Abstract
Plasma protein C is a serine protease zymogen that is transformed into the active, trypsin-like protease, activated protein C (APC), which can exert multiple activities. For its anticoagulant action, APC causes inactivation of the procoagulant cofactors, factors Va and VIIIa, by limited proteolysis, and APC’s anticoagulant activity is promoted by protein S, various lipids, high density lipoprotein, and factor V. Hereditary heterozygous deficiency of protein C or protein S is linked to moderately increased risk for venous thrombosis while a severe or total deficiency of either protein is linked to neonatal purpura fulminans. In recent years, the beneficial direct effects of APC on cells which are mediated by several specific receptors have become the focus of much attention. APC-induced signaling can promote multiple cytoprotective actions which can minimize injuries in various preclinical animal injury models. Remarkably, pharmacologic therapy using APC demonstrates substantial neuroprotective effects in various murine injury models, including ischemic stroke. This review summarizes the molecules that are central to the protein C pathways, the relationship of pathway deficiencies to venous thrombosis risk, and mechanisms for the beneficial effects of APC.
Keywords: Protein C, Protein S, Thrombomodulin, Endothelial protein C receptor, Neuroprotection
1 Introduction
Many studies show that the protein C pathways serve multiple purposes with the overall function of maintaining a regulated balance between hemostasis and host defense systems in response to vascular injury. Plasma protein C is a serine protease zymogen. Following its activation, activated protein C (APC) is capable of many different biologic activities. These include antithrombotic actions and diverse anti-inflammatory and cytoprotective activities, with the net effect of maintaining the health and integrity of the vasculature. This review provides an overview of the protein C pathways (see Figure 1) including the components and cofactors protein S, thrombomodulin (TM) and endothelial protein C receptor (EPCR). The reader seeking details that might not be included here due to space limitations is referred to additional sources [1–13], as well as to other reports that summarize multiple preclinical and clinical studies of APC and protein S [6, 9, 12, 14–16].
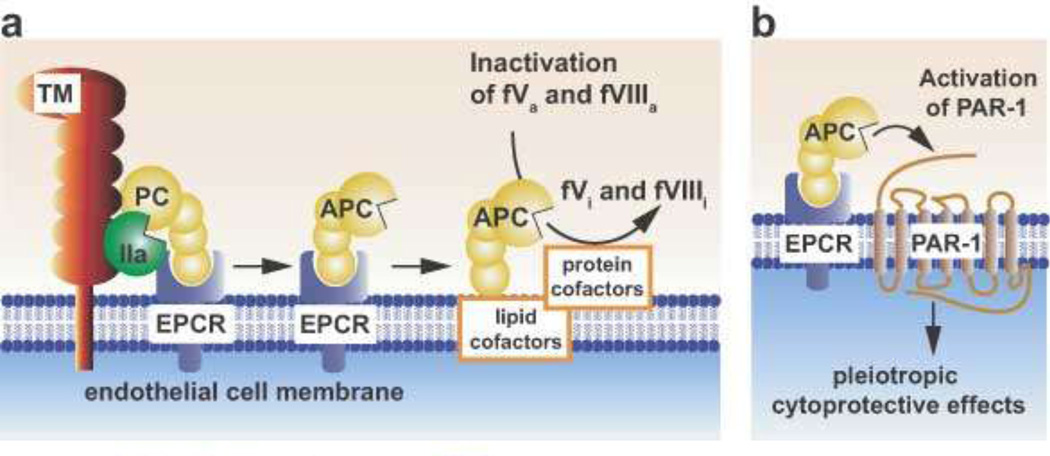
Figure 1. Schematic models of Protein C activation and expression of APC activities.
(a) Physiologic activation of protein C (PC) by the thrombin (IIa)-thrombomodulin (TM) complex occurs on the surface of endothelial cells. IIa bound to TM activates PC, especially when PC is bound to the endothelial receptor (EPCR). Since protein C and APC have a similar affinity for EPCR, after activation APC can either dissociate from EPCR to exert anticoagulant activity or remain bound to EPCR where it might express multiple direct cellular activities (b). Dissociation of APC from EPCR allows expression of APC’s anticoagulant activity on cell membrane surfaces, various microparticles, or lipoproteins (e.g., High Density Lipoprotein). As an anticoagulant, APC cleaves the activated cofactors Va (fVa) and VIIIa (fVIIIa) to yield inactivated cofactors, fVi and fVIIIi. This proteolytic inactivation is enhanced by protein cofactors (e.g., protein S, factor V) and lipids cofactors (e.g., phosphatidylserine, cardiolipin, glucosylceramide, or HDL).
(b) PAR1 mediates multiple cytoprotective effects of APC. In most, but not all, reported studies of APC’s beneficial effects of on endothelial cells, the cellular receptors EPCR and PAR1 are required. These cytoprotective effects include anti-apoptotic activities, anti-inflammatory activities, protection of endothelial barrier functions, and favorable alteration of gene expression profiles. This paradigm in which EPCR-bound APC activates PAR1 to initiate signaling is consistent with many in vitro and in vivo data. Localization of APC signaling in the caveolin-1 rich microdomains (caveolae) may help differentiate mechanisms for cytoprotective APC signaling versus proinflammatory thrombin signaling. Additional mechanisms for APC effects on cells may involve other receptors. These effects include APC anti-inflammatory effects on leukocytes or cytoprotective effects on dendritic cells and neurons. Other receptors may include PAR3, various integrins (e.g., Mac-1 (CD11b/CD18), β1 integrins, β3 integrins), S1P1, or apolipoprotein E receptor 2 (LRP8). This scheme is taken from Blood. (LO Mosnier et al. The cytoprotective protein C pathway. Blood 2007 109:3161–3172. © the American Society of Hematology)
2 Protein C pathway molecules
The molecules that are central participants in the protein C pathways include protein C, protein S, TM, EPCR, and protease activated receptor 1 (PAR1), and many biochemical and genetic details are known about these key components.
2.1 Protein C
The plasma protein C concentration is 70 nM (4 µ g/ml) with a half live of ~8 hours [17]. Plasma also contains a low level of APC (< 40 pM). Clearance of APC in plasma is significantly driven by serine protease inhibitors (SERPINs) which contribute to a remarkably long circulation half live of APC in man of ~ 20–25 min. The known inhibitors of APC in plasma include protein C inhibitor (PCI), α1-antitrypsin, α2-macroglobulin and α2-antiplasmin. Protein C is homologous to the vitamin K-dependent coagulation factors VII, IX, and X and the protein C gene (PROC) is comprised of nine exons and eight introns, located on chromosome 2q13–14 [18, 19]. The polypeptide structure of mature protein C includes an amino-terminal Gla domain (residues 1 to 37), an aromatic stack (residues 38 to 45), two epidermal growth factor (EGF)-like regions (EGF-1, residues 46 to 92 and EGF-2, residues 93 to 136), an N-terminal activation peptide (residues 158 to 169) on the heavy chain, and the serine protease domain (residues 170 to 419).
2.2 Protein S
Protein S, a vitamin K-dependent plasma glycoprotein, has a plasma level of 320 nM. Because it binds with high affinity to the complement system protein, C4b-binding protein, over half of plasma protein S circulates bound to C4BP while the concentration of free protein S is 130 nM [20, 21]. The protein S (PROS1) gene is located on chromosome 3 (3q11.2) and contains 15 exons. Mature protein S contains 635 amino acids comprising 5 distinct domains, including an N-terminal Gla domain (residues 1–37) and aromatic stack (residues 38–45), a so-called “thrombin-sensitive region” (TSR; residues 46–74), 4 EGF-like domains (EGF-1 (residues 75–115), EGF-2 (residues 116–159), EGF-3 (residues 160–201) and EGF-4 (residues 202–242)), and a large C-terminal region of 393 amino acids referred to as a sex-hormone binding globulin (SHBG)-like domain (residues 243–635) whose structure represents two laminin G-type domains (Figure 2).
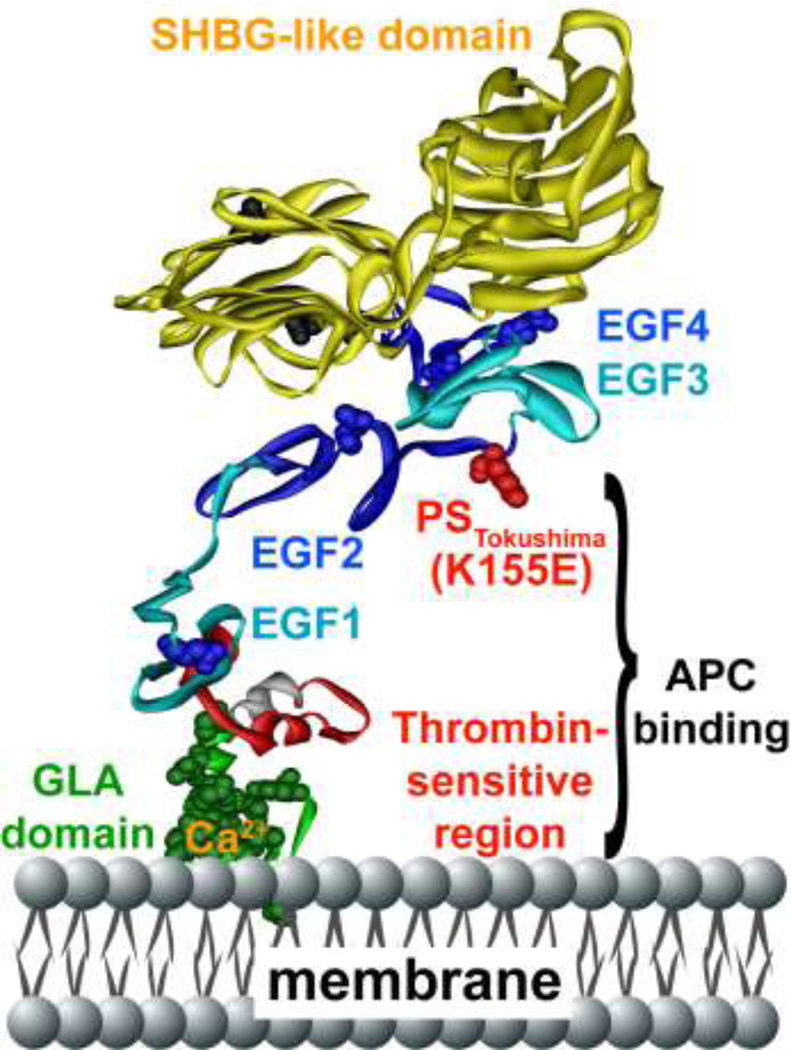
Figure 2. Protein S polypeptide scheme showing multiple domains and protein S Tokushima mutation.
Specific protein S domains are color coded and labeled. The N-terminal cluster of domains is responsible for binding to APC. The amino acid side chain of residue 155 is shown in red, representing the protein S Tokushima polymorphism, K155E (“155” is the mature protein S numbering which equates to K196E in full length precursor numbering). The schematic model of protein S was based on available models and structures of the individual GLA-TSR-EGF1 [167], EGF3–4 (1Z6C) [168], and the SHBG [169] domains. EGF2 was modeled by homology modeling using Swiss Model based on templates for extracellular domain of the LDL receptor (1N7DA), low-density lipoprotein receptor (1HZ8A), low-density lipoprotein receptor (1HJ7A) and EGF-like module-containing mucin-like hormone receptor-like 2 (2BO2A) as templates [170]. The individual domains were put together using Modeller [171], displayed and colored in Accelrys Discovery Studio and rendered in POV-Ray.
2.3 Thrombomodulin
TM (CD141) remarkably modulates the biologic specificity of thrombin [22–24]. TM reduces thrombin’s procoagulant actions (clotting fibrin and activating platelets) while it greatly increases the ability of thrombin to activate protein C (Figure 1a). TM also can promote inactivation of thrombin by PCI and antithrombin (AT). TM also enhances activation of the latent plasma carboxypeptidase B, thrombin activatable fibrinolysis inhibitor. The TM intron-less gene (THBD) is on chromosome 20 [25, 26]. TM contains 575 residues and contains a single transmembrane helix [26]. Low levels of soluble TM that come from proteolytic release of the TM ectodomain from the endothelium are present in plasma and are often taken as a marker of endothelial damage [22]. The ultimate significance of circulating TM is unknown.
2.4 EPCR
EPCR (CD201) is a membrane receptor that binds protein C [27]. It is homologous to the CD1/MHC superfamily. The gene for EPCR (PROCR) is on chromosome 20 [28]. EPCR's short cytoplasmic tail implies that EPCR plays no direct role in APC signaling, but rather plays an indirect role. Palmitoylation of the C-terminal Cys residue localizes EPCR to caveolae which may enable APC signaling [29, 30]. Although first recognized for binding protein C and APC with similar affinity (KD ~ 60 nM), EPCR also binds factor VII and factor VIIa, presumably via the Gla domain association with one end of an EPCR alpha helix. EPCR is also present on epithelial cells, monocytes, macrophages, neutrophils, eosinophils, natural killer cells and in mice on specific bone marrow derived dendritic cells [27, 31–34]. EPCR’s ectodomain is susceptible to being shed due to proteolysis in the region of residues 193–200, just above the trans-membrane domain, by tumor necrosis factor-alpha converting enzyme/ADAM17 (TACE). During inflammation, TNFα and Interleukin-1β can cause soluble EPCR formation in mice, and soluble EPCR is increased in plasmas of patients with a prothrombotic and proinflammatory tendency [35].
2.5 Protease activated receptor 1 (PAR1)
The search for the elusive platelet thrombin receptor led to discovery of PAR1 which is a major thrombin receptor on human platelets, though it is lacking on mouse platelets. PAR1 is the prototype of a four-member subfamily of protease activated G protein coupled receptors (GPCR) [36–39]. Typical of 7 transmembrane receptors, each PAR contains seven transmembrane helical domains and an extracellular N-terminal tail. The remarkable feature of PARs is that upon cleavage by various proteases the new amino terminus acts as a tethered agonist which triggers cell signaling [37, 40]. The PAR1 gene (F2R) is found on chromosome 5q13. This gene has two exons. PAR1 expression varies substantially among different cells and tissues [36–39]. Whereas thrombin activates human platelets via PAR1 and PAR4, murine platelet activation involves PAR3 and PAR4, but not PAR1 which is not present on murine platelets [41].
3 Protein C anticoagulant pathway and venous thrombosis
As demonstrated by the hereditary deficiencies of various components of the pathway in patients experiencing venous thrombosis, a physiologically essential function of the protein C pathway is reduction of risk for venous thrombosis. Anticoagulant activity of APC involves proteolytic inactivation of factors Va and VIIIa with enhancements from various lipid and protein cofactors (Figure 1a). Inactivation of factor Va by APC very effectively blunts thrombin generation due to one or more cleavages at Arg306, Arg506, and Arg679 in factor Va. Arg506 cleavage by APC occurs rapidly whereas cleavage at Arg306 is slower, but required for complete inactivation of factor Va. Mutagenesis studies show that positively charged residues in exosites on the APC protease domain surface are required for rapid inactivation of factor Va [42–47]. Physiological relevance of APC-mediated factor VIIIa inactivation has been debated due to the spontaneous inactivation of factor VIIIa. Nonetheless, several studies do support a role for APC for factor VIIIa inactivation [48, 49].
APC anticoagulant cofactors: Protein S
The APC-cofactor function of protein S involves a complex set of interactions of protein S with APC and with components of the prothrombinase complex, factor Xa and factor Va. Protein S binding to APC on phospholipid membranes brings APC’s active site closer to the membrane, presumably placing it in a better position to cleave Arg306. Protein S competes with factor Xa for binding to factor Va, such that protein S reduces the ability of factor Xa to block APC’s binding to and cleavage of factor Va [50, 51]. Areas of protein S that bind APC are located in the Gla domain, the thrombin-sensitive region, and in both EGF-1 and EGF-2 domains (Figure 2). Regions on the APC surface that bind protein S are the hydrophobic helix at the end of the Gla domain, several EGF-1 residues, and the C-terminus of the light chain, although this picture remains conjecture based on indirect evidence [52–54]. Protein S cofactor function is downregulated both in vitro and in vivo by at least two mechanisms. First, reversible binding of protein S to C4b binding protein (C4BP) results in the loss of its APC-cofactor activity for Arg506 cleavage in FVa. Protein S bound to C4BP can enhance cleavage at Arg306, but this cleavage in factor Va is not well reflected in typical coagulation assays for APC activity [55–58]. Second, protein S can be cleaved at Arg-49, Arg-60 and/or Arg-70 with a consequent loss of APC cofactor activity [59, 60].
Independent of its role as an APC cofactor, protein S can also inhibit coagulation reactions by its direct binding to factor Xa, factor Va, and factor VIIIa [61]. Additional factors affecting protein S anticoagulant actions include binding of protein S to tissue factor pathway inhibitor (TFPI) and to Zn2+ ions [13, 62–65]. More data are needed to determine the physiologic importance of APC-independent protein S anticoagulant activity.
APC anticoagulant cofactors: Lipids
In addition to procoagulant actions, negatively charged phospholipids, such as phosphatidylserine and cardiolipin, promote APC anticoagulant activity. Moreover, sphingolipids (e.g. glycosphingolipids and sphingosine), notably glucosylceramide, also enhance APC anticoagulant activity, and plasma glucosylceramide deficiency may be a marker for venous thrombosis risk [66–73].
APC anticoagulant cofactors: High Density Lipoprotein (HDL)
A very large amount of lipids circulate in lipoproteins. HDL stimulates APC’s cleavage at Arg306 in factor Va that is protein S-dependent [68]. Purified lipoprotein particles are chemically very complex with many components, making them challenging for functional coagulation studies. A recent challenge to this concept was based on faulty methodology because HDL was not properly handled or was frozen [74] and we have reproduced the original findings [68] showing HDL enhances APC’s anticoagulant actions. Consistent with the hypothesis that HDL is indirectly antithrombotic via APC, clinical data show that low HDL levels are found in venous thrombosis patients [69, 75–77].
APC anticoagulant cofactors: Factor V
Interestingly, factor V and protein S seem to be synergistic cofactors for factor VIIIa inactivation by APC [78]. It also appears that factor V promotes APC's inactivation of factor Va [79]. This APC-cofactor activity is defective in factor V Leiden, leading to the hypothesis that risk for thrombosis in factor V Leiden carriers is related, in part, to defective APC cofactor activity and, in part, to resistance to APC inactivation [80, 81].
3.1 Protein C deficiency
Heterozygous deficiency of protein C causes a moderately increased risk for venous thrombosis while severe homozygous or compound heterozygous deficiency causes neonatal purpura Fulminans [82]. Infants with severe deficiency present with massive thrombotic complications, and if they survive due to maintenance replacement therapy, they tend to present with mental retardation and/or visual impairment [82, 83]. Hundreds of mutations of protein C in thrombosis patients have been reported (see protein C mutation databases [84, 85]) and the basis for hereditary protein C defects can most often be rationalized based on the structure of protein C [86, 87]. Acquired protein C deficiency arises during initiation of Coumadin therapy due to the relatively short half life of protein C (T½ ~ 8 h) compared to most procoagulant vitamin K-dependent factors (T½ ~ 20 to 48 h), leading to the conventional wisdom of using heparin during initiation of therapy.
3.2 Protein S deficiency and protein S Tokushima
Heterozygous protein S deficiency conveys a significantly increased risk of thrombosis [88–90]. Homozygous or compound heterozygous severe protein S deficiency is rare and patients present with purpura fulminans in the neonatal period similar to severe protein C deficiency [91]. Testing for protein S deficiency is challenging due to the binding of protein S to C4b binding protein in plasma [90]. Classification of deficiencies is as follows: Type I (quantitative deficit), reduction of both free and total protein S antigen; type II or qualitative deficiency, functional protein S defect with a low functional activity but normal antigen level; and type III, with a low level of free protein S level but a normal level of total Protein S antigen. Protein S mutations are found in the majority of deficient patients (see protein S mutation databases) [90, 92]. Remarkably, in contrast to the absence of commonly found mutations in protein C deficient patients, two polymorphisms causing deficiencies are known as protein S Heerlen (Ser460Pro), a type III defect among subjects of European ancestry, and protein S Tokushima (K196E), a type II defect among Japanese [93–97]. Protein S Tokushima is found in ~ 10 % of venous thrombosis patients in Japan but not elsewhere [90, 93, 98–100]. Protein S Tokushima (K196E which corresponds to residue 155 in mature protein S, sometimes designated K155E) likely arose from a single founder in Japan. This represents a balanced polymorphism, like factor V Leiden or prothrombin 20210A, that carries a beneficial effect (e.g., prevention of excess fatal bleeding) and a mild risk factor for venous thrombosis.
3.3 APC-resistant factor V - factor V Leiden
APC resistance is defined as an abnormally reduced anticoagulant response of a plasma sample to APC and this condition conveys an increased risk for venous thrombosis. There are many potential molecular causes for this phenomenon, but a mutation in factor V which arose in a European ancestor involving an APC-cleavage site (Arg506Gln), known as factor V Leiden, is the most common cause for hereditary thrombosis [101–103].
3.4 TM and EPCR Genetic Variants
Although TM genetic mutations may be associated with increased risk of arterial thrombosis and myocardial infarction, there is little data for association with venous thrombosis risk. TM mutations were recently linked to atypical hemolytic-uremic syndrome, a syndrome that is strongly linked to excessive complement activation [104]. The lectin-like domain of TM can contribute to inhibition of complement activation and provide direct anti-inflammatory activity [22–24]. Association of EPCR defects with venous and arterial thrombosis appears to be controversial [105–109]. Four EPCR haplotypes have been characterized, and the H1 haplotype was associated with reduced venous thrombosis risk and with diminished risk of thrombosis in carriers of the factor V Leiden mutation [28, 110].
4 Protein C cytoprotective pathways
The PROWESS trial showed that recombinant APC reduced mortality in adult severe sepsis, although the recent PROWESS-SHOCK trial performed 10 years later with different intensive care unit standards of care failed to show benefit for APC when infused at low dose over 96 hours. The PROWESS trial stimulated new phases of APC research in many labs, leading to the elucidation of multiple cytoprotective activities of APC that are mediated by APC’s direct effects on cells via cell receptors [1–13]. These multiple activities include anti-apoptotic activity, anti-inflammatory activity, regulation of gene expression, and stabilization of endothelial barrier protection, and can be initiated when APC targets two key receptors, PAR1 and EPCR (Figure 1b) [6, 111–116]. Cells subject to cytoprotective signaling induced by APC are not limited to the endothelium but also include epithelial cells, various lymphocytes, dendritic cells and neurons. In addition to EPCR and PAR1, other receptors may also play key roles depending on the cell type.
A growing body of literature is addressing many questions related to APC's effects on cells, such as: 1) which signaling pathways are key for APC’s cytoprotective effects on different cell types; 2) what are the receptor target(s) for APC’s effects on cells; 3) what overlaps exist for molecular mechanisms for APC’s anti-apoptotic activity, anti-inflammatory activity, regulation of gene expression, and endothelial or epithelial barrier protection; and, perhaps most relevant for mechanistic insights that lead to clinical translation, 4) what are the relative contributions of APC anticoagulant activity versus APC’s cytoprotective activities for the observed reduction in morbidity and mortality in various in vivo injury preclinical animal models? APC and the protein C pathway components are central to the body's host-defense system, and, thus, they are ideal targets for translational medicine research. The basic and preclinical research on this system will certainly lead to more opportunities for therapeutic treatment of complex and challenging medical disorders, including thrombosis, severe sepsis and ischemic stroke among other maladies.
4.1 APC receptors – EPCR and protease activated receptor 1 (PAR1)
APC multiple cytoprotective effects, including (a) anti-apoptotic activity (b) anti-inflammatory activities, (c) protection of endothelial barrier function, and (d) alteration of gene expression profiles, are mediated by multiple receptors. At present, most but not all studies support the paradigm for the direct cytoprotective actions of APC shown in Figure 1b in which EPCR-bound-APC activates PAR1 to initiate signaling (see reviews [2, 5, 6, 9, 12, 14]). Whether initiated by thrombin, APC or other proteases, mechanisms for PAR1-stimualted signaling are complex and likely involve a spectrum of conformational states. Essentially each protease may be viewed as a different agonist whose detailed molecular mechanisms merit full description and comparisons to other agonists. As a complex GPCR, PAR1 has a diverse family of agonists, partial agonists, co-receptors, and allosteric effectors [117–119]. In addition to the fact that EPCR can act as an essential cofactor for PAR1 activation by APC, EPCR can also modulate PAR1 activation in vitro by factor VIIa and factor Xa [112–114, 120–124]. One must note that in addition to EPCR and PAR1, other receptors may mediate, directly or indirectly, APC-initiated signaling in various cells (see below).
Although the EPCR-APC-PAR1 paradigm is broadly useful and supported [6, 14, 29, 111–113, 125–130], there is a paradox of how PAR1 can be activated by both APC and thrombin with such contrasting outcomes [2, 29, 30, 130, 131]. A partial explanation is related to the localization of PAR1 and EPCR in membrane rafts, specifically in caveolae [29, 130, 132, 133]. Localization of PAR1 in caveolin-1 rafts is required for cytoprotective signaling on endothelial cells by APC but this is not the case for thrombin activation of PAR1 [30]. Moreover, thrombin-cleaved PAR1 is rapidly internalized whereas APC-activated PAR1 tends to remain on the cell surface, which could prolong signaling by APC compared to thrombin [30, 134]. Recent work indicates that PAR1 is a biased receptor because thrombin-initiated signaling proceeds via G-protein-signaling whereas APC-initiated signaling proceeds via β-arrestin-signaling which seems independent of a G-protein [135]. Finally, the mechanisms mediating APC-induced signaling will vary and depend on the cell type and on the temporal and spatial context of cells, tissues and organs.
4.2 Cytoprotective barrier stabilization on endothelial cells by APC
One of the remarkable actions of APC is its ability to stabilize endothelial barriers and to minimize vascular permeability. It is clear that thrombin-mediated PAR1 activation results in activation of barrier disruptive RhoA, Rho kinase-mediated inactivation of myosin light chain phosphatase, and actin-myosin contractility due to increased myosin light chain phosphorylation, whereas APC prevents such morphologic transformations and maintains endothelial barrier functions both in vitro and in vivo [115, 116, 128, 136–138]. Multiple studies show that APC requires EPCR, PAR1, S1P1, and caveolin-1 to activate barrier protective Rac1 via a β-arrestin, and the dishevelled-2 scaffold [30, 130, 131, 135, 139]. The effects of APC's barrier protective effects via Rac1 activation resemble endothelial barrier stabilization caused by sphingosine-1-phosphate, an endogenous bioactive lipid, in that APC causes EPCR-dependent clustering and transactivation of the S1P receptor 1 (S1P1) in membrane lipid rafts and with potential sphingosine kinase 1 (SphK1) “inside-out” signaling by S1P [115, 116, 138, 140].
4.3 Other APC receptors and other cells
Studies show that APC's cytoprotective effects in cells other than endothelial cells require additional receptors, including sphingosine-1-phosphate receptor 1 (S1P1), several integrins, PAR3, apolipoprotein E receptor 2 (ApoER2), glycoprotein Ib, Tie2, and epidermal growth factor receptor (EGFR) [115, 116, 127, 141–149].
Lymphocytes are a particularly important target for APC’s beneficial effects. APC's actions either directly or indirectly can reduce lymphocyte adhesion and tissue infiltration. Integrins appear to mediate APC's effects on lymphocytes. On macrophages, APC binds integrin CD11b/CD18 (αMβ2; Mac-1; CR3) where the integrin presumably replaces the role of EPCR in facilitating PAR1-dependent signaling by APC. APC-initiated production of barrier protective S1P and suppression of the proinflammatory response of activated macrophages depends on CD11b/CD18 but not on EPCR [149]. Human APC has an RGD (Arg-Gly-Asp) sequence which appears to mediate its binding β1 and β3 integrins and its inhibition of neutrophil migration [144].
PAR3 is the least studied member of the PAR four-membered family. However, it is clear that APC's cytoprotective effects in the brain, particularly on neurons, and in the kidney, particularly on podocytes, require PAR3 [127, 143, 150]. The mechanisms for APC's action on PAR3 remain to be clarified but likely involve PAR3 cleavage and activation.
ApoER2 (gene symbol LRP8 for low density lipoprotein receptor-related protein 8) binds APC with high affinity and this receptor can mediate APC-induced activation of signaling pathways in U937 cells [141, 148]. In this cell line, APC activates the PI3K-Akt survival pathway via Dab1-dependent activation of the Src family kinases, thereby promoting anti-inflammatory effects [148]. The in vivo significance of apoER2 as a physiologic or pharmacologic receptor for APC has not been assessed.
The ability of APC to reduce mortality in murine endotoxemia sepsis models requires EPCR on dendritic cells [34]. Given the growing list of APC receptors on vascular cells, it seems less and less likely that there is one unifying mechanism for each of APC’s cytoprotective effects on various cell types. Instead, APC cellular activities and their underlying mechanisms seem to be dictated by cell-type specific expression of APC receptor complexes.
It is noteworthy that APC proteolytic actions can also indirectly protect cells from the potentially damaging effects of histones or of neutrophil-derived histone-DNA complexes (NETs) because APC can cleave extracellular histones, and this activity appears to protect against sepsis in mice [151].
5 APC engineered mutants that distinguish anticoagulant from cytoprotective functions
Site-directed mutagenesis has provided many mutants of APC that help to clarify structure-function relationships for this protein. Initially, such studies mainly involved efforts to clarify why human mutations in protein C deficient patients caused thrombosis and to explore structure-function relationships for APC’s anticoagulant activity [2, 6, 152, 153]. Subsequently, stimulated by the PROWESS sepsis trial, by the bleeding risks identified in that trial, and by the discovery of APC's multiple cytoprotective actions, we and others undertook mutagenesis studies to discover APC mutants with selective loss of either anticoagulant or cytoprotective properties. The goal was initially to use APC mutants for proof of principle preclinical studies to clarify the relative importance of APC's various activities for its ability to reduce morbidity and mortality in various animal injury models. Our initial goal was also to prepare APC variants with a reduced risk of bleeding due to reduced anticoagulant activity but with normal direct effects on cells [154]. Moreover, our efforts to engineer cytoprotective-selective APC mutants were based on the assumptions that the factor Va exosites that mediate anticoagulant activity and the PAR1 exosites that mediate cytoprotective activities are, at least partially, non-overlapping. Thus, we attempted to alter factor Va exosites in APC without affecting exosites that might recognize PAR1, although nothing was known about exosites mediating PAR1 cleavage [6, 154]. Reported mutations that reduced anticoagulant activity involved a positively charged surface of the protease domain that includes loop 37 (protein C residues 190–193, equivalent to chymotrypsin (CHT) residues 36–39), the Ca++-binding loop (residues 225–235, CHT residues 70–80) and the autolysis loop (residues 301–316, CHT residues 142–153) (see Figure 3) [42, 44–47, 155, 156]. To our delightful surprise, when two APC variants with Ala mutations in two exosites defined by two APC surface loops involving Ala replacements of Arg229 and 230 and/or of Lys191, 192 and 193 were tested in assays of staurosporine-induced endothelial cell apoptosis, success was achieved. The two APC variants, 3K3A-APC and 5A-APC, had reduced anticoagulant activity but retained normal anti-apoptotic activity that requires PAR1 and EPCR and exhibited a normal ability to cleave a PAR1 Nterminal peptide at Arg41 [154]. When the anticoagulant and anti-apoptotic activities of these APC variants were normalized to wild-type APC, the two APC variants exhibited 25-times and 33-times greater anti-apoptotic activity relative to anticoagulant activity when compared to wild-type APC. Subsequently, additional APC mutants with altered activity profiles were made that include the cytoselective mutants, L38D-APC, an engineered disulphide APC mutant, as well as anticoagulant-selective mutants with mutations at residues E149, E330 or E333 (see Figure 3) [52, 157–160].
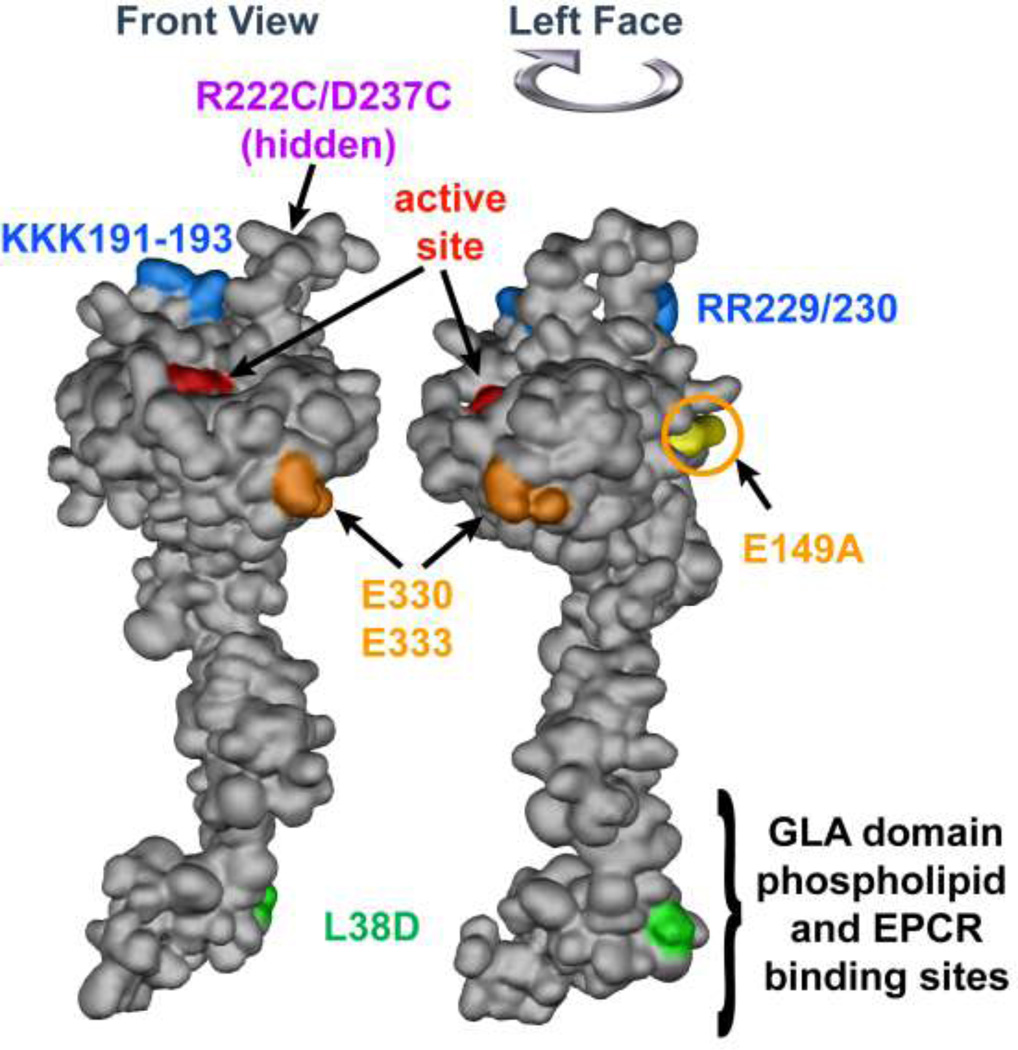
Figure 3. APC space-filling model showing mutations that differentiate amino acid residue requirements for anticoagulant versus cytoprotective activities.
The model of APC extending from bottom to top depicts the N-terminal Gla domain at the bottom which binds EPCR and phospholipids membranes. The protease domain is at at the top, with the EGF-1and EGF-2 domains in the middle section of the model. The "active site" triad of Ser, His and Asp residues is noted in red. Green highlights the L38D mutation that reduces anticoagulant activity due to reduced protein S enhancement. Gold highlights mutations of E330 and E333 to Ala that selectively reduce PAR1 signaling and the E149A mutation in the C-terminus of the light chain that causes loss of cytoprotective anti-inflammatory and anti-apoptotic activities but gain-of-function of anticoagulant activity due to enhanced protein S cofactor effects. On the top of the model, blue highlights five basic residues (KKK191–193 and RR229/230) which form a large positively charged exosite that recognizes factor Va. When these five basic residues are mutated to Ala, < 10 % anticoagulant activity remains whereas cytoprotective activities remain intact. Purple highlights two residues (R222 and D237 that are not in view) which when mutated to Cys can form a disulfide bond, causing loss of most anticoagulant activity but retention of cytoprotective activity. The model of full length APC [172] is based on the serine protease domain structure of APC (Protein Data Bank entry 1AUT [155]).
For proof of principle studies in preclinical research, murine cytoprotective-selective 3K3A-APC and 5A-APC mutants lacking most anticoagulant activity were compared in vivo to wt-APC for APC’s ability to reduce mortality in sepsis or to provide neuroprotection in ischemic stroke models. Remarkably, these cytoprotectiveselective APC mutants were fully active and appeared indistinguishable from wt-APC [34, 128, 142, 161]. In contrast, the murine anticoagulant-selective E149A-APC mutant lacking anti-apoptotic activity failed to reduce mortality in mice given lethal doses of endotoxin. Furthermore, murine 5A-APC also reduced death in murine pneumonia [138]. Thus, based on a growing set of data, it appears that APC’s cytoprotective actions are primarily responsible for APC’s in vivo benefits in sepsis and neuroprotection for stroke. These preclinical results help set the stage for developing novel APC mutants as second generation biologics with greatly reduced risk of bleeding. The PROWESS and PROWESS-SHOCK clinical trials were seriously limited in scope because they employed a 96 hour long, low-dose infusion regimen which was associated with serious bleeding risk in the former trial. In the future, cytoprotective-selective APC mutants should permit trials with altered dosing regimens, consistent with APC’s ability to alter cell signaling.
6 APC neuroprotective activities
The protein C system has particular relevance for the brain. Prospective epidemiology studies show that low levels of plasma protein C tend to be associated with increased risk for stroke, but not myocardial infarction. Infants with severe deficiency of protein C who survived the neonatal period tend to be blind or have cognitive impairments. Moreover, the organ for which the largest body of research exists for the cytoprotective actions of APC is the brain [12]. Based on the emerging concept that multiple cells contribute to acute and chronic brain maladies, disease-modifying agents should ideally target multiple cells via multiple actions. We believe that APC is such an agent as it has multiple actions and can act on multiple targets, as summarized above. Extensive data show that APC provides remarkable neuroprotective effects in vivo and increases survival in murine ischemic stroke studies [127, 162–164]. In vitro studies show that APC targets both brain endothelial cells and neurons in different injury model systems [12] with the net effect in vivo of inhibiting breakdown of the bloodbrain-barrier, neuronal damage, and inflammatory responses (Figure 4). Moreover, chronic, daily doses of APC showing beneficial effects in animal models of amyotrophic lateral sclerosis also appear promising in animal models of multiple sclerosis and Alzheimer’s disease [12, 146, 165]. Remarkably, APC conveys diseasemodifying effects in both acute neurological injury (e.g., stroke and nerve crush injury) and in chronic neurodegenerative disorders [12]. To a substantial extent, the neuroprotective actions are likely based on APC’s cytoprotective properties. In murine brain injury models, neuroprotection required PAR1, EPCR and PAR3 [113, 126, 127, 142, 143, 166]. For ischemic stroke, tPA is useful but is limited by its short window of 4.5 hours and its neurotoxicity. APC can block tPA’s neurotoxic effects [164, 166], prevent tPA-induced hemorrhagic conversion in stroke and reduce neuronal damage [126]. Thus, extensive data suggest that APC’s cytoprotective activities are very promising for providing neuroprotective acute and chronic therapies.
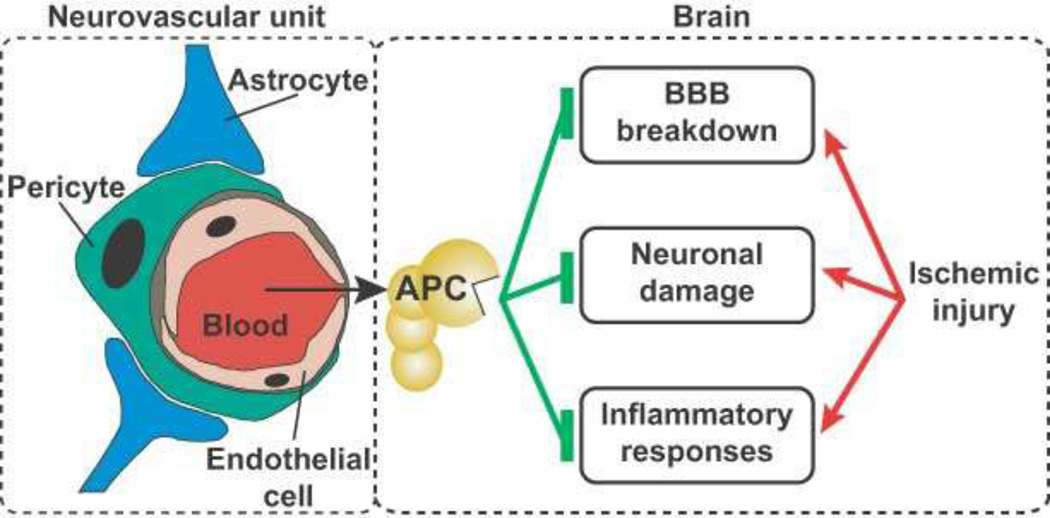
Figure 4. APC’s neuroprotective actions on the neurovascular unit following ischemic injury.
Ischemia promotes endothelial cell apoptosis, breakdown of the blood-brain-barrier (BBB), neuroinflammation, and damage to neurons. By its cytoprotective actions, APC protects vascular integrity and ameliorates post-ischemic BBB breakdown, thereby preventing secondary neuronal damage mediated by entry of several blood-derived neurotoxic and vasculotoxic molecules. APC can cross an intact BBB via EPCR-dependent transport to reach its neuronal targets in brain. APC can express direct neuronal protective activity to prevent neuron damage. APC also expresses anti-inflammatory activities by blocking early post-ischemic infiltration of brain by neutrophils and by suppressing microglia activation.
7 Conclusions
Further elucidation of APC’s cytoprotective pathways and receptors combined with site-directed mutagenesis studies of APC mutants is likely to lead to clinically interesting second generation APC biologics that go far beyond the classical anticoagulant properties of APC.
Acknowledgements
We gratefully acknowledge helpful discussions with members of the Griffin, Zlokovic and Mosnier laboratories. We apologize to our colleagues whose excellent work was not cited due to space limitations.
Contributor Information
John H Griffin, Department of Molecular & Experimental Medicine, The Scripps Research Institute, La Jolla CA 92037 USA.
Berislav V Zlokovic, Department of Physiology and Biophysics, University of Southern California, Los Angeles CA 90089-9142 USA.
Laurent O Mosnier, Department of Molecular & Experimental Medicine, The Scripps Research Institute, La Jolla CA 92037 USA.
Reference List
- 1.Esmon CT. Inflammation and the activated protein C anticoagulant pathway. Semin.Thromb.Hemost. 2006;32:49–60. doi: 10.1055/s-2006-939554. [DOI] [PubMed] [Google Scholar]
- 2.Rezaie AR. Regulation of the protein C anticoagulant and antiinflammatory pathways. Curr.Med.Chem. 2010;17:2059–2069. doi: 10.2174/092986710791233706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riewald M, Ruf W. Science review: Role of coagulation protease cascades in sepsis. Crit.Care. 2003;7:123–129. doi: 10.1186/cc1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiler H, Kerschen E. Modulation of sepsis outcome with variants of activated protein C. J Thromb Haemost. 2009;7:127–131. doi: 10.1111/j.1538-7836.2009.03377.x. [DOI] [PubMed] [Google Scholar]
- 5.Jackson CJ, Xue M. Activated protein C--an anticoagulant that does more than stop clots. Int.J.Biochem.Cell Biol. 2008;40:2692–2697. doi: 10.1016/j.biocel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Mosnier LO, Zlokovic BV, Griffin JH. The cytoprotective protein C pathway. Blood. 2007;109:3161–3172. doi: 10.1182/blood-2006-09-003004. [DOI] [PubMed] [Google Scholar]
- 7.van de Wouwer M, Collen D, Conway EM. Thrombomodulin-Protein C-EPCR System: Integrated to Regulate Coagulation and Inflammation. Arterioscler.Thromb.Vasc.Biol. 2004;24:1374–1383. doi: 10.1161/01.ATV.0000134298.25489.92. [DOI] [PubMed] [Google Scholar]
- 8.Rezende SM, Simmonds RE, Lane DA. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood. 2004;103:1192–1201. doi: 10.1182/blood-2003-05-1551. [DOI] [PubMed] [Google Scholar]
- 9.Danese S, Vetrano S, Zhang L, Poplis VA, Castellino FJ. The protein C pathway in tissue inflammation and injury: pathogenic role and therapeutic implications. Blood. 2010;115:1121–1130. doi: 10.1182/blood-2009-09-201616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castoldi E, Rosing J. APC resistance: biological basis and acquired influences. J Thromb Haemost. 2010;8:445–453. doi: 10.1111/j.1538-7836.2009.03711.x. [DOI] [PubMed] [Google Scholar]
- 11.Dahlbäck B. Advances in understanding pathogenic mechanisms of thrombophilic disorders. Blood. 2008;112:19–27. doi: 10.1182/blood-2008-01-077909. [DOI] [PubMed] [Google Scholar]
- 12.Zlokovic BV, Griffin JH. Cytoprotective protein C pathways and implications for stroke and neurological disorders. Trends Neurosci. 2011;34:198–209. doi: 10.1016/j.tins.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackeng TM, Rosing J. Protein S as cofactor for TFPI. Arterioscler.Thromb.Vasc.Biol. 2009;29:2015–2020. doi: 10.1161/ATVBAHA.108.177436. [DOI] [PubMed] [Google Scholar]
- 14.Jackson C, Whitmont K, Tritton S, March L, Sambrook P, Xue M. New therapeutic applications for the anticoagulant, activated protein C. Expert.Opin.Biol.Ther. 2008;8:1109–1122. doi: 10.1517/14712598.8.8.1109. [DOI] [PubMed] [Google Scholar]
- 15.van Sluis GL, Buller HR, Spek CA. The role of activated protein C in cancer progression. Thromb Res. 2010;125:S138–S142. doi: 10.1016/S0049-3848(10)70032-3. [DOI] [PubMed] [Google Scholar]
- 16.Gupta A, Williams MD, Macias WL, Molitoris BA, Grinnell BW. Activated protein C and acute kidney injury: Selective targeting of PAR-1. Curr.Drug Targets. 2009;10:1212–1226. doi: 10.2174/138945009789753291. [DOI] [PubMed] [Google Scholar]
- 17.Griffin JH, Evatt B, Zimmerman TS, Kleiss AJ, Wideman C. Deficiency of protein C in congenital thrombotic disease. J.Clin.Invest. 1981;68:1370–1373. doi: 10.1172/JCI110385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster DC, Yoshitake S, Davie EW. The nucleotide sequence of the gene for human protein C. Proc.Natl.Acad.Sci.U.S.A. 1985;82:4673–4677. doi: 10.1073/pnas.82.14.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patracchini P, Aiello V, Palazzi P, Calzolari E, Bernardi F. Sublocalization of the human protein C gene on chromosome 2q13-q14. Hum.Genet. 1989;81:191–192. doi: 10.1007/BF00293902. [DOI] [PubMed] [Google Scholar]
- 20.Griffin JH, Gruber A, Fernández JA. Reevaluation of total, free, and bound protein S and C4b-binding protein levels in plasma anticoagulated with citrate or hirudin. Blood. 1992;79:3203–3211. [PubMed] [Google Scholar]
- 21.Linse S, Hardig Y, Schultz DA, Dahlbäck B. A region of vitamin K-dependent protein S that binds to C4b binding protein (C4BP) identified using bacteriophage peptide display libraries. J.Biol.Chem. 1997;272:14658–14665. doi: 10.1074/jbc.272.23.14658. [DOI] [PubMed] [Google Scholar]
- 22.Conway EM. Thrombomodulin and its role in inflammation. Semin.Immunopathol. 2012;34:107–125. doi: 10.1007/s00281-011-0282-8. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Maruyama I. Thrombomodulin: protectorate God of the vasculature in thrombosis and inflammation. J Thromb Haemost. 2011;9(Suppl 1):168–173. doi: 10.1111/j.1538-7836.2011.04319.x. [DOI] [PubMed] [Google Scholar]
- 24.Esmon CT, Owen WG. The discovery of thrombomodulin. J.Thromb.Haemost. 2004;2:209–213. doi: 10.1046/j.1538-7933.2003.00537.x. [DOI] [PubMed] [Google Scholar]
- 25.Jackman RW, Beeler DL, Fritze L, Soff G, Rosenberg RD. Human thrombomodulin gene is intron depleted: nucleic acid sequences of the cDNA and gene predict protein structure and suggest sites of regulatory control. Proc.Natl.Acad.Sci.U.S.A. 1987;84:6425–6429. doi: 10.1073/pnas.84.18.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen DZ. Human thrombomodulin: complete cDNA sequence and chromosome localization of the gene. Biochemistry. 1987;26:4350–4357. doi: 10.1021/bi00388a025. [DOI] [PubMed] [Google Scholar]
- 27.Fukudome K, Esmon CT. Identification, cloning, and regulation of a novel endothelial cell protein C/activated protein C receptor. J.Biol.Chem. 1994;269:26486–26491. [PubMed] [Google Scholar]
- 28.Simmonds RE, Lane DA. Structural and functional implications of the intron/exon organization of the human endothelial cell protein C/activated protein C receptor (EPCR) gene: comparison with the structure of CD1/major histocompatibility complex alpha1 and alpha2 domains. Blood. 1999;94:632–641. [PubMed] [Google Scholar]
- 29.Bae JS, Yang L, Manithody C, Rezaie AR. The ligand occupancy of endothelial protein C receptor switches the protease-activated receptor 1-dependent signaling specificity of thrombin from a permeability-enhancing to a barrier-protective response in endothelial cells. Blood. 2007;110:3909–3916. doi: 10.1182/blood-2007-06-096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo A, Soh UJ, Paing MM, Arora P, Trejo J. Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc.Natl.Acad.Sci.U.S.A. 2009;106:6393–6397. doi: 10.1073/pnas.0810687106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturn DH, Kaneider NC, Feistritzer C, Djanani A, Fukudome K, Wiedermann CJ. Expression and function of the endothelial protein C receptor in human neutrophils. Blood. 2003;102:1499–1505. doi: 10.1182/blood-2002-12-3880. [DOI] [PubMed] [Google Scholar]
- 32.Yuksel M, Okajima K, Uchiba M, Horiuchi S, Okabe H. Activated protein C inhibits lipopolysaccharide-induced tumor necrosis factor-alpha production by inhibiting activation of both nuclear factor-kappa B and activator protein-1 in human monocytes. Thromb.Haemost. 2002;88:267–273. [PubMed] [Google Scholar]
- 33.Pereira C, Schaer DJ, Bachli EB, Kurrer MO, Schoedon G. Wnt5A/CaMKII signaling contributes to the inflammatory response of macrophages and is a target for the antiinflammatory action of activated protein C and interleukin-10. Arterioscler.Thromb.Vasc.Biol. 2008;28:504–510. doi: 10.1161/ATVBAHA.107.157438. [DOI] [PubMed] [Google Scholar]
- 34.Kerschen EJ, Hernandez I, Zogg M, Jia S, Hessner MJ, Fernandez J, et al. Activated protein C targets CD8+ dendritic cells to reduce the mortality of endotoxemia in mice. J Clin.Invest. 2010;120:3167–3178. doi: 10.1172/JCI42629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu D, Wang Y, Esmon NL, Esmon CT. Regulated endothelial protein C receptor shedding is mediated by tumor necrosis factor-alpha converting enzyme/ADAM17. J.Thromb.Haemost. 2007;5:395–402. doi: 10.1111/j.1538-7836.2007.02347.x. [DOI] [PubMed] [Google Scholar]
- 36.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 37.Coughlin SR. Thrombin signaling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 38.Leger AJ, Covic L, Kuliopulos A. Protease-activated receptors in cardiovascular diseases. Circulation. 2006;114:1070–1077. doi: 10.1161/CIRCULATIONAHA.105.574830. [DOI] [PubMed] [Google Scholar]
- 39.Traynelis SF, Trejo J. Protease-activated receptor signaling: new roles and regulatory mechanisms. Curr.Opin.Hematol. 2007;14:230–235. doi: 10.1097/MOH.0b013e3280dce568. [DOI] [PubMed] [Google Scholar]
- 40.Coughlin SR, Camerer E. PARticipation in inflammation. J.Clin.Invest. 2003;111:25–27. doi: 10.1172/JCI17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakanishi-Matsui M, Zheng YW, Sulciner DJ, Weiss EJ, Ludeman MJ, Coughlin SR. PAR3 is a cofactor for PAR4 activation by thrombin. Nature. 2000;404:609–613. doi: 10.1038/35007085. [DOI] [PubMed] [Google Scholar]
- 42.Gale AJ, Heeb MJ, Griffin JH. The autolysis loop of activated protein C interacts with factor Va and differentiates between the Arg506 and Arg306 cleavage sites. Blood. 2000;96:585–593. [PubMed] [Google Scholar]
- 43.Shen L, Villoutreix BO, Dahlbäck B. Interspecies loop grafting in the protease domain of human protein C yielding enhanced catalytic and anticoagulant activity. Thromb.Haemost. 1999;82:1078–1087. [PubMed] [Google Scholar]
- 44.Friedrich U, Nicolaes GAF, Villoutreix BO, Dahlbäck B. Secondary substrate-binding exosite in the serine protease domain of activated protein C important for cleavage at Arg-506 but not at Arg-306 in factor Va. J.Biol.Chem. 2001;276:23105–23108. doi: 10.1074/jbc.M103138200. [DOI] [PubMed] [Google Scholar]
- 45.Rezaie AR. Exosite-dependent regulation of the protein C anticoagulant pathway. Trends Cardiovasc.Med. 2003;13:8–15. doi: 10.1016/s1050-1738(02)00191-3. [DOI] [PubMed] [Google Scholar]
- 46.Gale AJ, Tsavaler A, Griffin JH. Molecular characterization of an extended binding site for coagulation factor Va in the positive exosite of activated protein C. J.Biol.Chem. 2002;277:28836–28840. doi: 10.1074/jbc.M204363200. [DOI] [PubMed] [Google Scholar]
- 47.Gale AJ, Griffin JH. Characterization of a thrombomodulin binding site on protein C and its comparison to an activated protein C binding site for factor Va. Proteins. 2004;54:433–441. doi: 10.1002/prot.10627. [DOI] [PubMed] [Google Scholar]
- 48.Gale AJ, Radtke KP, Cunningham MA, Chamberlain D, Pellequer JL, Griffin JH. Intrinsic stability and functional properties of disulfide bond-stabilized coagulation factor VIIIa variants. J.Thromb.Haemost. 2006;4:1315–1322. doi: 10.1111/j.1538-7836.2006.01951.x. [DOI] [PubMed] [Google Scholar]
- 49.Gale AJ, Cramer TJ, Rozenshteyn D, Cruz JR. Detailed mechanisms of the inactivation of factor VIIIa by activated protein C in the presence of its cofactors, protein S and factor V. J Biol.Chem. 2008;283:16355–16362. doi: 10.1074/jbc.M708985200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heeb MJ, Kojima Y, Rosing J, Tans G, Griffin JH. C-terminal residues 621-635 of protein S are essential for binding to factor Va. J.Biol.Chem. 1999;274:36187–36192. doi: 10.1074/jbc.274.51.36187. [DOI] [PubMed] [Google Scholar]
- 51.Heeb MJ, Kojima Y, Hackeng TM, Griffin JH. Binding sites for blood coagulation factor Xa and protein S involving residues 493-506 in factor Va. Protein Sci. 1996;5:1883–1889. doi: 10.1002/pro.5560050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harmon S, Preston RJ, Ainle FN, Johnson JA, Cunningham MS, Smith OP, et al. Dissociation of activated protein C functions by elimination of protein S cofactor enhancement. J.Biol.Chem. 2008;283:30531–30539. doi: 10.1074/jbc.M802338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosnier LO, Zampolli A, Kerschen EJ, Schuepbach RA, Banerjee Y, Fernandez JA, et al. Hyperantithrombotic, non-cytoprotective Glu149Ala-activated protein C mutant. Blood. 2009;113:5970–5978. doi: 10.1182/blood-2008-10-183327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersson HM, Arantes MJ, Crawley JT, Luken BM, Tran S, Dahlbäck B, et al. Activated protein C cofactor function of protein S: a critical role for Asp95 in the EGF1-like domain. Blood. 2010;115:4878–4885. doi: 10.1182/blood-2009-11-256610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishioka J, Suzuki K. Inhibition of cofactor activity of protein S by a complex of protein S and C4bbinding protein. Evidence for inactive ternary complex formation between protein S, C4b-binding protein, and activated protein C. J.Biol.Chem. 1990;265:9072–9076. [PubMed] [Google Scholar]
- 56.Fernández JA, Heeb MJ, Griffin JH. Identification of residues 413-433 of plasma protein S as essential for binding to C4b-binding protein. J.Biol.Chem. 1993;268:16788–16794. [PubMed] [Google Scholar]
- 57.Greengard JS, Fernández JA, Radtke KP, Griffin JH. Identification of candidate residues for interaction of protein S with C4b binding protein and activated protein C. Biochem.J. 1995;305:397–403. doi: 10.1042/bj3050397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández JA, Griffin JH, Chang GT, Stam J, Reitsma PH, Bertina RM, et al. Involvement of amino acid residues 423-429 of human protein S in binding to C4b-binding protein. Blood Cells Mol.Dis. 1998;24:101–112. doi: 10.1006/bcmd.1998.0175. [DOI] [PubMed] [Google Scholar]
- 59.Brinkman HJ, Mertens K, van Mourik JA. Proteolytic cleavage of protein S during the hemostatic response. J Thromb Haemost. 2005;3:2712–2720. doi: 10.1111/j.1538-7836.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- 60.Chang GT, Aaldering L, Hackeng TM, Reitsma PH, Bertina RM, Bouma BN. Construction and characterization of thrombin-resistant variants of recombinant human protein S. Thromb Haemost. 1994;72:693–697. [PubMed] [Google Scholar]
- 61.Heeb MJ, Mesters RM, Tans G, Rosing J, Griffin JH. Binding of protein S to factor Va associated with inhibition of prothrombinase that is independent of activated protein C. J.Biol.Chem. 1993;268:2872–2877. [PubMed] [Google Scholar]
- 62.Hackeng TM, Sere KM, Tans G, Rosing J. Protein S stimulates inhibition of the tissue factor pathway by tissue factor pathway inhibitor. Proc.Natl.Acad.Sci.U.S.A. 2006;103:3106–3111. doi: 10.1073/pnas.0504240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hackeng TM, Maurissen LF, Castoldi E, Rosing J. Regulation of TFPI function by protein S. J Thromb Haemost. 2009;7:165–168. doi: 10.1111/j.1538-7836.2009.03363.x. [DOI] [PubMed] [Google Scholar]
- 64.Fernandes N, Mosnier LO, Tonnu L, Heeb MJ. Zn2+-containing protein S inhibits extrinsic factor X activating complex independently of tissue factor pathway inhibitor (TFPI) J Thromb Haemost. 2010;8:1976–1985. doi: 10.1111/j.1538-7836.2010.03919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heeb MJ, Prashun D, Griffin JH, Bouma BN. Plasma protein S contains zinc essential for efficient activated protein C-independent anticoagulant activity and binding to factor Xa but not for efficient binding to tissue factor pathway inhibitor. FASEB J. 2009;23:2244–2253. doi: 10.1096/fj.08-123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernández JA, Kojima K, Petäjä J, Hackeng TM, Griffin JH. Cardiolipin enhances protein C pathway anticoagulant activity. Blood Cells Mol.Dis. 2000;26:115–123. doi: 10.1006/bcmd.2000.0285. [DOI] [PubMed] [Google Scholar]
- 67.Bakker HM, Tans G, Janssen-Claessen T, Thomassen MC, Hemker HC, Griffin JH, et al. The effect of phospholipids, calcium ions and protein S on rate constants of human factor Va inactivation by activated human protein C. Eur.J.Biochem. 1992;208:171–178. doi: 10.1111/j.1432-1033.1992.tb17171.x. [DOI] [PubMed] [Google Scholar]
- 68.Griffin JH, Kojima K, Banka CL, Curtiss LK, Fernández JA. High-density lipoprotein enhancement of anticoagulant activities of plasma protein S and activated protein C. J. Clin.Invest. 1999;103:219–227. doi: 10.1172/JCI5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffin JH, Fernández JA, Deguchi H. Plasma lipoproteins, hemostasis and thrombosis. Thromb.Haemost. 2001;86:386–394. [PubMed] [Google Scholar]
- 70.Deguchi H, Fernández JA, Pabinger I, Heit JA, Griffin JH. Plasma glucosylceramide deficiency as potential risk factor for venous thrombosis and modulator of anticoagulant protein C pathway. Blood. 2001;97:1907–1914. doi: 10.1182/blood.v97.7.1907. [DOI] [PubMed] [Google Scholar]
- 71.Deguchi H, Fernández JA, Griffin JH. Neutral glycosphingolipid-dependent inactivation of coagulation factor Va by activated protein C and protein S. J.Biol.Chem. 2002;277:8861–8865. doi: 10.1074/jbc.M110252200. [DOI] [PubMed] [Google Scholar]
- 72.Yegneswaran S, Deguchi H, Griffin JH. Glucosylceramide, a neutral glycosphingolipid anticoagulant cofactor, enhances the interaction of human- and bovine-activated protein C with negatively charged phospholipid vesicles. J.Biol.Chem. 2003;278:14614–14621. doi: 10.1074/jbc.M206746200. [DOI] [PubMed] [Google Scholar]
- 73.Deguchi H, Yegneswaran S, Griffin JH. Sphingolipids as bioactive regulators of thrombin generation. J Biol.Chem. 2004;279:12036–12042. doi: 10.1074/jbc.M302531200. [DOI] [PubMed] [Google Scholar]
- 74.Oslakovic C, Norstrom E, Dahlbäck B. Reevaluation of the role of HDL in the anticoagulant activated protein C system in humans. J Clin.Invest. 2010;120:1396–1399. doi: 10.1172/JCI42260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mineo C, Deguchi H, Griffin JH, Shaul PW. Endothelial and antithrombotic actions of HDL. Circ.Res. 2006;98:1352–1364. doi: 10.1161/01.RES.0000225982.01988.93. [DOI] [PubMed] [Google Scholar]
- 76.Deguchi H, Pecheniuk NM, Elias DJ, Averell PM, Griffin JH. High-density lipoprotein deficiency and dyslipoproteinemia associated with venous thrombosis in men. Circulation. 2005;112:893–899. doi: 10.1161/CIRCULATIONAHA.104.521344. [DOI] [PubMed] [Google Scholar]
- 77.Eichinger S, Pecheniuk NM, Hron G, Deguchi H, Schemper M, Kyrle PA, et al. High-Density Lipoprotein and the Risk of Recurrent Venous Thromboembolism. Circulation. 2007;115:1609–1614. doi: 10.1161/CIRCULATIONAHA.106.649954. [DOI] [PubMed] [Google Scholar]
- 78.Shen L, Dahlbäck B. Factor V and protein S as synergistic cofactors to activated protein C in degradation of factor VIIIa. J.Biol.Chem. 1994;269:18735–18738. [PubMed] [Google Scholar]
- 79.Cramer TJ, Griffin JH, Gale AJ. Factor V Is an Anticoagulant Cofactor for Activated Protein C during Inactivation of Factor Va. Pathophysiol.Haemost.Thromb. 2010;37:17–23. doi: 10.1159/000315141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castoldi E, Rosing J. Factor V Leiden: a disorder of factor V anticoagulant function. Curr.Opin.Hematol. 2004;11:176–181. doi: 10.1097/01.moh.0000130315.41033.32. [DOI] [PubMed] [Google Scholar]
- 81.Nicolaes GAF, Dahlbäck B. Factor V and thrombotic disease: description of a janus-faced protein. Arterioscler.Thromb.Vasc.Biol. 2002;22:530–538. doi: 10.1161/01.atv.0000012665.51263.b7. [DOI] [PubMed] [Google Scholar]
- 82.Chalmers E, Cooper P, Forman K, Grimley C, Khair K, Minford A, et al. Purpura fulminans: recognition, diagnosis and management. Arch.Dis.Child. 2011;96:1066–1071. doi: 10.1136/adc.2010.199919. [DOI] [PubMed] [Google Scholar]
- 83.Gladson CL, Groncy P, Griffin JH. Coumarin necrosis, neonatal purpura fulminans, and protein C deficiency. Arch.Dermatol. 1987;123:1701a–1706a. [PubMed] [Google Scholar]
- 84.D'Ursi P, Marino F, Caprera A, Milanesi L, Faioni EM, Rovida E. ProCMD: a database and 3D web resource for protein C mutants. BMC.Bioinformatics. 2007;8:S11. doi: 10.1186/1471-2105-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saunders RE, Perkins SJ. CoagMDB: a database analysis of missense mutations within four conserved domains in five vitamin K-dependent coagulation serine proteases using a text-mining tool. Hum.Mutat. 2008;29:333–344. doi: 10.1002/humu.20629. [DOI] [PubMed] [Google Scholar]
- 86.Rovida E, Merati G, D'Ursi P, Zanardelli S, Marino F, Fontana G, et al. Identification and computationally-based structural interpretation of naturally occurring variants of human protein C. Hum.Mutat. 2006;28:345–355. doi: 10.1002/humu.20445. [DOI] [PubMed] [Google Scholar]
- 87.Greengard JS, Fisher CL, Villoutreix B, Griffin JH. Structural basis for type I and type II deficiencies of antithrombotic plasma protein C: patterns revealed by three-dimensional molecular modelling of mutations of the protease domain. Proteins. 1994;18:367–380. doi: 10.1002/prot.340180407. [DOI] [PubMed] [Google Scholar]
- 88.Comp PC, Esmon CT. Recurrent venous thromboembolism in patients with a partial deficiency of protein S. N.Engl.J.Med. 1984;311:1525–1528. doi: 10.1056/NEJM198412133112401. [DOI] [PubMed] [Google Scholar]
- 89.Schwarz HP, Fischer M, Hopmeier P, Batard MA, Griffin JH. Plasma protein S deficiency in familial thrombotic disease. Blood. 1984;64:1297–1300. [PubMed] [Google Scholar]
- 90.Garcia de Frutos P, Fuentes-Prior P, Hurtado B, Sala N. Molecular basis of protein S deficiency. Thromb Haemost. 2007;98:543–556. [PubMed] [Google Scholar]
- 91.Mahasandana C, Suvatte V, Marlar RA, Manco-Johnson MJ, Jacobson LJ, Hathaway WE. Neonatal purpura fulminans associated with homozygous protein S deficiency. Lancet. 1990;335:61–62. doi: 10.1016/0140-6736(90)90201-f. [DOI] [PubMed] [Google Scholar]
- 92.Gandrille S, Borgel D, Ireland H, Lane DA, Simmonds R, Reitsma PH, et al. Protein S deficiency: a database of mutations. For the Plasma Coagulation Inhibitors Subcommittee of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 1997;77:1201–1214. [PubMed] [Google Scholar]
- 93.Hayashi T, Nishioka J, Shigekiyo T, Saito S, Suzuki K. Protein S Tokushima: abnormal molecule with a substitution of Glu for Lys-155 in the second epidermal growth factor-like domain of protein S. Blood. 1994;83:683–690. [PubMed] [Google Scholar]
- 94.Hayashi T, Nishioka J, Suzuki K. Characterization of dysfunctional protein S-Tokushima (K155-->E) in relation to the molecular interactions required for the regulation of blood coagulation. Pol.J Pharmacol. 1996;48:221–223. [PubMed] [Google Scholar]
- 95.Ikejiri M, Wada H, Sakamoto Y, Ito N, Nishioka J, Nakatani K, et al. The association of protein S Tokushima-K196E with a risk of deep vein thrombosis. Int.J Hematol. 2010;92:302–305. doi: 10.1007/s12185-010-0671-0. [DOI] [PubMed] [Google Scholar]
- 96.Nicolaes GA, Hackeng TM, Segers K, Rosing J. A structural model of the SHBG domain of human variant protein S Heerlen. Thromb Haemost. 2006;96:538–540. [PubMed] [Google Scholar]
- 97.Heeb MJ, Koenen RR, Fernandez JA, Hackeng TM. Direct anticoagulant activity of protein S-C4b binding protein complex in Heerlen heterozygotes and normals. J Thromb Haemost. 2004;2:1766–1773. doi: 10.1111/j.1538-7836.2004.00901.x. [DOI] [PubMed] [Google Scholar]
- 98.Miyata T, Kimura R, Kokubo Y, Sakata T. Genetic risk factors for deep vein thrombosis among Japanese: importance of protein S K196E mutation. Int.J Hematol. 2006;83:217–223. doi: 10.1532/IJH97.A20514. [DOI] [PubMed] [Google Scholar]
- 99.Kimura R, Kokubo Y, Miyashita K, Otsubo R, Nagatsuka K, Otsuki T, et al. Polymorphisms in vitamin K-dependent gamma-carboxylation-related genes influence interindividual variability in plasma protein C and protein S activities in the general population. Int.J Hematol. 2006;84:387–397. doi: 10.1532/IJH97.06082. [DOI] [PubMed] [Google Scholar]
- 100.Hayashi T, Nishioka J, Suzuki K. Molecular mechanism of the dysfunction of protein S(Tokushima) (Lys155-->Glu) for the regulation of the blood coagulation system. Biochim.Biophys.Acta. 1995;1272:159–167. doi: 10.1016/0925-4439(95)00081-x. [DOI] [PubMed] [Google Scholar]
- 101.Dahlbäck B, Carlsson M, Svensson PJ. Familial thrombophilia due to a previously unrecognized mechanism characterized by poor anticoagulant response to activated protein C: prediction of a cofactor to activated protein C. Proc.Natl.Acad.Sci.U.S.A. 1993;90:1004–1008. doi: 10.1073/pnas.90.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Greengard JS, Sun X, Xu X, Fernández JA, Griffin JH, Evatt B. Activated protein C resistance caused by Arg506Gln mutation in factor Va. Lancet. 1994;343:1361–1362. doi: 10.1016/s0140-6736(94)92497-x. [DOI] [PubMed] [Google Scholar]
- 103.Bertina RM, Koeleman BPC, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, et al. Mutations in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 104.Delvaeye M, Noris M, de Vriese A, Esmon CT, Esmon NL, Ferrell G, et al. Thrombomodulin mutations in atypical hemolytic-uremic syndrome. N.Engl.J.Med. 2009;361:345–357. doi: 10.1056/NEJMoa0810739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poort SR, Vos HL, Rosendaal FR, Bertina RM. The endothelial protein C receptor (EPCR) 23 bp insert mutation and the risk of venous thrombosis. Thromb Haemost. 2002;88:160–162. [PubMed] [Google Scholar]
- 106.Saposnik B, Lesteven E, Lokajczyk A, Esmon CT, Aiach M, Gandrille S. Alternative mRNA splicing is favored by the A3 haplotype of the EPCR gene PROCR and generates a novel soluble form of EPCR in plasma. Blood. 2008;111:3442–3451. doi: 10.1182/blood-2007-08-104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Biguzzi E, Merati G, Liaw PCY, Bucciarelli P, Oganesyan N, Qu D, et al. A 23bp insertion in the endothelial protein C receptor (EPCR) gene impairs EPCR function. Thromb.Haemost. 2001;86:945–948. [PubMed] [Google Scholar]
- 108.von Depka M, Czwalinna A, Eisert R, Wermes C, Scharrer I, Ganser A, et al. Prevalence of a 23bp insertion in exon 3 of the endothelial cell protein C receptor gene in venous thrombophilia. Thromb Haemost. 2001;86:1360–1362. [PubMed] [Google Scholar]
- 109.Medina P, Navarro S, Estelles A, Espana F. Polymorphisms in the endothelial protein C receptor gene and thrombophilia. Thromb.Haemost. 2007;98:564–569. [PubMed] [Google Scholar]
- 110.Medina P, Navarro S, Estelles A, Vaya A, Bertina RM, Espana F. Influence of the 4600A/G and 4678G/C polymorphisms in the endothelial protein C receptor (EPCR) gene on the risk of venous thromboembolism in carriers of factor V Leiden. Thromb.Haemost. 2005;94:389–394. doi: 10.1160/TH05-02-0089. [DOI] [PubMed] [Google Scholar]
- 111.Joyce DE, Gelbert L, Ciaccia A, DeHoff B, Grinnell BW. Gene expression profile of antithrombotic protein C defines new mechanisms modulating inflammation and apoptosis. J.Biol.Chem. 2001;276:11199–11203. doi: 10.1074/jbc.C100017200. [DOI] [PubMed] [Google Scholar]
- 112.Riewald M, Petrovan RJ, Donner A, Mueller BM, Ruf W. Activation of endothelial cell protease activated receptor 1 by the protein C pathway. Science. 2002;296:1880–1882. doi: 10.1126/science.1071699. [DOI] [PubMed] [Google Scholar]
- 113.Cheng T, Liu D, Griffin JH, Fernández JA, Castellino FJ, Rosen ED, et al. Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat.Med. 2003;9:338–342. doi: 10.1038/nm826. [DOI] [PubMed] [Google Scholar]
- 114.Mosnier LO, Griffin JH. Inhibition of staurosporine-induced apoptosis of endothelial cells by activated protein C requires protease activated receptor-1 and endothelial cell protein C receptor. Biochem.J. 2003;373:65–70. doi: 10.1042/BJ20030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feistritzer C, Riewald M. Endothelial barrier protection by activated protein C through PAR1- dependent sphingosine 1-phosphate receptor-1 crossactivation. Blood. 2005;105:3178–3184. doi: 10.1182/blood-2004-10-3985. [DOI] [PubMed] [Google Scholar]
- 116.Finigan JH, Dudek SM, Singleton PA, Chiang ET, Jacobson JR, Camp SM, et al. Activated protein C mediates novel lung endothelial barrier enhancement: Role of sphingosine 1-phosphate receptor transactivation. J.Biol.Chem. 2005;280:17286–17293. doi: 10.1074/jbc.M412427200. [DOI] [PubMed] [Google Scholar]
- 117.Kenakin T. New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat.Rev.Drug Discov. 2005;4:919–927. doi: 10.1038/nrd1875. [DOI] [PubMed] [Google Scholar]
- 118.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular Mechanism of β-Arrestin-Biased Agonism at Seven-Transmembrane Receptors. Annu.Rev.Pharmacol.Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr.Opin.Struct.Biol. 2011;21:541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 120.Jalbert LR, Rosen ED, Moons L, Chan JCY, Carmeliet P, Collen D, et al. Inactivation of the gene for anticoagulant protein C causes lethal perinatal consumptive coagulopathy in mice. J.Clin.Invest. 1998;102:1481–1488. doi: 10.1172/JCI3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pendurthi UR, Rao LV. Factor VIIa interaction with endothelial cells and endothelial cell protein C receptor. Thromb.Res. 2010;125:S19–S22. doi: 10.1016/j.thromres.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sen P, Gopalakrishnan R, Kothari H, Keshava S, Clark CA, Esmon CT, et al. Factor VIIa bound to endothelial cell protein C receptor activates protease activated receptor-1 and mediates cell signaling and barrier protection. Blood. 2011;117:3199–3208. doi: 10.1182/blood-2010-09-310706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Disse J, Petersen HH, Larsen KS, Persson E, Esmon N, Esmon CT, et al. The endothelial protein C receptor supports tissue factor ternary coagulation initiation complex signaling through proteaseactivated receptors. J Biol.Chem. 2011;286:5756–5767. doi: 10.1074/jbc.M110.201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schuepbach RA, Riewald M. Coagulation factor Xa cleaves PAR1 and mediates signaling dependent on binding to the endothelial protein C receptor. J.Thromb.Haemost. 2010;8:379–388. doi: 10.1111/j.1538-7836.2009.03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Domotor E, Benzakour O, Griffin JH, Yule D, Fukudome K, Zlokovic BV. Activated protein C alters cytosolic calcium flux in human brain endothelium via binding to endothelial protein C receptor and activation of Protease Activated Receptor-1. Blood. 2003;101:4797–4801. doi: 10.1182/blood-2002-12-3680. [DOI] [PubMed] [Google Scholar]
- 126.Cheng T, Petraglia AL, Li Z, Thiyagarajan M, Zhong Z, Wu Z, et al. Activated protein C inhibits tissue plasminogen activator-induced brain hemorrhage. Nat.Med. 2006;12:1278–1285. doi: 10.1038/nm1498. [DOI] [PubMed] [Google Scholar]
- 127.Guo H, Liu D, Gelbard H, Cheng T, Insalaco R, Fernández JA, et al. Activated protein C prevents neuronal apoptosis via protease activated receptors 1 and 3. Neuron. 2004;41:563–572. doi: 10.1016/s0896-6273(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 128.Kerschen EJ, Fernandez JA, Cooley BC, Yang XV, Sood R, Mosnier LO, et al. Endotoxemia and sepsis mortality reduction by non-anticoagulant activated protein C. J.Exp.Med. 2007;204:2439–2448. doi: 10.1084/jem.20070404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mosnier LO, Yang XV, Griffin JH. Activated protein C mutant with minimal anticoagulant activity, normal cytoprotective activity, and preservation of thrombin activable fibrinolysis inhibitor-dependent cytoprotective functions. J.Biol.Chem. 2007;282:33022–33033. doi: 10.1074/jbc.M705824200. [DOI] [PubMed] [Google Scholar]
- 130.Bae JS, Yang L, Rezaie AR. Receptors of the protein C activation and activated protein C signaling pathways are colocalized in lipid rafts of endothelial cells. Proc.Natl.Acad.Sci.USA. 2007;104:2867–2872. doi: 10.1073/pnas.0611493104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Russo A, Soh UJ, Trejo J. Proteases Display Biased Agonism at Protease-Activated Receptors: Location matters! Mol.Interv. 2009;9:87–96. doi: 10.1124/mi.9.2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bae JS, Yang L, Rezaie AR. Lipid raft localization regulates the cleavage specificity of protease activated receptor 1 in endothelial cells. J.Thromb.Haemost. 2008;6:954–961. doi: 10.1111/j.1538-7836.2008.02924.x. [DOI] [PubMed] [Google Scholar]
- 133.Bae JS, Rezaie AR. Protease activated receptor 1 (PAR-1) activation by thrombin is protective in human pulmonary artery endothelial cells if endothelial protein C receptor is occupied by its natural ligand. Thromb.Haemost. 2008;100:101–109. doi: 10.1160/TH08-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schuepbach RA, Feistritzer C, Brass LF, Riewald M. Activated protein C-cleaved protease activated receptor-1 is retained on the endothelial cell surface even in the presence of thrombin. Blood. 2008;111:2667–2673. doi: 10.1182/blood-2007-09-113076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Soh UJ, Trejo J. Activated protein C promotes protease-activated receptor-1 cytoprotective signaling through beta-arrestin and disheveled-2 scaffolds. Proc.Natl.Acad.Sci.U.S.A. 2011;108:E1372–E1380. doi: 10.1073/pnas.1112482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baumer Y, Spindler V, Werthmann RC, Bunemann M, Waschke J. Role of Rac 1 and cAMP in endothelial barrier stabilization and thrombin-induced breakdown. J.Cell Phys. 2009;220:716–726. doi: 10.1002/jcp.21819. [DOI] [PubMed] [Google Scholar]
- 137.Spindler V, Schlegel N, Waschke J. Role of GTPases in control of microvascular permeability. Cardiovasc.Res. 2010;87:243–253. doi: 10.1093/cvr/cvq086. [DOI] [PubMed] [Google Scholar]
- 138.Bir N, Lafargue M, Howard M, Goolaerts A, Roux J, Carles M, et al. Cytoprotective-selective activated protein C attenuates Pseudomonas aeruginosa-induced lung injury in mice. Am.J Respir.Cell Mol.Biol. 2011;45:632–641. doi: 10.1165/rcmb.2010-0397OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bae JS, Rezaie AR. Thrombin inhibits nuclear factor kappaB and RhoA pathways in cytokinestimulated vascular endothelial cells when EPCR is occupied by protein C. Thromb.Haemost. 2009;101:513–520. [PMC free article] [PubMed] [Google Scholar]
- 140.Wang L, Dudek SM. Regulation of vascular permeability by sphingosine 1-phosphate. Microvasc.Res. 2009;77:39–45. doi: 10.1016/j.mvr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.White TC, Berny MA, Tucker EI, Urbanus RT, de Groot PG, Fernandez JA, et al. Protein C supports platelet binding and activation under flow: role of glycoprotein Ib and apolipoprotein E receptor 2. J.Thromb.Haemost. 2008;6:995–1002. doi: 10.1111/j.1538-7836.2008.02979.x. [DOI] [PubMed] [Google Scholar]
- 142.Guo H, Singh I, Wang Y, Deane R, Barrett T, Fernandez JA, et al. Neuroprotective activities of activated protein C mutant with reduced anticoagulant activity. Eur.J.Neurosci. 2009;29:1119–1130. doi: 10.1111/j.1460-9568.2009.06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 143.Madhusudhan T, Wang H, Straub BK, Grone E, Zhou Q, Shahzad K, et al. Cytoprotective signaling by activated protein C requires protease activated receptor-3 in podocytes. Blood. 2012;119:874–883. doi: 10.1182/blood-2011-07-365973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Elphick GF, Sarangi PP, Hyun YM, Hollenbaugh JA, Ayala A, Biffl WL, et al. Recombinant human activated protein C inhibits integrin-mediated neutrophil migration. Blood. 2009;113:4078–4085. doi: 10.1182/blood-2008-09-180968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minhas N, Xue M, Fukudome K, Jackson CJ. Activated protein C utilizes the angiopoietin/Tie2 axis to promote endothelial barrier function. FASEB J. 2010;24:873–881. doi: 10.1096/fj.09-134445. [DOI] [PubMed] [Google Scholar]
- 146.Zhong Z, Ilieva H, Hallagan L, Bell R, Singh I, Paquette N, et al. Activated protein C therapy slows ALS-like disease in mice by transcriptionally inhibiting SOD1 in motor neurons and microglia cells. J Clin.Invest. 2009;119:3437–3449. doi: 10.1172/JCI38476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xue M, Chow SO, Dervish S, Chan YK, Julovi S, Jackson CJ. Activated protein C enhances human keratinocyte barrier integrity via sequential activation of epidermal growth factor receptor and tie2. J Biol.Chem. 2011;286:6742–6750. doi: 10.1074/jbc.M110.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang XV, Banerjee Y, Fernandez JA, Deguchi H, Xu X, Mosnier LO, et al. Activated protein C ligation of ApoER2 (LRP8) causes Dab1-dependent signaling in U937 cells. Proc.Natl.Acad.Sci.U.S.A. 2009;106:274–279. doi: 10.1073/pnas.0807594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cao C, Gao Y, Li Y, Antalis TM, Castellino FJ, Zhang L. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J Clin.Invest. 2010;120:1971–1980. doi: 10.1172/JCI40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Isermann B, Vinnikov IA, Madhusudhan T, Herzog S, Kashif M, Blautzik J, et al. Activated protein C protects against diabetic nephropathy by inhibiting endothelial and podocyte apoptosis. Nat.Med. 2007;13:1349–1358. doi: 10.1038/nm1667. [DOI] [PubMed] [Google Scholar]
- 151.Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, et al. Extracellular histones are major mediators of death in sepsis. Nat.Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wildhagen KC, Lutgens E, Loubele ST, Ten CH, Nicolaes GA. The structure-function relationship of activated protein C. Thromb Haemost. 2011;106:1034–1045. doi: 10.1160/TH11-08-0522. [DOI] [PubMed] [Google Scholar]
- 153.Weiler H. Multiple receptor-mediated functions of activated protein C. Hamostaseol. 2011;31:185–195. doi: 10.5482/ha-1166. [DOI] [PubMed] [Google Scholar]
- 154.Mosnier LO, Gale AJ, Yegneswaran S, Griffin JH. Activated protein C variants with normal cytoprotective but reduced anticoagulant activity. Blood. 2004;104:1740–1745. doi: 10.1182/blood-2004-01-0110. [DOI] [PubMed] [Google Scholar]
- 155.Mather T, Oganessyan V, Hof P, Huber R, Foundling S, Esmon CT, et al. The 2.8 Å crystal structure of Gla-domainless activated protein C. EMBO J. 1996;15:6822–6831. [PMC free article] [PubMed] [Google Scholar]
- 156.Shen L, Villoutreix BO, Dahlbäck B. Involvement of lys 62(217) and lys 63(218) of human anticoagulant protein C in heparin stimulation of inhibition by the protein C inhibitor. Thromb.Haemost. 1999;82:72–79. [PubMed] [Google Scholar]
- 157.Bae JS, Yang L, Manithody C, Rezaie AR. Engineering a disulfide bond to stabilize the calcium binding loop of activated protein C eliminates its anticoagulant but not protective signaling properties. J.Biol.Chem. 2007;282:9251–9259. doi: 10.1074/jbc.M610547200. [DOI] [PubMed] [Google Scholar]
- 158.Yang L, Bae JS, Manithody C, Rezaie AR. Identification of a specific exosite on activated protein C for interaction with protease activated receptor 1. J.Biol.Chem. 2007;282:25493–25500. doi: 10.1074/jbc.M702131200. [DOI] [PubMed] [Google Scholar]
- 159.Preston RJ, Villegas-Mendez A, Sun YH, Hermida J, Simioni P, Philippou H, et al. Selective modulation of protein C affinity for EPCR and phospholipids by Gla domain mutation. FEBS J. 2005;272:97–108. doi: 10.1111/j.1432-1033.2004.04401.x. [DOI] [PubMed] [Google Scholar]
- 160.Ni Ainle F, O'Donnell JS, Johnson JA, Brown L, Gleeson EM, Smith OP, et al. Activated protein C Nlinked glycans modulate cytoprotective signaling function on endothelial cells. J Biol.Chem. 2011;286:1323–1330. doi: 10.1074/jbc.M110.159475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guo H, Wang Y, Singh I, Liu D, Fernandez JA, Chow N, et al. Species-dependent neuroprotection by activated protein C mutants with reduced anticoagulant activity. J.Neurochem. 2009;109:116–124. doi: 10.1111/j.1471-4159.2009.05921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fernández JA, Xu X, Liu D, Zlokovic BV, Griffin JH. Recombinant murine-activated protein C is neuroprotective in a murine ischemic stroke model. Blood Cells Mol.Dis. 2003;30:271–276. doi: 10.1016/s1079-9796(03)00034-2. [DOI] [PubMed] [Google Scholar]
- 163.Shibata M, Kumar SR, Amar A, Fernández JA, Hofman F, Griffin JH, et al. Anti-inflammatory, antithrombotic, and neuroprotective effects of activated protein C in a murine model of focal ischemic stroke. Circulation. 2001;103:1799–1805. doi: 10.1161/01.cir.103.13.1799. [DOI] [PubMed] [Google Scholar]
- 164.Zlokovic BV, Zhang C, Liu D, Fernández JA, Griffin JH, Chopp M. Functional recovery after embolic stroke in rodents by activated protein C. Ann.Neurol. 2005;58:474–477. doi: 10.1002/ana.20602. [DOI] [PubMed] [Google Scholar]
- 165.Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, et al. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 2008;451:1076–1081. doi: 10.1038/nature06559. [DOI] [PubMed] [Google Scholar]
- 166.Liu D, Cheng T, Guo H, Fernández JA, Griffin JH, Song X, et al. Tissue plasminogen activator neurovascular toxicity is controlled by activated protein C. Nat.Med. 2004;10:1379–1383. doi: 10.1038/nm1122. [DOI] [PubMed] [Google Scholar]
- 167.Villoutreix BO, Teleman O, Dahlback B. A theoretical model for the Gla-TSR-EGF-1 region of the anticoagulant cofactor protein S: from biostructural pathology to species-specific cofactor activity. J.Comput.Aided Mol.Des. 1997;11:293–304. doi: 10.1023/a:1007912929828. [DOI] [PubMed] [Google Scholar]
- 168.Drakenberg T, Ghasriani H, Thulin E, Thamlitz AM, Muranyi A, Annila A, et al. Solution structure of the Ca2+-Binding EGF3-4 pair from vitamin K-dependent protein S: identification of an unusual fold in EGF3. Biochemistry. 2005;44:8782–8789. doi: 10.1021/bi050101f. [DOI] [PubMed] [Google Scholar]
- 169.Villoutreix BO, Dahlback B, Borgel D, Gandrille S, Muller YA. Three-dimensional model of SHBGlike region of anticoagulant protein S: new structure-function insights. Proteins. 2001;43:203–216. [PubMed] [Google Scholar]
- 170.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 171.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol.Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 172.Pellequer JL, Gale AJ, Getzoff ED, Griffin JH. Three-dimensional model of coagulation factor Va bound to activated protein C. Thromb.Haemost. 2000;84:849–857. [PubMed] [Google Scholar]