Abstract
This report details the lipid composition of nascent HDL (nHDL) particles formed by the action of the ATP binding cassette transporter A1 (ABCA1) on apolipoprotein A-I (apoA-I). nHDL particles of different size (average diameters of ∼12, 10, 7.5, and <6 nm) and composition were purified by size-exclusion chromatography. Electron microscopy suggested that the nHDL were mostly spheroidal. The proportions of the principal nHDL lipids, free cholesterol, glycerophosphocholine, and sphingomyelin were similar to that of lipid rafts, suggesting that the lipid originated from a raft-like region of the cell. Smaller amounts of glucosylceramides, cholesteryl esters, and other glycerophospholipid classes were also present. The largest particles, ∼12 nm and 10 nm diameter, contained ∼43% free cholesterol, 2–3% cholesteryl ester, and three apoA-I molecules. Using chemical cross-linking chemistry combined with mass spectrometry, we found that three molecules of apoA-I in the ∼9–14 nm nHDL adopted a belt-like conformation. The smaller (7.5 nm diameter) spheroidal nHDL particles carried 30% free cholesterol and two molecules of apoA-I in a twisted, antiparallel, double-belt conformation. Overall, these new data offer fresh insights into the biogenesis and structural constraints involved in forming nascent HDL from ABCA1.
Keywords: ATP binding cassette transporter A1, nascent HDL, cholesterol, apolipoprotein A-I, mass spectrometry
In humans, the plasma concentration of HDLs has been repeatedly shown to be inversely correlated with the risk of developing coronary heart disease (1, 2). The concentrations of plasma HDL has been shown to be largely dependent on hepatic ATP-binding cassette transporter A1 (ABCA1) (3–5), which transports and promotes the efflux of glycerophosphocholine (PC), free cholesterol (FC), and sphingomyelin (SM) to wild-type apolipoprotein A-I (apoA-I), resulting in the formation of nascent HDLs (nHDLs). Functional mutations in human ABCA1 cause Tangier disease (6, 7), which is characterized by very low levels of plasma HDL apoA-I. Tangier disease is believed to alter a process termed “reverse cholesterol transport.” Although defects in ABCA1 function have been identified by decreases in production of plasma HDL, ABCA1 dysfunction can also significantly alter cell signaling by modulating membrane cholesterol levels, leading to hyperactive immune cells (8–10). Most recently, studies have shown that ABCA1 functions to maintain cellular cholesterol content within membrane microdomains, such as lipid rafts (8, 11). Therefore, it is tempting to speculate that transport of cholesterol from cellular membranes to apoA-I-forming nHDL particles is foremost a means of maintaining membrane lipid raft integrity and is the first step in the well-known reverse cholesterol transport pathway.
ABCA1-mediated efflux of cholesterol and PC leading to the biogenesis of nHDL has been extensively studied in a wide variety of cell types and systems (12, 13). Although these studies have yielded a better understanding of ABCA1 function, there remain important questions that require clarification. It is generally agreed that apoA-I binds to ABCA1 and to high-capacity binding sites on the plasma membrane (14–16), which in turn stimulate the assembly of lipids into a nHDL particle. Although previous studies have shown that ABCA1 is not associated with cholesterol/sphingomyelin-rich raft regions (17), the dependence of ABCA1 function on maintenance of microdomain cholesterol composition is clear (18–21). The following questions still need to be addressed: (1) Which cellular compartments do the lipids that initially comprise nHDL particles derive? and (2) How do three molecules of apoA-I come together and conformationally adapt on the cell surface to acquire these lipids? The current studies were undertaken to answer these and other important questions. We report from our investigations that the largest spheroidal nHDL particle has an average diameter of ∼11 nm, carries up to 45% of the total particles’ lipid as cholesterol, and is bound by three apoA-I molecules organized in a belt-like conformation. We conclude that these studies demonstrate the importance of nHDL composition and protein conformation in understanding how ABCA1 effluxes cellular cholesterol and PC as part of the reverse cholesterol transport pathway and maintenance of the plasma membrane microdomain cholesterol content.
MATERIALS AND METHODS
Reagents
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was from Avanti Polar Lipids Inc. Bis(sulfosuccinimidyl)suberate, dithiobis(succinimidylpropionate) (DSP), bis-(N-succinimidy) penta(ethyleneglycol), and bis-(N-succinimidyl)nona(ethyleneglycol) were from ThermoFisher. Me2SO and formic acid was from Sigma-Aldrich. Sequencing grade modified trypsin and restriction enzymes were from Promega. RapiGest SF™ was from Waters Inc. Sodium desoxycholate, potassium chloride, optima grade methanol, chloroform, acetonitrile, and glacial acetic acid were from Fisher Scientific. Mark 12 molecular weight standards and Simply Blue™ Safestain were from Invitrogen. Ultrafree-15 centrifugal and Biomax™ 10K membranes were from Millipore Corp.. All other solvents and routine reagents were of the highest available commercial grades.
Cell culture
Human embryonic kidney (HEK) 293 cells expressing ABCA1 were a generous gift from Dr. Michael Hayden, University of British Columbia, Canada and supplied by Dr. John Parks (22, 23). HEK293 Flp-In™ cells from Invitrogen, which do not express ABCA1, were used for controls to determine the contribution of cellular sloughing during 18 to 24 h incubations or artifacts during concentration and fast protein liquid chromatography (FPLC) of postcellular media. All cells were maintained in DMEM containing 4.5 g/l glucose, 50 µg/ml hygromycin, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM L-glutamine, and 10% FBS. Cells were maintained at 37°C in an atmosphere of 5% CO2. ABCA1-expressing cells were plated in 100 mm dishes until cells reached ∼90% confluence, washed four times with balanced salt solution, and incubated for 18–24 h with 10 µg/ml of lipid-free human 125I-apoA-I in serum-free DMEM. In experiments designed to analyze cholesterol incorporation, 1 µCi of 3H-cholesterol per ml media was added to the cell monolayer when cells were ∼80% confluent and incubated for 24 h before incubating with unlabeled apoA-I, as described above.
In experiments to quantify the mass of nHDL lipids or to determine protein structure, 40 to 100 mm plates of confluent cells were grown. To remove traces of contaminating fetal calf serum proteins, cells were washed three times with balanced salt solution, and then 10 ml of serum-free was added and the cells were incubated for 1 h at 37°C. The media was removed, and another 10 ml of serum-free DMEM was added for second 1 h incubation. Then 10 ml of fresh DMEM containing 10 µg/ml of lipid-free human apoA-I was added, and the cells were incubated overnight at 37°C. The next day the culture media was harvested, dialyzed against 10 mM ammonium bicarbonate (pH 7.4) at 4°C, and concentrated using Amicon Ultra-15 Filter Devices (Millipore). In parallel experiments, HEK 293 Flp-In™ cells were treated with lipid-free apoA-I in the same manner as described above, and essentially no cholesterol and only trace amounts of phospholipids were detected, which were subtracted from composition data.
In companion experiments, bone marrow-derived macrophages (BMDMs) were obtained from the femurs and tibias of chow-fed adult mice. Briefly, bones were cut at one end, and the marrow was collected by centrifugation into 1.5 ml tubes and then plated in low-glucose DMEM containing 30% L929 cell-conditioned medium (LCM), 20% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 µg/ml streptomycin for 7 days until the cells reached confluence. Approximately 40 dishes of confluent 100 mm dishes of BMDMs were then used for nHDL production and lipid raft isolation. LCM was obtained from L929 cells cultured in RMPI media containing 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 µg/ml streptomycin, which had been plated at a density of 5.3 × 105 cells per T-150 flask and grown until confluence. No other additions were made to increase expression of ABCA1. After incubation for 24 h with 10 µg/ml human apoA-I, the conditioned media was collected, passed through a 0.2 µm filter, and stored at −80°C until needed.
Preparation of lipid-free human ApoA-I
ApoA-I was purified from human plasma by sequential ultracentrifugation as previously described (24). Mass spectrometry and 12% SDS-PAGE were used to assess protein purity before use. Protein concentration was determined using the Lowry assay (25).
Purification of nHDL
Dialyzed, concentrated culture media was adjusted to 1.225 g/ml with KBr. The d < 1.25 g/ml fraction after centrifugation was isolated, dialyzed, and then concentrated and fractionated by FPLC using three Superdex-200 10/3000GL (GE Healthcare) columns connected in series at a flow rate of 0.3 ml/min. Fractions were collected, and, for experiments using radioactive tracers, fractions were counted for 125I-apoA-I or 3H-cholesterol (Perkin Elmer). FPLC Peaks 1–4 were identified from plots of radioactivity versus fraction number using Prism™ (GraphPad Software). Aliquots from each of the four peak fractions were analyzed by 4–30% nondenaturing gradient gel electrophoresis (NDGGE) to determine the particle diameter size homogeneity by comparison to high-molecular-weight standards (Amersham Biosciences) using either radioactivity or mass. Radioactive protein bands were visualized on the gel using a BAS 5000 phosphorimager (Fujifilm). Protein mass gels was determined by staining with Simply Blue. FPLC peaks in mass studies were pooled according to regions identified from radioactive tracer studies: Peak 1 = fractions #110–125, Peak 2 = fractions #126–139, Peak 3 = fractions #140–157, and Peak 4 = fractions #158–175. Pooled peaks were dialyzed against 10 mM ammonium bicarbonate (pH 7.4) at 4°C in a 6 liter container that had never been exposed to phosphate detergent. An aliquot from each pooled peak fraction was assayed for apoA-I content using ELISA as previously described (26).
Chemical cross-linking
To determine the number of apoA-I molecules per particle, purified nHDL particles were cross-linked with DSP as previously reported (27). Cross-linking for structural analysis was conducted with several different cross-linkers having different arm lengths as previously reported (24, 28). Cross-linkers were dissolved in Me2SO and added to the particles at a molar ratio of 100:1 cross-linker to apoA-I to determine the number of molecules per particle. For structure determination, the cross-linker to apoA-I molar ratio was 25:1. After addition of freshly made cross-linker, the tube was capped and mixed. After 15 min at room temperature (RT), 50 mM Tris was added to stop the cross-linking reaction. Aliquots containing 15 µg apoA-I were analyzed by nonreducing Novex 4–12% Tris-Glycine SDS-PAGE (Invitrogen).
SDS-PAGE and trypsin digestion
Products from chemical cross-linking of nHDL were separated on 12% SDS-PAGE. Protein bands were excised from the gel, minced, and repeatedly dehydrated with acetonitrile. Gel pieces were rehydrated with a cold, freshly prepared solution of 20 ng/µl trypsin in 10 mM ammonium bicarbonate (pH 7.8), 0.1% (w/v) RapiGest SF™, and 1 mM CaCl2, as previously described (24, 28). The final trypsin to apoA-I mass ratio was 1:20. After incubation on ice for 10 min, the digests were incubated for 18 h at 37°C.
Peptide isolation and liquid chromatography mass spectrometry
After digestion, gel pieces were covered with 200 µl of extraction solvent (acetonitrile-formic acid-water, v/v/v, 50:5:45) as previously reported (24, 28). The supernatant was transferred to a fresh tube before mass spectrometry. To identify candidate cross-linked peptides for MS/MS sequencing, survey scans were performed on each peptide mixture using a Waters quadrupole-time-of-flight API-US mass spectrometer equipped with a Waters CapLC as previously reported (24, 28). Positive ion electrospray survey scans were recorded in the continuum mode using a scan window from 300 to 1500 m/z with an accumulation time of 2 s. The source temperature was 80°C, and the cone and capillary voltages were 45 V and 3.5 kV, respectively. The experimental m/z was corrected for the +2 charge state using apoA-I tryptic fragment 7 (m/z = 806.8969) and for the +1 charge state using apoA-I tryptic fragment 12 (m/z = 831.4365). The survey ion scan was deconvoluted to give a list of +1 charge states that eluted from the column. This experimental list was sorted against a list of all possible lysine-to-lysine cross-links for the particular chemical cross-linker used in an experiment. Product ion MS/MS spectra were acquired in the continuum mode from 50 to 1800 m/z using a data–directed, charge-state selective collision energy and an accumulation time of 2 s. Sequence analysis was performed with a fragment ion tolerance of ± 0.05 m/z.
Molecular modeling
The molecular modeling of apoA-I bound to recombinant HDL particles has been described in previous publications using the coordinates for lipid-free Δ43-apoA-I (29) that were joined with 1–43 amino acids of the N-terminal end (24, 28). Unmodified full-length lipid-free apoA-I has been studied by others and the results were used to guide the conformation (30–34). We used the number of apoA-I per particle and particle size as constraints for each nHDL. Cross-linked positions on the individual apoA-I molecules were oriented to their correct distances. To do this, we used the maximum distance of Cα-Lysine-(cross-linker)-Cα-Lysine calculated for each of the cross-linkers: 26.0 Å, 26.6 Å, 30.7 Å, and 36.3Å for bis(sulfosuccinimidyl)suberate, DSP, bis-(N-succinimidy) penta(ethyleneglycol), and bis-(N-succinimidy) nona(ethyleneglycol), respectively. ApoA-I was bent only at the proline or glycine-glycine sites between the amphipathic segments of apoA-I. Tools available in Swiss-PdbViewer OSX version 4.0.4 (http://www.expasy.org/spdbv/) were used to optimize the conformations, and pdb files were manipulated using PyMOL version 1.5 (http:/www.pymol.org). Swiss-PdbViewer, PyMOL, and Visual Molecular Dynamics for Mac OSX, version 1.9 (35) (www.ks.uiuc.edu/Research/vmd) were used to generate the molecular figures shown in the manuscript.
Lipid extraction
Lipids were extracted from the dried nHDL Peaks #1–4 according to Bligh and Dyer (36) after adding 4 µg of [3,4-14C2]cholesterol as the internal standard for cholesterol analyses (9). Lipid extracts were evaporated under a stream of argon, dissolved in 1 ml of chloroform-methanol (1:1), and stored at −80°C until analyzed. In all cases, parallel experiments were conducted using control (non-ABCA1 expressing) cells, and those results were subtracted from the values obtained from ABCA1-expressing cells.
Cholesterol analysis by GC-MS
An aliquot from each of the Peak #1–4 lipid extracts was evaporated under argon, dissolved in toluene, and analyzed on a Finnigan Trace 2000 mass spectrometer or a Finnigan TSQ Quantum XLS tandem mass spectrometer interfaced to a Trace gas chromatograph (9). A second 50 µl aliquot was saponified at room temperature for 2 h with methanolic-KOH (similar to a reported procedure using ethanol), cooled, and then extracted with hexane.
Cholesteryl ester molecular species
An aliquot of the lipid extract was mixed with 95 µl of a methanol solution containing 500 pg/µl cholesterol heptadecanoate (Nu-Chek Prep) and 1 ng/µl sodium formate. After standing for 30 min, the solution was analyzed by direct infusion at 4.3 µl/min into a Waters Quattro II tandem mass spectrometer operated in the positive ion mode. Analysis settings were: collision energy (MSCE), 3.8kV; cone, 65 V; source temp, 80°C; desolvation nitrogen was introduced at 300 l/h and heated to 150°C; nebulizing nitrogen was set to 10 l/h; collision gas, argon at 1 mTorr. SRM detected the neutral loss of 368.4 Da over a scan range of m/z 640–740 with a scan time of 0.5 s. The data were quantitated with a response curve from 0.1 µM to 6.4 µM of each cholesteryl ester against 0.78 µM internal standard.
Phospholipid analysis
Aliquots of the lipid extract were separated and analyzed by normal-phase HPLC-tandem mass spectrometry (LC-MS/MS) using modifications of previously described methods (37). Samples were dried down in a stream of argon, and the residue was dissolved in 100 µl of chloroform-methanol (2:1) and 60 µl injected onto a 3.9 × 300 mm Waters µPorasil column (10 µm particle size). The components were separated by gradient elution: 100% solvent A for 1 min, 100% A to 100% solvent B over 59 min, hold at 100%B for 10 min, and then regenerating the column at 100% A for 20 min. Solvent A was composed of hexane-isopropanol-water-ammonium hydroxide (150:200:5:2, v/v), and solvent B contained hexane-isopropanol-water-ammonium hydroxide (150:200:35:2, v/v). A splitting tee directed 7% of the column effluent to the electrospray source of a Finnigan TSQ Quantum Discovery Max triple-quadrupole mass spectrometer for monitoring the elution of nonpolar lipids, glycerophospholipid (GP) classes (38), SM, and hexosylceramides (HexCer). The remaining 95% of the column effluent was collected for quantitative analysis of the individual lipid fractions. The amount of phosphorus in the GP and SM classes was quantified using the method of Rouser (39). HexCer and triglyceride fractions were collected and analyzed as described below. The following GP classes were detected as [M+H]+ ions by scanning in positive-ion mode: PE by the common neutral loss of 141 Da at 25 eV MSCE; PS by the common neutral loss of 185 Da at 22 eV MSCE; and choline-containing lipids, PC, lyso-glycerophosphocholine (LPC), and sphingomyelin (SM), as precursors of m/z 184.1 at 35 eV MSCE. Scanning in negative-ion mode for [M-H]− ions gave glycerophosphoinositol (PI) as precursors of m/z 241 at 48 eV MSCE and PA/PG as precursors of m/z 153 at 40 eV MSCE. Data were collected throughout the LC run using the Scan Events function of the Xcalibur™ 2.0 instrument control software (ThermoFisher). The spray potential was 3400 V in positive-ion mode and −2800 V in negative-ion mode. The nitrogen sheath gas pressure was 10 psi. Argon at 0.8 mTorr was used as the collision gas. The ion transfer capillary temperature was 270°C. Molecular species profiles for each GP class were obtained by averaging all the spectra acquired in the scan filter of interest during the elution time of the peak of interest. Molecular species were reported as mole-percent (mol%) of total lipid within each class after isotope correction and mass-dependent throughput correction.
Sphingomyelin FA distribution
Acyl composition was investigated by treating the SM fractions eluted from the LC column with methanolic-BF3 (Sigma-Aldrich) and then analyzing the methyl esters by GC/MS (40) using a Finnigan Trace 2000 mass spectrometer. Heptadecanoic (Nu-Chek Prep) acid was added as the internal standard.
Hexosylceramide analysis
The hexosylceramide-containing HPLC fraction was evaporated under a stream of nitrogen, redissolved in 100 µl of 100:2 chloroform-acetic acid, and then analyzed by LC-MS/MS using a modification of the method of Farwanah et al. (41). A 1 × 150 mm Phenomenex Luna Silica2 column containing 3 µm-diameter particles was eluted at a flow rate of 65 µl/min using the following gradient: 100% Solvent A (chloroform-acetic acid, 100:2) for 1 min switching to 100% Solvent B (chloroform-isopropanol-acetic acid, 80:20:2) over 0.1 min held for 5 min, followed by a step to 100% Solvent C (isopropanol-methanol-water-acetic acid, 50:50:25:2.5) over 0.1 min, held for 12 min, step to 100% Solvent B over 0.1 min, held for 20 min, followed by column regeneration for 20 min with 95% Solvent A plus 5% hexane. The spray potential was 3700 V, and data were collected in the positive ion mode. Sheath gas pressure was 25, and the ion transfer capillary temperature was 270°C. Hexosyl-ceramides were detected by scanning for precursors of m/z 264.2 using argon as the collision gas at 0.8 mTorr and MSCE = 27 eV. d3-Glucosylceramide (Matreya, LLC) was added as the internal standard.
Triglyceride analysis
Approximately 100 pmol of trilinolein (Hormel Institute) was added to each triglyceride fraction before it was evaporated with a nitrogen stream. The residue was dissolved in 400 µl of 1:1 chloroform-methanol containing 1 ng/µl of sodium formate. Each sample was infused at 5 µl/min with positive ion mass spectra acquired from m/z 700 to 1,200 by LC-MS/MS. Triglycerides were quantified by comparison of the sodiated triglyceride peak intensities to that of the sodiated internal standard. The spray potential was 3700 V with a capillary offset voltage of −35V. The sweep gas was on, but no sheath gas was used. The transfer capillary temperature was set to 270°C. A scan time of 0.5 s was used with spectral accumulation of 1 min.
Electron microscopy
For negative staining of HDL particles, a formvar-coated, 300-mesh, copper grid was floated on top of a 40 μl drop of sample for 30 s. The grid was then removed, and excess fluid was gently wicked away using the edge of a piece of filter paper, carefully avoiding completely drying out the sample. The grid, with adsorbed particles, was then floated (sample side down) on a 40 µl drop of 2% aqueous phosphotungstic acid (pH 5.1) for 30 s. The grid was removed, and excess fluid was wicked away until the sample was thoroughly dried. Negative stained preparations were viewed at 80 KeV using an FEI CM-12 transmission electron microscope. To quantify particle diameters, more than 200 particles were arbitrarily chosen by placing a grid of lines over a series of micrographs. Particles transected by a line were measured. The diameter was taken as the length along the longest axis of the identified particle. An average of 10 particles were measured from each field, and three separate samples were investigated for each condition.
Detergent-free membrane isolation
Subcellular fractions were purified from HEK cells and BMDMs using the method of Smart et al. (42). Briefly, a plasma membrane fraction was prepared from 16 T-75 flasks of confluent ABCA1-expressing HEK cells and 40 to 150 mm dishes of confluent BMDMs. To remove HEK cells, the flasks were trypsinized, and the cells pooled into two 50 ml Falcon tubes. From this point, all steps were carried out at 4°C. The cells were washed once with 30 ml PBS, followed by two washes with 30 ml Buffer A (0.25 M sucrose, 1 mM EDTA, and 20 mM Tricine [pH 7.8]). The cells were then resuspended in 1 ml Buffer A containing cOmplete Ultra, Mini, EDTA-free protease inhibitor cocktail according to the manufacturers recommendations (Roche Applied Science) and transferred to 2 ml Eppendorf microcentrifuge tubes. In the case of the BMDMs, the cell monolayer was washed once with 5 ml PBS and then twice with 5 ml Buffer A. The cells were collected by scraping using 1 ml Buffer A containing protease inhibitor cocktail, centrifuged at 1,000 rpm for 10 min, and then resuspended in 1 ml Buffer A before being transferred to 2 ml Eppendorf microcentrifuge tubes. Both cell types underwent the same steps after this point. Cells were homogenized by passing through a 21G needle at least 20 times and then centrifuged at 5,000 rpm for 10 min. Postnuclear supernate was removed and placed on ice. Cell pellets were resuspended in 1 ml Buffer A with protease inhibitors, homogenized as before, and centrifuged again. Postnuclear supernate (2 ml) fractions were combined and layered on top of 30% Percoll (Sigma-Aldrich) in Buffer A and centrifuged for 30 min at 84,000 g in a Beckman Ti60 rotor. The plasma membrane fraction, a visibly cloudy band ∼5.7 cm from the bottom of the centrifuge tube, was collected using a Pasteur pipette. Its volume adjusted to 2 ml with Buffer D (0.25 M sucrose, 1 mM EDTA, 20 mM Tricine [pH 7.6]). It was sonicated twice for 6 s at 50J and then placed on ice for 2 min. This step was repeated two more times for a total of six sonication steps. The sonicate was mixed with 1.84 ml Buffer C (50% OptiPrep (Sigma-Aldrich) in 0.25 M sucrose, 1 mM EDTA, 20 mM Tricine [pH 7.8]) and 0.16 ml Buffer D, bringing its final OptiPrep concentration to 23% before centrifugation. The sonicated plasma membrane was transferred to a 15 ml centrifuge tube, 9 ml of a 20% to 10% linear OptiPrep gradient was overlayered on the sample, and the sample was centrifuged for 90 min at 52,000 g in a Beckman SW40 rotor. In preliminary studies, 1 ml samples were collected, and aliquots were analyzed for transferrin receptor (non-raft) and flotillin-1 (raft) by Western blotting showing that the raft fraction was the top 5 ml of the gradient. The top 5 ml of the gradient was transferred to a new 15 ml centrifuge tube and mixed with 4 ml Buffer C. The remaining 8 ml was saved in 1 ml fractions as “non-raft” material. The top 5 ml was overlaid with 1 ml 15% OptiPrep, followed by 0.5 ml 5% OptiPrep (both prepared by mixing Buffer C with Buffer D) then centrifuged for 90 min at 52,000 g in a Beckman SW40 rotor. A visibly cloudy band was present at the interface between the 15% and 5% OptiPrep overlays. This band was collected as the lipid raft fraction.
Cholesterol oxidase assay
A total of 200 mU of recombinant cholesterol oxidase from Streptomyces sp. (Sigma-Aldrich) was used for each analysis. Analyses were conducted in tubes containing 6 µg of FC from mouse HDL, nHDL Peak 1 (12–14 nm diameter), nHDL Peak 2 (9–11 nm diameter), or Peak 3 (7–8 nm diameter) in 1 ml of PBS. For the zero time point, 250 µl were withdrawn. Samples were incubated at 37°C, and cholesterol oxidase was added to give 200 mU/ml. Aliquots (250 µl) were withdrawn at intervals, quenched with 250 µl of chloroform-methanol 2:1 (v:v), and extracted as described above. Samples were stored at −20°C. FC was quantified by GC/MS/MS as described above.
Statistical analyses
Results were compared statistically using the Student's t-test in Prism™ 4 (GraphPad Software). Two-tailed P values < 0.05 were considered statistically significant.
RESULTS
Nascent HDL particle isolation and characterization
A purification strategy was investigated for nHDL particles formed from the incubation of lipid-free 125I-apoA-I and ABCA1-expressing HEK cells in serum-free media for 18–24 h. After incubation with cells, the medium was fractionated using FPLC and the 125I-apoA-I nHDL distribution profile shown in Fig. 1A (filled circles) as percent of total radioactivity. Also shown is the elution profile of 3H-cholesterol contained in nHDL (Fig. 1A, open circles) from ABCA1-expressing cells preloaded with 3H-cholesterol and incubated with unlabeled apoA-I. The majority of the labeled cholesterol was in the first three peaks with very little, if any, in Peak 4, closely following the radiolabeled apoA-I profile. In addition, control cells (HEK 293 Flp-In™) were treated with radiolabeled apoA-I as described for ABCA1-expressing cells. In control cells, the bulk of the radiolabeled apoA-I eluted (Fig. 1A, filled squares) in Peak 4, which corresponds to lipid-free apoA-I. As expected, there was no radiolabeled cholesterol associated with any FPLC peak from control cells (Fig. 1A, open squares). In control cell incubations, a small peak of radiolabeled apoA-I eluted in Peak 3 (Fig. 1A, filled squares). This small peak was subjected to cross-linking followed by SDS-PAGE and was found to represent a small amount of dimeric apoA-I, containing no detectable lipids, and was present in the starting radiolabeled apoA-I preparation (data not shown) (23). Further characterization by 4–30% NDGGE using standards of known Stokes diameter (Fig. 1A, inset) showed particles with diameters corresponding to specific ranges: Peak 1, 12–14 nm, which is a shoulder of Peak 2, 9–11 nm; Peak 3, 7–8 nm; and Peak 4, <6 nm as detected by 125I-apoA-I (Fig. 1A, inset).
Fig. 1.
Nascent HDL particle diameter distribution. A: Distribution of nHDL particles from cells incubated for 18–24 h with 10 µg/ml of lipid-free human 125I-apoA-I in serumfree media. After incubation, the medium was dialyzed and concentrated, and nHDL particles were purified based on size using FPLC as described in Materials and Methods. The radioactive content of each fraction was determined and plotted as the percentage of total 125I-apoA-I (filled circles) per fraction. As a control, HEK cells not expressing ABCA1 (Flp-In™) were incubated in the same manner as ABCA1-expressing cells and plotted as the percentage of total 125I-apoA-I (filled squares) per fraction. In other experiments, cells were preincubated with 1 µCi/ml of 3H-cholesterol overnight and incubated with 10 µg/ml unlabeled lipid-free apoA-I. The radioactive content of each fraction was determined and plotted as the percentage of total 3H-cholesterol from ABCA1-expressing cells (open circles) or nonABCA1-expressing cells (open squares) per fraction. Aliquots of peak fractions (Fx) were taken and separated using 4–30% NDGE and are shown in the inset. Radioactive protein bands were visualized on the gel using phosphorimaging. From these experiments, Peaks 1–4 were defined: Peak 1 = fractions 110–125, Peak 2 = fractions 126–139, Peak 3 = fractions 140–157, and Peak 4 = fractions 158–175. B: Analysis of nHDL from ABCA1-expressing cells. In these experiments, ABCA1-expressing cells were incubated with 10 µg/ml of unlabeled apoA-I for 24 h as outlined above and in Materials and Methods. Fractions were pooled as described in A, and aliquots containing 10 µg of apoA-I mass were analyzed by 4–30% NDGE using Coomassie Blue G for visualization: Lane 1, pooled Peak 1 nHDL; Lane 2, pooled Peak 2 nHDL; Lane 3, pooled Peak 3 nHDL; Lane 4, pooled Peak 4 nHDL. Lane MW shows the Stoke's diameter for high molecular weight standards. C: To determine the number of apoA-I molecules per particle, aliquots of pooled nHDL fractions were subjected to cross-linking using a 100:1 molar ratio of DSP to apoA-I. Cross-linked nHDL were separated using nonreducing 4–12% SDS-PAGE. The lanes are as follows: ApoA-I Ladder; Pk1 = cross-linked Peak 1 nHDL; Pk2 = cross-linked Peak 2 nHDL; Pk3 = cross-linked Peak 3 nHDL; Dimer Mix = unreduced mixture of Q109C apoA-I homodimer and A176C apoA-I homodimer (24). These figures represent typical analyses from four independent experiments.
To address concerns related to potential artifacts associated with nHDL production using our ABCA1 expressing cell system beyond that demonstrated using control cells (HEK 293 Flp-In™), we measured nHDL size and composition when radiolabeled apoA-I mutant proteins with known defects in conformational adaptation to lipid binding were used (supplementary Fig. I). Supplementary Figure I shows that when dimeric radiolabeled apoA-I R173C (28) (supplementary Fig. IB, B′) or radiolabeled monomeric apoA-I L159R (43) (supplementary Fig. IC, C′) were incubated with ABCA1-expressing cells, lesser amounts of radiolabeled nHDL particles were produced (supplementary Fig. IB, C) compared with radiolabeled wild-type apoA-I (supplementary Fig. IA). Note the ∼3-fold axis scale difference in supplementary Fig. IA versus IB and IC. Molecular diameters, as shown in insets to supplementary Fig. IA–C, were determined by NDGGE of FPLC fractions after comparison to standards of known Stokes diameter. Furthermore, when unlabeled mutant apoA-I proteins were incubated with cells preloaded with 3H-cholesterol, supplementary Fig. IA′–C′ show that, for the same incubation time, ABCA1 transferred significantly less cholesterol to apoA-I R173C or apoA-I L159R than it did to wild-type apoA-I, as would be expected based on studies of these mutant proteins in human plasma (43, 44). In support of these results, previous studies showed that the amount of nHDL expressed as total nmol of glycerolphosphatidylcholine, total cholesterol, and apoA-I were significantly reduced when mutant apoA-I was used for nHDL assembly compared with nHDL formed using wild-type apoA-I (43). These results confirm the trends observed with radioactive tracer studies and clearly show that nHDL particle biogenesis by ABCA1-overexpressing HEK cells reflects properties associated with apoA-I conformation and lipid cargo assembly. Using information derived from radioactive tracer studies, we scaled-up this system to obtain mg amounts of nHDL particles without the use of radiolabeled apoA-I. Finally, to address concerns that HEK cell-derived nHDL particles do not constitute a valid model of physiologically derived nHDL particles, we prepared nHDL from mouse BMDMs and purified these nHDL using the same procedures used with our HEK cell system. As previously demonstrated, nHDL particles from BMDMs show an nHDL particle diameter distribution that was very similar to that obtained by incubating apoA-I with ABCA1-overexpressing HEK cells and numerous other cell lines (16).
ApoA-I Content of nHDL Particles
To begin our analyses of nascent HDL composition, large-scale experiments were initiated using HEK cells. Fig. 1B shows the particle diameter distribution of nHDL fractions from Peaks 1–4 after FPLC. Approximately 10 µg of apoA-I were run per lane on a 4–30% NDGGE and then stained with Coomassie Blue. We next subjected nHDL particles to cross-linking analysis to determine the number of apoA-I molecules per particle for each of the peak fractions, and the results are shown in Fig. 1C. Aliquots were cross-linked with 100:1 molar ratio of dithiobis(succinimidylpropionate) (DSP) and then separated on a nonreducing 4–12% SDS-PAGE and stained with Simply Blue™. The first lane in Fig. 1C shows an apoA-I ladder generated by cross-linking lipid-free apoA-I at high concentration. Lanes labeled Pk1 and Pk2 in Fig. 1C show that trimers of apoA-I were obtained from cross-linking nHDL with diameters of 12–14 nm and 9–11 nm (Peaks 1 and 2). The lane labeled Pk3 shows an apoA-I doublet demonstrating that 7.5 nm nHDL particles (Peak 3) carry two molecules of apoA-I. The doublet of dimeric apoA-I shows similar electrophoretic migration as the unreduced mixture of Q109C apoA-I homodimer and A176C apoA-I homodimer (24). Previous studies have demonstrated that dimers of apoA-I having similar sizes display different or aberrant electrophoretic mobility, which was found to depend on the location of the cross-link (24). When an aliquot of <6 nm nHDL was cross-linked, only monomer was detected (data not shown).
Electron microscopy studies of nHDL particles
Nascent HDL shape and size were further evaluated by negative stain electron microscopy (EM). Fig. 2A–D show representative images of nHDL from each of the peak fractions, and Fig. 2A′–D′ show the corresponding histograms of particle sizes for Peaks 1–4. A large fraction of particles appeared as round en face with a width to length ratio of >0.87, suggesting a spheroid. To confirm that particles seen by negative stain EM were indeed spheroidal, samples were tilted 45 degrees on either side of the zero tilt, and the change in particle dimensions was recorded. Analysis was performed on particles that did not show interference from surrounding particles. If the round samples represent discoidal particles, the tilt should dramatically change the length to width ratio with a distinct shortening along the dimension parallel to the tilt axis and no change in the dimension perpendicular to the tilt axis. On the other hand, if the particles are spheroidal, there should be little or no change in either dimension with tilting. We found that tilting made less than 2% change in the width to length ratio, suggesting that the round particles viewed by EM were indeed spherical in shape.
Fig. 2.
Electron microscopy evaluation of nascent HDL. Aliquots of Peaks 1–4 prepared and purified as described in Materials and Methods were adjusted to 0.1 µg/ml of protein and evaluated by electron microscopy. A–D: Representative images of nHDL. A′–D′: The corresponding histogram distribution of particle diameters in nm. A and A′, Peak 1 nHDL; B and B′, Peak 2 nHDL; C and C′, Peak 3 nHDL; D and D′, Peak 4 nHDL. Results shown are representative of data obtained from the analysis of particles from three independent experiments.
We also analyzed the changes in individual particles that had a length to width ratio greater than 2. If these were discoidal particles viewed on end, then tilting when the particles were oriented along the axis of tilt should produce an increase in width and thus lessen the length to width ratio. However, we found that tilting the sample along the long axis did not make much change in the length to width ratio, but if we tilted perpendicular to the long axis of the particle there was a shortening of the particle length. This suggests that the elongated particles were not true discs but were egg shaped to elongated egg shaped (i.e., no flat side). In contrast to the single elongate particles, tilting rouleaux produced changes in the width-length ratio that would be expected of stacks of discoidal particles. Thus, overall, most of the particles were spheroidal rather than oval. Using the above criteria, we find that ∼12–14 nm nHDL contained mostly spheroidal particles and a few isolated, single discoidal particles but no rouleaux. The ∼9–11 nm nHDL contained many more spherical particles than discoidal particles in an 8:1 ratio. Particles in 7.5 nm nHDL were predominantly spherical with a few discoidal particles and rouleaux, and <6 nm nHDL particles appeared to be spheroidal with no appreciable rouleaux formation.
Major nHDL particle lipids
To investigate the types and amounts of lipids within nHDL particles, lipids were extracted using the procedure of Bligh and Dyer (36) and then analyzed by mass spectrometry. Table 1 shows the mol% lipid composition while Table 2 shows the per particle molar lipid composition for nHDL Peaks 1 through 4. Experiments using Flp-In (non-ABCA1-expressing cells) were carried out in parallel, and only traces of lipid were detected in the volume where Peaks 1–4 would have eluted. However, the small amount detected was subtracted from data obtained from ABCA1-expressing cells (Tables 1 and 2). No microparticles of material eluting in the void volume during FPLC separation were found in any preparations of nHDL. Supplementary Table I shows the data as molar lipid composition per apoA-I. The data demonstrate that the total amount of lipid and the molar ratio of lipid to apoA-I decreased as the diameter of the nHDL particle decreased. For example, the total number of lipids per particle went from ∼240 lipid molecules for the largest nHDL (Peak 1 and Peak 2 nHDL were similar in composition) to 26 lipid molecules in 7.5 nm nHDL but only four lipid molecules in <6 nm nHDL. These data show that by far the most abundant lipids in the all the nHDL particles are PC, FC, and SM.
TABLE 1.
Mole Percent Lipid Composition of Nascent HDL Particle
| Peak ID, FPLC Fraction # | Approx. Diameter* | Total GP | PC | LPC | PS | PE | PI | PG | SM | FC | CE | GC | TG |
| nm | mol% | ||||||||||||
| Peak 1 (#97–116) | ∼12 | 39.1a ± 0.6 | 34.9a ± 2.2 | 1.1a ± 0.4 | 0.8a ± 0.5 | 1.3a ± 0.7 | 1.0a ± 0.7 | nd | 10.3a ± 1.9 | 45.7a ± 1.6 | 3.0a ± 0.6 | 1.8a ± 0.8 | 0.3a ± 0.1 |
| Peak 2 (#117–131) | ∼10 | 44.1a ± 1.5 | 39.5a ± 2.5 | 1.5a ± 1.0 | 0.9a ± 0.5 | 0.6a ± 0.7 | 1.6a,b ± 0.3 | nd | 10.2a ± 0.1 | 40.7a ± 2.5 | 2.5a ± 0.7 | 2.4a ± 0.6 | 0.2a ± 0.1 |
| Peak 3 (#132–146) | ∼7.5 | 55.9b ± 2.6 | 44.9b ± 1.8 | 2.1a ± 0.5 | 2.5b ± 0.9 | 2.7a ± 0.9 | 3.5a,b ± 2.7 | nd | 10.2a ± 0.2 | 29.4b ± 2.3 | 2.3a ± 0.7 | 2.3a ± 0.1 | 0.5a ± 0.1 |
| HEK cells | - | 59.6b ± 3.3 | 31.3a,c ± 3.0 | 1.6a ± 1.1 | 4.0d ± 0.3 | 16.9c ± 0.4 | 5.5b,c ± 1.1 | 0.8 ± 0.2 | 5.0b ± 0.6 | 20.5b ± 3.0 | 9.6b ± 1.0 | 0.8b ± 0.1 | 3.9a ± 0.3 |
Nascent HDL were analyzed for different lipid classes as described in Materials and Methods. Statistical differences in each column are indicated by different letters at P < 0.05.
* Particle size was calculated by comparison to standards of known diameter and by electron microscopy as described.
† ApoA-I was measured by ELISA, and the number of apoA-I molecules per particle was determined by cross-linking with DSP followed by separation using nonreducing 2–12% SDS-PAGE. Results shown are the mean ± SD of three independent experiments.
TABLE 2.
Per Particle Molecular Lipid Composition of Nascent HDL Particles
| Peak ID, FPLC Fraction # | Approx.Diameter* | ApoA-I† | (GP+SL) | TC | Total Lipid | PC | Other GP | SM | FC | CE | GC | TG |
| nm | per particle | molecules/particle | ||||||||||
| Peak 1 (#97–116) | ∼12 | 3 | 109 | 103 | 212 | 74 | 9 | 22 | 97 | 6 | 4 | 1 |
| Peak 2 (#117–131) | ∼10 | 3 | 150 | 115 | 266 | 105 | 12 | 27 | 108 | 7 | 6 | 1 |
| Peak 3 (#132–146) | ∼7.5 | 2 | 18 | 9 | 26 | 12 | 3 | 3 | 8 | 1 | 1 | 0 |
| Peak 4 (#147–170) | ∼6 | 1 | 3 | 1 | 4 | 1 | 2 | 0 | 1 | 0 | 0 | 0 |
Nascent HDL were analyzed for different lipid classes as described in Materials and Methods.
* Particle size was calculated by comparison to standards of known diameter and by electron microscopy as described.
† ApoA-I was measured by ELISA, and the number of apoA-I molecules per particle was determined by cross-linking with DSP followed by separation using nonreducing 2–12% SDS-PAGE. Results shown are the mean ± SD of three independent experiments.
Fig. 3A shows the mol% for the three principal classes of nHDL lipids, total cholesterol (TC), GP, and sphingolipid (SL) compared with total HEK cell membrane lipids. The mol% of total SLs was relatively constant for all nHDL. TC decreased steadily from 14 nm to <6 nm nHDL and was inversely related to the mol% of GP. The mol% of SLs in HEK cell membranes was about half that measured in nHDL Peaks 1–4, whereas the mol% of cholesterol was similar to that in <6 nm nHDL.
Fig. 3.
Distribution of major lipid components in nHDL. A: Total lipid (mol%) for each of the three major classes of lipids. Compositional analysis for FPLC Peaks 1–4 showed that the major lipids comprising nHDL particles were cholesterol (hatched columns), GP (open columns), and SM (filled columns). B: The nmole content of free (open columns) and CE (filled columns) for Peaks 1–4. C: the GP class distribution for FPLC Peaks 1–4 are expressed as nmol total lipid per particle. D: GP class distribution for FPLC Peaks 1–4 expressed as mol%. GP accounts for 40–70% of the total nHDL lipids. Analyses were carried out as described in Materials and Methods. For each class, the GP was separated by HPLC, and individual fractions collected were analyzed for lipid phosphorus. Values represent total cell lipid extract. Total cholesterol and FC were measured using GC/MS methodology; GP, SM, and GC were analyzed using LC/MS/MS. Values are from three or more independent experiments. Statistical differences are indicated by different letters at P < 0.05.
Fig. 3B shows that within the TC fraction approximately 90% was FC; however, there was a significant amount of cholesteryl ester (CE) in the 12–14 nm and 9–11 nm nHDL but only a trace in the 7.5 nm nHDL. To confirm that the particles contained CE, LC-MS/MS analysis of sodiated samples showed that the CE contained predominantly fatty acids (FAs) corresponding to 16:1, 16:0, 18:2, and 18:1, which comprised 16, 21, 23, and 26%, respectively, of the total (data not shown). The CE-FA acyl compositions were similar to those reported by Schifferer et al. (45) for fibroblast and macrophage nHDL. Because HEK cells do not express lecithin cholesterol acyl transferase (LCAT) but do express acyl-CoA cholesterol acyltransferase (ACAT) mRNA (S. Bhat, unpublished data), it seems likely that ACAT may be the source of the CE. The CE acyl composition in this study (16:0 ≈ 18:2 ≈ 18:1) was similar to that reported for ACAT-1 acyl chain preferences (16:0 ≈ 18:2 ≈ 18:1) (46) but different from that reported for LCAT (18:2 > 16:1 > 18:1 > 16:0) (47).
To assess the availability of FC in nHDL particles as compared with spherical plasma HDL, we treated nHDL with cholesterol oxidase and compared the results to purified mouse plasma HDL. The results are plotted as percent FC versus time in supplementary Fig. II. Overall, nHDL FC was oxidized slightly slower than plasma HDL at 2 min but then appeared to oxidize slightly faster by 10 min.
Fig. 3C and show the distribution of GP and SM for each of the peaks as total moles/particle and mol%, respectively. PC was the predominant GP in all nHDL, with no significant difference between Peaks 1, 2, and 3. The <6 nm nHDL showed a significant decrease in the mole fraction of PC and a significant increase in the percentages of LPC, glycerophosphoserine (PS), glycerophosphoethanolamine (PE), and PI. The composition of 7.5 nm nHDL was intermediate between that of 9–14 nm nHDL (Peaks 1 and 2) and the <6 nm nHDL. SM was the next most abundant phospholipid in nHDL, with diameters from 12 nm to 7.5 nm. All other GP classes were relatively minor components. HEK cell lipid distribution was different from that of nHDL (Fig. 3C, D). The predominant GP in total HEK cell lipids were PC and PE, followed by PI and PS, with very little LPC.
Lipid enrichment of nHDL particles
To investigate the source of nHDL lipids, we compared the plasma membrane (PM) and lipid raft composition from two different cell sources to that of nHDL particles obtained from its source. To do this, we used previously described nondetergent methods to isolate plasma membrane lipid rafts from ABCA1 overexpressing cells and mouse BMDMs, and nHDL particles were obtained as previously described. Fig. 4A shows the TC to PC content for PM and lipid rafts for both cells types when compared with that for purified ∼9–11 nm diameter nHDL particles (Peak 2). As expected, HEK and BMDM cell lipid rafts showed an approximately 2- to 3-fold higher TC/PC ratio compared with the PM (48, 49). The ∼9–11 nm diameter nHDL particles (Peak 2) from ABCA1-expressing HEK cells had a higher TC-PC ratio than the PM or lipid raft fractions. The same diameter particles from BMDM had a TC-PC ratio higher than the PM but slightly lower than the lipid raft fraction. The same trends were observed when SM was measured and compared with the PC content and are shown as the SM-PC ratio in Fig. 4B. To confirm fractionation, Fig. 4C shows the purity of the lipid raft and nonraft fractions as determined by Western blot analysis using antibodies to flotillin-1 and the transferrin receptor, respectively, as previously described (50).
Fig. 4.
Comparison of lipid raft composition and HEK293 cells and BMDM nHDL. A: The ratio of total cholesterol to PC for isolated plasma membrane, lipid rafts, and nHDL particles derived from HEK 293 cells overexpressing ABCA1 or the same fractions isolated from BMDMs. B: SM-PC ratio for the same fractions listed in A. Fractions from HEK cells or BMDM were obtained using the nondetergent procedure as described in Materials and Methods. C: Purity of the raft and nonraft fractions after Western blotting for Flotillin-1 and Transferrin receptor after 12% SDS-PAGE. Values are from two or three independent experiments for each cell type. FC and total cholesterol was measured using GC/MS methodology; PC, and SM were analyzed using LC/MS/MS.
Sphingomyelin and hexosylceramides
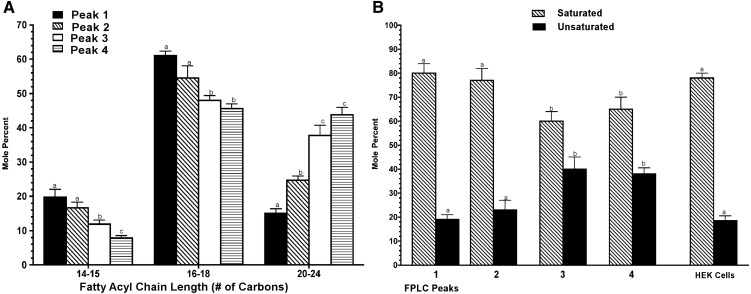
LC-MS/MS analysis for SM showed that there was not a uniform distribution of FA across the different nHDL diameters. Fig. 5A shows that higher-molecular-weight FA predominated in the smallest-diameter particles, whereas lower-molecular-weight species predominated in the larger-diameter nHDL. The sphingosine portion of all SM molecular species showed a singly charged ion at m/z = 264.27, suggesting that the sphingosine base for these species was d18:1Δ4t sphingosine (51). A higher degree of unsaturation was associated with an SM species that carried longer-chain FA (Fig. 5B), usually C22 and C24 FA having one or two double bonds.
Fig. 5.
Sphingomyelin fatty acyl chain length and saturation in nHDL. A: SM fatty acyl chain length shown as the total number of carbons for FPLC Peaks 1–4. B: Saturated and unsaturated fatty acyl chains (mol%) carried by PC for FPLC Peaks 1–4. All samples were analyzed by LC-MS/MS. Molar abundance for each SM chain length was calculated from the SM mass spectra after correction for isotope and the mass dependent instrument response. Results shown are the mean ± SD of four independent experiments. Within a specific class, statistically significant differences are indicated by different letters at P < 0.05.
To determine whether glucosylceramide (GCer) or galactosylceramide was the principal component, nHDL samples were doped with 16:0 GCer or 16:0 galactosylceramide (52). LC-MS/MS analysis showed that 16:0 GCer was the only detectable product, suggesting that nHDL HexCer were exclusively GCer. FA analysis of GCer showed that the 24 carbon FA 24:1 and 24:0 were the principal FAs in 7.5 nm to 12 nm nHDL, comprising about 39% of the total. The remaining FA were 16:0 at 32% and 22:0 at 23%, and 14:0, 18:0, and 20:0 were 6% of the total. The FA distribution was confirmed by GC/MS. For 12 nm to 7.5 nm nHDL, the total amount of GCer was about 5 ± 1% of the total GP and 17 ± 4% of the total ceramide. GCer from ABCA1-expressing HEK cells had FA distributions of 16:0, 18:0, 22:0, 24:1, and 24:0 with 28, 10, 23, 19, and 20%, respectively. The principal FA in nHDL GCer were similar to that reported for HEK cells by Lahiri et al. (53) and human plasma (54). However, the C18 ceramide content of nHDL was lower than in HEK cells.
Saturated FAs in GP
We further characterized the differences in acyl saturation between nHDL lipids and HEK cell lipids. Supplementary Table II shows the distribution of diacyl and ether-acyl GP (columns 2 and 3). The mol% values of saturated, mono-, di-, and polyunsaturated FA for diacyl and ether-acyl GP are given in columns 4–11. nHDL PC was enriched in saturated FA compared with HEK cells. The principal saturated FAs were 14:0 and 16:0, whereas the principle unsaturated FA was 18:1. The other GP classes—PE, PI, and PS—contained a much higher fraction of unsaturated FA compared with PC. Thus, these results show that nHDL particles have PC with ∼2-fold enrichment in saturated FA compared with total HEK cell lipids. All nHDL were analyzed for triglyceride (TG) content using mass spectrometric methods. Only small amounts of TG were detected (Tables 1 and 2).
Conformation of apoA-I on nHDL particles
nHDL particles were cross-linked at low cross-linker to protein ratios to minimize alterations in protein conformation due to over cross-linking. This material was separated using SDS-PAGE and subjected to in-gel digestion as previously described (24). Using four lysine-specific chemical cross-linkers, several intermolecular, interpeptide, and intrapeptide cross-links were identified in nHDL with diameters of 9–11 nm and 7.5 nm nHDL. The overall structures deduced from these studies are shown in Figs. 6 and 7 for the 7.5 nm and 11 nm nHDL, respectively. Cross-linked peptides that were used in these analyses are listed in supplementary Tables III and IV, along with the Cα-link-Cα distance for the proposed conformations. Intrapeptide connotes peptides that have cross-links in the same peptide (e.g., adjacent lysines were linked). Interpeptide means that the two cross-linked peptides were close together within a single apoA-I strand but that there was an intervening tryptic cut site. Intrapeptide and interpeptide cross-linked peptides were distinguished from intermolecular cross-linked peptides because they were found in the monomer fraction. An intermolecular cross-linked peptide is one that was only found in tryptic digests of dimeric or trimeric protein bands from SDS-PAGE and was not present in the digests of monomeric protein. Because separation of cross-linked apoA-I products is excellent using SDS-PAGE (Fig. 1C), there is little in the way of cross contamination. The ∼7.5 nm nHDL showed two cross-linked products, one running with the monomeric apoA-I standard and the second running at the same level as an apoA-I dimers. Given the low abundance of Peak 1 nHDL (12–14 nm nHDL), we only solved the structure for the 9–11 nm nHDL, which showed three cross-link products, one running as apoA-I monomer, a second as an apoA-I dimer (24), and a third that ran with trimeric apoA-I (Fig. 1C). Each band was excised from the gel and then subjected to in-gel tryptic digestion using previously reported procedures (24, 28). A total of 14 cross-linked peptides were identified for each particle based on predicted m/z with an error of ±0.03 Da and MS/MS sequencing.
Fig. 6.
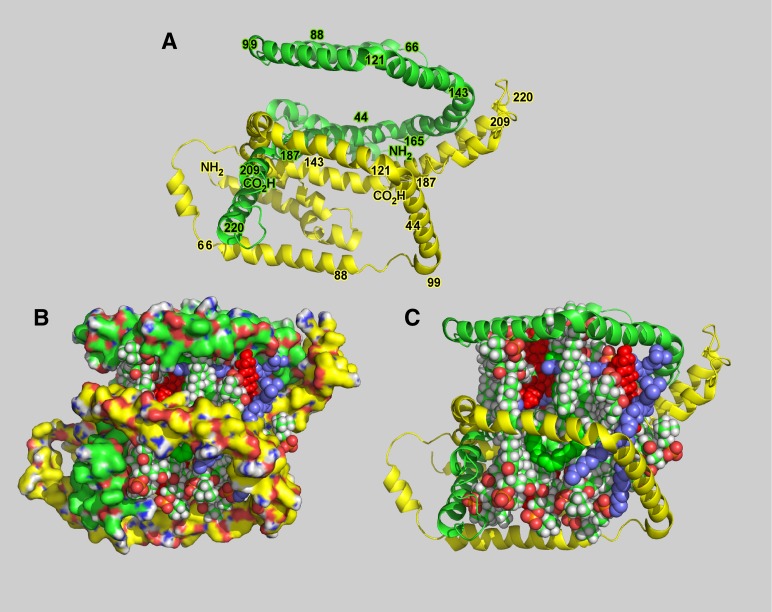
Conformation of two ApoA-I molecules on 7.5 nm nascent HDL particles. Molecular models of 7–8 nm nHDL particles (Peak 3) containing 26 molecules of total lipid (Table 2). The lipids include representative PC, SM, GCer, CE, and FC in the same ratio reported in Table 1. A: Two apoA-I molecules arranged in an antiparallel orientation. The regions of the two strands are indicated by the residue number. Note that the configuration of the protein is helical. B: The two strands shown in A, including 26 lipids. C: The particle in B with the apoA-I displayed as a space-filling surface. Glycerophospholipid is indicated by chains having a green backbone and white hydrogen atoms, ceramides are in blue, cholesteryl ester is in green, and free cholesterol is in solid red.
Fig. 7.
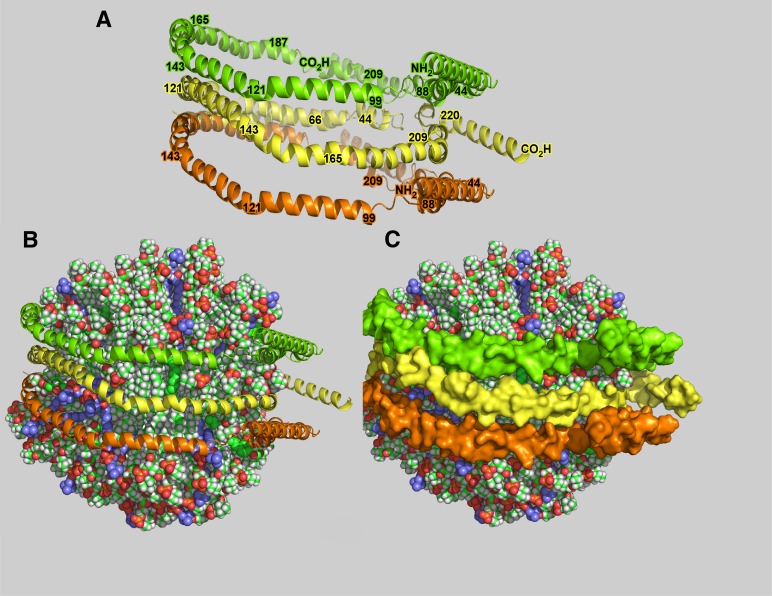
Conformation of three ApoA-I molecules on ∼9–11 nm nascent HDL particles. Molecular models of 9–11 nm diameter nHDL particles (Peak 2) containing 266 molecules of total lipid. The lipids include representative PC, SM, GCer, CE, and FC in the same ratio reported in Table 2. A: Three apoA-I strands in an antiparallel orientation. The two outer strands are in the same orientation but are antiparallel to the center strand. Regions of the three strands are indicated by residue numbers. Note that apoA-I assumes a belt configuration. B: The three strands shown in A wrapped around 239 lipids. C: B with the apoA-I displayed as a space-filling surface. Glycerophospholipid is indicated by chains having a green backbone and white hydrogen atoms, ceramides are in blue, CE in green, and FC in solid red.
Constraints on assessing apoA-I conformation were the cross-links and the size of the particle. Overall particle dimensions were obtained from EM and by comparison to molecular weight standards on NDGE. Particle volumes were calculated from the particle composition using volumes for each component reported in the literature. Particle diameters calculated from the volume suggested that the particles could be disk like or spherical. EM assessment showed that the particles were predominantly spheroidal. Fig. 6 shows the structure proposed for the 7.5 nm particle that carries two apoA-I molecules and about 26 lipids (Table 2). Size, volume, and compositional constraints suggest that these particles have a smaller radius than the 80 Å diameter rHDL particles (24). The intermolecular cross-links suggest that the two apoA-Is were twisted around one another in a helical fashion, with an antiparallel orientation of the two apoA-I monomers (55).
The lipid–rich, 9 to 11 nm diameter nHDL particle was also investigated using chemical cross-linking and mass spectrometry and is shown in Fig. 7. The figure shows the accommodation of 239 molecules of lipid, the average composition of Peak 1 and Peak 2, packaged with three apoA-I monomers. Particle diameter and particle volume suggest that apoA-I was a belt of antiparallel apoA-I molecules with overlap of the central regions (helix 5 amino acids 121–142), an orientation similar to that reported for 9.6 nm diameter rHDL (24). We have used an arrangement with the inner apoA-I molecule antiparallel to the two outer molecules. Structural features of several reports on apoA-I structure were incorporated into our analysis (34, 56–58).
DISCUSSION
nHDL lipids
We have characterized apoA-I containing nHDL particles purified from ABCA1-expressing HEK cells. The size distribution as well as the preβ mobility of these nHDL particles are similar to particles reported from numerous other cell lines and sources (12, 16, 23, 59, 60). There were modest differences among the reports regarding the amounts of each particle, and we find small differences in particle distribution and lipid composition between the different cell types used in this study. Small changes in particle distribution may reflect differences in apoA-I availability, ABCA1 activity, and lipid availability. Overall, our data support the concept that to maintain cellular membrane cholesterol levels, ABCA1 transports and displaces lipids to apoA-I, yielding thermodynamically stable lipid-raft-like protein complexes, which are rapidly remodeled once released into plasma. Our results show that the most abundant lipids in the nHDL were PC, FC, and SM, with smaller contributions from CE and GCer. The other classes of GP make up a small percentage of the total. This finding is different from other studies that reported a greater contribution of PE and PI to the nHDL particle (61), similar to what we found for the total cell extract (Fig. 4C, D). The reason for these differences is not clear, but the source of the nHDL particles does not appear to play a role because both mouse BMDM and HEK cells showed similar overall lipid compositions, with only acyl chain composition of the phospholipids being variable (B. Fulp, unpublished observations). There is some controversy in the literature regarding the overall composition of nHDL. We find that the percentage distributions for GP, FC, and SL were similar to several reports that used different cell expression systems (45, 59, 62–65) but very different from other reports (23, 61, 66). Work by Duong et al. (61, 66) was characterized by the formation of microparticles that were not detected in these studies, suggesting that Duong et al. were studying a mechanistically different process. The amount of PC per particle reported here was similar to that of Mulya et al. (23), but the FC concentration reported in this study was substantially greater.
nHDL cholesterol
A significant finding in this report was that ∼43% of the lipid packaged in large nHDL particles was FC. These results suggest that formation and release of nHDL is a highly efficient packaging mechanism primarily functioning as a means to finely tune cellular membrane cholesterol content, using apoA-I as an abundant and flexible means of organizing and solubilizing large amounts of cholesterol. However, the conformation of apoA-I is critical for cholesterol transfer and packaging because we have demonstrated that mutant forms of apoA-I associated with decreased HDL plasma concentrations (44) yield lower amounts of nHDL cholesterol (43) compared with those packaged with wild-type apoA-I (supplementary Fig. I). The unusually high level of cholesterol enrichment contained in nHDL has previously been reported only for small plasma HDLs isolated from LCAT-deficient patients (67, 68). Because HEK cells do not express LCAT, remodeling of cholesterol to CE in nHDL particles would not be anticipated. CE in nHDL was consistent with the known expression of ACAT in these cells and the distribution of incorporated FAs.
A subfraction of human plasma HDL, 9.8 nm diameter HDL2α, that is reported to carry three or four molecules of apoA-I, had a total molar lipid content (69, 70) that was similar to that of the larger-diameter nHDL we report in this study. What is striking was the similarity in total cholesterol carried by plasma-derived HDL2α and the 9–11 nm nHDL reported here (49% and 43%, respectively). However, HDL2α is a mature particle carrying most of its cholesterol as CE and not FC (38% and 11% vs. 2.5% and 41%, respectively) (69).
Besides an abundance of FC in the 9–11 nm nHDL, they also carried about seven molecules of CE, unlike HEK cell PM and the lipid raft fraction, which contained no measurable CE (B. Fulp, unpublished observations). Small amounts of CE have previously been reported in fibroblast- and macrophage-derived nHDL (45). Because cholesteryl esters are much more hydrophobic than cholesterol, it is very likely sequestered in the core of nHDL particle and may constitute the beginnings of a hydrophobic core that yields spheroidal particles (71). Previous biophysical studies have demonstrated that CE was soluble in phospholipid up to about 3 mol%, whereas above that concentration there was phase separation (72). rHDL particles containing more than six CE have been routinely classified as spheroidal (73, 74), and small amounts of cholesteryl ester have been included in dynamic computer simulations to generate spheroidal particles (75). Our EM data (Fig. 3) was consistent with a spheroidal shape for 9–11 nm diameter ABCA1-derived nHDL particles.
Conformation of apoA-I on nHDL
To assess the conformation of apoA-I on nHDL, we used features that were common to other HDL particles that we previously reported. The intermolecular cross-link Lys118-Lys140 was common to ∼7.5 nm nHDL, ∼9–11 nm nHDL, and to 8.0 nm and 9.6 nm rHDL (24, 76, 77). Previous studies of rHDL suggested that helical regions 4 and 5 of each protein strand overlap with one another and that the nHDL cross-link Lys118-Lys140 was consistent with nHDL having the same protein-to-protein interaction found in rHDL, otherwise referred to as the “belt-buckle” (24, 76, 77). Other cross-links common to ∼9–11 nm nHDL and 9.6 nm rHDL include Lys40-Lys239 and Lys133-Lys140, suggesting similar conformational constraints for apoA-I on the two particles and an antiparallel arrangement for the two apoA-I molecules. However, 7.5 nm nHDL appears to be unique, having only the central region cross-link in common, suggesting a conformation that was substantially different from that of ∼9–11 nm nHDL, 8.0 nm rHDL, or 9.6 nm rHDL.
Our analysis of the cross-linking data suggests two very different conformations for apoA-I associated with particles of 7.5 nm diameter compared with that for particles of ∼9–11 nm diameter (Figs. 6 and 7, respectively). nHDL of 7.5 nm diameter were only slightly smaller than the 8.0 nm diameter rHDL particles made with POPC but had half of the total lipid molecules (26 vs. 54) (24). The 8.0 nm diameter rHDL were discs with the two apoA-I molecules encircling the periphery as an antiparallel belt. The N-terminal end was folded back onto the protein belt to a greater extent than reported for the 9.6 nm diameter discs (24). A recent report on the crystal structure of a truncated apoA-I, Δ(185-243), suggests that that the N-terminal region folds back onto the body, to about helical region 4, of the belt (34). Because the quantity of lipid in the 7.5 nm diameter nHDL was only half that of 8.0 nm diameter rHDL, the circumference of the belt was, by necessity, smaller to accommodate contact between apoA-I and lipid. However, to accommodate the smaller lipid volume, more of the protein folded over itself in a helix, yielding a particle that was more spherical than 8 nm diameter rHDL. Assuming tight bending of the apoA-I strands to accommodate lipid the intermolecular cross-links then tightly lock the protein conformation.
Previous studies of 9.6 nm diameter rHDL carrying 150 molecules of POPC suggested that two antiparallel apoA-I strands formed a belt around the periphery of a disc of PC, with N- and C-terminal ends folded back onto the belt of protein that covered the periphery of the lipid disc (24, 78). Because ∼9–11 nm diameter nHDL carried ∼266 lipids, 150 of which were GP and SM, we presumed that the apoA-I conformation and particle structure of nHDL would be significantly different compared with synthetic rHDL particles. Consistent with anticipated results, the particles were spherical, in contrast to the 9.6 nm diameter rHDL, but cross-link analysis suggested that the three apoA-I particles carrying apoA-I have a belt-like conformation similar to that of 9.6 nm rHDL.
Two recent studies have suggested conformational models for three apoA-I molecules on mature HDL, one described as a trefoil (69, 79) and the second a helical dimer with hairpin, HdHp (80). Silva et al. (79) showed that there was a similarity between the cross-linking pattern of a 9.6 nm diameter rHDL disc and a 9.3 nm diameter rHDL sphere; however, the first carried two molecules of apoA-I, whereas the second carried three apoA-Is. In a three-strand belt, interstrand ion pairs that stabilized the two-belt model could only be partially shared between the other two strands. This would suggest that the central belt has a conformation that is different from the other two strands. This can be more easily seen by assuming that each “belt” has two surfaces, L1 and L2. The only arrangements for antiparallel belts are then L1L1 or L2L2.
All but one of the cross-links listed in supplementary Table IV fit both the belt and the trefoil model (79); however, the intermolecular Lys94-Lys96 cross-link reported here can only be accommodated by a belt, which led us to propose a belt instead of a trefoil. A belt conformation for these nHDL particles does not preclude rearrangement of apoA-I into a trefoil conformation because the size and composition changes during maturation into CE-filled HDL, as occurs in plasma in the presence of LCAT. When the cross-links reported herein were superimposed on the helical dimer called the double superhelix (80), five of the nine cross-links could not be reasonably matched. It is possible that the hairpin conformation “floating” on the particle surface may allow for the unmatched cross-links, but that seems unlikely. The shape, but not the proportions, of the double superhelix seems to resemble the conformation of the smaller, 7.5 nm diameter nHDL particle (Fig. 6) rather than that of apoA-I on larger nHDL (Fig. 7).
Raft-like lipid composition and lipid packaging
Larger nHDL particles had a strikingly high TC-PC ratio, reminiscent of ratios normally found in lipid rafts rather than the PM (81). FC and SM are suggested to associate in rafts (82, 83), and studies have shown that high mole fractions of FC can be incorporated into bilayer and monolayer phospholipids (84, 85). Biophysical studies of miscible mixtures of PC and SM with FC suggested that there is no preferential affinity of cholesterol for PC or SM but that a phase separation must occur before preferential association of FC with SM (86). The FC content of lipid rafts range from about 24% to 47% of the total lipid or roughly twice the amount of FC that was in the PM (48, 49, 87). The 9–14 nm diameter nHDLs had a TC/PC ratio, 1.4, that was similar to the average TC/PC ratio, 1.3, of rafts from both HEK cells and BMDM. These TC/PC ratios were roughly twice that of the PM, 0.6. The TC-PC ratio for the smallest particle (0.65) was close to that of the PM.
Not only were FC levels greater in rafts compared with the PM, but raft membranes also have higher levels of SM (48, 87, 88) and contain a higher percentage of saturated FA (89). The 9–14 nm nHDL had an average SM-PC ratio of 0.32, similar to the average ratio of 0.35 for the lipid raft fraction but larger than the average ratio of 0.1 for the PM and carried more saturated FA. The similarity of the SM-PC and TC-PC ratios between nHDL and PM raft fractions and increased levels of saturated FA in larger nHDL suggest that the lipid pool used to populate large nHDL had a raft-like composition.
Lipidation by ABCA1 takes place at the surface of the cell (13). Lipid compositions measured from the smallest particle were consistent with the composition of the nonraft regions of the PM. Proportionally much more FC than SM were transferred to the larger particles, suggesting a lipid source from a raft-like region. Small, but significant, amounts of CE were also present in larger nHDL particles. In these cells, ACAT-1, localized in the rough ER (90), was the most likely source of CE, suggesting that recycling endosomes, the primary source of PM lipids, were involved in the transfer of CE from the ER to the PM. Because nHDL do not carry significant amounts of TG, lipid droplets were not considered to be a likely source of CE (91, 92).
Because previous studies of nHDL particles from ABCA1-overexpressing cells contained FC that was easily converted to CE by LCAT (93), the current studies examined the accessibility of FC at the nHDL surface by treating the particles with cholesterol oxidase. These studies showed that the FC was oxidized in a kinetically distinct manner but was similar to plasma HDL FC (supplementary Fig. II). Previous studies with cholesterol oxidase had shown that most of the FC in HDL and LDL (>85–95%) was susceptible to oxidation (94). The oxidation profile for these particles showed a biphasic oxidation profile, suggesting that there might be two separate regions in slow equilibrium contributing to FC oxidation.
Mechanism of lipidation
nHDL were generated as three relatively homogenous populations containing one, two, or three molecules of apoA-I. Lipid per apoA-I increased exponentially (Fig. 8A). For simplicity, the 10 nm and 12 nm diameter particles are averaged to ∼9–14 nm because their composition and number of apoA-Is per particle were similar. The total lipids per apoA-I monomer were ∼9–14 nm nHDL carrying three apoA-I molecules with about 80 lipid molecules per apoA-I, 7.5 nm nHDL carrying two molecules of apoA-I with 12 molecules of lipid per apoA-I, and <6 nm nHDL carrying a single apoA-I and four lipid molecules. Because isolated nHDL maintained their size on storage, particle fusion was not a major factor in particle assembly. Previous studies have shown that there was no precursor-product relationship between the smallest and largest particle (23); therefore, each particle appears to have been derived from a separate step in the lipidation process.
The change in protein conformation going from lipid-free apoA-I to lipidated apoA-I suggests that the opening of lipid-free apoA-I to accept lipid takes place in a concerted fashion that can be simplified to discrete steps, each of which entails sequential opening of helical pairs of the 4-helix bundle (Fig. 8B). Therefore, it is interesting to speculate on a mechanism that might explain the continuity of nHDL particle size and composition regardless of the cell type from which they are derived. There may be three points in the lipidation process at which further lipidation is hindered and nHDL was released from ABCA1. Given that the particles contain one, two, or three molecules of apoA-I, these steps appear to involve the sequential addition of apoA-I molecules during the lipidation process. If the next apoA-I is unavailable, if it was not properly opened, or if there is insufficient lipid, the nHDL particle is released from the ABCA1/apoA-I complex. This process also suggests the possible assistance of accessory protein(s) or chaperones that may aid in the assembly of three apoA-I molecules at the membrane surface as well as in opening of lipid-poor apoA-Is for assembly of the larger, more apoA-I lipid-rich particles. In plasma, the 3-apoA-I nHDL would be rapidly converted to a mature HDL in which cholesteryl ester would replace most of the free cholesterol, forming a core and transforming the particle to a true sphere.
Fig. 8.
Formation of nHDL Particles by ABCA1. Lipidation of nascent HDL particles by ABCA1. A: The large jump in total lipid content per apoA-I between the lipid rich 9–11 nm diameter nHDL and the lipid-poor 7.5 nm diameter nHDL. These data suggest that accessory or chaperone protein(s) may aid in coordination and opening of lipid-poor apoA-I for assembly of the 3-apoA-I lipid-rich particles. B: Mechanism for the ABCA1-mediated lipidation of apoA-I. apoA-I binds at the membrane surface, possibly to accessory protein(s), and then moves to ABCA1. These accessory protein(s) are probably involved in how the apoA-I /ABCA1 complex adds a second apoA-I or how lipid-poor apoA-I is released. ABCA1 is not located in lipid rafts (regions of high cholesterol and sphingomyelin content), but these regions participate as the source of lipids transferred to nHDL. When a second molecule of apoA-I binds, then the two apoA-I/ABCA1 complex can package small amounts of cholesterol and then be released as the 7.5 nm nHDL particle. However, to package maximal amounts of cholesterol, a third molecule of apoA-I must be present to adapt to the proper conformation and generate the ∼9–11 nm diameter nHDL, thus allowing ABCA1 to coordinate the removal of excess membrane cholesterol and maintain lipid raft integrity.
Low-temperature Triton X100 extracts do not contain active ABCA1. However, ABCA1 and mutants with a single amino acid mutation that prevented ATPase activity were found to colocalize in the plasma membrane (95), but further study indicated that the mutant slightly favored the raft region (21). Landry et al. (20) suggested that ABCA1 disrupts raft membrane microdomains present on the cell surface. Therefore, we assume that our studies, along with the work of others, suggest that 1) ABCA1 initially binds to raft regions of the plasma membrane but then remodels these regions by transferring lipids to apoA-I or 2) that ABCA1 in the plasma membrane associates with the lipid disordered phase adjacent islands of raft lipids and uses raft lipids to populate nHDL.
The previous hypothesis had suggested that nHDL was discoidal and probably contained two molecules of apoA-I per particle. Considerable effort was expended to develop methods to prepare discoidal particles of defined composition and size. The maximum cholesterol content reported in these synthetic particles never exceeded about 14% of total lipid (96). However, the smaller nHDL particle, having a diameter of 7.5 nm with two apoA-Is per particle, was spherical like the larger particles. A unique feature was that FC constituted 30% of the total lipid. Because none of the nHDL was discoidal, these results suggest that rHDL prepared by cholate dialysis methods do not duplicate the nHDL endpoint of lipids packaged through the action of ABCA1 on lipid-free apoA-I (97).
In conclusion, large, cholesterol-rich nHDL particles are spheroidal and contain three apoA-I molecules. Their lipid composition has been shown to be similar to that of the membrane lipid rafts, suggesting that the lipids transferred to apoA-I may be derived from raft-like regions of the PM. Because lipid rafts are regions of the plasma membrane that have key roles in receptor signaling and cell activation, these studies support the idea that ABCA1 functions to coordinate or maintain surface concentrations of cholesterol in membrane lipid rafts by apoA-I and therefore in forming nHDL particles. Overall, the process of nascent HDL formation represents a mechanism designed to preserve cellular membrane lipid-raft function by initiating the first step of a process known as reverse cholesterol transport.
Supplementary Material
Acknowledgments
The authors thank Dr. Michael Hayden for the use of the ABCA1 expressing HEK293 cell line.
Footnotes
Abbreviations:
- BMDM
- bone marrow-derived macrophage
- CE
- cholesteryl ester
- DSP
- dithiobis(succinimidylpropionate)
- EM
- electron microscopy
- FC
- free cholesterol
- FPLC
- fast protein liquid chromatography
- GP
- glycerophospholipids
- HEK
- human embryonic kidney
- HexCer
- hexosylceramides
- LPC
- lyso-glycerophosphocholine
- MSCE
- mass spectrometer collision energy
- NDGGE
- nondenaturing gradient gel electrophoresis
- nHDL
- nascent HDL
- PC
- glycerophosphocholine
- PE
- glycerophosphoethanolamine
- PG
- glycerophosphoglycerol
- PI
- glyerophosphoinositol
- PM
- plasma membrane
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PS
- glycerophosphoserine
- rHDL
- recombinant HDL
- RT
- room temperature
- SL
- sphingolipid
- TC
- total cholesterol
These studies were supported by grants from the National Institutes of Health grants HL-49373 and HL-64163 (M.S.T.) and HL-48148 (W.G.J.) and by American Heart Association grant 09GRNT2280053 (M.J.T.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. Electron microscopy was carried out in the Vanderbilt University Research Electron Microscopy Core of the Cell Imaging Resource. This resource is partially supported by National Institutes of Health grants CA68485, DK20593, and DK58404. Mass spectrometers used for these studies were acquired with funds from several agencies (MJT): TSQ Discovery Max LC-MS/MS and Advion Nanomate from the North Carolina Biotechnology Grant 2007-IDG-1021; TRACE MS from the North Carolina Biotechnology Grant 9903-IDG-1002 and the Winston-Salem Foundation; Finnigan TSQ Quantum XLS GC/MS/MS from NIH Shared Instrumentation Grant 1S10RR027940. The Waters Q-TOF mass spectrometer was from National Institutes of Health Shared Instrumentation Grant 1S10RR17846. MS analyses were performed in the Mass Spectrometer Facility of the Comprehensive Cancer Center of Wake Forest University School of Medicine, supported in part by NCI center grant 5P30CA12197.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and four tables.
REFERENCES
- 1.Castelli W. P., Doyle J. T., Gordon T., Hames C. G., Hjortland M. C., Hulley S. B., Kagan A., Zukel W. J. 1977. HDL cholesterol and other lipids in coronary heart disease. The Cooperative Lipoprotein Phenotyping Study. Circulation. 55: 767–772. [DOI] [PubMed] [Google Scholar]
- 2.Duffy D., Rader D. J. 2009. Update on strategies to increase HDL quantity and function. Nat Rev Cardiol. 6: 455–463. [DOI] [PubMed] [Google Scholar]
- 3.Francis G. A. 2010. The complexity of HDL. Biochim. Biophys. Acta. 1801: 1286–1293. [DOI] [PubMed] [Google Scholar]
- 4.Oram J. F., Heinecke J. W. 2005. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 85: 1343–1372. [DOI] [PubMed] [Google Scholar]
- 5.Timmins J. M., Lee J. Y., Boudyguina E., Kluckman K. D., Brunham L. R., Mulya A., Gebre A. K., Coutinho J. M., Colvin P. L., Smith T. L., et al. 2005. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Invest. 115: 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodzioch M., Orsó E., Klucken J., Langmann T., Böttcher A., Diederich W., Drobnik W., Barlage S., Büchler C., Porsch-Özcürümez M., et al. 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22: 347–351. [DOI] [PubMed] [Google Scholar]
- 7.Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., et al. 1999. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22: 336–345. [DOI] [PubMed] [Google Scholar]
- 8.Yvan-Charvet L., Wang N., Tall A. R. 2010. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30: 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm A. J., Zabalawi M., Grayson J. M., Weant A. E., Major A. S., Owen J., Bharadwaj M., Walzem R., Chan L., Oka K., et al. 2009. Apolipoprotein A-I and its role in lymphocyte cholesterol homeostasis and autoimmunity. Arterioscler. Thromb. Vasc. Biol. 29: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm A. J., Zabalawi M., Owen J. S., Shah D., Grayson J. M., Major A. S., Bhat S., Gibbs D. P., Jr., Thomas M. J., Sorci-Thomas M. G. 2010. Apolipoprotein A-I modulates regulatory T cells in autoimmune LDLr−/−, ApoA-I−/− mice. J. Biol. Chem. 285: 36158–36169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fessler M. B., Parks J. S. 2011. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J. Immunol. 187: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vedhachalam C., Duong P. T., Nickel M., Nguyen D., Dhanasekaran P., Saito H., Rothblat G. H., Lund-Katz S., Phillips M. C. 2007. Mechanism of ATP-binding cassette transporter AI (ABCA1)-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 282: 25123–25130. [DOI] [PubMed] [Google Scholar]
- 13.Denis M., Landry Y. D., Zha X. 2008. ATP-binding cassette A1-mediated lipidation of apolipoprotein A-I occurs at the plasma membrane and not in the endocytic compartments. J. Biol. Chem. 283: 16178–16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan H. H., Denis M., Lee D. Y., Iatan I., Nyholt D., Ruel I., Krimbou L., Genest J. 2007. Identification of an ABCA1-dependent phospholipid-rich plasma membrane apolipoprotein A-I binding site for nascent HDL formation: implications for current models of HDL biogenesis. J. Lipid Res. 48: 2428–2442. [DOI] [PubMed] [Google Scholar]
- 15.Vedhachalam C., Ghering A. B., Davidson W. S., Lund-Katz S., Rothblat G. H., Phillips M. C. 2007. ABCA1-induced cell surface binding sites for ApoA-I. Arterioscler. Thromb. Vasc. Biol. 27: 1603–1609. [DOI] [PubMed] [Google Scholar]
- 16.Krimbou L., Hassan H. H., Blain S., Rashid S., Denis M., Marcil M., Genest J. 2005. Biogenesis and speciation of nascent apoA-I-containing particles in various cell lines. J. Lipid Res. 46: 1668–1677. [DOI] [PubMed] [Google Scholar]
- 17.Mendez A. J., Lin G. R., Wade D. P., Lawn R. M., Oram J. F. 2001. Membrane lipid domains distinct from cholesterol/sphingomyelin-rich rafts are involved in the ABCA1-mediated lipid secretory pathway. J. Biol. Chem. 276: 3158–3166. [DOI] [PubMed] [Google Scholar]
- 18.Fielding P. E., Fielding C. J. 1995. Plasma membrane caveolae mediate the efflux of cellular free cholesterol. Biochemistry. 34: 14288–14292. [DOI] [PubMed] [Google Scholar]
- 19.Iatan I., Bailey D., Ruel I., Hafiane A., Campbell S., Krimbou L., Genest J. 2011. Membrane microdomains modulate oligomeric ABCA1 function: impact on apoAI-mediated lipid removal and phosphatidylcholine biosynthesis. J. Lipid Res. 52: 2043–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry Y. D., Denis M., Nandi S., Bell S., Vaughan A. M., Zha X. 2006. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J. Biol. Chem. 281: 36091–36101. [DOI] [PubMed] [Google Scholar]
- 21.Zarubica A., Plazzo A. P., Stockl M., Trombik T., Hamon Y., Muller P., Pomorski T., Herrmann A., Chimini G. 2009. Functional implications of the influence of ABCA1 on lipid microenvironment at the plasma membrane: a biophysical study. FASEB J. 23: 1775–1785. [DOI] [PubMed] [Google Scholar]
- 22.See R. H., Caday-Malcolm R. A., Singaraja R. R., Zhou S., Silverston A., Huber M. T., Moran J., James E. R., Janoo R., Savill J. M., et al. 2002. Protein kinase A site-specific phosphorylation regulates ATP-binding cassette A1 (ABCA1)-mediated phospholipid efflux. J. Biol. Chem. 277: 41835–41842. [DOI] [PubMed] [Google Scholar]
- 23.Mulya A., Lee J. Y., Gebre A. K., Thomas M. J., Colvin P. L., Parks J. S. 2007. Minimal lipidation of pre-beta HDL by ABCA1 results in reduced ability to interact with ABCA1. Arterioscler. Thromb. Vasc. Biol. 27: 1828–1836. [DOI] [PubMed] [Google Scholar]
- 24.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry O. J., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 26.Sorci-Thomas M. G., Parks J. S., Kearns M. W., Pate G. N., Zhang C., Thomas M. J. 1996. High level secretion of wild-type and mutant forms of human proapoA-I using baculovirus-mediated Sf-9 cell expression. J. Lipid Res. 37: 673–683. [PubMed] [Google Scholar]
- 27.Li H. H., Lyles D. S., Pan W., Alexander E., Thomas M. J., Sorci-Thomas M. G. 2002. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J. Biol. Chem. 277: 39093–39101. [DOI] [PubMed] [Google Scholar]
- 28.Bhat S., Sorci-Thomas M. G., Calabresi L., Samuel M. P., Thomas M. J. 2010. Conformation of dimeric apolipoprotein A-I milano on recombinant lipoprotein particles. Biochemistry. 49: 5213–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. 2007. Electron paramagnetic resonance spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apoA-I in solution. J. Biol. Chem. 282: 9143–9149. [DOI] [PubMed] [Google Scholar]
- 31.Okon M., Frank P. G., Marcel Y. L., Cushley R. J. 2002. Heteronuclear NMR studies of human serum apolipoprotein A-I. Part I. Secondary structure in lipid-mimetic solution. FEBS Lett. 517: 139–143. [DOI] [PubMed] [Google Scholar]
- 32.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166. [PubMed] [Google Scholar]
- 33.Silva R. A., Hilliard G. M., Fang J., Macha S., Davidson W. S. 2005. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 44: 2759–2769. [DOI] [PubMed] [Google Scholar]
- 34.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of HDL by dimerization. J. Biol. Chem. 286: 38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. 1996. VMD - Visual Molecular Dynamics. J. Mol. Graph. 14: 33–38. [DOI] [PubMed] [Google Scholar]
- 36.Bligh E. G., Dyer W. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 37.Van Kessel W. S., Hax W. M., Demel R. A., De Gier J. 1977. High performance liquid chromatographic separation and direct ultraviolet detection of phospholipids. Biochim. Biophys. Acta. 486: 524–530. [DOI] [PubMed] [Google Scholar]
- 38.DeLong C. J., Baker P. R., Samuel M., Cui Z., Thomas M. J. 2001. Molecular species composition of rat liver phospholipids by ESI-MS/MS: the effect of chromatography. J. Lipid Res. 42: 1959–1968. [PubMed] [Google Scholar]
- 39.Rouser G., Siakotos A. N., Fleischer S. 1966. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1: 85–86. [DOI] [PubMed] [Google Scholar]
- 40.Thomas M. J., Chen Q., Sorci-Thomas M. G., Rudel L. L. 2001. Isoprostane levels in lipids extracted from atherosclerotic arteries of nonhuman primates. Free Radic. Biol. Med. 30: 1337–1346. [DOI] [PubMed] [Google Scholar]
- 41.Farwanah H., Wirtz J., Kolter T., Raith K., Neubert R. H., Sandhoff K. 2009. Normal phase liquid chromatography coupled to quadrupole time of flight atmospheric pressure chemical ionization mass spectrometry for separation, detection and mass spectrometric profiling of neutral sphingolipids and cholesterol. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2976–2982. [DOI] [PubMed] [Google Scholar]
- 42.Smart E. J., Ying Y. S., Mineo C., Anderson R. G. 1995. A detergent-free method for purifying caveolae membrane from tissue culture cells. Proc. Natl. Acad. Sci. USA. 92: 10104–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorci-Thomas M. G., Zabalawi M., Bharadwaj M. S., Wilhelm A. J., Owen J. S., Asztalos B. F., Bhat S., Thomas M. J. 2012. Dysfunctional HDL containing L159R ApoA-I leads to exacerbation of atherosclerosis in hyperlipidemic mice. Biochim. Biophys. Acta. 1821: 502–512. [Google Scholar]
- 44.Sorci-Thomas M. G., Thomas M. J. 2002. The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc. Med. 12: 121–128. [DOI] [PubMed] [Google Scholar]
- 45.Schifferer R., Liebisch G., Bandulik S., Langmann T., Dada A., Schmitz G. 2007. ApoA-I induces a preferential efflux of monounsaturated phosphatidylcholine and medium chain sphingomyelin species from a cellular pool distinct from HDL(3) mediated phospholipid efflux. Biochim. Biophys. Acta. 1771: 853–863. [DOI] [PubMed] [Google Scholar]
- 46.Cases S., Novak S., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Welch C. B., Lusis A. J., Spencer T. A., Krause B. R., et al. 1998. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J. Biol. Chem. 273: 26755–26764. [DOI] [PubMed] [Google Scholar]
- 47.Sgoutas D. S. 1972. Fatty acid specificity of plasma phosphatidylcholine: cholesterol acyltransferase. Biochemistry. 11: 293–296. [DOI] [PubMed] [Google Scholar]
- 48.Pike L. J., Han X., Gross R. W. 2005. Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: a shotgun lipidomics study. J. Biol. Chem. 280: 26796–26804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown D. A., Rose J. K. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68: 533–544. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X., Owen J. S., Wilson M. D., Li H., Griffiths G. L., Thomas M. J., Hiltbold E. M., Fessler M. B., Parks J. S. 2010. Macrophage ABCA1 reduces MyD88-dependent Toll-like receptor trafficking to lipid rafts by reduction of lipid raft cholesterol. J. Lipid Res. 51: 3196–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu F. F., Turk J. 2000. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 11: 437–449. [DOI] [PubMed] [Google Scholar]
- 52.Zama K., Hayashi Y., Ito S., Hirabayashi Y., Inoue T., Ohno K., Okino N., Ito M. 2009. Simultaneous quantification of glucosylceramide and galactosylceramide by normal-phase HPLC using O-phtalaldehyde derivatives prepared with sphingolipid ceramide N-deacylase. Glycobiology. 19: 767–775. [DOI] [PubMed] [Google Scholar]
- 53.Lahiri S., Park H., Laviad E. L., Lu X., Bittman R., Futerman A. H. 2009. Ceramide synthesis is modulated by the sphingosine analog FTY720 via a mixture of uncompetitive and noncompetitive inhibition in an Acyl-CoA chain length-dependent manner. J. Biol. Chem. 284: 16090–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scherer M., Bottcher A., Schmitz G., Liebisch G. 2011. Sphingolipid profiling of human plasma and FPLC-separated lipoprotein fractions by hydrophilic interaction chromatography tandem mass spectrometry. Biochim. Biophys. Acta. 1811: 68–75. [DOI] [PubMed] [Google Scholar]
- 55.Klon A. E., Segrest J. P., Harvey S. C. 2002. Molecular dynamics simulations on discoidal HDL particles suggest a mechanism for rotation in the apo A-I belt model. J. Mol. Biol. 324: 703–721. [DOI] [PubMed] [Google Scholar]
- 56.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. 2007 doi: 10.1038/nsmb1284. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868. [Erratum. Nat Struct Mol Biol. 2015: 2330. ] [DOI] [PubMed] [Google Scholar]
- 57.Chetty P. S., Mayne L., Lund-Katz S., Stranz D., Englander S. W., Phillips M. C. 2009. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones M. K., Zhang L., Catte A., Li L., Oda M., Ren G., Segrest J. P. 2010. Assessment of the validity of the double super helix model for reconstituted high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem.285: 41161–41171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Bortnick A. E., Nickel M., Dhanasekaran P., Subbaiah P. V., Lund-Katz S., Rothblat G. H., Phillips M. C. 2003. Effects of apolipoprotein A-I on ATP-binding cassette transporter A1-mediated efflux of macrophage phospholipid and cholesterol: formation of nascent high density lipoprotein particles. J. Biol. Chem. 278: 42976–42984. [DOI] [PubMed] [Google Scholar]
- 60.Asztalos B., Zhang W., Roheim P. S., Wong L. 1997. Role of free apolipoprotein A-I in cholesterol efflux. Formation of pre-alpha-migrating high-density lipoprotein particles. Arterioscler. Thromb. Vasc. Biol. 17: 1630–1636. [DOI] [PubMed] [Google Scholar]
- 61.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. 2008. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindholm E. M., Bielicki J. K., Curtiss L. K., Rubin E. M., Forte T. M. 1998. Deletion of amino acids Glu146→Arg160 in human apolipoprotein A-I (ApoA-ISeattle) alters lecithin:cholesterol acyltransferase activity and recruitment of cell phospholipid. Biochemistry. 37: 4863–4868. [DOI] [PubMed] [Google Scholar]
- 63.Zhang W., Asztalos B., Roheim P. S., Wong L. 1998. Characterization of phospholipids in pre-alpha HDL: selective phospholipid efflux with apolipoprotein A-I. J. Lipid Res. 39: 1601–1607. [PubMed] [Google Scholar]
- 64.Forte T. M., Bielicki J. K., Goth-Goldstein R., Selmek J., McCall M. R. 1995. Recruitment of cell phospholipids and cholesterol by apolipoproteins A-II and A-I: formation of nascent apolipoprotein-specific HDL that differ in size, phospholipid composition, and reactivity with LCAT. J. Lipid Res. 36: 148–157. [PubMed] [Google Scholar]
- 65.Lee C. Y., Lesimple A., Larsen A., Mamer O., Genest J. 2005. ESI-MS quantitation of increased sphingomyelin in Niemann-Pick disease type B HDL. J. Lipid Res. 46: 1213–1228. [DOI] [PubMed] [Google Scholar]
- 66.Duong P. T., Collins H. L., Nickel M., Lund-Katz S., Rothblat G. H., Phillips M. C. 2006. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J. Lipid Res. 47: 832–843. [DOI] [PubMed] [Google Scholar]
- 67.Norum K. R., Glomset J. A., Nichols A. V., Forte T. 1971. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: physical and chemical studies of low and high density lipoproteins. J. Clin. Invest. 50: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell C. D., King W. C., Applegate K. R., Forte T., Glomset J. A., Norum K. R., Gjone E. 1980. Characterization of apolipoprotein E-rich high density lipoproteins in familial lecithin:cholesterol acyltransferase deficiency. J. Lipid Res. 21: 625–634. [PubMed] [Google Scholar]
- 69.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Souza J. A., Vindis C., Hansel B., Negre-Salvayre A., Therond P., Serrano C. V., Jr., Chantepie S., Salvayre R., Bruckert E., Chapman M. J., et al. 2008. Metabolic syndrome features small, apolipoprotein A-I-poor, triglyceride-rich HDL3 particles with defective anti-apoptotic activity. Atherosclerosis. 197: 84–94. [DOI] [PubMed] [Google Scholar]
- 71.Peters-Libeu C. A., Newhouse Y., Hall S. C., Witkowska H. E., Weisgraber K. H. 2007. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J. Lipid Res. 48: 1035–1044. [DOI] [PubMed] [Google Scholar]
- 72.Hamilton J. A., Miller K. W., Small D. M. 1983. Solubilization of triolein and cholesteryl oleate in egg phosphatidylcholine vesicles. J. Biol. Chem. 258: 12821–12826. [PubMed] [Google Scholar]
- 73.Jonas A., Wald J. H., Toohill K. L., Krul E. S., Kezdy K. E. 1990. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J. Biol. Chem. 265: 22123–22129. [PubMed] [Google Scholar]
- 74.Sparks D. L., Lund-Katz S., Phillips M. C. 1992. The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 267: 25839–25847. [PubMed] [Google Scholar]
- 75.Catte A., Patterson J. C., Bashtovyy D., Jones M. K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemela P., et al. 2008. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 94: 2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas M. J., Bhat S., Sorci-Thomas M. G. 2008. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J. Lipid Res. 49: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. 2005. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280: 33015–33025. [DOI] [PubMed] [Google Scholar]
- 78.Li H., Lyles D. S., Thomas M. J., Pan W., Sorci-Thomas M. G. 2000. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J. Biol. Chem. 275: 37048–37054. [DOI] [PubMed] [Google Scholar]
- 79.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Z., Gogonea V., Lee X., May R. P., Pipich V., Wagner M. A., Undurti A., Tallant T. C., Baleanu-Gogonea C., Charlton F., et al. 2011. The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering. J. Biol. Chem. 286: 12495–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 82.Simons K., Ikonen E. 1997. Functional rafts in cell membranes. Nature. 387: 569–572. [DOI] [PubMed] [Google Scholar]
- 83.Rietveld A., Simons K. 1998. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta. 1376: 467–479. [DOI] [PubMed] [Google Scholar]
- 84.Huang J., Feigenson G. W. 1999. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys. J. 76: 2142–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Veatch S. L., Keller S. L. 2002. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 89: 268101. [DOI] [PubMed] [Google Scholar]
- 86.Calhoun W. I., Shipley G. G. 1979. Sphingomyelin–lecithin bilayers and their interaction with cholesterol. Biochemistry. 18: 1717–1722. [DOI] [PubMed] [Google Scholar]
- 87.Pike L. J., Han X., Chung K. N., Gross R. W. 2002. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 41: 2075–2088. [DOI] [PubMed] [Google Scholar]
- 88.Fridriksson E. K., Shipkova P. A., Sheets E. D., Holowka D., Baird B., McLafferty F. W. 1999. Quantitative analysis of phospholipids in functionally important membrane domains from RBL-2H3 mast cells using tandem high-resolution mass spectrometry. Biochemistry. 38: 8056–8063. [DOI] [PubMed] [Google Scholar]
- 89.Zech T., Ejsing C. S., Gaus K., de Wet B., Shevchenko A., Simons K., Harder T. 2009. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 28: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Underwood K. W., Jacobs N. L., Howley A., Liscum L. 1998. Evidence for a cholesterol transport pathway from lysosomes to endoplasmic reticulum that is independent of the plasma membrane. J. Biol. Chem. 273: 4266–4274. [DOI] [PubMed] [Google Scholar]
- 91.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277: 44507–44512. [DOI] [PubMed] [Google Scholar]
- 92.Atshaves B. P., Storey S. M., McIntosh A. L., Petrescu A. D., Lyuksyutova O. I., Greenberg A. S., Schroeder F. 2001. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J. Biol. Chem. 276: 25324–25335. [DOI] [PubMed] [Google Scholar]
- 93.Mulya A., Seo J., Brown A. L., Gebre A. K., Boudyguina E., Shelness G. S., Parks J. S. 2010. Apolipoprotein M expression increases the size of nascent pre beta HDL formed by ATP binding cassette transporter A1. J. Lipid Res. 51: 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Slotte J. P., Gronberg L. 1990. Oxidation of cholesterol in low density and high density lipoproteins by cholesterol oxidase. J. Lipid Res. 31: 2235–2242. [PubMed] [Google Scholar]
- 95.Hamon Y., Broccardo C., Chambenoit O., Luciani M. F., Toti F., Chaslin S., Freyssinet J. M., Devaux P. F., McNeish J., Marguet D., et al. 2000. ABC1 promotes engulfment of apoptotic cells and transbilayer redistribution of phosphatidylserine. Nat. Cell Biol. 2: 399–406. [DOI] [PubMed] [Google Scholar]
- 96.Sparks D. L., Frank P. G., Neville T. A. 1998. Effect of the surface lipid composition of reconstituted LPA-I on apolipoprotein A-I structure and lecithin: cholesterol acyltransferase activity. Biochim. Biophys. Acta. 1390: 160–172. [DOI] [PubMed] [Google Scholar]
- 97.Cavigiolio G., Shao B., Geier E. G., Ren G., Heinecke J. W., Oda M. N. 2008. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry. 47: 4770–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.