Abstract
Multiphoton fluorescence recovery after photobleaching (MP-FRAP) is a laser microscopy technique used to measure diffusion coefficients of macromolecules in biological systems. The three-dimensional resolution and superior depth penetration within scattering samples offered by MP-FRAP make it an important tool for investigating both in vitro and in vivo systems. However, biological systems frequently confine diffusion within solid barriers, and to date the effect of such barriers on the measurement of absolute diffusion coefficients via MP-FRAP has not been studied. We have used Monte Carlo simulations of diffusion and MP-FRAP to understand the effect of barriers of varying geometries and positions relative to the two-photon focal volume. Furthermore, we supply ranges of barrier positions within which MP-FRAP can confidently be employed to measure accurate diffusion coefficients. Finally, we produce two new MP-FRAP models that can produce accurate diffusion coefficients in the presence of a single plane boundary or parallel infinite plane boundaries positioned parallel to the optical axis, up to the resolution limit of the multiphoton laser scanning microscope.
I. INTRODUCTION
Multiphoton fluorescence recovery after photobleaching (MP-FRAP) is a laser microscopy technique typically employed to measure diffusion coefficients within biological systems [1]. MP-FRAP is performed by using a brief, high-intensity, laser flash to generate photobleaching within a region of interest in a fluorescent sample. The laser is then attenuated and the region of interest is monitored as still-fluorescent molecules from outside the region diffuse in to replace the outwardly diffusing bleached molecules. The resulting fluorescence versus time curve can be fitted to an analytical formula to produce the diffusion coefficient of the mobile fluorophore. In an MP-FRAP experiment, fluorescence and photobleaching are both generated via multiphoton excitation [1]. The intrinsic spatial confinement of multiphoton excitation results in a three-dimensionally (3D) resolved bleaching/monitoring volume [2] and allows the use of MP-FRAP to measure three-dimensionally resolved diffusion coefficients within intact samples. This intrinsic spatial confinement obviates the need for a confocal pinhole and allows MP-FRAP, as well as the multiphoton laser-scanning microscope upon which it is based, to probe living tissue down to depths of several hundred micrometers. In its original formulation [1] MP-FRAP was demonstrated with a parked bleaching and monitoring beam. Several variants of MP-FRAP have since been demonstrated with patterned photobleaching accomplished by interfering beams [3], beam scanning [4], and other methods. In this work we refer to the experiment performed in its classical parked beam configuration as MP-FRAP.
Other techniques employed to measure biological diffusion include (single-photon) fluorescence recovery after photobleaching (FRAP), FRAP with spatial Fourier analysis (SFA-FRAP), single particle tracking (SPT), and fluorescence correlation spectroscopy (FCS). FRAP is the precursor to MPFRAP and was developed in the 1970s [5–7] to probe transport parameters in biological systems. Due to the lack of spatial confinement of the one-photon excitation process, FRAP is applied to two-dimensional samples, defined as having an axial extent that is significantly smaller than the Rayleigh length of the focused bleach and monitor beam. High-resolution measurements with high numerical aperture (NA) lenses are therefore limited to thin samples (<1 μm). FRAP can be used on thicker samples by using cylindrical beams (i.e., a low NA lens) with longer Rayleigh lengths. This again provides a two-dimensional measurement of the diffusion coefficient, averaged over the axial extent of the sample. Finally, FRAP with a confocal pinhole can be used to achieve 3D resolved measurements [8], but rely on numerical analysis rather than analytical formulas. The use of spatial Fourier analysis allows SFA-FRAP [9] to probe diffusion in intact thick tissue, but the technique also has low spatial resolution (~40 μm) and is limited to the depth penetration of epifluorescence microscopy (~50 μm). SPT tracks individual molecules via high spatiotemporal imaging to determine their transport properties, with two-dimensional (2D) imaging techniques (i.e., epifluorescence) producing 2D resolved measurements of diffusion coefficients [10,11] and 3D imaging techniques producing 3D resolved measurements of diffusion coefficients. FCS comes in both one-photon [12] and two-photon [13,14] varieties and like MP-FRAP it can be used to measure diffusion with high, 3D , resolution. SPT and FCS rely on low fluorophore concentrations (with accompanying low signals), while MP-FRAP relies on high fluorophore concentrations (with accompanying large signals), making the techniques complementary, especially in the difficult optical environment of scattering tissue.
The effect of barriers to diffusion is an important and relevant topic regarding the study of both biological and nonbiological systems. MP-FRAP has been employed to measure absolute diffusion coefficients in the cytoplasm of cells [1,15], in cartilage [16], in optically fabricated gels [17], and in blood plasma [18]. In these cases, the volume surrounding the bleached spot was assumed to be “open,” with barriers to diffusion at infinity. However, many biological systems inherently confine measurements to regions within solid barriers to diffusion, such as cell walls and organelles. In vivo systems in particular offer few free volumes for diffusion measurements. Clearly, when the focal volume is positioned within a cell there is concern that the confined geometry will affect the measured diffusion coefficient. However, even when the bleach and monitor volume is positioned outside of individual cells, the tissue interstitium exhibits a complex structure that can interfere with free diffusion due to adjacent cellular structures. Determining the effects of these barriers, and the distances over which these effects become significant, is absolutely necessary for MP-FRAP to be performed accurately in vivo.
In the limit that these barriers become extremely close, their impact is relatively straightforward to model because they simply change the dimensionality of the system. For example, it has been shown that an MP-FRAP experiment in microvilli (an extremely narrow tube) can be modeled by one-dimensional diffusion [19,20]. However, there has been no analysis of MP-FRAP applied in systems with boundaries to diffusion positioned at intermediate distances, i.e., neither approaching zero nor at infinity, in order to determine their effects on the reported diffusion coefficient.
There have been several recent studies investigating the various effects caused by barriers to diffusion in relation to micro- and nanostructures [21,22], nuclear magnetic resonance [23], diffusion-convection processes [24,25], anomalous diffusion [26], and diffusion in channels [27]. However, the results of these studies are not presented in the context of experimental biological diffusion measurement techniques. In recognition of the prevalence of small volumes presented by many biological systems, work has been done to apply FCS [28–32], SPT [33,34], and FRAP [35] in systems where the focal volume is confined. While MP-FRAP has been applied to both living cells and in vivo, to date the effect of barriers on the measurement of absolute diffusion coefficients via MP-FRAP has been largely neglected (although in the development of one patterned photobleaching variant of MP-FRAP the effect of axial boundaries was considered [4]).
Barriers to diffusion are omnipresent in vivo, and their effects must be considered when attempting to perform accurate MP-FRAP. In this work we explore the effects of different barrier geometries on the diffusion coefficient reported by MP-FRAP, discuss the mechanism by which these barriers affect the reported diffusion coefficient, and determine the appropriate distances at which the effects of barriers can be neglected. To do this we simulate the diffusive spread of a distribution of bleached molecules in the presence of various barriers to diffusion via Monte Carlo simulation and then calculate the resultant (two-photon) fluorescence signal, producing an artificial fluorescence versus time curve. We then fit many such curves to the classical MP-FRAP equation, which assumes all barriers are at infinity, and explore how the presence of differing barriers produces errors in the reported diffusion coefficient. In the case of both a single plane boundary and two parallel infinite plane boundaries oriented parallel to the optical axis, we introduce new models of MP-FRAP that explicitly account for the presence of one or more boundaries and explore how these new models improve the accuracy of the reported diffusion coefficients. This work is essential to the application of MP-FRAP in vivo.
II. MONTE CARLO MODEL OF MP-FRAP
A. Initial fluorophore distribution
The initial concentration distribution of unbleached fluorophore immediately after the photobleaching pulse, in the limit that the boundaries to diffusion are at infinity, is given by Brown et al. [1]:
| (1) |
where co is the initial equilibrium concentration of fluorophore, b is the number of photons absorbed per photobleaching event, qb is the quantum efficiency for b-photon photobleaching, δb is the multiphoton fluorescence action cross-section of the fluorophore for the order of excitation required for photobleaching, is the time average of the bleach intensity raised to the bth power, and Δt is the duration of the bleaching pulse.
The bleach intensity can be approximated as a 3D Gaussian [1]:
| (2) |
where ωr and ωz are the 1/e2 radial and axial dimensions of the two-photon focal volume, respectively, and is the time average of the intensity at the two-photon focal volume center raised to the bth power.
For the purpose of simulation, it is more efficient and effective to follow the bleached fluorophores [36–38]. Substituting Eq. (2) into Eq. (1), setting b = 2 for a two-photon bleaching process and co = 1 in anticipation of populating nodes later to determine the amplitude, and noting that the bleach depth parameter is defined as , we find the initial distribution of bleached fluorophore:
| (3) |
The bleach depth parameter was chosen to be β = 0.25, a value typical of experimental in vivo MP-FRAP recovery curves [1,18]. The axial and radial extents of the two-photon focal volume were defined as ωr ≡ 2.6λ/(2πNA) and ωz ≡ 8.8nλ/[2π(NA)2], respectively, where λ is the wavelength of the excitation laser, n is the index of refraction of the immersion medium, and NA is the numerical aperture of the lens [39]. Our simulations represent the NA extremes of typical water-immersion lenses (λ = 780 nm, n = 1.33, NA = 0.5 or 1.2). Space was discretized into a regular lattice with spacing defined by the expected diffusion properties (see “Diffusion” below). One thousand bleached fluorophores were placed at lattice points using Eq. (3) as the probability distribution and with the caveat that no fluorophores were allowed outside any diffusive barriers introduced into the system. Multiple occupancy on a single node was permitted, though rarely occurred.
It is important to note that the size of the bleaching distribution is not necessarily the same as that of the optical focal volume. As explained in Brown et al. [1], the shape of the photobleaching volume will depend upon the photochemical mechanism of photobleaching, the bleach depth parameter, and the characteristic radii of the optical point spread function (PSF). In order to explore a tractable parameter space, we are assuming the simple first-order model of photobleaching photochemistry described within Brown et al., an experimentally relevant value for the bleach depth parameter of β = 0.25, and two values of microscope NA (and hence two values for each of the characteristic radii of the optical PSF) of 0.5 and 1.2. Due to the relatively small value of β, this produces a characteristic size for the bleaching volume that is within 3% of the size of the optical PSF; hence we plot the effects of barrier proximity as a function of distance from the center of the focal volume in units of ωr and ωz, the radial and axial e–2 extents of the PSF, respectively. To extrapolate these results to other photobleaching chemistries or greater bleach depths, one can calculate the characteristic radial and axial dimensions of that photobleaching distribution and treat those as ωr and ωz for comparison to our results.
B. Diffusion
Diffusion was modeled as a random walk on a 3D lattice [37,38,40,41]. Lattice spacing was determined by the 3D diffusion equation, 〈r2〉 = 6Dt, where the diffusion coefficient, D, was chosen a priori and the time step, t, was chosen to be approximately 1/1000 of the typical diffusive recovery time for a system with a diffusion coefficient D and with radial and axial focal volume widths ωr and ωz. For both the low and high NA cases, D was chosen as 10 μm2/s, approximately the experimental diffusion coefficient for 2000 kD fluorescein dextran [18]. The corresponding time steps were chosen as 3.12 and 0.376 μs, respectively, and the lattice spacings were calculated to be 13.7 and 4.75 nm, respectively.
C. Boundary conditions
Four boundary models were applied to the diffusing system: a single infinite plane boundary parallel and perpendicular to the optical axis, two parallel infinite plane boundaries parallel and perpendicular to the optical axis, a hollow infinite cylindrical boundary parallel and perpendicular to the optical axis, and a hollow spherical boundary. In the context of our simulations a barrier is considered infinite if a particle cannot cross the barrier at any time during the simulation. The parallel, cylindrical, and spherical boundaries were positioned symmetrically about the focal volume center, and the positions of all of the boundaries were defined as fractions of ωr or ωz relative to the focal volume center. All boundaries were assumed to be perfectly reflecting; i.e., any particle attempting to cross a boundary was returned to the node it was occupying when the step began. There were no bleached or unbleached molecules beyond the boundaries at t = 0 or at any subsequent point.
D. MP-FRAP
The fluorescence intensity generated by a weak monitoring beam that is held stationary in the sample and produces fluorescence through an m-photon process is given by
| (4) |
where δm is the multiphoton fluorescence action cross-section of the fluorophore for the order of excitation required to produce fluorescence, E is the collective efficiency of the detection system, and m is the number of photons absorbed per excitation event.
We can calculate the “missing fluorescence” that would be generated by the bleached fluorophores were they not bleached by re-expressing the integral as a sum of the monitor intensity [given by Eq. (2) with b → m] over all bleached fluorophore locations (xi,yi,zi). We can also let (1/m)Eδm → 1, as it will be divided out when the fluorescence is normalized for fitting:
| (5) |
To obtain the normalized fluorescence of the unbleached molecules, F(t)/Fo, we first normalize the missing fluorescence of the bleached molecules by the prebleach fluorescence of all the fluorophores in the system, Fo, and then subtract from one: F(t)/Fo = 1 – Fbl(t)/Fo. Fo was determined by first setting t = 0 and β = 0.25 in Eq. (6), below, truncating the sum to the first ten terms, and solving for F(0)/Fo. This value was then substituted into F(0)/Fo = 1 – Fbl(0)/Fo to deduce F0 from Fbl(0).
The natural variation of a Monte Carlo simulated random walk introduced a small amount of noise into the resulting F(t) recovery curves. On top of this, we added Poisson distributed noise to mimic the typical distribution of noise arising from photon counting experiments, and in an amount typical of in vitro MP-FRAP experiments [1,18]. Fluorescence recoveries were terminated when the change in the recovered fluorescence was less than 1% over a time equivalent to the half-time for complete recovery of a freely diffusing system with diffusion coefficient D.
Unless otherwise stated, all simulated F(t) curves were fit to the accepted diffusive recovery model [1] for a stationary bleaching and monitoring beam:
| (6) |
where τD is the characteristic diffusion time and R is the square of the ratio of the axial and radial dimensions of the focal volume. The diffusion coefficient is given by .
III. RESULTS AND DISCUSSION
A. Single plane boundary
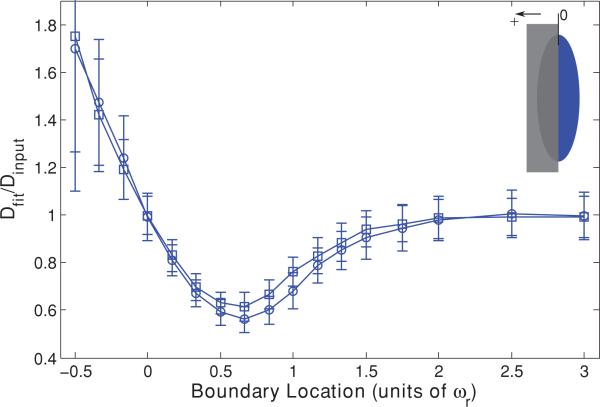
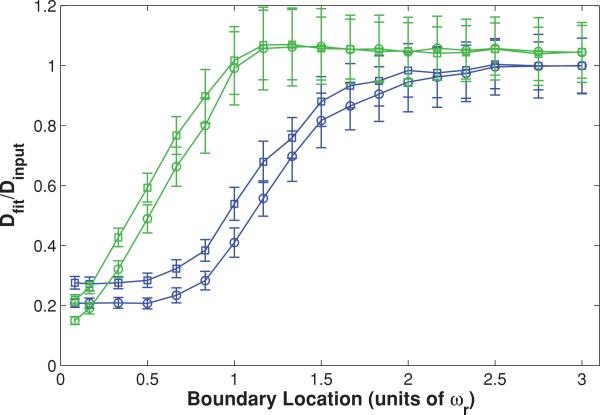
We begin our Monte Carlo investigation by introducing a single infinite plane reflective boundary parallel to the optical axis, at a range of distances measured in units of ωr relative to the focal volume center. This models diffusion measurements adjacent to cell walls [42–44]. We then generate an initial distribution of bleached molecules according to Eq. (1), with the caveat that no molecules are located beyond the boundary. Then we simulate the random diffusion of those molecules and produce an F(t) curve as described above. The resultant curve is fit to Eq. (6), the MP-FRAP formula that assumes all boundaries are at infinity. The fit diffusion coefficient is then divided by the true diffusion coefficient (defined a priori in setting up the diffusion random walk); hence errors due to the presence of a boundary are readily identified by a deviation of this ratio from one. Note that we ceased our simulations at a boundary location of –0.5ωr because at this point the average fluorescence from the focal volume in steady state is <10% of the value of the unobstructed focal volume.
The resultant data are presented in Fig. 1 and show that MP-FRAP begins to yield diffusion coefficients significantly different from the input diffusion coefficient (defined hereafter as when the mean fit diffusion coefficient is more than 1 standard deviation (SD) different from the input diffusion coefficient) when the boundary passes a distance of 1.3ωr from the focal volume center for a high NA lens and a distance of 1.5ωr for a low NA lens. In each of these cases, the fit diffusion coefficient, Dfit, becomes significantly different from the input coefficient, Dinput, before the boundary crosses the focal volume center (0ωr), and the deviation is biphasic, with Dfit initially smaller than Dinput, then becoming significantly larger than Dinput as the boundary crosses the focal volume center. The erroneously low value of Dfit is most pronounced when the boundary is in the range of ~0ωr–1.5ωr, and we hypothesized that this occurs because the boundary hinders the complete escape of bleached molecules from the focal volume, forcing a selection of fluorophores to reside longer in the neighborhood of the focal volume, thereby lengthening the recovery time. We further hypothesized that as a growing portion of the focal volume becomes “hidden” behind the boundary its characteristic radial size will become smaller than ωr and fitting of the resultant recovery curves to Eq. (6), which assumes that ωr is the relevant radial length scale, will produce the erroneously large values of Dfit that become apparent as the wall approaches –0.5ωr.
FIG. 1.
(Color) Single boundary parallel to the optical axis at a range of positions, presented as fractions of ωr from the focal volume center, for both a high NA (squares) lens and a low NA (circles) lens. Negative position values indicate that the boundary has crossed the focal volume center (i.e., more than half the focal volume is hidden beyond the boundary). Fluorescence recovery curves were generated via Monte Carlo simulation and fit to the standard MPFRAP model [Eq. (6)]. Fit diffusion coefficients were normalized to the input diffusion coefficient; hence an accurate fit produces a ratio of one.
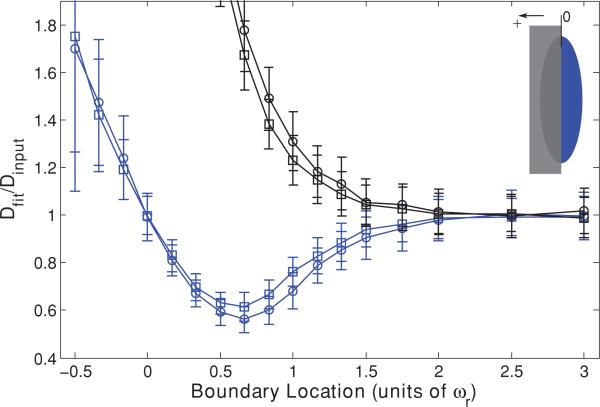
To test these hypotheses we first repeated the series of Monte Carlo simulations, now using a “destructive” boundary instead of a “reflective” one, such that each bleached molecule that attempted to cross the boundary was removed from the simulation. As shown in Fig. 2, removal of the reflected fluorophores eliminated the initially low values of Dfit but retained the later large values, suggesting that it is indeed reflection of bleached molecules off of the boundary and back into the focal volume that lengthens the recovery time and leads to the initial erroneously small values of Dfit. One may note that the curves describing the destructive case begin to upswing while those for the reflective case are still in their initial downturn. At this point in the reflective case the effect on recovery is dominated by the fluorophores reflecting back into the region of the focal center, and only as the boundary crosses the focal center does the effect on shortening ωr begin to overcome particle deflection and reverse the trend, eventually leading to too-large values of Dfit.
FIG. 2.
(Color) Single “destructive” boundary (black) and “reflective” boundary (blue) parallel to the optical axis at a range of positions, presented as fractions of ωr from the focal volume center for a high NA (squares) lens and a low NA (circles) lens. Negative position values indicate that the boundary has crossed the focal volume center. Fluorescence recovery curves were generated via Monte Carlo simulation and fit to the standard MP-FRAP model. Fit diffusion coefficients were normalized to the input diffusion coefficient.
Next, to demonstrate the effect that changing the focal volume dimensions has on fluorescence recovery and fitting, we generated data assuming an unobstructed focal volume and free diffusion, but with ωr reduced to mimic the influence of the barriers reducing the focal volume as introduced in the simulations. We then fit the resulting fluorescence curves assuming a focal volume with the original ωr. As ωr was reduced to successively smaller values, the fit diffusion coefficient became increasingly large, beyond the value of Dinput (data not shown). This reproduces the trend seen in Fig. 2 and suggests that the erroneously large values of Dfit are indeed due solely to a reduction in the bleaching distribution and monitoring volume, and hence are an overestimate of their characteristic size during the fitting process.
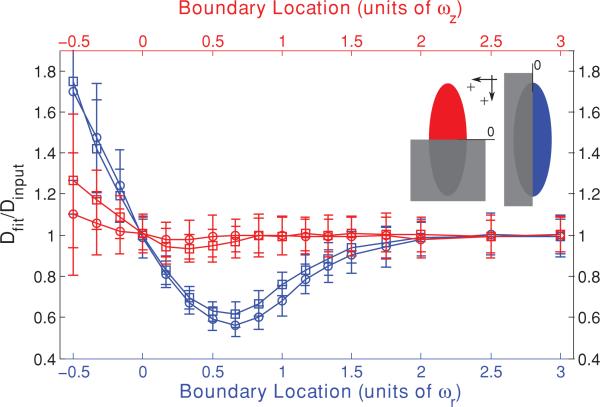
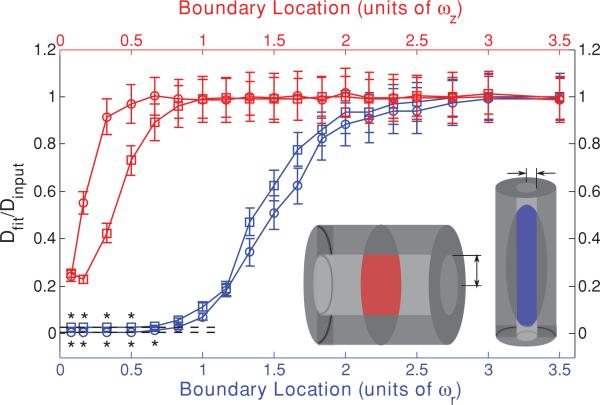
Inspection of Fig. 1 also reveals that the low NA curve is more affected by the approach of the barrier than is the high NA curve, with more significantly low initial values for Dfit. We hypothesized that this is due to the different aspect ratios of the focal volumes (ωz/ωr = 3.75 for the high NA case and ωz/ωr = 9 for the low NA case). The fastest route for diffusive escape from an initial bleached distribution will be along the shortest dimension of the initial distribution, and the higher aspect ratio of the low NA focal volume means that the diffusive transport in the direction of the approaching barrier is a more significant contributor to fluorescence recovery for that objective lens than for a higher NA objective, resulting in a more significant effect of the barrier. To test this hypothesis we repeated the series of Monte Carlo simulations, but brought in a barrier to diffusion that was perpendicular to the optical axis. The fastest route for diffusive escape in this geometry is now parallel to the surface of the approaching barrier and thus unhindered by it, leading us to predict that the initially low values of Dfit should deviate from Dinput far less than for the case of boundaries parallel to the optical axis. Furthermore, the low values of Dfit should now be least significant for the low NA case, as that has the highest aspect ratio. As shown in Fig. 3, the initially low values of Dfit for the boundary perpendicular to the optical axis do indeed deviate less from Dinput and are now least significant for the low NA case, thus confirming our hypothesis. Figure 3 also reveals that MP-FRAP begins to yield diffusion coefficients significantly different from the input diffusion when a boundary perpendicular to the optical axis passes –0.3ωz for a high NA lens. For the low NA case, Dfit does not deviate significantly for any of the boundary locations assessed.
FIG. 3.
(Color) Single boundary parallel (blue) and perpendicular (red) to the optical axis at a range of positions, presented as fractions of ωr or ωz, respectively, from the focal volume center for a high NA (squares) lens and a low NA (circles) lens. Negative position values indicate that the boundary has crossed the focal volume center. Fluorescence recovery curves were generated via Monte Carlo simulation and fit to the standard MP-FRAP model. Fit diffusion coefficients were normalized to the input diffusion coefficient.
To improve the accuracy of the reported diffusion coefficient for the case of a single barrier to diffusion, we have derived an alternative analytical model of the fluorescence recovery (see Appendix) that takes into account the presence of the barrier. For a barrier lying parallel to the optical axis, the new model is
| (7) |
where μn(t) = 1 + 2nt/τD, νn(t) = 1 + 2nt/RτD, and u is the x or y position of the bleached molecule distribution center relative to the boundary. For a barrier perpendicular to the optical axis, the form is the same but μn → νn and ωr → ωz in the exponential and complementary error functions.
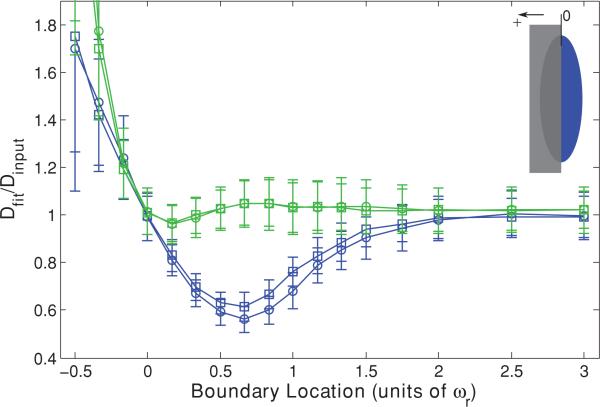
When this new MP-FRAP “single boundary” model is used to fit simulated diffusion curves produced in the presence of a single barrier parallel to the optical axis at a known distance, the fit diffusion coefficients improve dramatically over a wide range of barrier distances (see Fig. 4). For the case of a barrier perpendicular to the optical axis, Dfit does not improve significantly over the already generally accurate results using the standard model (data not shown). Figure 4 shows that Dfit remains accurate until after the boundary has crossed the center of the focal volume (0ωr), but becomes significantly different from Dinput as a boundary parallel to the optical axis passes –0.15ωr from the focal volume center for both a high NA lens and a low NA lens. If the position of the boundary is not known, we can allow u to be a free fitting parameter. However, doing so yields erroneous values for the diffusion coefficient for barrier positions <1ωr (data not shown).
FIG. 4.
(Color) Single boundary parallel to the optical axis at a range of positions, presented as fractions of ωr from the focal volume center for a high NA (squares) lens and a low NA (circles) lens. Negative position values indicate that the boundary has crossed the focal volume center. Fluorescence recovery curves were generated via Monte Carlo simulation and fit to the standard MP-FRAP model (blue) and the new MP-FRAP model (green) designed for use near a single barrier. Fit diffusion coefficients were normalized to the input diffusion coefficient.
B. Parallel plane boundaries
Two parallel infinite plane reflective boundaries mimic systems such as the regions between cell walls found in tumor and brain extracellular space [42–44]. To model this system we introduce parallel plane barriers symmetrically about the focal volume center, positioned parallel or perpendicular to the optical axis at a range of distances measured in units of ωr or ωz relative to the focal volume center. As before, the data are presented as Dfit/Dinput as a function of boundary location. In each case, Fig. 5 shows that as the boundaries approach the focal volume, the fit diffusion coefficient begins to drop compared to the input diffusion coefficient. Dfit becomes significantly different from Dinput as boundaries parallel to the optical axis pass 1.5ωr from the focal volume center for a high NA lens and 1.8ωr for a low NA lens. For boundaries perpendicular to the optical axis, Dfit becomes statistically significantly different from Dinput as the boundaries pass 0.5ωz from the focal center for a high NA lens. For the low NA case, Dfit does not deviate significantly for any of the boundary locations assessed.
FIG. 5.
(Color) Two parallel boundaries parallel (blue) or perpendicular (red) to the optical axis at a range of positions symmetric about the focal volume center and presented as fractions of ωr or ωz for a high NA (squares) lens and a low NA (circles) lens. Fluorescence recovery curves were generated through Monte Carlo simulation and fit to the standard MP-FRAP model. Fit diffusion coefficients were normalized to the input diffusion coefficient. Dashed lines mark the limit of 2D diffusion as indicated by generating data with the 2D MP-FRAP recovery equation and fitting it to the 3D MP-FRAP recovery equation. For data points marked with an asterisk (*), the accurate diffusion coefficient was recovered when the data were fit with the 2D MP-FRAP model.
As in the case of the single boundary, the effect on the diffusion coefficient is more significant for the case of two parallel plane boundaries running parallel to the optical axis. As demonstrated previously, this arises because boundaries parallel to the optical axis reduce the opportunity for diffusing molecules to leave the focal volume via the shorter radial dimension, which predominantly determines the duration of recovery for a freely diffusing sample. This is also shown by the opposite behaviors of low and high NA lenses in the two geometries. When the boundaries are parallel to the optical axis, the low NA lens is the most affected because radial diffusion is more significant in this high aspect ratio focal volume. Conversely, when the boundaries are perpendicular to the optical axis, the low NA lens is the least affected, for the same reason. Unlike the single boundary case, however, as the boundaries significantly reduce the focal volume Dfit does not rise but levels off. The characteristic length of the focal volume is reduced in the direction normal to the plane surfaces, suggesting that the characteristic recovery time should become shorter, as in the single boundary case. However, as the distance between the planes approaches zero, diffusion is effectively confined to two dimensions, and this effect dominates.
By letting ωz → ∞ in Eq. (6) we obtain a two-dimensional form of the MP-FRAP model, which is valid in the limit of a 2D system perpendicular to the optical axis and identical to the original one-photon FRAP model [5]:
| (8) |
By generating data using this 2D formula and then fitting the data to the standard 3D model we find that in the limit of 2D diffusion the 3D model should yield a value of the diffusion coefficient that is 0.84 ± 0.04 times the accepted value for a high NA lens and 0.96 ± 0.03 times the accepted value for a low NA lens. These limits are plotted in Fig. 5 as the dashed lines and coincide with the values of the normalized diffusion coefficients at small values of ωz as determined by the Monte Carlo simulations of MP-FRAP. The asterisks (*) mark data sets that, when refit with the 2D MP-FRAP model given by Eq. (8), recovered the input diffusion coefficient to within 1 SD.
To improve the accuracy of the reported diffusion coefficient for the case of parallel infinite plane barriers to diffusion, we have derived an alternative analytical model of the fluorescence recovery (see the Appendix) that takes into account the presence of the barriers. For barriers lying parallel to the optical axis, the new model is
| (9) |
For barriers perpendicular to the optical axis, the form is the same but μn → νn and ωr → ωz in the exponential and error functions.
When this new MP-FRAP “parallel boundary” model is used to fit simulated diffusion curves produced assuming the presence of two parallel plane barriers parallel to the optical axis at a known distance, the fit diffusion coefficients improve dramatically over a wide range of barrier distances (see Fig. 6). For the case of barriers perpendicular to the optical axis the values for Dfit do not improve significantly over the already widely accurate results using the standard model (data not shown). Figure 6 shows that Dfit becomes significantly different from Dinput as boundaries parallel to the optical axis pass ωr from the focal volume center for both a high NA lens and a low NA lens. If the position of the boundary is not known, we can allow u to be a free fitting parameter. However, doing so yields poor fits for barrier positions <1ωr (data not shown).
FIG. 6.
(Color) Two parallel plane boundaries parallel to the optical axis at a range of positions, presented as fractions of ωr from the focal volume center for a high NA (squares) lens and a low NA (circles) lens. Fluorescence recovery curves were generated via Monte Carlo simulation and fit to the standard MP-FRAP model (blue) and the new MP-FRAP model (green) designed for use between two parallel plane barriers. Fit diffusion coefficients were normalized to the input diffusion coefficient.
C. Cylindrical boundary
An infinite hollow cylindrical boundary provides an excellent approximation for neuronal dendrites and axons, in which transport measurements are of interest in neurobiological research [45,46]. In our simulations, we introduce a cylindrical boundary both parallel and perpendicular to the optical axis, positioned symmetrically about the focal volume center with a range of radii measured in units of ωr and ωz. Again, the data are presented as Dfit/Dinput as a function of boundary location. As with the case of the parallel plane boundaries, Fig. 7 shows that as the cylindrical boundary approaches the focal volume, Dfit begins to drop compared to Dinput, and these effects occur at values of ωr for the case of a cylindrical boundary parallel to the optical axis larger than for corresponding values of ωz in the perpendicular case. Specifically, Dfit becomes significantly smaller than Dinput as the radius of a cylinder parallel to the optical axis becomes smaller than 1.8ωr for a high NA lens and 2ωr for a low NA lens. Dfit becomes statistically significantly smaller than Dinput as the radius of a cylinder perpendicular to the optical axis becomes smaller than 0.7ωz for a high NA lens and 0.3ωz for a low NA lens.
FIG. 7.
(Color) Cylindrical boundary parallel (blue) and perpendicular (red) to the optical axis at a range of positions, symmetric about the focal volume center and presented as fractions of ωr or ωz, for a high NA (squares) lens and a low NA (circles) lens. Fluorescence recovery curves were generated through Monte Carlo simulation and fit to the standard MP-FRAP model. Fit diffusion coefficients were normalized to the input diffusion coefficient. Dashed lines mark the limit of 1D diffusion as indicated by generating data with the 1D MP-FRAP recovery equation and fitting it to the 3D MP-FRAP recovery equation. For data points marked with an asterisk (*), the accurate diffusion coefficient was recovered when the data were fit with the 1D MP-FRAP model.
Similar to the parallel plane boundaries, we find that when the cylindrical boundary is sufficiently constricting the diffusion effectively becomes one dimensional (1D). Consequently, although the size of the available volume decreases Dfit does not increase above Dinput after initially dropping significantly below Dinput. In the limit of 1D diffusion, the MP-FRAP model can be altered to account for the dimensional change. By letting ωr → ∞ in Eq. (6) we obtain a 1D form of the MP-FRAP model for diffusion along the optical axis:
| (10) |
By generating data using this 1D model and fitting it to the 3D model we find that in the limit of 1D diffusion the 3D model should yield a value of the diffusion coefficient that is 0.026 ± 0.001 times the accepted value for a high NA lens and 0.006 ± 0.001 times the accepted value for a low NA lens. These limits are plotted in Fig. 7 as the dashed lines and coincide with the values of the normalized diffusion coefficients at small values of ωr as determined by the Monte Carlo simulations of MP-FRAP. The asterisks (*) mark data sets that, when refit with the 1D MP-FRAP model, recovered the input diffusion coefficient, within 1 SD.
D. Spherical boundary
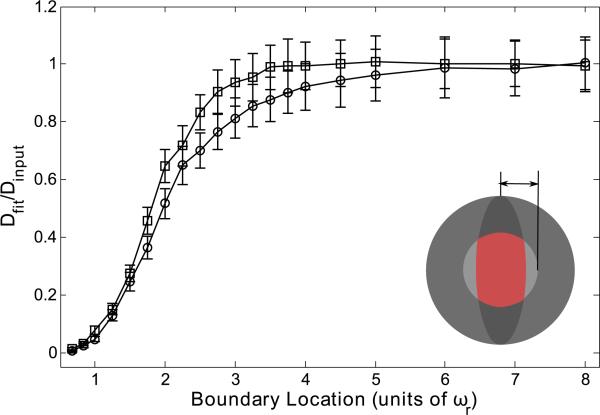
A hollow spherical boundary well approximates a cell body, cell nucleus, or dendritic spine [47–49]. We introduce the spherical boundary symmetrically about the focal volume center with a range of radii measured in units of ωr. The data are presented as Dfit/Dinput as a function of boundary location. Figure 8 shows that the fit diffusion coefficient drops rapidly relative to the input diffusion coefficient as the boundaries narrow in on the focal volume. Specifically, Dfit becomes significantly smaller than Dinput as the radius of the sphere becomes smaller than 2.75ωr for a high NA lens and 3.75ωr for a low NA lens.
FIG. 8.
(Color) Spherical boundary at a range of positions, symmetric about the focal volume center and presented as fractions of ωr, for a high NA (squares) lens and a low NA (circles) lens. Fluorescence recovery curves were generated through Monte Carlo simulation and fit to the standard MP-FRAP model. Fit diffusion coefficients were normalized to the input diffusion coefficient.
As the sphere becomes sufficiently small, many (and eventually all) of the bleached molecules will be unable to leave the focal volume. This suggests that the standard 3D MP-FRAP model [Eq. (6)], which assumes free diffusion of the entire fluorophore population, may be inappropriate to fit the recovery. The concept of an “immobile fraction” has been discussed previously [1,50] in the context of fluorophore populations attached to the extracellular matrix or cell cytoskeleton and involves an additional fitting parameter to account for the incomplete recovery caused by a subset of immobile fluorophores. We refit the data generated for free diffusion within a spherical boundary using standard 3D MP-FRAP with a fitting parameter for an immobile fraction, but did not achieve significantly more accurate values for the diffusion coefficient (data not shown). This arises because the case of an immobile fluorophore population and our case of a trapped but mobile population are only superficially similar. In the case of an immobile fraction, a subset of fluorophores are permanently (relative to the duration of the experiment) fixed in space. The remaining fluorophores, however, are assumed to diffuse freely. In the case of fluorophores confined within a spherical boundary, however, while many fluorophores remain within the focal volume they are always free to move. As the bleached molecules spread from their initial center-heavy distribution to a more uniform distribution, the fluorescence of the sample changes. The shape of the recovery is subtly, yet significantly, different from the case of immobile fluorophores, and fitting with this added parameter does not significantly improve the resultant diffusion coefficient.
E. Summary
Using Monte Carlo simulations, we have defined the range of applicability of MP-FRAP to measure diffusion in the presence of barriers of various geometries and positions relative to the focal volume center. Table I summarizes the results of our simulations. Using the standard MP-FRAP model, the onset of deviations in the fit diffusion coefficient is at a distance of 1.3ωr–1.5ωr for a single plane barrier parallel to the optical axis and ~–0.3ωz for a barrier perpendicular to the optical axis. Using the new MP-FRAP single boundary model (Eq. (7)) for a barrier parallel to the optical axis, the initially low values of Dfit are avoided and the point of onset of significant deviations greatly improves to ~–0.15ωr. For two parallel plane boundaries, the onset of deviations is at a distance of 1.5ωr–1.8ωr for barriers parallel to the optical axis and <0.1ωz to ~0.5ωz for barriers perpendicular to the optical axis. To improve the range of applicability for barriers parallel to the optical axis, the user could perform MP-FRAP analysis using the new two-boundary model [Eq. (9)], enabling accurate diffusion coefficients to be produced down to ωr. The onset of deviations within a hollow cylindrical boundary is at a radius of ~1.8ωr–2ωr for a cylinder parallel to the optical axis and ~0.3ωz–0.7ωz for a cylinder perpendicular to the optical axis. In each of these geometries, the high-aspect ratio of two-photon focal volumes leads to greater deviations when the boundaries are brought in parallel to the optical axis. For the sphere, the onset of deviations occurs at a radius of ~2.75ωr–3.75ωr and is not improved by fitting with an immobile fraction term.
TABLE I.
Summary of results of fitting fluorescence recovery curves in the presence of reflective boundaries to diffusion with the standard MP-FRAP diffusive-recovery model. Boundaries are located symmetrically about the focal volume center at positions measured in units of ωr or ωz. The boundary locations for a single parallel-plane boundary and two parallel-plane boundaries laying parallel to the optical axis within which the standard MP-FRAP model yields erroneous diffusion coefficients can be narrowed by using the new models presented in the text.
| Boundary geometry | Orientation with respect to optical axis | Boundary limit with standard MP-FRAP | Boundary limit with new model |
|---|---|---|---|
| Single plane | parallel | 1.3ωr–1.5ωr | ~–0.15ωr |
| perpendicular | ~–0.3ωz | ||
| Parallel-plane | parallel | 1.5ωr–1.8ωr | 1ωr |
| perpendicular | <0.1ωz–~0.5ωz | ||
| Cylinder | parallel | 1.8ωr–2ωr | |
| perpendicular | 0.3ωz –0.7ωz | ||
| Sphere | 2.75ωr –3.75ωr |
For all geometries, the error in the fit diffusion coefficient as the boundary approaches the focal volume is initially presented as a deviation below the input diffusion coefficient. This is caused by a hindrance of the ability of bleached molecules to leave the environment of the focal volume as a result of reflection off of the boundary wall(s) and back into the focal volume. For the case of a single plane barrier to diffusion, after initially dipping below Dinput, Dfit begins to rise, eventually leading to deviations greater than Dinput. This is caused by truncation of the bleach distribution and monitor volume as the focal volume crosses into the boundary wall. This same trend is not seen for the other geometries, however, even though our simulations bring the boundaries inside the focal volume, thus also truncating the bleach distribution and monitor volumes. Instead, Dfit levels off in all cases. For the parallel plane and cylindrical boundaries, this is because the confinement mimics the approach to a 2D (parallel plane) or 1D (cylindrical) system. When data at this limit for barriers parallel to the optical axis are refit with the corresponding 1D or 2D MP-FRAP models, Dfit returns the same value as Dinput. For the case of the spherically confining boundary, Dfit continues to get smaller as the boundaries move inward.
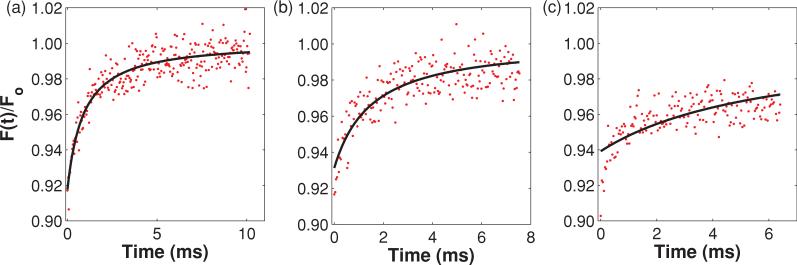
Given the significant errors evident in the fit diffusion coefficient when the various boundaries closely confine the focal volume, it is reasonable to expect that the corresponding recovery curves would be significantly altered compared to their unbounded counterparts and that the fits may be obviously poor by simple inspection. In the case of a single plane barrier or two parallel-plane barriers, while the recovery curves do lengthen as the boundaries are brought in (hence the drop in Dfit), they retain the classic recovery curve shape and fits to recovery curves for all boundary locations assessed look good by inspection (data not shown). It is not until a cylindrical barrier is brought in to narrowly confine the focal volume that any obvious error is detected. Figure 9 shows representative data sets and fits for recovery curves generated within cylindrical boundaries of three different radii. To simulate the data shown in Fig. 9(a) the cylinder radius was 3.5ωr. In this case the boundary was far enough away to achieve a result consistent with free diffusion, and Dfit/Dinput = 1.02. To generate the data in Fig. 9(b) the cylinder radius was 1.5ωr. Although by direct comparison with Fig. 9(a) the recovery curve does appear altered, the shape is typical of MP-FRAP recovery curves and the fit looks good by eye. However, in agreement with Fig. 7, Dfit/Dinput = 0.59; the fit diffusion coefficient is not accurate. Finally, to produce the data in Fig. 9(c) the cylinder radius was 1.0ωr, very nearly at the limit of 1D diffusion. In this case the curve is further altered and the fit now looks poor to the eye. Again in agreement with Fig. 7, Dfit/Dinput = 0.15. Clearly, the experimental researcher cannot rely on inspection of recovery curves and fits to indicate an error in the measurement, except in the extreme case of a narrowly constrained cylinder. Instead, she or he should follow the guidelines described here.
FIG. 9.
(Color) Representative recovery curves and fits for the case of a cylindrical barrier of three different radii (3.5ωr, 1.5ωr, and 1.0ωr, respectively), oriented parallel to the optical axis. In (a) the shape of the recovery curve matches that for free diffusion, and Dfit/Dinput = 1.02. By direct comparison with (a) the recovery curve in (b) appears altered, but the shape is typical of MP-FRAP recovery curves and the fit looks good by eye. However, Dfit/Dinput = 0.59; Dfit is not accurate. In (c) the curve is significantly altered in shape and the fit both looks poor to the eye and yields an inaccurate diffusion coefficient, Dfit/Dinput = 0.15.
Most MP-FRAP experiments are performed using a multi-photon laser scanning microscopy (MPLSM) platform, with a radial and axial resolution of imaging of ~ωr and ~ωz, respectively [51], although improved resolution can be achieved with deconvolution techniques [52]. Furthermore, MP-FRAP relies on large fluorophore concentrations, typically high enough to allow in vivo imaging of the diffusing fluorophore population. Based upon our results, the user can therefore image the spatial distribution of fluorophores and avoid MP-FRAP analysis using the standard model [Eq. (6)] when the boundaries move within the locations presented in the third column of Table I. In the cases of planar barriers and a cylindrical barrier, all oriented perpendicular to the optical axis, one cannot take advantage of the full range of safe barrier distances because the resolution limit of the microscope (~1ωz) prevents us from accurately determining our position relative to the barrier with accuracy less than the resolution limit. In those cases, MP-FRAP should only be performed when the barrier is at a resolvable distance from the focal volume center (typically ≥1ωz) to ensure that unsafe regions are not interrogated. In the case of a single barrier or two parallel-plane barriers lying parallel to the optical axis, the range of safe barrier distances has been extended by the new models [Eqs. (7) and (9)] down to or below the resolution limit.
In some experiments, the diffusing population may be concentrated enough to use for MP-FRAP but too dilute to allow imaging of the diffusive barriers. In this case, a second fluorophore could be employed to highlight the boundaries within and between which the measurement will be taken, for example, by labeling the cell walls or by introducing a second diffusible fluorophore into the region of tissue interstitium under study. By using an appropriately chosen color for the second population, the spectral overlap in fluorescence is minimized and the effects on the MP-FRAP results will be negligible.
Our results provide assurance to the experimental researcher that if she or he can successfully image the region of interest of the intended measurement, then MP-FRAP can now be relied upon to yield an accurate measurement of the absolute diffusion coefficient within the constrained space. This opens up a wide range of in vivo systems within which MP-FRAP can now be confidently applied.
IV. CONCLUSION
In this paper, we have used Monte Carlo simulations to model multiphoton fluorescence recovery after photo-bleaching in the presence of reflecting boundaries of various geometries and sizes. Our results show that MP-FRAP can produce erroneous values of the diffusion coefficient even when the boundaries are significantly larger than the focal volume. The size limit at which the boundaries begin affecting the MP-FRAP measurement varies with the geometry of the boundary, with the two extremes being a single plane perpendicular to the optical axis (Dfit becomes erroneous at ~–0.3ωz) and a sphere (Dfit becomes erroneous at 3.75ωr). The significance of the error is a function of the aspect ratio of the focal volume (i.e., the NA) and the orientation of the barriers, with barriers to diffusion in the radial direction having the greatest effect. Using our guidelines, a researcher can first image a sample using two-photon fluorescence and then measure and locate a region with the appropriate dimensions to allow an accurate measurement of the diffusion coefficient using the appropriate model. For both a single barrier and paired barriers parallel to the optical axis, we present a new model of MP-FRAP that can be used to produce accurate diffusion coefficients for boundary distances much closer than is possible with the standard MP-FRAP model. Measurements of diffusion via MP-FRAP can now be completed with confidence in an array of in vivo systems previously believed to be inaccessible.
ACKNOWLEDGMENTS
The authors would like to thank Matthew C. Sullivan for several useful conversations. This work was supported by the Department of Defense Era of Hope Scholar Research Award Program (No. W81XWH-09-1-0405), the NIH Directors New Innovator Award Program (No. 1DP2OD006501-01), and the Pew Scholars Program in the Biomedical Sciences (Edward Brown).
APPENDIX
The time-dependent concentration distribution of un-bleached fluorophore following the bleach pulse is given by Brown et al. [1]. When written in Cartesian coordinates for a concentration distribution centered at the origin, the expression is
| (A1) |
where
| (A2) |
| (A3) |
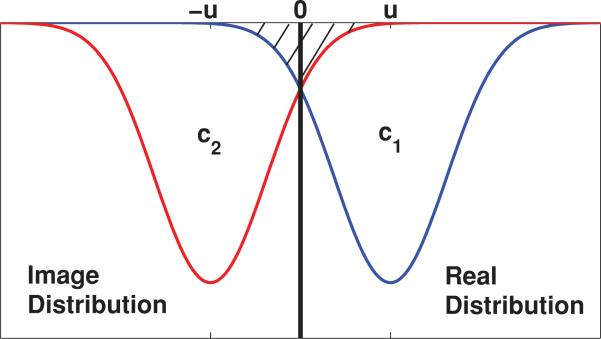
For the case of a single infinite plane boundary at the origin we can develop an approximate analytical model by replacing the barrier with the real distribution and an “image” distribution placed symmetrically about the origin at positions u and –u, which represent the distance of the real distribution from the barrier (see Fig. 10). In the region to the right of the origin, this closely models the behavior of the system: as the distributions spread due to diffusion, the overlap of the image distribution with the real distribution mimics the behavior of those fluorophores that bounce off the barrier and back into the space to the right of the barrier. With this approximation, there is some overcounting of the initial fluorophore population (hatched region in Fig. 10) when the two distributions are oriented close to the barrier. This “extra” distribution of bleached molecules will evolve over time, producing an error in the fluorescence as a function of time, and hence in the diffusion coefficient, D. However, this error remains small until the peak of the image distribution approaches the origin. As the two distributions perfectly overlap the error in D disappears and then grows again as the distribution centers continue past one another.
FIG. 10.
(Color) Placement of real and “image” bleached molecule concentration distributions for the case of a single infinite plane boundary. The overlap of the image distribution in the region of the real distribution as the two distributions spread (t > 0) mimics the behavior of diffusing molecules that bounce off the barrier and back into the focal volume. At t = 0, there will be overcounting of bleached molecules when the two distributions are close to the barrier. This error will remain small until the image distribution peak approaches the barrier.
The combined concentration distribution, assuming a barrier parallel to the optical axis in the x dimension, is given by
| (A4) |
where c1 and c2 are produced by introducing a coordinate shift into Eq. (A1), and the 1/2 accounts for the fact that we have introduced an image distribution that doubles the true concentration of fluorophore.
The fluorescence recovery is monitored by a low-intensity laser beam centered on the real concentration distribution and is given by
| (A5) |
where δm is the multiphoton fluorescence action cross-section, E is the collection efficiency of the system, m is the number of photons required to produce fluorescence from a single fluorophore, and is the time-average of the bleach intensity raised to the mth power, given by
| (A6) |
It is important to note that while for the standard model derivation the integral in F(t) is taken over all space, in the presence of a single barrier the integral along the dimension interrupted by the barrier (in this case x) is taken only from 0 → ∞.
When Eqs. (A4) and (A6) are substituted into Eq. (A5) and the integral is performed, the simplified expression for the fluorescence recovery, letting m = b = 2, is
| (A7) |
where is the equilibrium value of the fluorescence before the photobleaching pulse. We can compare Eq. (A7) with the standard MP-FRAP model [Eq. (6)] to gain some insight into the new form. As noted earlier, the 1/2 arises from our introduction of the image distribution. The first complementary error function comes from the calculation of , which is evaluated over the limits 0 → ∞. The first part of the summation, appearing on the first line, is the standard MP-FRAP equation. Finally, the contribution to the fluorescence recovery is shared between the real concentration distribution, represented by the second complementary error function, and the image concentration distribution, represented by the exponential × complementary error function term. In the limit u → ∞,
| (A8) |
With the image distribution center pushed to negative infinity, the distribution of unbleached molecules in the region of the focal volume (at positive infinity) is at equilibrium and contributes a constant 1/2 to the normalized fluorescence. Meanwhile, the real distribution center translates to infinity with the focal volume and produces a fluorescence recovery with the same form as a standard fluorescence recovery. At full recovery (t → ∞), , as expected.
This derivation can be repeated for a wall perpendicular to the optical axis by introducing the appropriate coordinate shifts in z, rather than x or y. The result has the same form as Eq. (A7), but with ωr → ωz and μn(t) → νn(t) in the exponential and complimentary error functions.
Following similar logic leading to the derivation of the “single boundary” MP-FRAP model, we can also produce an approximate analytical model for use in the presence of two parallel infinite-plane boundaries. In this case, we place the real concentration distribution at the origin and model two boundaries placed symmetrically about the distribution center (at u and –u) with an image distribution on the opposite side of each barrier (at –2u and 2u). The concentration distribution for this configuration has three parts, c″ (x,y,z; t) = (1/3)[c1(x – 2u,y,z,; t) + c2(x,y,z; t), + c3(x + 2u,y,z; t)], and the integration to determine the fluorescence is limited to –u → u along the dimension in which the barriers appear. The resulting normalized fluorescence recovery, for boundaries parallel to the optical axis, is given by
| (A9) |
Similar to the single boundary formula, the 1/3 arises from the introduction of the two image distributions. The first error function comes from the calculation of , which is evaluated from –u → u. The standard MP-FRAP model appears again and is weighted by contributions from the real distribution, represented by the second error function, and the image distributions, represented by the two exponential × error function terms. In the limit u → ∞,
| (A10) |
Here, the image distributions contribute a constant 1/3 each to the normalized fluorescence, while the real distribution recovers as would a standard MP-FRAP curve. At full recovery (t → ∞), , as expected.
As with the one-boundary model, this derivation can be repeated for walls perpendicular to the optical axis by introducing the appropriate coordinate shifts in z, rather than x or y. The result has the same form as Eq. (A9), but with ωr → ωz and μn(t) → νn(t) in the exponential and error functions.
Contributor Information
Kelley D. Sullivan, Department of Physics and Astronomy, University of Rochester, Rochester, New York 14627, USA
Edward B. Brown, Department of Biomedical Engineering, University of Rochester, Rochester, New York 14627, USA.
References
- 1.Brown EB, Wu ES, Zipfel W, Webb WW. Biophys. J. 1999;77:2837. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denk W, Strickler JH, Webb WW. Science. 1990;248:73. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 3.Davis SK, Bardeen CJ. Rev. Sci. Instrum. 2002;73:2128. [Google Scholar]
- 4.Mazza D, Braeckmans K, Cella F, Testa I, Vercauteren D, Demeester J, De Smedt SS, Diaspro A. Biophys. J. 2008;95:3457. doi: 10.1529/biophysj.108.133637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Biophys. J. 1976;16:1055. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edidin M, Zagyansky M, Lardner T. Science. 1976;191:466. doi: 10.1126/science.1246629. [DOI] [PubMed] [Google Scholar]
- 7.Koppel DE, Axelrod D, Schlessinger J, Elson EL, Webb WW. Biophys. J. 1976;16:1315. doi: 10.1016/S0006-3495(76)85776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubitscheck U, Wedekind P, Peters R. J. Microsc. 1998;192:126. doi: 10.1111/j.1365-2818.1994.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 9.Berk DA, Yuan F, Leunig M, Jain RK. Biophys. J. 1993;65:2428. doi: 10.1016/S0006-3495(93)81326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geerts H, Brabander MD, Nuydens R, Geuens S, Moeremans M, Mey JD, Hollenbeck P. Biophys. J. 1987;52:775. doi: 10.1016/S0006-3495(87)83271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian H, Sheetz MP, Elson EL. Biophys. J. 1991;60:910. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magde D, Elson EL, Webb WW. Biopolymers. 1974;13:29. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 13.Berland KM, So PTC, Gratton E. Biophys. J. 1995;68:694. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mertz J, Xu C, Webb WW. Opt. Lett. 1995;20:2532. doi: 10.1364/ol.20.002532. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe W, Matsunaga S, Higashi T, Fukui K, Itoh K. J. Biomed. Opt. 2008;13:031213. doi: 10.1117/1.2939401. [DOI] [PubMed] [Google Scholar]
- 16.Williams RM, Zipfel WR, Tinsley ML, Farnum CE. Biophys. J. 2007;93:1039. doi: 10.1529/biophysj.106.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu S, Rodionov V, Terasaki M, Campagnola PJ. Opt. Lett. 2005;30:159. doi: 10.1364/ol.30.000159. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan KD, Sipprell WH, III, Brown EB, Jr., Brown EB., III Biophys. J. 2009;96:5082. doi: 10.1016/j.bpj.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coscoy S, Waharte F, Gautreau A, Martin M, Louvard D, Mangeat P, Arpin M, Amblard F. Proc. Natl. Acad. Sci. USA. 2002;99:12813. doi: 10.1073/pnas.192084599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waharte F, Brown CM, Coscoy S, Coudrier E, Amblard F. Biophys. J. 2005;88:1467. doi: 10.1529/biophysj.104.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grebenkov DS. Phys. Rev. E. 2007;76:041139. doi: 10.1103/PhysRevE.76.041139. [DOI] [PubMed] [Google Scholar]
- 22.Burada PS, Hänggi P, Marchesoni F, Schmid G, Talkner P. Chem. Phys. Chem. 2009;10:45. doi: 10.1002/cphc.200800526. [DOI] [PubMed] [Google Scholar]
- 23.Frøhlich AF, Jespersen SN, Ostergaard L, Kiselev VG. J. Magn. Reson. 2008;194:128. doi: 10.1016/j.jmr.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Krepysheva N, Di Pietro L, Neel MC. Phys. Rev. E. 2006;73:021104. doi: 10.1103/PhysRevE.73.021104. [DOI] [PubMed] [Google Scholar]
- 25.Szymczak P, Ladd AJC. Phys. Rev. E. 2003;68:036704. doi: 10.1103/PhysRevE.68.036704. [DOI] [PubMed] [Google Scholar]
- 26.Yudson VI, Reineker P. Phys. Rev. E. 2001;64:031108. doi: 10.1103/PhysRevE.64.031108. [DOI] [PubMed] [Google Scholar]
- 27.Ribrault C, Triller A, Sekimoto K. Phys. Rev. E. 2007;75:021112. doi: 10.1103/PhysRevE.75.021112. [DOI] [PubMed] [Google Scholar]
- 28.Gennerich A, Schild D. Biophys. J. 2000;79:3294. doi: 10.1016/S0006-3495(00)76561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gennerich A, Schild D. Biophys. J. 2002;83:510. doi: 10.1016/S0006-3495(02)75187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foquet M, Korlach J, Zipfel WR, Webb WW, Craighead HG. Anal. Chem. 2004;76:1618. doi: 10.1021/ac035088o. [DOI] [PubMed] [Google Scholar]
- 31.Etienne E, Lenne P-F, Sturgis JN, Rigneault H. Appl. Opt. 2006;45:4497. doi: 10.1364/ao.45.004497. [DOI] [PubMed] [Google Scholar]
- 32.von der Hocht I, Enderlein J. Exp. Mol. Pathol. 2007;82:142. doi: 10.1016/j.yexmp.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Saxton MJ. Biophys. J. 1995;69:389. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Görisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Proc. Natl. Acad. Sci. USA. 2004;101:13221. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sbalzarini IF, Mezzacasa A, Helenius A, Koumoutsakos P. Biophys. J. 2005;89:1482. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soumpasis DM. Biophys. J. 1983;41:95. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coelho FP, Vaz WL, Melo E. Biophys. J. 1997;72:1501. doi: 10.1016/S0006-3495(97)78798-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxton MJ. Biophys. J. 2001;81:2226. doi: 10.1016/S0006-3495(01)75870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown E, Webb W. Methods in Enzymology. In: Marriott Gerard., editor. Caged Compounds. Vol. 291. Academic Press; San Diego: 1998. pp. 356–380. [DOI] [PubMed] [Google Scholar]
- 40.Tsourkas PK, Longo ML, Raychaudhuri S. Biophys. J. 2008;95:1118. doi: 10.1529/biophysj.107.122564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieser S, Axmann M, Schütz GJ. Biophys. J. 2008;95:5988. doi: 10.1529/biophysj.108.141655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexandrakis G, Brown EB, Tong RT, McKee TD, Campbell RB, Boucher Y, Jain RK. Nat. Med. 2004;10:203. doi: 10.1038/nm981. [DOI] [PubMed] [Google Scholar]
- 43.Binder DK, Papadopoulos MC, Haggie PM, Verkman AS. J. Neurosci. 2004;24:8049. doi: 10.1523/JNEUROSCI.2294-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadopoulos MC, Kim JK, Verkman AS. Biophys. J. 2005;89:3660. doi: 10.1529/biophysj.105.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabso M, Neher E, Spira ME. Neuron. 1997;18:473. doi: 10.1016/s0896-6273(00)81247-7. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt H, Arendt O, Brown EB, Schwaller B, Eilers J. J. Neurochem. 2007;100:727. doi: 10.1111/j.1471-4159.2006.04231.x. [DOI] [PubMed] [Google Scholar]
- 47.Svoboda K, Tank DW, Denk W. Science. 1996;272:716. doi: 10.1126/science.272.5262.716. [DOI] [PubMed] [Google Scholar]
- 48.Majewska A, Brown E, Ross J, Yuste R. J. Neurosci. 2000;20:1722. doi: 10.1523/JNEUROSCI.20-05-01722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holthoff K, Tsay D, Yuste R. Neuron. 2002;33:425. doi: 10.1016/s0896-6273(02)00576-7. [DOI] [PubMed] [Google Scholar]
- 50.Feder TJ, Brust-Mascher I, Slattery JP, Baird B, Webb WW. Biophys. J. 1996;70:2767. doi: 10.1016/S0006-3495(96)79846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zipfel WR, Williams RM, Webb WW. Nat. Biotechnol. 2003;21:1369. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 52.Crivaro M, Enjieu-Kadji H, Hatanaka R, Nakauchi S, Bosch J, Judin J, Riera J, Kawashima R. J. Microsc. 2010 doi: 10.1111/j.1365-2818.2010.03473.x. doi: 10.1111/j.1365-2818.2010.03473.x. [DOI] [PubMed] [Google Scholar]