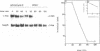Abstract
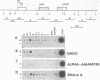
c-myc RNA rapidly decreases to barely detectable levels in Friend erythroleukemia cells induced to differentiate upon the addition of dimethylsulfoxide. We show here that c-myc gene is down-regulated both at the transcriptional level presumably by a block in the elongation of primary transcripts and at the post-transcriptional level by an increase in the degradation of its mRNA.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985 Dec 12;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981 Dec;27(2 Pt 1):381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Armelin H. A., Armelin M. C., Kelly K., Stewart T., Leder P., Cochran B. H., Stiles C. D. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984 Aug 23;310(5979):655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- Aviv H., Voloch Z., Bastos R., Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976 Aug;8(4):495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- Bastos R. N., Volloch Z., Aviv H. Messenger RNA population analysis during erythroid differentiation: a kinetical approach. J Mol Biol. 1977 Feb 25;110(2):191–203. doi: 10.1016/s0022-2836(77)80068-5. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Campisi J. c-myc regulation during retinoic acid-induced differentiation of F9 cells is posttranscriptional and associated with growth arrest. Mol Cell Biol. 1986 Feb;6(2):518–524. doi: 10.1128/mcb.6.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Dyson P. J., Littlewood T. D., Forster A., Rabbitts T. H. Chromatin structure of transcriptionally active and inactive human c-myc alleles. EMBO J. 1985 Nov;4(11):2885–2891. doi: 10.1002/j.1460-2075.1985.tb04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einat M., Resnitzky D., Kimchi A. Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature. 1985 Feb 14;313(6003):597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- Endo T., Nadal-Ginard B. Transcriptional and posttranscriptional control of c-myc during myogenesis: its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol Cell Biol. 1986 May;6(5):1412–1421. doi: 10.1128/mcb.6.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Marty L., Piechaczyk M., el Sabrouty S., Dani C., Jeanteur P., Blanchard J. M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985 Mar 11;13(5):1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser N. W., Nevins J. R., Ziff E., Darnell J. E., Jr The major late adenovirus type-2 transcription unit: termination is downstream from the last poly(A) site. J Mol Biol. 1979 Apr 25;129(4):643–656. doi: 10.1016/0022-2836(79)90474-1. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Harris L. J., Remmers E. F., Brodeur P., Riblet R., D'Eustachio P., Marcu K. B. c-myc Gene rearrangements involving gamma immunoglobulin heavy chain gene switch regions in murine plasmacytomas. Nucleic Acids Res. 1983 Dec 10;11(23):8303–8315. doi: 10.1093/nar/11.23.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation of late simian virus 40 mRNA synthesis is enhanced by the agnoprotein and is temporally regulated in isolated nuclear systems. Mol Cell Biol. 1985 Jun;5(6):1327–1334. doi: 10.1128/mcb.5.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Keath E. J., Caimi P. G., Cole M. D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984 Dec;39(2 Pt 1):339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Klein G., Klein E. Evolution of tumours and the impact of molecular oncology. Nature. 1985 May 16;315(6016):190–195. doi: 10.1038/315190a0. [DOI] [PubMed] [Google Scholar]
- Knight E., Jr, Anton E. D., Fahey D., Friedland B. K., Jonak G. J. Interferon regulates c-myc gene expression in Daudi cells at the post-transcriptional level. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1151–1154. doi: 10.1073/pnas.82.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska A., Yenofsky R., Brawerman G. Regulation of messenger RNA stability in mouse erythroleukemia cells. J Mol Biol. 1985 Jan 20;181(2):231–239. doi: 10.1016/0022-2836(85)90087-7. [DOI] [PubMed] [Google Scholar]
- Lachman H. M., Skoultchi A. I. Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature. 1984 Aug 16;310(5978):592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Cellular oncogenes and multistep carcinogenesis. Science. 1983 Nov 18;222(4625):771–778. doi: 10.1126/science.6356358. [DOI] [PubMed] [Google Scholar]
- Leder A., Pattengale P. K., Kuo A., Stewart T. A., Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986 May 23;45(4):485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Lindell T. J., Weinberg F., Morris P. W., Roeder R. G., Rutter W. J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970 Oct 23;170(3956):447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Marks P. A., Rifkind R. A. Erythroleukemic differentiation. Annu Rev Biochem. 1978;47:419–448. doi: 10.1146/annurev.bi.47.070178.002223. [DOI] [PubMed] [Google Scholar]
- Mechti N., Affabris E., Romeo G., Lebleu B., Rossi G. B. Role of interferon and 2',5'-oligoadenylate synthetase in erythroid differentiation of Friend leukemia cells. Studies with interferon-sensitive and -resistant variants. J Biol Chem. 1984 Mar 10;259(5):3261–3265. [PubMed] [Google Scholar]
- Nevins J. R., Blanchard J. M., Darnell J. E., Jr Transcription units of adenovirus type 2. Termination of transcription beyond the poly(A) addition site in early regions 2 and 4. J Mol Biol. 1980 Dec 15;144(3):377–386. doi: 10.1016/0022-2836(80)90096-0. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Blanchard J. M., Marty L., Dani C., Panabieres F., El Sabouty S., Fort P., Jeanteur P. Post-transcriptional regulation of glyceraldehyde-3-phosphate-dehydrogenase gene expression in rat tissues. Nucleic Acids Res. 1984 Sep 25;12(18):6951–6963. doi: 10.1093/nar/12.18.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Prochownik E. V., Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. 1986 Aug 28-Sep 3Nature. 322(6082):848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- Rapp U. R., Cleveland J. L., Brightman K., Scott A., Ihle J. N. Abrogation of IL-3 and IL-2 dependence by recombinant murine retroviruses expressing v-myc oncogenes. Nature. 1985 Oct 3;317(6036):434–438. doi: 10.1038/317434a0. [DOI] [PubMed] [Google Scholar]
- Rifkind R. A., Sheffery M., Profous-Juchelka H. R., Reuben R. C., Marks P. A. Induction of globin gene expression during erythroid cell differentiation. Ann N Y Acad Sci. 1983;411:141–149. doi: 10.1111/j.1749-6632.1983.tb47296.x. [DOI] [PubMed] [Google Scholar]
- Rougeon F., Kourilsky P., Mach B. Insertion of a rabbit beta-globin gene sequence into an E. coli plasmid. Nucleic Acids Res. 1975 Dec;2(12):2365–2378. doi: 10.1093/nar/2.12.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Hagenbüchle O., Wellauer P. K., Pittet A. C. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 1983 Jun;33(2):501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Sariban E., Mitchell T., Kufe D. Human c-myc and N-ras expression during induction of HL-60 cellular differentiation. Biochem Biophys Res Commun. 1985 Feb 15;126(3):999–1005. doi: 10.1016/0006-291x(85)90284-0. [DOI] [PubMed] [Google Scholar]
- Westin E. H., Wong-Staal F., Gelmann E. P., Dalla-Favera R., Papas T. S., Lautenberger J. A., Eva A., Reddy E. P., Tronick S. R., Aaronson S. A. Expression of cellular homologues of retroviral onc genes in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2490–2494. doi: 10.1073/pnas.79.8.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]