Abstract
Oncolytic adenoviruses (Ads) are an emerging alternative therapy for cancer; however, clinical trial have not yet demonstrated sufficient efficacy. When oncolytic Ads are used in combination with taxoids a synergistic increase in both cytotoxicity and viral replication is observed. In order to generate a next generation oncolytic adenovirus, virion were physically conjugated to a highly potent taxoid, SB-T-1214, and a folate targeting motif. Conjugation was enabled via the metabolic incorporation of non-canonical monosaccharides (O-GlcNAz) and amino acids (homopropargylglycine), which served as sites for chemoselective modification.
Despite substantial progress in understanding the molecular underpinnings of cancer, current chemotherapeutic options are limited and often unsuccessful. On promising alternative strategy is the use of conditionally replicative oncolytic vectors. Such viruses are designed to preferentially replicate in cancerous cells, such as those lacking common tumor suppressors (e.g. p53), leading to partially selective tumor toxicity. In addition, many carry a toxic transgene designed to amplify the inherent cytotoxic nature, which results from viral protein expression and immune stimulation.1 Despite this multifaceted cytotoxicity, the major limitation for oncolytic viruses in clinical trials has been efficacy.2 In an effort to increase potency, a number of oncolytic viruses have been used in combination with traditional chemotherapeutics.3-5 In particular, conditionally replicative adenoviruses (Ads) have demonstrated significant synergism when used in combination with a number of different chemotherapeutics including doxorubicin, paclitaxel/docetaxel, cisplatin and histone deacetylase inhibitors.6 In the case of taxoid/oncolytic Ad combination therapy, an increase in viral replication is seen in addition to synergistic cytotoxicity. 7-12 While the mechanistic origin of synergism is not well understood, it is clearly a general and significant phenomenon.
Paclitaxel treatment of cancer cells results in the upregulation of TNF related apoptosis inducing ligand (TRAIL) receptors.13 Notably, one of the most promising oncolytic Ads in clinical trials bears the cytotoxic TRAIL transgene, which induces apoptosis in the infected cell and mediates substantial bystander cytotoxicity. As a result, taxoid/AdTRAIL would be expected to have an additional source of synergism. SB-T-1214 is a next generation taxoid that exhibits significantly improved cytotoxicity, against a number of drug resistant cancer cell lines.14-16 In addition, this taxoid exhibited substantial inhibition of cancer stem cell related genes (Oct4, Sox2, Nanog, and c-Myc) when screened against 3 unrelated invasive colon cancer cell lines.17 These results indicate that SB-T-1214 has significant potential, particularly with respect to cancer stem cells and cancers that are resistant to traditional chemotherapeutics.
While combination therapy demonstrates significant promise, it holds that efficiently targeted Ad particles bearing a therapeutic payload would provide an additional boost in efficacy. This would be a result of spatially and temporally concerted delivery of cytotoxicity, and may have the added benefit of reducing systemic toxicity. In order to achieve this goal, selective chemical modification routes for adenovirus are required, particularly those that allow the generation of multifunctional particles. Previously we reported the incorporation and modification of a non-canonical sugar residue, O-GlcNAz on serine 109 of the fiber protein, as a means of chemoselectively tailoring Ad particles.18 The specificity of this strategy, derived from the fidelity of the biosynthetic machinery and the highly selective chemistries developed for azide modification, allowed folate modification without compromising virus infectivity. Folate decorated Ad exhibited substantial (~20 fold) increase transgene delivery to breast cancer cells.18
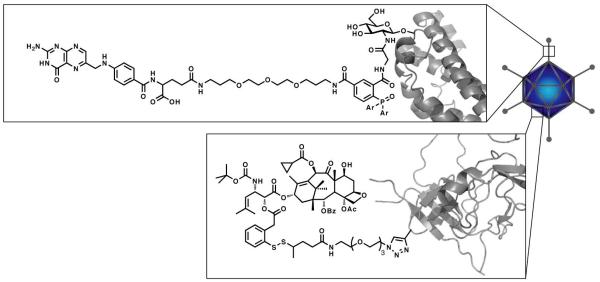
Here, we extend this approach towards multimodal adenovirus particles via the simultaneous metabolic labeling with O-GlcNAz and an alkyne bearing non-canonical amino acid, homopropargylglycine (HPG)(Fig. 1). Introduction of these surrogates into Ad particles was envisioned to allow sequential Staudinger ligation of O-GlcNAz followed by copper assisted “click” modification of homopropargylglycine (HPG).
Figure 1.
A cartoon illustrating adenovirus particles chemically modified with both a folate moiety, via Staudinger conjugation with an introduced O-GlcNAz, and SB-T-1214, via “click” modification of metabolically introduced homopropargylglycine.
In order to explore this potential, adenovirus type 5 particles were produced in the presence of a metabolic precursor of GlcNAz, peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), and HPG.19,20 Azido-sugar incorporation was accomplished by supplementing media with 50 μM Ac4GalNAz for the entire duration of virus production (48 hours). Introduction of the alkyne-amino acid was mediated by exposure of producer cells to 4 mM HPG during a six-hour window (18-24 hours post infection), in a pulse chase format with methionine containing media. 48 hours post infection, the cells were harvested, lysed and viruses were purified via a standard two-step ultracentrifugation procedure in CsCl gradients.21
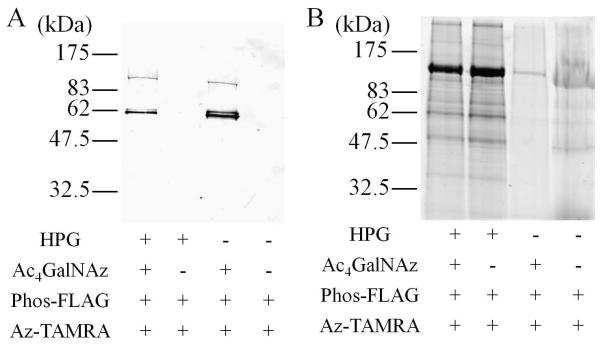
Purified O-GlcNAc and HPG bearing virion were treated with 300 μM of Staudinger probe bearing a FLAG epitope (PhosFLAG) (3 hours, RT).22 Reaction mixtures were subsequently exposed to tetramethylrhodamine 5-carboxamido-(6-azidohexanyl) (az-TAMRA) dye (500 μM) using copper assisted “click” reaction conditions under de-oxygenating conditions in the presence of bathophenanthroline disodium salt (3 mM) and CuBr (1 mM) (RT, 12 hours).23 Particles were purified by size exclusion (Sephadex G-25) and interrogated by western blot and fluorescent gel imaging. Western analysis demonstrated that virus particles produced in the presence of both Ac4GalNAz and HPG and only Ac4GalNAz are labeled on a single coat protein occurring at 62 kD by PhosFLAG (Fig. 2a). No PhosFLAG labeling is seen on particles produced in the absence of Ac4GalNAz, consistent with previous studies demonstrating the specific labeling of the fiber protein via O-GlcNAz. Fluorescent gel imaging of az-TAMRA labeled HPG-Ad and HPG/O-GlcNAz-Ad demonstrated labeling of a number of different proteins, consistent with the presence of solvent exposed methionine sites (Fig. 2b).24
Figure 2.
Chemoselective modification of GalNAz and HPG labeled adenovirus with Phosphine-FLAG and az-TAMRA. A) Anti-FLAG western blot of peptide and dye labeled Ad5, demonstrating incorporation of azido sugar onto adenoviral fiber. B) Same samples from A, analyzed for fluorescence as a reporter of HPG incorporation onto virus capsid.
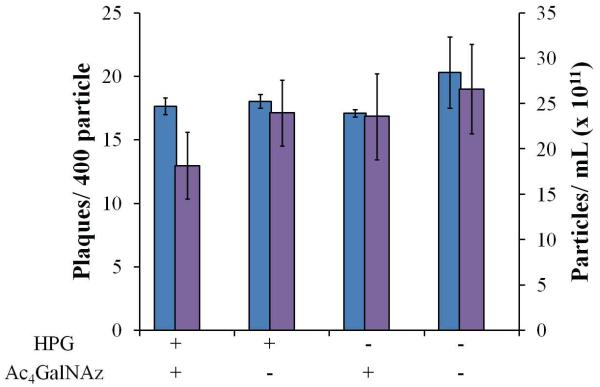
Previous characterization of O-GlcNAz labeled Ad particles demonstrated 22 ± 1.5 chemically addressable azides per particle.18 In order to quantitate HPG incorporation, TAMRA labeled virion were quantified via fluorescent gel imaging against a free az-TAMRA standard addition curve (supplementary figure 2), demonstrating an attachment of 193 ± 12 dyes per virion (supplementary table 1). Vector production and infectivity are often compromised during genetic engineering of Ad particles, which has slowed the pace of vector development and effectively limited the production of multifunctional particles. While previous results indicate that O-GlcNAz incorporation does not impact either particle production or infectivity, the incorporation of HPG into the protein backbone at significantly higher incorporation levels was a concern. Surprisingly, no significant loss in either particle production or infectivity was observable for either of the singly modified Ad particles or particle bearing both O-GlcNAz and HPG (Fig 3).
Figure 3.
Viral fitness analysis. Adenoviral particle count was assessed after purification (purple bars) as assayed by UV detection showing efficient particle generation in presence of different non-natural substrates. Virus plaque assay (blue bars) showing infectivity of modified virus particles.
In order to generate a chemotherapeutically “armed” Ad particle, an azido derivative of SB-T-1214 (az-SB-T-1214) was synthesized that included a reductively self-immolative linker, designed to release the taxoid after Ad particle endocytosis (Scheme 1). This linker has demonstrated efficient endosomal release of SBT-1214 in cell culture studies.16,25 Modification of AdTRAIL with az-SB-T-1214 was accomplished in an identical manner to az-TAMRA modification of Ad described above. Chemically modified virion (SB-T-1214AdTRAIL) were purified by size exclusion and used to infect ovarian cancer cells, ID8, at multiplicities of infection (MOIs) that were expected to be subtoxic for AdTRAIL alone. Levels of free SB-T-1214 equivalent to that loaded on virion, as well as unmodified AdTRAIL were used for comparison. Importantly, the only difference between the processing of AdTRAIL and SB-T-1214AdTRAIL was the addition of HPG during production of the latter. Specifically, AdTRAIL was exposed to identical “click” conditions as SB-T-1214AdTRAIL, however due to the absence of HPG was presumably unmodified. Five days post infection, cytotoxicity was assessed via MTT assay (Roche, KitI). TRAIL expressing viruses that were covalently linked with az-SB-T-1214 demonstrated an increase in cytotoxicity compared to free azSB-T-1214 and AdTRAIL alone (Fig. 4), consistent with the proposed synergistic effect. Unmodified SB-T-1214, both alone and co-administered with AdTRAIL demonstrated higher levels of cell cytotoxicity. Presumably, this effect is due to a difference in the kinetics of cellular penetration of the free drug versus covalently linked SB-T-1214 and AdTRAIL.
Scheme1.
Structures of az-SBT1214 and phosphine-folate used to chemoselectively modify O-GlcNAz and HPG labeled Ad-Luc.
Figure 4.

Comparison of cytoxicity profiles of metabolically unlabeled TRAIL-Ad5(AdTRAIL), free drug (SBT-1214), free azide-drug conjugate (az-SBT-1214), and co-administered AdTRAIL and SBT-1214 (SBT-1214 + AdTRAIL) with drug labeled TRAIL expressing adenovirus (SBT-AdTRAIL) on ovarian carcinoma cells (ID8). Cells seeded on 96 well plates were infected with shown concentrations of virus and/or drug. After 5 days MTT assay was performed to determine cell viability.
As many cancers demonstrate significantly higher levels of folate receptor (FR), folate conjugates demonstrate selectivity for this receptor and folate conjugates are efficiently internalized, folate has been widely used for cancer targeting.26,27 Although we have previously the gene delivery of folate-Ad particles, those modified with both az-SB-T-1214 and folate may demonstrate altered uptake profiles. In order to explore these effects, dually modified Ad bearing a luciferase transgene (AdLuc) were screened against a murine breast cancer cell line, 4T1.28 Cells grown under folate free media for 2 weeks were seeded on 96 well plates at concentrations of 1 × 104 cells/ well. Infection was accomplished at a MOI of 50 and cells were examined for luciferase activity 24 hours post infection (Luc-Bright-Glow). Folate targeted SB-T-1214AdTRAIL demonstrated (Fig. 5) a ~30 fold increase in transgene expression on breast cancer cell type (4T1) compared to virion lacking O-GlcNAz. In addition, treatment of cells with free folate prior to infection led to a dose dependent loss in virus infection (Fig. 5).
Figure 5.
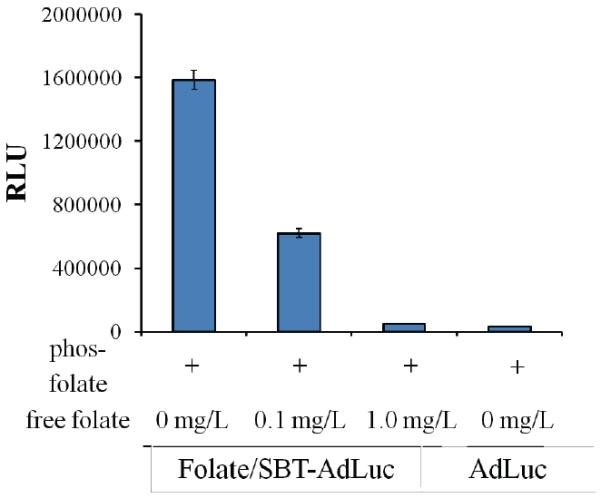
Targeting analysis of SB-T-1214 and folate modified AdLuc. Luciferase analysis of 4T1 cells 24 hours post infection with modified virions both in the presence and absence of external folic acid showing dose dependent gene expression.
In summary we report the development of a novel multi-functional adenoviral platform via non-canonical substrate incorporation and chemoselective modification. We utilized the platform to develop a combination vector targeted towards the folate receptor and armed with a next generation taxoid. The described system allowed the efficient modification with both functionalities without impact on viral fitness. Further, initial studies indicate significant synergistic cell toxicity. Ongoing studies will evaluate the construct in the context of different cancers, both in cell culture and within in vivo xenograft model systems. In principal the described dual modification methodology can be utilized to append targeting, imaging, diagnostic and chemotherapeutic modules to replication selective Ad, potentially accelerating vector development and allowing the evaluation of alternative combination therapies.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- (1).Yamamoto M, Curiel DT. Mol. Ther. 2009;18:243. doi: 10.1038/mt.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Pesonen S, Kangasniemi L, Hemminki A. Mol Pharm. 2010;8:12. doi: 10.1021/mp100219n. [DOI] [PubMed] [Google Scholar]
- (3).Passer BJ, Castelo-Branco P, Buhrman JS, Varghese S, Rabkin SD, Martuza RL. Cancer Gene Ther. 2009;16:551. doi: 10.1038/cgt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Huang B, Sikorski R, Kirn DH, Thorne SH. Gene Ther. 2011;18:164. doi: 10.1038/gt.2010.121. [DOI] [PubMed] [Google Scholar]
- (5).Tseng JC, Granot T, DiGiacomo V, Levin B, Meruelo D. Cancer Gene Ther. 2010;17:244. doi: 10.1038/cgt.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ottolino-Perry K, Diallo J-S, Lichty BD, Bell JC, McCart J. Andrea. Mol Ther. 2009;18:251. doi: 10.1038/mt.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yu DC, Chen Y, Dilley J, Li YH, Embry M, Zhang H, Nguyen N, Amin P, Oh J, Henderson DR. Cancer Res. 2001;61:517. [PubMed] [Google Scholar]
- (8).Zhang J, Ramesh N, Chen Y, Li YH, Dilley J, Working P, Yu DC. Cancer Res. 2002;62:3743. [PubMed] [Google Scholar]
- (9).Cheong SC, Wang Y, Meng JH, Hill R, Sweeney K, Kirn D, Lemoine NR, Hallden G. Cancer Gene Ther. 2008;15:40. doi: 10.1038/sj.cgt.7701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hassan M, Braam SR, Kruyt FAE. Cancer Gene Ther. 2006;13:1105. doi: 10.1038/sj.cgt.7700984. [DOI] [PubMed] [Google Scholar]
- (11).Li YM, Song ST, Jiang ZF, Zhang Q, Qu YM, Su CQ, Zhao CH, Li ZQ, Ge FJ, Qian QJ. Chin. J. Canc. Res. 2007;19:76. [Google Scholar]
- (12).Radhakrishnan S, Miranda E, Ekblad M, Holford A, Pizarro MT, Lemoine NR, Hallden G. Human Gene Ther. 2010;21:1311. doi: 10.1089/hum.2010.019. [DOI] [PubMed] [Google Scholar]
- (13).Nagano S, Perentes JY, Jain RK, Boucher Y. Cancer Res. 2008;68:3795. doi: 10.1158/0008-5472.CAN-07-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ojima I, Chen J, Sun L, Borella CP, Wang T, Miller ML, Lin SN, Geng XD, Kuznetsova LR, Qu CX, Gallager D, Zhao XR, Zanardi I, Xia SJ, Horwitz SB, Mallen-St Clair J, Guerriero JL, Bar-Sagi D, Veith JM, Pera P, Bernacki RJ. J. Med. Chem. 2008;51:3203. doi: 10.1021/jm800086e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kuznetsova L, Chen J, Sun L, Wu XY, Pepe A, Veith JA, Pera P, Bernacki RJ, Ojima I. Bioorg. Med. Chem. Lett. 2006;16:974. doi: 10.1016/j.bmcl.2005.10.089. [DOI] [PubMed] [Google Scholar]
- (16).Chen SY, Zhao XR, Chen JY, Chen J, Kuznetsova L, Wong SS, Ojima I. Bioconj. Chem. 2010;21:979. doi: 10.1021/bc9005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Botchkina G, Zuniga E, Das M, Wang Y, Wang H, Zhu S, Savitt A, Rowehl R, Leyfman Y, Ju J, Shroyer K, Ojima I. Mol Cancer. 2010;9:192. doi: 10.1186/1476-4598-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Banerjee PS, Ostapchuk P, Hearing P, Carrico I. J. Am. Chem. Soc. 2010;132:13615. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Boyce M, Carrico IS, Ganguli AS, Yu S-H, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2011;108:3141. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Curr. Op. Chem. Biol. 2010;14:774. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Tollefson AE, Hermiston TW, Wold WS. 1998;Vol. 21:1. [Google Scholar]
- (22).Luchansky SJ, Hang HC, Saxon E, Grunwell JR, Danielle CY, Dube DH, Bertozzi CR. Recognition of Carbohydrates in Biological Systems Pt A: General Procedures. Vol. 362. Academic Press Inc; San Diego: 2003. p. 249. [Google Scholar]
- (23).Gupta SS, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Bioconj. Chem. 2005;16:1572. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- (24).Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Science. 2010;329:1071. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ojima I. Acc. Chem. Res. 2007;41:108. doi: 10.1021/ar700093f. [DOI] [PubMed] [Google Scholar]
- (26).Low PS, Henne WA, Doorneweerd DD. Acc. Chem. Res. 2007;41:120. doi: 10.1021/ar7000815. [DOI] [PubMed] [Google Scholar]
- (27).Xia W, Low PS. J. Med. Chem. 2010;53:6811. doi: 10.1021/jm100509v. [DOI] [PubMed] [Google Scholar]
- (28).Jogler C, Hoffmann D, Theegarten D, Grunwald T, Uberla K, Wildner O. J. Virol. 2006;80:3549. doi: 10.1128/JVI.80.7.3549-3558.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.