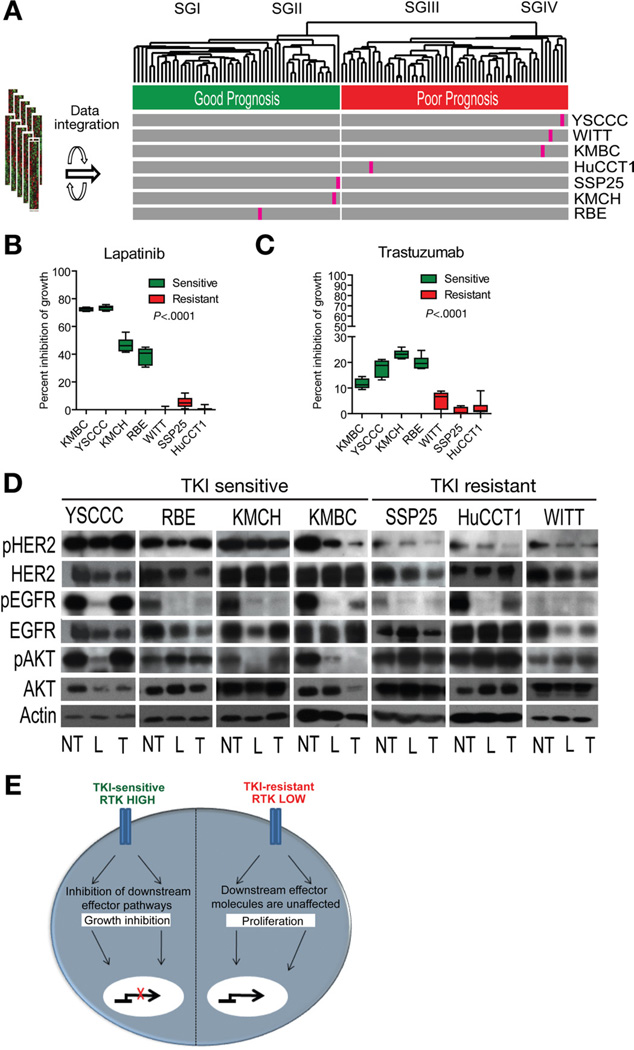
To assess RTKs as potential drug targets against CCA, we integrated 7 human CCA cell lines with a patient cohort (Figure 6A). All cell lines were treated for 7 days with known RTK inhibitors trastuzumab and lapatinib (Figure 6B and C and Supplementary Figure 6). Lapatinib, a dual inhibitor of EGFR and HER2, was significantly more effective than trastuzumab, which selectively targets HER2, as evaluated by growth inhibition and Western blot analysis of HER2, EGFR, and downstream AKT (Figure 6B–D). Furthermore, lapatinib was efficient at the median lethal dose (50%–80% growth inhibition), which varied between 1.1 and 5.9 µmol/L among the examined CCA cell lines, whereas the inhibitory effect of trastuzumab did not exceed 20% even at the maximum upper limit dose (500 µg/mL) (Supplementary Figure 6). The growth inhibition directly correlated with the extent of HER2 and EGFR activation, which was considerably higher in the drug-sensitive cell lines as compared with the drug-resistant cell lines. It is noteworthy that 2 of the TKI-resistant cell lines (HuCCT1 and WITT) that integrated with the poor prognosis subclass (Figure 6A) had mutations in KRAS codon 12, supporting a link between resistance to EGFR-based therapies and activation of KRAS, a downstream effector of RTKs.
Figure 6.
Effect of TKIs in CCA. (A) Integration of 7 human CCA cell lines with the patient cohort using the 238-gene classifier. (B and C) Effect of a 7-day treatment with (B) lapatinib and (C) trastuzumab on the viability of CCA cell lines using an estimated 50% lethal dose for lapatinib or 500 µg/mL trastuzumab, respectively, for each cell line. Bars represent 8 experiments as mean ± 95% CI viability expressed as percent versus corresponding controls. The statistical significance was determined by one-way analysis of variance with Tukey’s multiple comparison tests (α = .05). (D) Western blot analysis of drug-target EGFR, HER2, and downstream AKT following treatment with lapatinib (L), trastuzumab (T), and untreated control (NT), respectively. (E) Schematic representation of TKI response. TKI-sensitive CCA cell lines have high level and activity of EGFR and HER2 expression. Downstream AKT signaling is unaffected in TKI-resistant CCA cell lines.
Meta-analysis of the CCA Classifier
Meta-analysis of the classifier showed a prognostic value and its ability to predict clinical outcome for numerous types of human cancer (eg, lung adenocarcinoma, kidney, and breast) (odds ratio, >2, P < .0001) (Supplementary Figure 7A). We also showed a significant association of the classifier with high-grade and advanced-stage HCC (Supplementary Figure 7B and C). GSEA confirmed a significant enrichment (NES = 1.78; P < .007) and association with poor subtype A HCCs10 (Supplementary Figure 7D). Among the characteristics of HCCs with poor survival is activation of the mTOR pathway (odds ratio, 2.5; P < .0001).22 Similarly, our immunohistochemical analysis revealed frequent overexpression of pRPS6, a marker of mTOR signaling, in patients with poor outcome and early recurrence (Supplementary Figures 5A and 7E and F). Tumors from the poor subclass also displayed stronger Ki67 staining (Supplementary Figure 7G and H), in agreement with the prognostic significance of increased proliferation in resectable CCA.23
Discussion
We performed a comprehensive molecular and genomic characterization of 104 surgically resected CCAs. Our 238-gene classifier identified a high-risk group of patients with CCA, significantly differentiating patients according to overall and recurrence-free survival independent of specific clinical subtypes. Reflecting the strength of the classifier, it could be further reduced to 36 genes, which differentiated individuals in outcome-linked categories with greater accuracy. A comparison with our recent genomic data obtained from a limited number of CCAs24 confirmed a significant cholangio-specific association of our 238-gene classifier as well as the 36 survival genes identified in this study (Supplementary Figure 8). Furthermore, in a meta-analysis, our classifier revealed a strong capacity to predict clinical outcome for other types of cancer, including HCC (Supplementary Figure 7B–D). The close genomic relationships found between HCC and CCA suggest that acquisition of CCA-like expression traits may play a role in HCC heterogeneity.
Combining laser microdissection with transcriptomics allowed us to identify the core biological processes in tumor epithelium and stroma, which drive CCA disease progression and outcome. The most malignant tumor phenotype was characterized by a strong up-regulation of HER2 signaling in the epithelial cell compartment and concomitant overexpression of proinflammatory cytokines in tumor stroma, including interleukin-625 and CXCR4.26 A recently described 26-gene stromal-derived prognostic predictor in breast cancer16 was significantly enriched in the stromal compartment of CCA.
Aberrant HER2 expression has been described in many cancers (eg, ovarian, gastric) and most prominently in breast cancer, where it has a significant role in malignant transformation27 and choice of therapy. In CCA, overexpression of HER2 was reported in ~30% of tumors.20 In our study, HER2 up-regulation was found only in tumors from patients with poor prognosis, who were also characterized by a frequent coactivation of ERBB3 and EGFR, 2 other members of the ErbB receptor family, as well as MET and mTOR. Multiple oncogenic pathways were frequently coactivated within a single tumor from the poor prognosis group (Supplementary Figure 5A), indicating that oncogenic addiction may be a hallmark of CCA progression. To explore the effects of available drugs targeting RTKs for treatment of CCA, we used an integrated in vitro/in vivo approach to identify CCA cell lines that closely mimic the genomic phenotypes of the identified subclasses of patients with CCA (Figure 6A). The results showed that our newly recognized subclass of patients with poor outcome CCA with increased EGFR and HER2 signaling may benefit from dual-target TKIs, whereas KRAS mutations may confer resistance to this treatment. Although TKIs may present a therapeutic strategy to target CCA, a secondary target downstream of KRAS may be required to sensitize TKI-resistant cancer cells to TKIs. Indeed, we found a significant association of activating KRAS mutations (24.6%) in the cohort with outcome when integrated with the classifier. The presence of KRAS mutations is predictive of resistance to EGFR therapy in colorectal cancer. Clinical trials with TKIs in non–small cell lung cancer show that patients responding to therapy typically have activating mutations in EGFR.28,29 Although a low frequency of EGFR mutations (13.6%) was described in CCA,30 we found no EGFR-specific mutations or amplification of EGFR in our cohort. A recent phase 2 study with erlotinib in advanced biliary cancers7 showed a therapeutic benefit against tumors overexpressing EGFR. Also, given a strong activation of the downstream mTOR pathway in tumors from patients with poor outcome, targeting this pathway may present an alternative treatment option for CCA. A more in-depth analysis of the prognostic subclasses by class comparison identified a group of patients (SGIII) characterized by overrepresentation of genes involved in proteasomal activity, suggesting a potential therapeutic benefit of proteasome and antiinflammatory inhibitors.
In conclusion, we identified 2 prognostic categories of patients with CCA, each containing 2 subclasses (SGI–IV) characterized by distinct gene expression profiles. A prognostic 36-gene classifier either alone or in combination with other molecular predictors (ie, mutations, coactivation of multiple oncogenic pathways) improved the molecular classification and outcome prediction in CCA. The study also shows the therapeutic potential for dual-target TKIs (eg, lapatinib) in CCA. Taken together, the present findings establish the foundation for future directions in the development of diagnostic and therapeutic modalities for CCA.
Supplementary Material
Acknowledgments
The authors thank Tanya Hoang (Laboratory of Experimental Carcinogenesis, National Cancer Institute) for laboratory assistance and Teresa Mettler (Mayo Clinic) for abstracting clinical data.
Funding
Supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health; National Institutes of Health grants CA100882 and CA128633 (to L.R.R.) and P30DK084567 (Mayo Clinic Center for Cell Signaling in Gastroenterology); and grant 271-070712 from the Danish Medical Research Council (to J.B.A.).
Abbreviations used in this paper
- CCA
cholangiocarcinoma
- CI
confidence interval
- EGFR
epidermal growth factor receptor
- ES
enrichment score
- GSEA
gene set enrichment analysis
- NES
normalized enrichment score
- RTK
receptor tyrosine kinase
- SG
subgroup
- TGF
transforming growth factor
- TKI
tyrosine kinase inhibitor
- TNF
tumor necrosis factor
Footnotes
Supplementary Materials
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at 10.1053/j.gastro.2011.12.005.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.DeOliveira ML, Cunningham SC, Cameron JL, et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg. 2007;245:755–762. doi: 10.1097/01.sla.0000251366.62632.d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deoliveira ML, Schulick RD, Nimura Y, et al. New staging system and a registry for perihilar cholangiocarcinoma. Hepatology. 2011;53:1363–1371. doi: 10.1002/hep.24227. [DOI] [PubMed] [Google Scholar]
- 4.Aljiffry M, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of cholangiocarcinoma, 1990–2009. World J Gastroenterol. 2009;15:4240–4262. doi: 10.3748/wjg.15.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glimelius B, Hoffman K, Sjoden PO, et al. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Ann Oncol. 1996;7:593–600. doi: 10.1093/oxfordjournals.annonc.a010676. [DOI] [PubMed] [Google Scholar]
- 6.Bengala C, Bertolini F, Malavasi N, et al. Sorafenib in patients with advanced biliary tract carcinoma: a phase II trial. Br J Cancer. 2010;102:68–72. doi: 10.1038/sj.bjc.6605458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philip PA, Mahoney MR, Allmer C, et al. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol. 2006;24:3069–3074. doi: 10.1200/JCO.2005.05.3579. [DOI] [PubMed] [Google Scholar]
- 8.Lubner SJ, Mahoney MR, Kolesar JL, et al. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol. 2010;28:3491–3497. doi: 10.1200/JCO.2010.28.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 10.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Harris T. Does large scale DNA sequencing of patient and tumor DNA yet provide clinically actionable information? Discov Med. 2010;10:144–150. [PubMed] [Google Scholar]
- 13.Harris T. Gene and drug matrix for personalized cancer therapy. Nat Rev Drug Discov. 2010;9:660. doi: 10.1038/nrd3181-c1. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 17.Kipp BR, Barr Fritcher EG, Clayton AC, et al. Comparison of KRAS mutation analysis and FISH for detecting pancreatobiliary tract cancer in cytology specimens collected during endoscopic retrograde cholangio-pancreatography. J Mol Diagn. 2010;12:780–786. doi: 10.2353/jmoldx.2010.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin Liver Dis. 2004;24:139–154. doi: 10.1055/s-2004-828891. [DOI] [PubMed] [Google Scholar]
- 19.Tannapfel A, Benicke M, Katalinic A, et al. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000;47:721–727. doi: 10.1136/gut.47.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirica AE. Role of ErbB family receptor tyrosine kinases in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2008;14:7033–7058. doi: 10.3748/wjg.14.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang SA, Lee ES, Yoon HY, et al. PTK6 inhibits down-regulation of EGF receptor through phosphorylation of ARAP1. J Biol Chem. 2010;285:26013–26021. doi: 10.1074/jbc.M109.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. 1983, e1–e11. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briggs CD, Neal CP, Mann CD, et al. Prognostic molecular markers in cholangiocarcinoma: a systematic review. Eur J Cancer. 2009;45:33–47. doi: 10.1016/j.ejca.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma- like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott JL, Gores GJ. Targeting IL-6 in cholangiocarcinoma therapy. Am J Gastroenterol. 2007;102:2171–2172. doi: 10.1111/j.1572-0241.2007.01394.x. [DOI] [PubMed] [Google Scholar]
- 26.Ohira S, Sasaki M, Harada K, et al. Possible regulation of migration of intrahepatic cholangiocarcinoma cells by interaction of CXCR4 expressed in carcinoma cells with tumor necrosis factor-alpha and stromal-derived factor-1 released in stroma. Am J Pathol. 2006;168:1155–1168. doi: 10.2353/ajpath.2006.050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 28.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 29.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 30.Gwak GY, Yoon JH, Shin CM, et al. Detection of response-predicting mutations in the kinase domain of the epidermal growth factor receptor gene in cholangiocarcinomas. J Cancer Res Clin Oncol. 2005;131:649–652. doi: 10.1007/s00432-005-0016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.