Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a secreted protein that promotes degradation of cell surface LDL receptors (LDLRs) in selected cell types. Here we used genetic and pharmacological inhibitors to define the pathways involved in PCSK9-mediated LDLR degradation. Inactivating mutations in autosomal recessive hypercholesterolemia (ARH), an endocytic adaptor, blocked PCSK9-mediated LDLR degradation in lymphocytes but not in fibroblasts. Thus, ARH is not specifically required for PCSK9-mediated LDLR degradation. Knockdown of clathrin heavy chain with siRNAs prevented LDLR degradation. In contrast, prevention of ubiquitination of the LDLR cytoplasmic tail, inhibition of proteasomal activity, or disruption of proteins required for lysosomal targeting via macroautophagy (autophagy related 5 and 7) or the endosomal sorting complex required for trafficking (ESCRT) pathway (hepatocyte growth factor-regulated Tyr-kinase substrate and tumor suppressor gene 101) failed to block PCSK9-mediated LDLR degradation. These findings are consistent with a model in which the LDLR-PCSK9 complex is internalized via clathrin-mediated endocytosis and then routed to lysosomes via a mechanism that does not require ubiquitination and is distinct from the autophagy and proteosomal degradation pathways. Finally, the PCSK9-LDLR complex appears not to be transported by the canonical ESCRT pathway.
Keywords: cholesterol, lysosomes, autophagy, endosomes, low density lipoprotein receptor
Elevated plasma levels of LDL-cholesterol (LDL-C) are a cardinal risk factor for coronary heart disease. LDL particles are formed in the circulation as a catabolic product of triglyceride-rich lipoprotein metabolism and are removed by LDL receptor (LDLR)-mediated endocytosis in the liver. Circulating levels of LDL are exquisitely sensitive to changes in LDLR activity (1). LDLR activity is controlled at the transcriptional level by feedback inhibition and at the post-translational level by targeted degradation through the action of proprotein convertase subtilisin/kexin type 9 (PCSK9) (2–5). The importance of PCSK9 in LDLR catabolism is illustrated by the impact of naturally occurring PCSK9 mutations on plasma levels of LDL-C (6). Gain-of-function mutations in PCSK9 are associated with 2- to 3-fold increases in plasma levels of LDL-C (6, 7), whereas PCSK9 deficiency results in very low plasma LDL-C levels (8).
PCSK9 is a 692-amino acid glycoprotein that contains a 22-residue signal sequence followed by a prodomain and a catalytic domain that shares structural homology with the proteinase K family of subtilisin-like serine proteases (9). The C-terminal portion of the protein comprises a cysteine- and histidine-rich domain. PCSK9 undergoes autocatalytic cleavage in the endoplasmic reticulum that severs the covalent attachment of the prodomain and the catalytic domain (9). Nonetheless, the prodomain remains tightly associated with the catalytic domain and shields the catalytic triad as PCSK9 moves through the secretory pathway. After secretion, PCSK9 binds the extracellular domain of the LDLR on the cell surface, triggering receptor degradation (10).
The mechanism by which PCSK9 binding to the LDLR targets the receptor for degradation is not understood. After LDL binds the LDLR on the cell surface, the LDL:LDLR complex is internalized via clathrin-coated pits and delivered to endosomes (11). In the low pH environment of the endosome, the LDLR undergoes a conformational change that promotes the release of bound LDL (12). The receptor is then recycled to the cell surface, whereas the LDL is delivered to lysosomes (1). Usually, each LDLR undergoes multiple rounds of internalization and recycling.
Direct binding of PCSK9 to the LDLR at the cell surface is essential for PCSK9-mediated degradation of the receptor (13). PCSK9 binds specifically to epidermal growth factor-like repeat (EGF)-A, the first of three EGF-like repeats in the EGF precursor domain of the LDLR (13, 14). In addition to the EGFA-like repeat, the β-propeller domain and at least three copies of the ligand binding repeats of the LDLR are required for PCSK9-mediated degradation of the receptor (13, 14). Unlike LDL, the binding affinity of PCSK9 for the LDLR increases with a reduction in pH (13, 15, 16). Although the C-terminal domain of PCSK9 is not required for LDLR binding, it is required for LDLR degradation (14). A recent study indicates that the domain can be replaced by a heterologous protein (dsRED) of comparable size and charge (17). The roles of the β-propeller domain and the ligand binding repeats of the LDLR and of the C-terminal domain of PCSK9 in LDLR degradation are not known. Recently, it was reported that the C-terminal domain of PCSK9 associates with the ligand-binding domain of the LDLR in a pH-dependent manner (18, 19).
The present study was undertaken to elucidate the pathways and proteins involved in PCSK9-mediated degradation of the LDLR. We used genetic and pharmacological inhibitors to block protein degradation by the proteasome, autophagy, and lysosome and traced the effect of these agents on the intracellular itinerary of the PCSK9-LDLR complex. Our data are consistent with a model in which PCSK9-bound LDLRs are internalized via clathrin-mediated endocytosis and routed to lysosomes via a pathway that does not require ubiquitination of the receptor or the major components of the endosomal sorting complex required for trafficking (ESCRT) pathway and does not involve the core components of the autophagocytic machinery.
MATERIALS AND METHODS
Materials
Cell culture medium and PBS were obtained from Meditech, Inc. (Herndon, VA), and FBS was obtained from Atlanta Biologicals (Lawrenceville, GA). EDTA-free protein inhibitor cocktail was purchased from Roche Applied Science (Indianapolis, IN), and MG132 was purchased from Calbiochem (Billerica, MA). Lipofectamine 2000 and LipofectamineTM RNAiMax were obtained from Invitrogen(Grand Island, NY). All other chemicals and reagents were obtained from Sigma (St Louis, MO) unless otherwise specified.
The following antibodies were used in the experiments described: HL-1, a mouse monoclonal antibody (Ab) to the linker sequence between the 4th and 5th ligand binding repeat of human LDLR (20); 3143, a rabbit polyclonal Ab against a peptide comprising the 14 C-terminal residues of mouse LDLR (21); rabbit polyclonal antisera against full-length human PCSK9 (172C) (14) and against the C-terminal 15 residues of autosomal recessive hypercholesterolemia (ARH) (22); and a polyclonal antibody (804c) against HMG-CoA reductase, IgG-A9, a mouse MAb against the catalytic domain of hamster HMG-CoA reductase (amino acids 450–887) (23). In addition, antibodies were purchased to detect calnexin (StressGen, Farmingdale, NY), LC3B (Cell Signaling, Danvers, MA), clathrin heavy chain (BD Transduction Laboratory, San Jose, CA), GM130 (Sigma), cathepsin D and actin (Sigma), TSG101 and hepatocyte growth factor-regulated Tyr-kinase substrate (HRS) (Abcam, Cambridge, MA), epidermal growth factor receptor (EGFR) (Millipore, Billerica, MA), Atg5 (Sigma), ubiquitin (Santa Cruz Biotechnology, Inc. Santa Cruz, CA), and EEA1 (BD Bioscience, San Jose, CA). Alexa Fluor® 488 Donkey Anti-Mouse IgG was obtained from Invitrogen.
Biotinylation and immunoblot analysis
Cell surface proteins were biotinylated as described (13). Cell lysates were prepared in 150 μl of lysis buffer (1% v/v Triton-X100, 50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 5 mM EDTA plus protease inhibitor). A total of 90 μl of cell lysate was added to 50 μl of a 1:1 mixture of Neutravidin-agarose (Pierce) and lysis buffer (410 μl). The mixture was rotated overnight at 4°C. After centrifugation at 3,000 g for 5 min, the pellets were washed three times in lysis buffer for 10 min at 4°C. Cell surface proteins were eluted from the beads in 1× SDS loading buffer (31 mM TrisCl [pH 6.8], 1% SDS, 12.5% glycerol, 0.0025% bromophenol) for 5 min at 95°C. Proteins were analyzed by immunoblotting as described (13). Briefly, protein samples were separated on 8% SDS-PAGE gels and transferred to nitrocellulose membranes (GE Healthcare). Antibody binding was detected using HRP-conjugated rabbit anti-mouse IgG or goat anti-rabbit IgG (GE Healthcare) followed by enhanced chemiluminescence detection (Pierce). The membranes were then exposed to F-BX810TM Blue X-Ray films (Phoenix Research Products, Hayward, CA).
To quantify the immunoblot signals, films were scanned using a HP Scanjet 5590 and quantified using ImageJ (http://rsbweb.nih.gov/ij/). The intensity of each band was corrected for background using a blank from the same film and then normalized using a loading control (Calnexin or Actin) run on the same gel. The value of the control (Lane 1) on each gel was set to one.
Cell culture
Lymphocytes were isolated from circulating blood, immortalized using the Epstein-Barr virus, and cultured as described (24). Immortalized lymphocytes were maintained in medium A (RPMI 1640 medium supplemented with 10% [v/v] FCS, 100 units/ml penicillin G, and streptomycin). To up-regulate LDLR expression, lymphocytes (5 × 105 cells/ml) were grown for 2 days in medium B (medium A with 10% [v/v] human lipoprotein-poor serum in place of FCS). Cells were resuspended in a 35 mm dish at a density of 1 × 106 cells/ml.
HuH7 cells were cultured in medium C (high glucose DMEM medium [hDMEM] plus 10% FCS and 100 units/ml penicillin G/streptomycin). The medium was switched to medium D (hDMEM plus 10% newborn calf lipoprotein-poor serum [NCLPPS]) with the addition of PCSK9. Mouse hepatoma (Hepa1c1c7) cells were cultured in MEMα medium (Invitrogen), fibroblasts and HepG2 cells in DMEM medium (25), HeLa cells in MEM containing nonessential amino acids (1:100) and sodium pyruvate (1:100), and human embryonic kidney (HEK)-293 cells in high-glucose DMEM medium. All culture medium contained 10% FCS plus 100 units/ml penicillin G/streptomycin.
Purification of recombinant human PCSK9
C-terminal FLAG-tagged full-length PCSK9 fusion proteins (wild-type and D374Y) were purified using anti-FLAG M2 beads and size exclusion chromatography (Superdex 200 10/300 fast performance liquid chromatography; GE Healthcare, Piscataway, NJ) (10).
Site-directed mutagenesis
A pShuttle- RSV vector containing wild-type human LDLR (14) was used as a template for mutagenesis. Mutagenesis was performed using QuickChangeTM site-directed mutagenesis kits (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Oligonucleotides containing the residues to be mutated (K709R, K795R, K809R, and C818S) were synthesized by IDT, Inc. (Coralville, IA). The presence of the desired mutation and the integrity of each construct were verified by Sanger DNA sequencing.
Small interfering RNA knockdown
Small interfering (si)RNAs targeting ARH, clathrin heavy chain, ATG7, TSG101, and HRS and control siRNAs were obtained from Thermo Scientific.
qRT-PCR
ATG7 mRNA levels were quantified by RT-PCR using oligonucleotides specific to human ATG7 and PCR Master Mix (Applied Biosystems) according to the manufacturer's instructions. All samples were analyzed in triplicate. GAPDH was used as an internal control.
Cell fractionation
HuH7 cells were washed and collected in ice-cold PBS, resuspended in 1 ml of 1× SE buffer (0.25 M sucrose, 1 mM EDTA), and homogenized in a ball-bearing homogenizer with a clearance of 16 μm. The homogenized suspension was transferred to a 15 ml centrifuge tube, and 850 μl of 1× SE buffer was added. The suspension was centrifuged for 10 min at 370 g at 4°C in a swinging-bucket rotor. The postnuclear supernatant was removed and transferred to an ice-cold, 1.5 ml microcentrifuge tube. A Percoll gradient was prepared by layering 8.5 ml of 1× SE buffer/Percoll (16%) over 10× SE buffer (1.2 ml) in a 16 × 76 mm Beckman Ultra-Clear centrifuge tube. The postnuclear supernatant was layered on top of the Percoll gradient, and the tube was capped before centrifugation for 30 min at 26,500 g at 4°C. After centrifugation, eight 1.2 ml samples were collected from the bottom of the tube. SDS was added to a final concentration of 1%, and the solution was vortexed for 30 min at 4°C. Proteins were precipitated with TCA, resuspended in 2% SDS-PAGE loading buffer, and subjected to immunoblot analysis.
Isolation of primary hepatocytes
Animal experiments were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee and were performed in accordance with federal animal welfare policies and regulations. Atg5−/− mice were provided by Beth Levine (UT Southwestern) (26).
Livers were isolated from fetal (∼E16) Atg5−/− mice, rinsed in 5 ml ice-cold wash buffer (HBSS [Invitrogen], 10 mM Hepes [pH 7.4], 100 μg/ml gentamycin sulfate, 0.5 mM EGTA), and transferred to 60 mm dishes. The liver tissue was diced into small pieces (1–2 mm3) with sterile scissors, transferred to a 50 ml tube, and incubated at 37°C for 10 min on a gently rotating platform. After removing the supernatant, 10 ml of digestion buffer (HBSS with 0.3 mg/ml collagenase 2, 25 μl 1 M CaCl2, and 100 μg/ml gentamycin sulfate) was added to liver tissue and allowed to incubate for 30 min at 37°C. The supernatant was transferred to a 50 ml tube containing 40 ml cold medium (DMEM, 5% FBS, 10 mM Hepes [pH 7.4], 100 μg/ml gentamycin sulfate). Digestion buffer (10 ml) was added to the cells and gently rotated 37°C for 30 min. The supernatant was collected and transferred to a 50 ml tube containing 40 ml cold culture medium. The particulate material was resuspended in 5 ml digestion buffer, passed through a 27.5 gauge needle, and filtered through a 70 μm filter. The filtrate was added to 45 ml of ice-cold culture medium. The supernatants were centrifuged for 5 min at 70 g, and each pellet was washed twice with 50 ml ice-cold culture medium. Pellets were resuspended, pooled, subjected to centrifugation, resuspended in 6 ml culture medium (37°C), and plated on collagen-coated 6-well plates. Plates were incubated overnight at 37°C in 5% CO2. After 48 h, cells were treated with PCSK9 in DMEM plus 5% NCLPPS for 4 h at 37°C .
Imaging
The lysosome marker DND-26, transferrin-Alexa488, and the Alexa-546 protein labeling kit were obtained from Invitrogen. PCSK9 was labeled with Alexa-546 dye according to the manufacturer's instructions. Cells were treated with labeled PCSK9 and DND-26 or transferrin-Alex488 and imaged at ambient temperature using a confocal microscope. The imaging data were analyzed using ImageJ.
For immunofluorescence experiments, cells grown on cover slips were rinsed with PBS and buffer A (PBS+10 mm HEPES [pH 7.3]) and fixed with freshly prepared 3% formaldehyde in buffer A for 30 min at room temperature. Cells were rinsed with PBS, incubated in 50 mM NH4Cl for 30 min, rinsed twice with PBS, and permeabilized with 0.1% (v/v) Triton X-100 in PBS for 7 min at 4°C. The permeabilized cells were conditioned with buffer B (1% [w/v] BSA in PBS) for 30 min at 4°C and incubated with primary antibody in buffer B. Sixteen hours later, cells were washed three times (5 min per wash) with buffer C (0.1% BSA in PBS) at room temperature. After washing, the cells were incubated with secondary antibody in buffer B for 1 h at room temperature in muted light. Cells were then washed three times with buffer C at room temperature (5 min per wash) and rinsed three times with distilled water. The washed cover slips were mounted on the micro slide and left to dry overnight.
RESULTS
PCSK9 promotes LDLR degradation in primary fibroblasts lacking ARH
PCSK9 promotes LDLR degradation in some cell types (hepatocytes and lymphocytes) but not in others (fibroblasts) (10). The cell-type-specific pattern of PCSK9 activity parallels the cell-type-specific requirement of ARH for LDLR-mediated endocytosis of LDL (27). ARH is an adaptor protein that binds the cytoplasmic tail of the LDLR and links it to the endocytic machinery for uptake of LDL (27). ARH is required for LDL uptake by hepatocytes and lymphocytes but not by fibroblasts, suggesting that ARH may be specifically required for PCSK9-mediated LDLR degradation (28).
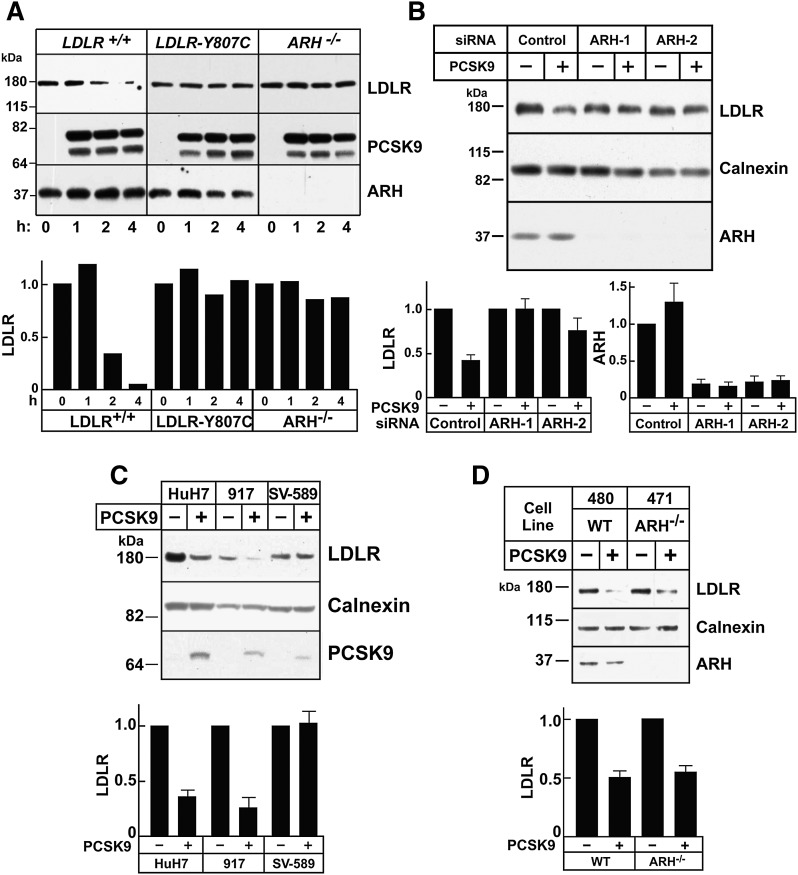
To test this hypothesis, we examined the effect of ARH on LDLR protein levels in transformed lymphocytes. The addition of PCSK9 to control lymphocytes resulted in degradation of LDLR within 4 h (Fig. 1A). Conversely, PCSK9 treatment of cells from an individual who lacks ARH failed to promote receptor degradation. PCSK9 treatment also failed to decrease LDLR levels in cells from a patient with familial hypercholesterolemia who is homozygous for a mutation (Y807C) that prevents ARH binding to the in the internalization motif (NPVY) of the receptor (29–31). Inactivation of ARH in cultured hepatocytes (HuH7 cells) using two different siRNAs also interfered with PCSK9-stimulated LDLR degradation (Fig. 1B), which is consistent with previous findings (10).
Fig. 1.
PCSK9-mediated LDLR degradation requires ARH in lymphocytes and hepatocytes but not in fibroblasts. A: Immortalized lymphocytes (5 × 105 cells/ml) from a normal subject, a patient homozygous for a mutation in the LDLR that blocks LDL internalization (LDLR-Y807C) (61), and a patient with ARH deficiency were cultured at 37°C. PCSK9 (5 μg/ml) was added to the medium for the indicated times. After cells were washed in ice-cold PBS, lysates were prepared and subjected to immunoblotting. The graph represents quantification of the data shown. B: HuH7 cells (1 × 105 cells/60 mm dish) were grown for 24 h and transfected with two separate anti-ARH siRNAs (ARH-1 and ARH-2) or a control siRNA (Control). After 3 days, PCSK9 (10 μg/ml) was added to the medium. After 4 h at 37°C, cells were processed as described in A. C: Primary human fibroblasts (917), immortalized fibroblasts (SV-589), and hepatocytes (HuH7) were treated with PCSK9 (10 μg/ml) for 4 h, and cell lysates were processed as described in A. D: Primary human fibroblasts from normal (480) or ARH-deficient (471) individuals were treated with PCSK9 and whole cell lysate were analyzed as described in A. All experiments were performed at least twice with similar results. Proteins were quantified as described in Materials and Methods. Each graph represents the mean ± SEM for three independent experiments, except for A, which represents the data shown.
Previous studies have reported that PCSK9 does not promote LDLR degradation in fibroblasts (10), a cell type that does not require ARH for LDL uptake (32). It has been proposed that DAB2, another related adaptor protein, substitutes for ARH in fibroblasts (33). Surprisingly, we found that PCSK9 treatment resulted in robust degradation of the LDLR in primary human fibroblasts (917 cells) (Fig. 1C). We also examined another primary fibroblast cell line (480) as well as fibroblasts from a patient with ARH deficiency (Fig. 1D), and identical results were obtained. In contrast to these results, PCSK9 did not promote LDLR degradation in SV40-transformed human fibroblasts (SV-589 cells) (Fig. 1C).
Thus, ARH is not specifically required for PCSK9-mediated LDLR degradation. The reason why PCSK9 fails to stimulate receptor degradation in transformed fibroblasts remains to be elucidated.
PCSK9-mediated LDLR degradation requires clathrin-mediated endocytosis
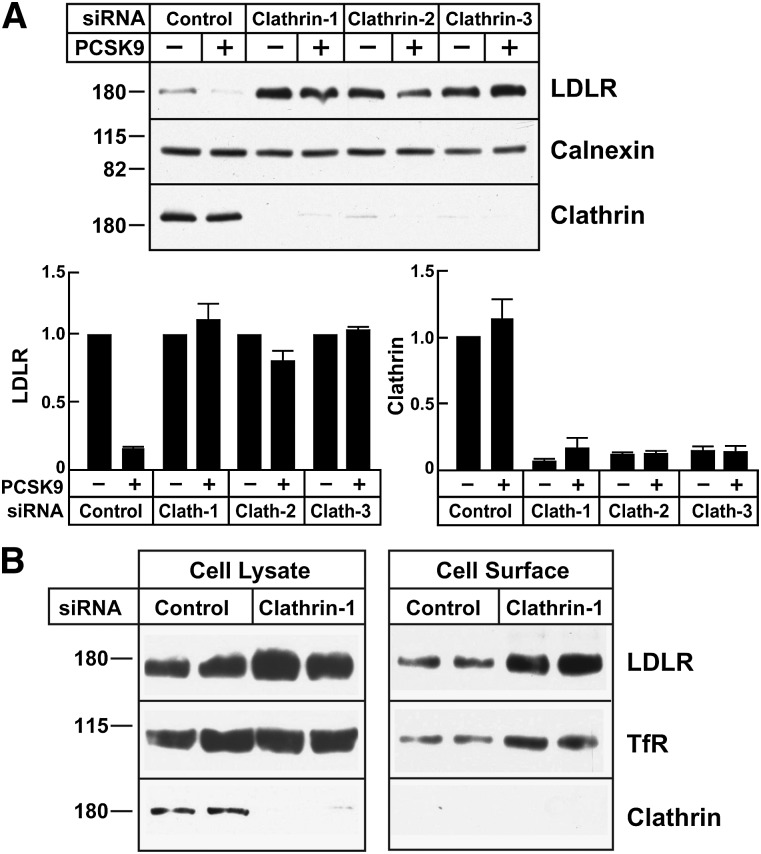
PCSK9 engages the LDLR at the cell surface (10), but the mechanisms responsible for its effects on LDLR trafficking have not been molecularly defined. Knockdown of the mRNA encoding clathrin heavy chain in cultured hepatocytes (HuH7) blocked PCSK9-mediated degradation of the LDLR (Fig. 2). The level of LDLR was increased in the clathrin siRNA-treated cells. Clathrin also participates in the delivery of proteins from the Golgi to the cell surface (34). To ensure that clathrin knockdown did not prevent the delivery of newly synthesized LDLRs from the Golgi to the plasma membrane, we examined the effect of clathrin depletion on the amount of cell surface LDLR by using biotinylation to label cell surface proteins. The transferrin receptor and clathrin served as positive and negative controls, respectively, for biotinylation of cell surface proteins. The enrichment of LDLR on the cell surface of clathrin-depleted cells was comparable to that observed in clathrin-replete cells (Fig. 2B).
Fig. 2.
PCSK9-mediated LDLR degradation in clathrin-depleted HuH7 cells. A: HuH7 cells were transfected with three different anti-clathrin heavy chain (Clathrin-1, Clathrin-2, and Clathrin-3) or control siRNAs as indicated in Fig. 1. After 3 days, PCSK9 (10 μg/ml) was added to the medium for 4 h. B: HuH7 cells were transfected with control siRNA or siRNA targeting clathrin heavy chain for 72 h as indicated in Fig. 1. Cell surface proteins were isolated by biotinylation as described in Materials and Methods. Whole cell lysates and streptavidin-precipitated proteins were analyzed by immunoblotting. The immunoreactive proteins were quantified as described in Materials and Methods. Graphs represent the means ± SEM from three independent experiments. TfR, transferrin receptor.
These data indicate that PCSK9 binding at the cell surface does not reroute the LDLR to an alternative uptake pathway, as occurs for some other receptors, such as the EGFR (35–37). Rather, the PCSK9-LDLR complex is internalized by the same endocytic machinery that imports LDL.
PCSK9-mediated LDLR degradation does not require ubiquitination of the LDL receptor tail or proteasome function
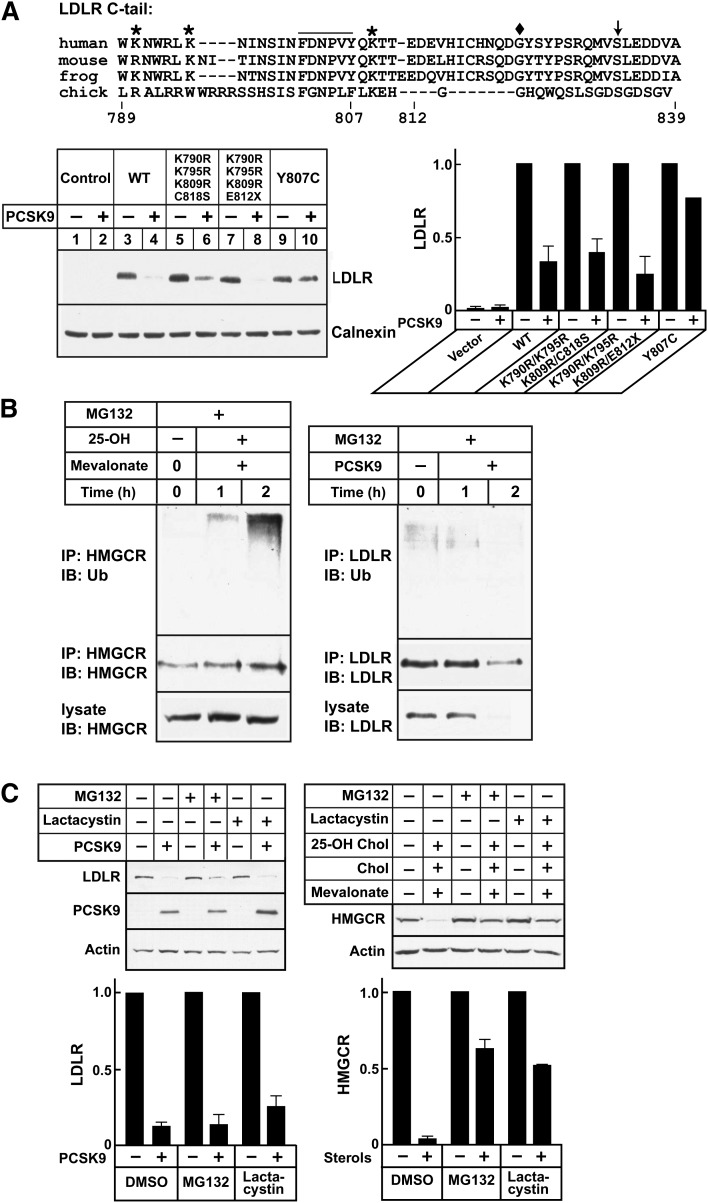
Ubiquitin functions as a targeting signal for protein degradation by proteasomes and lysosomes and is essential for degradation of LDLRs by the inducible degrader of the LDLR (IDOL) (38). To determine if ubiquitination of LDLR is required for PCSK9-mediated LDLR degradation, we substituted all of the lysines in the cytoplasmic tail of the LDLR with arginine residues and mutated a cysteine residue (C818) that is ubiquitinated by IDOL (38) to serine or introduced a mutation that truncates the protein at residue 812 (E812×). A construct in which the tyrosine in the internalization motif was mutated to cytosine (Y807C, the so-called “JD mutation”) was used as a positive control (29, 30) (Fig. 3A). Substitution of the lysine residues and cysteine residue of the LDLR cytoplasmic tail had no discernible effect on PCSK9-mediated degradation of the receptor, whereas the Y807C substitution, which disrupts the internalization sequence, protected the receptor from degradation. Thus, ubiquitination of the LDLR is not required for PCSK9-mediated degradation of the receptor under these conditions.
Fig. 3.
PCSK9-mediated LDLR degradation and the ubiquitin-proteosome pathway. A: Alignment of the amino acid sequence of the LDLR cytoplasmic tail from four species is shown. Mouse Hepalclc7 cells (plated at 4 × 105 cells/well in 6-well plates) were grown in 2 ml MEMα containing 10% FCS (Day 0). On Day 1, cells were transfected with empty vector or with plasmids expressing wild-type (WT) or mutant LDLR using Lipofectamine 2000. On Day 2, the medium was switched to MEMα plus 10% NCLPPS, cholesterol (10 μg/ml), and 25-hydroxycholesterol (1 μg/ml). On Day 3, PCSK9-D374Y (2.0 μg/ml) was added. After 4 h at 37°C, lysates were prepared and immunoblotted with HL1, a monoclonal antibody against human LDLR that does not recognize mouse LDLR. B: HuH7 cells were cultured as described and treated as indicated in B. HMGCR and LDLR were immunoprecipitated from cell lysates using antibodies 804c and 3143, respectively. Immunoprecipitates were size-fractionated on SDS-PAGE gels and probed with anti-ubiquitin (P4D1), anti-HMGCR (IgG-A9), and anti-LDLR (HL1) antibodies. C: HuH7 cells (plated at 3 × 105/well in 6-well plates) were grown in medium C (Day 0). On Day 1, medium was switched to hDMEM containing 10% NCLPPS plus compactin (10 µM), mevalonate (50 µM) for HMGCR degradation assays, and hDMEM containing 10% NCLPPS for LDLR degradation. After 16 h, medium containing MG132 (10 μM), lactacystin(10 μM), or DMSO alone was added. After 1 h, cells were treated with 25-hydroxycholesterol (1 μg/ml), cholesterol (10 μg/ml), and mevalonate (10 mM) or with PCSK9 (5 μg/ml) for 4 h. Cell lysates were subjected to immunoblot analysis. All experiments were performed twice with similar results. Proteins were quantified as described in Materials and Methods. Graphs represent the means ± SEM for two independent experiments.
We also examined the effect of pharmacological inhibition of proteosomes on PCSK9-stimulated LDLR degradation. PCSK9 was added to HuH7 cells in the presence of MG132, a proteosome inhibitor. Sterol-induced ubiquitination of hydroxymethylglutaryl-CoA reductase (HMGCR) served as a positive control in this experiment (39). 25-Hydroxycholesterol markedly increased the amount of ubiquitin associated with HMGCR (Fig. 3B, left). In contrast, very low levels of ubiquitinated LDLR were observed in cells treated with MG132 alone, and the levels did not increase when PCSK9 was added to the medium (Fig. 3B, right). The addition of proteosomal inhibitors MG132 or lactacystin failed to block PCSK9-mediated LDLR degradation, whereas both inhibited sterol- and mevalonate-stimulated degradation of HMGCR (Fig. 3C) by ∼50%.
Taken together, these data show that PCSK9-mediated degradation of LDLRs in hepatocytes does not require proteasome function or ubiquitination of the cytoplasmic tail of the LDLR.
Lysosome function is required for PCSK9-mediated LDLR degradation
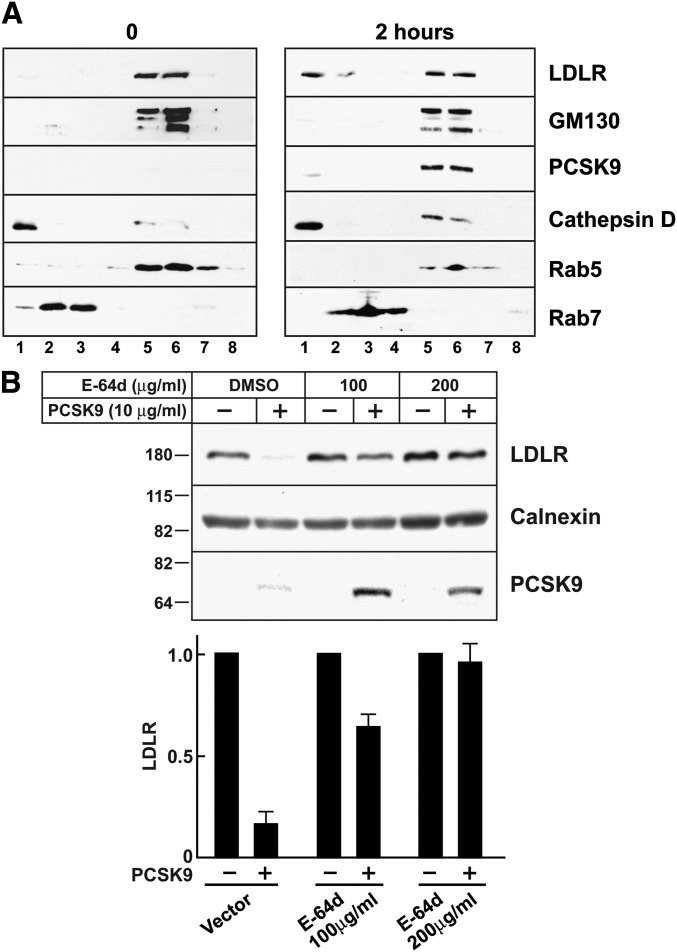
The notion that PCSK9 targets LDLRs for degradation in lysosomes is based on colocalization of PCSK9 and LDLRs with lysosome markers by fluorescence microscopy (10). However, Maxwell et al. (40) reported that inhibition of lysosomal proteases failed to inhibit the degradation of the LDLR in HepG2 cells expressing PCSK9. Live cell imaging revealed that PCSK9 colocalized with the lysosome marker lysotracker, whereas very little PCSK9 colocalized with transferrin, which is located in recycling endosomes (supplementary Fig. I). To confirm these microscopy studies, we used cell fractionation and immunoblot analysis to follow PCSK9-induced migration of the LDLR in HuH7 cells. At the zero time point, the bulk of the LDLR localized to fractions containing Rab5 and GM130 (Fig. 4A), consistent with partitioning of the receptor between the Golgi and early endosomes (41). No LDLR was detectable in the lysosome fraction, a dense fraction containing the lysosome marker cathepsin D but not markers of Golgi or other endosomal compartments. Two hours after the addition of PCSK9 to the cells, LDLRs were readily detectable in the lysosome fraction.
Fig. 4.
PCSK9-mediated LDLR degradation and lysosome function. A: Confluent HuH7 cells were treated with PCSK9 (10 μg/ml) for 0 or 2 h. Cell lysates were fractionated on Percoll gradients as described in Materials and Methods. Proteins were precipitated from the gradient fractions, size-fractionated on 4–12% gradient SDS-polyacrylamide gels, and visualized by immunoblot analysis. B: Confluent HuH7 cells were treated with the lysosome inhibitor E-64d at the doses indicated or with DMSO alone for 30 min at 37°C in medium D. PCSK9 (10 μg/ml) was added to the medium, and cells were allowed to grow for another 4 h. Cells were harvested, and cell lysates were subjected to immunoblot analysis. Proteins were quantified as described in Materials and Methods. Graphs represent the means ± SEM for three independent experiments.
Consistent with the microscopy and cell fractionation studies, PCSK9-mediated LDLR degradation was almost completely blocked by the lysosomal protease inhibitor E-64d (Fig. 4B). Treatment with E-64d also increased cellular levels of PCSK9 (Fig. 4B). Taken together, these data are consistent with a model in which PCSK9 binds to the LDLR and reroutes the receptor to lysosomes where both proteins are degraded.
Autophagy is not required for PCSK9-mediated LDLR degradation
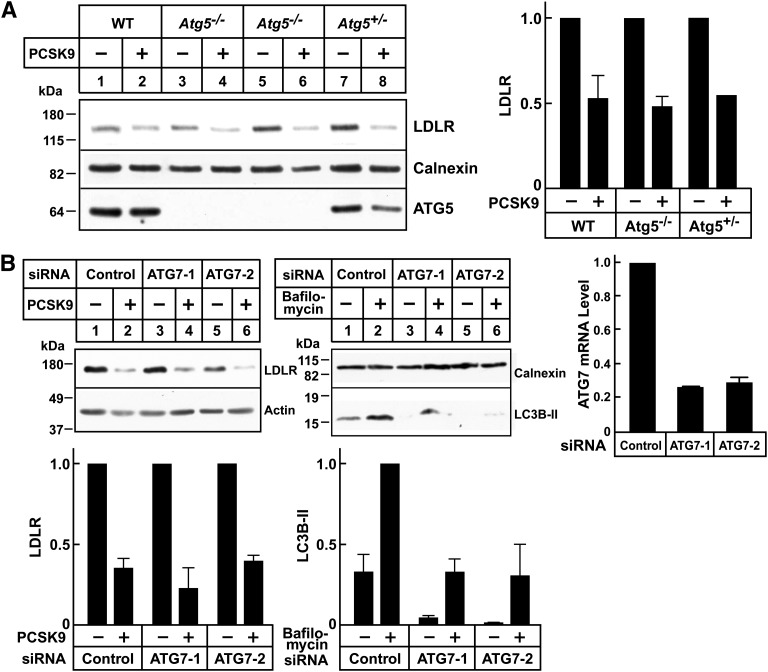
Autophagy is a highly conserved pathway by which proteins and organelles are routed to lysosomes for degradation (42). To determine whether autophagy is involved in PCSK9-mediated LDLR degradation, we evaluated PCSK9 function in hepatocytes lacking two proteins considered essential for the pathway, Atg5 and Atg7 (26, 42). PCSK9-mediated LDLR degradation was preserved in cultured primary hepatocytes from Atg5−/− mice (Fig. 5A). Knockdown of Atg7 using siRNAs had no effect on PCSK9-mediated LDLR degradation. To ensure that we had effectively inactivated ATG7, we examined the relative levels of LC3B-I and LC3B-II. Atg7 is essential for the covalent conjugation of LC3B-I with phosphatidylethanolamine, which forms LC3B-II. The conversion of LC3-I to LC3-II is indicative of autophagic activity. Bafilomycin 1A was added to the cells to block the degradation of LC3B-II (43). As expected with inactivation of the autophagocytic pathway, the levels of LC3B-II were decreased (Fig. 5B); the LC3B-I bands were only apparent on long exposures (not shown). Taken together, these data indicated that PCSK9-mediated LDLR degradation does not involve the autophagy pathway.
Fig. 5.
PCSK9-mediated LDLR degradation in autophagy-deficient hepatocytes. A: Primary hepatocytes were isolated from WT Atg5−/− or Atg5+/− mouse embryos. After 48 h, cells were treated with PCSK9 (10 μg/ml) for 4 h at 37°C. Cell lysates were subjected to immunoblot analysis. B: HuH7 cells were transfected with control or anti-ATG7 siRNA oligos as described in Fig. 1. After 72 h, cells were treated with PCSK9 (10 μg/ml) for 4 h or with bafilomycin A1 (100 nM) for 2 h at 37°C. To visualize LC3B protein, cell lysates were fractionated on 15% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes for immunoblot analysis. The efficiency of ATG7 knockdown was evaluated by qRT-PCR as described in Materials and Methods. Data were normalized to control dishes, and GAPDH was used as an internal standard. Similar results were obtained in at least two independent experiments. Proteins were quantified as described in Materials and Methods. Graphs represent the means ± SEM from two independent experiments.
Knockdown of the key ESCRT pathway proteins fails to block PCSK9-mediated LDLR degradation
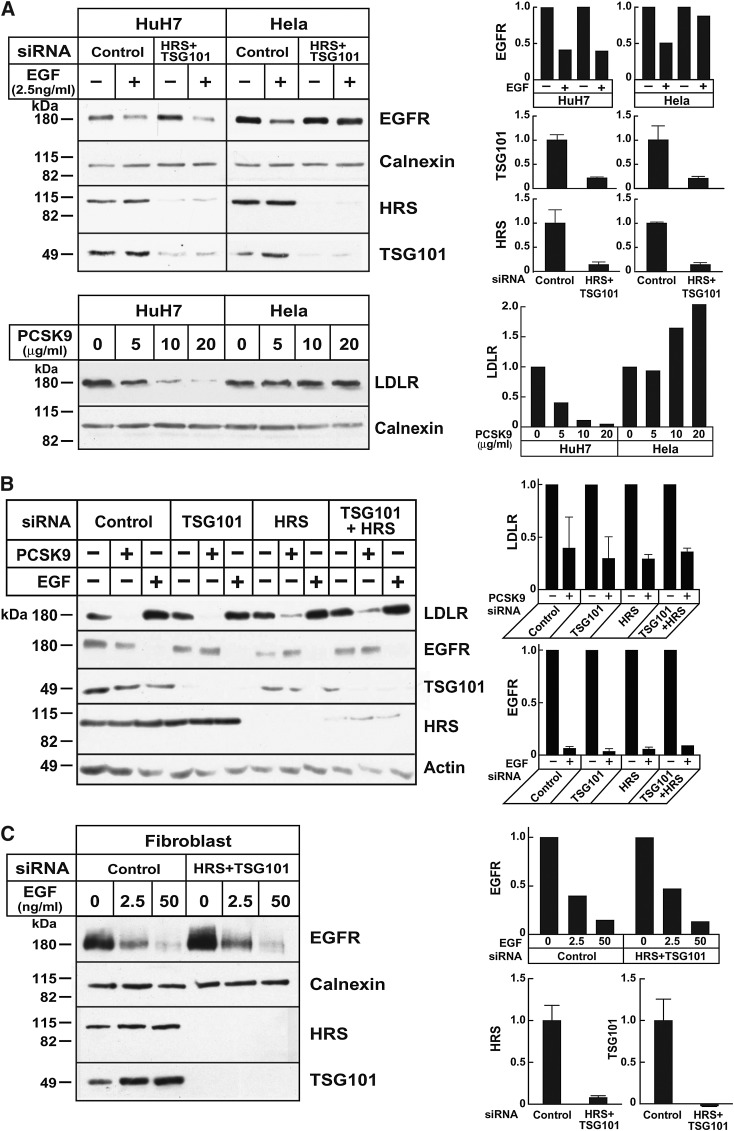
The ESCRT pathway plays a key role in sorting cell membrane proteins to lysosomes (44). To determine whether the ESCRT machinery routes the PCSK9-LDLR complex to lysosomes, we used siRNAs to knock down two components of the pathway: HRS, which is part of ESCRT-0 complex that participates in the initial selection of ubiquitinated cargo at the endosomal membrane (45), and tumor susceptibility gene 101 (TSG101), a component of ESCRT-I that couples HRS to other elements of the ESCRT machinery (44). Depleting essential components of the ESCRT pathway has been shown to inhibit EGF-stimulated degradation of the EGFR in Hela cells (45, 46). Silencing RNAs were used to knockdown TGS101 and HRS singly or together in HuH7 cells (Fig. 6A, B) and Hela cells (Fig. 6A). As expected, inactivation of the initial steps in the ESCRT pathway inhibited EGF-stimulated EGFR degradation in Hela cells (Fig. 6A). In contrast, inactivating TSG101 and HRS cells failed to block EGFR degradation in HuH7 (Fig. 6A, B), HepG2 cells (data not shown), and fibroblasts (Fig. 6C). Conversely, PCSK9 promoted degradation of LDLR in HepG2 cells and in fibroblasts but not in Hela cells. Knockdown of HRS and TSG101 did not affect PCSK9-stimulated degradation in any of these cell types. Thus, in cell types in which inactivation of the ESCRT pathway inhibited EGF-stimulated EGFR degradation, PCSK9 failed to promote LDLR degradation (i.e., Hela cells) (Fig. 6A), whereas in cell types where PCSK9 was active (hepatocytes and fibroblasts), interrupting the ESCRT pathway did not interfere with EGFR degradation by its ligand.
Fig. 6.
The ESCRT pathway and PCSK9-mediated LDLR degradation. A, upper panel: HuH7 and Hela cells were cultured as indicated in Materials and Methods, and siRNA transfection was performed as described in Fig. 1. The medium was changed to serum-free medium 72 h after transfection, and after 2 h, EGF was added to the medium. Cells were collected after another 4 h. A, lower panel: Confluent HuH7 and Hela cells were treated with the indicated amounts of PCSK9 at 37°C for 4 h. Cell lysates were subjected to immunoblot analysis. Graphs represent the mean ± SEM for two independent experiments. B: HuH7 cells were transfected with control siRNA or with siRNAs against TSG101, HRS, or both. After 72 h, cells were switched to hDMEM without serum. Cells were incubated for 24 h before the addition of PCSK9 (10 μg/ml) or EGF (50 ng/ml) for 4 h at 37°C. Cells lysates were subjected to immunoblot analysis. Data represent the mean ± SEM for three independent experiments. C: Human skin fibroblasts (917) were plated at a density of 2 × 104 cells per 60 mm dish in DMEM with 10% FCS and 100 units/ml penicillin G/streptomycin. After 24 h (day 1), cells were transfected with siRNA targeting HRS and TSG101 or with a control siRNA. On day 3, cells were split (1:3) and plated in new 60 mm dishes. On day 4, cells were transfected with the same siRNAs. On day 7, cells were incubated in serum-free medium for 2 h and treated with EGF. After treatment, cell lysates were subjected to immunoblot analysis as indicated. All experiments were performed at least twice with similar results. Proteins were quantified as described in Materials and Methods. Graphs represent the data from the experiment shown.
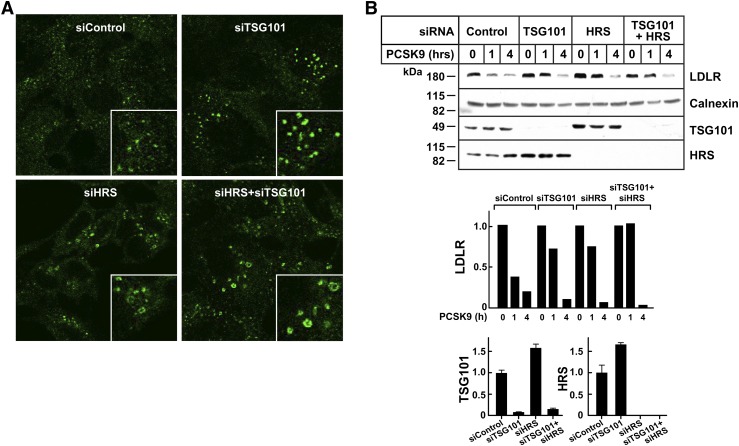
To develop another system in which to examine the role of the ESCRT pathway in PCSK9-mediated LDLR degradation, we performed the same assays in cultured HEK 293 cells. Inactivation of HRS and TGS101 is associated with an increase in the size of the early endosome and a reduced rate of EGF degradation in this cell type (47–49). Depletion of HRS or TGS101, singly or together, resulted in enlargement of early endosomes (Fig. 7A), as detected using EEA1, an early endosome marker (47). In parallel experiments, siRNA treatment of HEK293 cells decreased cellular levels of HRS and TSG101 by more than 80%. Knockdown of either protein appeared to delay LDLR-degradation at the 1 h time point (Fig. 7C), but the effect was not consistent across experiments. Knockdown of HRS and TSG101 had no detectable effect on PCSK9-mediated LDLR degradation at 4 h (Fig. 7C).
Fig. 7.
The ESCRT pathway and PCSK9-mediated LDLR degradation in HEK293 cells. A: HEK293 cells were cultured on cover slides as indicated in Materials and Methods, and siRNA transfection was performed as described in Fig. 1. After 72 h, cells were fixed and stained with anti-EEA1 antibody as indicated in Materials and Methods. B: HEK293 cells were cultured, and siRNA transfection was performed as described in Fig. 1. After 72 h, cells were treated with PCSK9 (10 μg/ml) for the indicated times. Cell lysates were subjected to immunoblot analysis. Graphs represent the quantitative results from this experiment. The experiments were performed twice with similar results. Graphs represent the data shown in the experiment.
These results, when taken together, strongly suggest that PCSK9-mediated LDLR degradation does not require the first components of the ESCRT pathway. It remains possible that knockdown of the ESCRT pathway was not as complete in the cultured hepatocytes and fibroblasts as it was in the Hela cells. Alternatively, PCSK9-LDLR complex may enter the ESCRT pathway downstream of the initial components, as has been described previously for another cell surface protein, protease-activated receptor 1 (PAR1) (50, 51).
DISCUSSION
In the present study, we used pharmacological inhibition, siRNA-mediated knockdown, and targeted gene disruption to investigate the pathway by which PCSK9 routes LDLRs to lysosomes. Our data are consistent with a model in which LDLRs that engage PCSK9 at the cell membrane are internalized via the canonical clathrin-dependent endocytic machinery but fail to enter the recycling pathway. Rather, the PCSK9-LDLR complex is routed to lysosomes via a pathway that does not require ubiquitination of the cytoplasmic tail of the receptor and does not involve the proteasomal or autophagy pathways.
To rule out the possibility that PCSK9-binding redirects LDLRs to an alternative internalization pathway that culminates in degradation, in a fashion similar to that described for EGFR (35) and FGFR3 (52), we examined the effect of inactivating clathrin heavy chain on PCSK9-mediated LDLR degradation. Addition of PCSK9 to the medium of cultured hepatocytes in which clathrin heavy chain was depleted failed to reduce cellular LDLR content. Biotin-tagging of cell-surface proteins revealed that clathrin knockdown increased cell surface expression of LDLRs, thus confirming that clathrin is required for internalization of the LDLR-PCSK9 complex that forms on the cell surface.
It has been shown that PCSK9 promotes LDLR degradation in hepatocytes (10), and here we show that PCSK9 also promotes receptor degradation in lymphocytes (Fig. 1A). Both of these cell lines require ARH for LDLR-mediated endocytosis of LDL (27, 53). In contrast, ARH is not required for LDLR-mediated endocytosis in fibroblasts. The addition of PCSK9 to the medium of transformed fibroblasts failed to stimulate LDLR degradation (Fig. 1). Hepatocytes and lymphocytes, but not fibroblasts, require ARH for LDL receptor-mediated endocytosis (22, 54). These findings, when taken together, suggested that ARH may be specifically required for PCSK9 action. Here we show that PCSK9 promotes LDLR degradation in several different primary cell lines of human fibroblasts, even cells that are deficient in ARH. Thus, ARH is not specifically required as the endocytic adaptor for PCSK9-mediated LDLR degradation.
Several cell surface receptors undergo ligand-induced degradation via ligand-stimulated ubiquitination of lysine residues in their cytoplasmic tails. For some receptors, ubiquitination of the cytoplasmic tail routes the receptor to lysosomes. For other receptors, such as the VEGF receptor 2, ligand-induced ubiquitination targets the receptor to the proteasome for degradation (55). Recent studies have shown that the LDLR is ubiquitinated by the LXR-responsive ubiquitin ligase IDOL (38), which promotes ubiquitination of lysine and cysteine residues in the cytoplasmic tail of the receptor (Fig. 3, top) and targets it for lysosomal degradation (38). Substitution of arginines for the lysines at residues 790, 795, and 809 and a serine for cysteine 818 in the cytoplasmic tail of the LDLR protects the receptor from IDOL-induced degradation. In the present study, PCSK9 did not promote ubiquitination of LDLRs and mutation of the lysine, and cysteine residues in the cytoplasmic tail failed to protect the receptor from PCSK9-mediated degradation (Fig. 3). Moreover, the proteosome inhibitors MG132 and lactacystin failed to maintain LDLR expression in PCSK9-treated cells. Therefore, PCSK9 does not function in hepatocytes by promoting ubiquitination of the cytoplasmic tail of the LDLR and acts independently of IDOL and the proteasome.
The pathway by which PCSK9 directs LDLRs from endosomes to lysosomes has not been molecularly characterized. Several integral membrane proteins, including receptors such as EGFR, MHC class 1 molecules (56), and transporters such as ABCA1 (57), are delivered from endosomes to the multivesicular bodies and then lysosomes by ESCRTs (58). Studies in yeast and selected mammalian cells have identified several components of the ESCRT machinery (58). Cargo recognition is mediated by the HRS/STAM complex (ESCRT-0) as well as ESCRT-I, which includes TSG101, VPS28, Vps37, and Mvb12. In the present study, simultaneous depletion of HRS and TSG101 disrupted the ESCRT pathway, as indicated by the inhibition of EGF-stimulated EGFR degradation in Hela cells (Fig. 6) (59) and the enlargement of early endosomes in HEK293 cells (Fig. 7) (47), but failed to block PCSK9-mediated LDLR degradation in multiple cell types, including cultured hepatocytes (HuH7, HepG2), embryonic kidney cells (HEK293), and primary fibroblasts. The discordant responses of inactivation of the ESCRT pathway on EGFR degradation by EGF and LDLR degradation by PCSK9 is most consistent with the notion that the PCSK9-LDLR complex traffics to lysosomes independently of the ESCRT pathway, or at least ESCRT-0 or ESCRT-I. Recently, Dores et al. (51) reported that PAR1, a G protein-coupled receptor, is trafficked to lysosomes via multivesicular bodies. Lysosomal trafficking of PAR1 did not require ubiquitination of the protein or the presence of HRS or TSG101. Knockdown of components of the ESCRT-III complex did prevent degradation of PAR1 (51). Additional experiments are required to determine if PCSK9-LDLR complexes bypass ESCRT-0 and ESCRT-I and use ESCRT-III to reach lysosomes.
Macroautophagy constitutes an alternative pathway for the lysosomal sorting of PCSK9-LDLR complexes. Initially considered a bulk degradation mechanism, there is mounting evidence that autophagy can also target proteins in a highly selective manner (60). Genetic deletion of Atg5 in mice and siRNA knockdown of ATG7 blocked maturation of LC3B-I to LC3B-II, a well-characterized marker of autophagy, but failed to interrupt PCSK9-mediated degradation of LDLR. This finding indicates that PCSK9 does not use the core molecular machinery required for autophagosome formation to effect degradation of the LDLR.
Although depletion of key components of the known lysosomal sorting machinery failed to block PCSK9-mediated degradation of LDLRs in HuH7 cells, cell fractionation studies confirmed that PCSK9 routed the receptor to lysosomes in these cells. Furthermore, in contrast to an earlier report (5), treatment with the cysteine protease inhibitor E64d effectively blocked PCSK9-mediated degradation of LDLR in HuH7 cells (Fig. 4B) and in primary fibroblasts (data not shown).
Like many cell-surface receptors, LDLR is a long-lived protein, with a half-life in fibroblasts of ∼12 h. The factors that determine basal turnover of the LDLR have not been defined. Accordingly, it is not known whether PCSK9 accelerates the basal degradation pathway of the receptor or uses an entirely different mechanism. In the present study, knockdown of clathrin heavy chain in HuH7 cells increased baseline levels of LDLR protein. Thus clathrin is involved in basal and PCSK9-mediated degradation of LDLRs. PCSK9 does not promote degradation of LDLRs in ARH-deficient lymphocytes (Fig. 1), yet basal levels of LDLR were not increased in these cells. Taken together, these findings suggest that basal and PCSK9-mediated degradation of LDLRs occur via distinct cellular pathways.
Supplementary Material
Acknowledgments
The authors thank Christina Zhao and Stephanie Blankenship for excellent technical assistance, Beth Levine for providing the genetically modified mice used in the studies described, Jay Horton for helpful discussions, and Eran Leitersdorf (Hebrew University, Hadassah Medical Center, Jerusalem, Israel) for cells from a patients homozygous for the Y807C substitution in the LDLR.
Footnotes
Abbreviations:
- ARH
- autosomal recessive hypercholesterolemia
- EGFR
- epidermal growth factor receptor
- ESCRT
- endosomal sorting complex required for trafficking
- HEK
- human embryonic kidney
- HRS
- hepatocyte growth factor-regulated Tyr-kinase substrate
- IDOL
- inducer of LDLR degradation
- LDLR
- LDL receptor
- PAR1
- protease-activated receptor 1
- PCSK9
- proprotein convertase subtilisin/kexin type 9
This work was supported by grants from the National Institutes of Health grants PO1HL20948 and RO1HL092550. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., Brown M. S. 2009. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29: 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park S. W., Moon Y. A., Horton J. D. 2004. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 279: 50630–50638. [DOI] [PubMed] [Google Scholar]
- 4.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the LDLR and LDL-cholesterol. J. Biol. Chem. 279: 48865–48875. [DOI] [PubMed] [Google Scholar]
- 5.Maxwell K. N., Breslow J. L. 2004. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA. 101: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 7.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. 2004. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum. Genet. 114: 349–353. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 9.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., Chretien M. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. 2006. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 116: 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton J. D., Cohen J. C., Hobbs H. H. 2009. PCSK9: a convertase that coordinates LDL catabolism. J. Lipid Res. 50(Suppl): S172–S177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 298: 2353–2358. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. 2008. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 105: 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., et al. 2007. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 282: 20502–20512. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., et al. 2007. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat. Struct. Mol. Biol. 14: 413–419. [DOI] [PubMed] [Google Scholar]
- 17.Holla O. L., Cameron J., Tveten K., Strom T. B., Berge K. E., Laerdahl J. K., Leren T. P. 2011. Role of the C-terminal domain of PCSK9 in degradation of the LDL receptors. J. Lipid Res. 52: 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tveten K., Holla O. L., Cameron J., Strom T. B., Berge K. E., Laerdahl J. K., Leren T. P. 2012. Interaction between the ligand-binding domain of the LDL receptor and the C-terminal domain of PCSK9 is required for PCSK9 to remain bound to the LDL receptor during endosomal acidification. Hum. Mol. Genet. 21: 1402–1409. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T., Lu C., Ryan R. O. 2011. A two-step binding model of PCSK9 interaction with the low density lipoprotein receptor. J. Biol. Chem. 286: 5464–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Driel I. R., Goldstein J. L., Südhof T. C., Brown M. S. 1987. First cysteine-rich repeat in ligand-binding domain of low density lipoprotein receptor binds Ca2+ and monoclonal antiobodies, but not lipoproteins. J. Biol. Chem. 262: 17443–17449. [PubMed] [Google Scholar]
- 21.Russell D. W., Schneider W. J., Yamamamoto T., Luskey K. L., Brown M. S., Goldstein J. L. 1984. Domain map of the LDL receptor: sequence homology with the epidermal growth factor precursor. Cell. 37: 577–585. [DOI] [PubMed] [Google Scholar]
- 22.Wilund K. R., Yi M., Campagna F., Arca M., Zuliani G., Fellin R., Ho Y. K., Garcia J. V., Hobbs H. H., Cohen J. C. 2002. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum. Mol. Genet. 11: 3019–3030. [DOI] [PubMed] [Google Scholar]
- 23.Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. 1983. Regulation of 3-hydroxy-3-methylglutaryl Coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J. Biol. Chem. 258: 8450–8455. [PubMed] [Google Scholar]
- 24.Michaely P., Li W. P., Anderson R. G., Cohen J. C., Hobbs H. H. 2004. The modular adaptor protein ARH Is required for low density lipoprotein (LDL) binding and internalization but not for LDL receptor clustering in coated pits. J. Biol. Chem. 279: 34023–34031. [DOI] [PubMed] [Google Scholar]
- 25.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. 2007. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 128: 931–946. [DOI] [PubMed] [Google Scholar]
- 27.Garcia C. K., Wilund K., Arca M., Zuliani G., Fellin R., Maioli M., Calandra S., Bertolini S., Cossu F., Grishin N., et al. 2001. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 292: 1394–1398. [DOI] [PubMed] [Google Scholar]
- 28.Horton J. D., Cohen J. C., Hobbs H. H. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown M. S., Goldstein J. L. 1976. Analysis of a mutant strain of human fibroblasts with a defect in the internalization of receptor-bound low density lipoprotein. Cell. 9: 663–674. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein J. L., Brown M. S., Stone N. J. 1977. Genetics of the LDL receptor: evidence that the mutations affecting binding and internalization are allelic. Cell. 12: 629–641. [DOI] [PubMed] [Google Scholar]
- 31.He G., Gupta S., Yi M., Michaely P., Hobbs H. H., Cohen J. C. 2002. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277: 44044–44049. [DOI] [PubMed] [Google Scholar]
- 32.Garcia C. K., Wilund K., Arca M., Zuliani G., Fellin R., Maioli M., Calandra S., Bertolini S., Cossu F., Grishin N., Barnes R., Cohen J. C., Hobbs H. H. 2001. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 292: 1394–1398. [DOI] [PubMed] [Google Scholar]
- 33.Maurer M. E., Cooper J. A. 2006. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J. Cell Sci. 119: 4235–4246. [DOI] [PubMed] [Google Scholar]
- 34.Pearse B. M., Robinson M. S. 1990. Clathrin, adaptors, and sorting. Annu. Rev. Cell Biol. 6: 151–171. [DOI] [PubMed] [Google Scholar]
- 35.Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 102: 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. 2008. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 15: 209–219. [DOI] [PubMed] [Google Scholar]
- 37.Mayor S., Pagano R. E. 2007. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8: 603–612. [DOI] [PubMed] [Google Scholar]
- 38.Zelcer N., Hong C., Boyadjian R., Tontonoz P. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 325: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo Y., Lee P. C., Sguigna P. V., DeBose-Boyd R. A. 2011. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. USA. 108: 20503–20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxwell K. N., Fisher E. A., Breslow J. L. 2005. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA. 102: 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancino J., Torrealba C., Soza A., Yuseff M. I., Gravotta D., Henklein P., Rodriguez-Boulan E., Gonzalez A. 2007. Antibody to AP1B adaptor blocks biosynthetic and recycling routes of basolateral proteins at recycling endosomes. Mol. Biol. Cell. 18: 4872–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He C., Klionsky D. J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barth S., Glick D., Macleod K. F. 2010. Autophagy: assays and artifacts. J. Pathol. 221: 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458: 445–452. [DOI] [PubMed] [Google Scholar]
- 45.Berlin I., Schwartz H., Nash P. D. 2010. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J. Biol. Chem. 285: 34909–34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raiborg C., Malerod L., Pedersen N. M., Stenmark H. 2008. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 314: 801–813. [DOI] [PubMed] [Google Scholar]
- 47.Razi M., Futter C. E. 2006. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 17: 3469–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bache K. G., Raiborg C., Mehlum A., Stenmark H. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278: 12513–12521. [DOI] [PubMed] [Google Scholar]
- 49.Komada M., Soriano P. 1999. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13: 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gullapalli A., Wolfe B. L., Griffin C. T., Magnuson T., Trejo J. 2006. An essential role for SNX1 in lysosomal sorting of protease-activated receptor-1: evidence for retromer-, Hrs-, and Tsg101-independent functions of sorting nexins. Mol. Biol. Cell. 17: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dores M. R., Chen B., Lin H., Soh U. J., Paing M. M., Montagne W. A., Meerloo T., Trejo J. 2012. ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 197: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haugsten E. M., Zakrzewska M., Brech A., Pust S., Olsnes S., Sandvig K., Wesche J. 2011. Clathrin- and dynamin-independent endocytosis of FGFR3–implications for signalling. PLoS ONE. 6: e21708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eden E. R., Patel D. D., Sun X. M., Burden J. J., Themis M., Edwards M., Lee P., Neuwirth C., Naoumova R. P., Soutar A. K. 2002. Restoration of LDL receptor function in cells from patients with autosomal recessive hypercholesterolemia by retroviral expression of ARH1. J. Clin. Invest. 110: 1695–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norman D., Sun X. M., Bourbon M., Knight B. L., Naoumova R. P., Soutar A. K. 1999. Characterization of a novel cellular defect in patients with phenotypic homozygous familial hypercholesterolemia. J. Clin. Invest. 104: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer R. D., Srinivasan S., Singh A. J., Mahoney J. E., Gharahassanlou K. R., Rahimi N. 2011. PEST motif serine and tyrosine phosphorylation controls vascular endothelial growth factor receptor 2 stability and downregulation. Mol. Cell. Biol. 31: 2010–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hewitt E. W., Duncan L., Mufti D., Baker J., Stevenson P. G., Lehner P. J. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21: 2418–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizuno T., Hayashi H., Naoi S., Sugiyama Y. 2011. Ubiquitination is associated with lysosomal degradation of cell surface-resident ATP-binding cassette transporter A1 (ABCA1) through the endosomal sorting complex required for transport (ESCRT) pathway. Hepatology. 54: 631–643. [DOI] [PubMed] [Google Scholar]
- 58.Williams R. L., Urbe S. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8: 355–368. [DOI] [PubMed] [Google Scholar]
- 59.Berlin I., Schwartz H., Nash P. D. 2010. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J. Biol. Chem. 285: 34909–34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroemer G., Marino G., Levine B. 2010. Autophagy and the integrated stress response. Mol. Cell. 40: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knoblauch H., Muller-Myhsok B., Busjahn A., Ben A. L., Bahring S., Baron H., Heath S. C., Uhlmann R., Faulhaber H. B., Shpitzen S., et al. 2000. A cholesterol-lowering gene maps to chromosome 13q. Am. J. Hum. Genet. 66: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.