Abstract
Computer-aided drug design (CADD) methodologies have made great advances and contributed significantly to the discovery and/or optimization of many clinically used drugs in recent years. CADD tools have likewise been applied to the discovery of inhibitors of HIV-I integrase, a difficult and worthwhile target for the development of efficient anti-HIV drugs. This article reviews the application of CADD tools, including pharmacophore search, quantitative structure–activity relationships, model building of integrase complexed with viral DNA and quantum-chemical studies in the discovery of HIV-I integrase inhibitors. Different structurally diverse integrase inhibitors have been identified by, or with significant help from, various CADD tools.
HIV-I integrase & anti-AIDS drug development
Integration of viral DNA into the host DNA is a crucial step in the replication of HIV-1. This reaction is catalyzed by integrase (IN), one of the three essential enzymes encoded by the pol gene of HIV-1 [1]. This enzyme has three domains: the N-terminal domain, which contains HHCC motif that coordinates zinc binding; the catalytic [CAT] core domain, which contains the catalytic triad Asp-64, Asp-116 and Glu-152 (known as the DDE motif), and the C-terminal domain, which is involved in host DNA binding through a mechanism that is not completely clear at present. For full catalytic activity in vivo, all three domains are required.
Integrase inserts a double-stranded DNA copy of the viral genome into the chromosomes of an infected cell through a multistep process that involves two distinct reactions known as 3′-end processing (3′-P) and strand transfer (ST), respectively [2,3]. Following reverse transcription of the HIV-1 genome, IN assembled on the newly synthesized transcript removes two bases from both 3′-ends of the double-stranded viral DNA (the 3′-P reaction). The complex of IN and the preprocessed viral DNA (plus possibly additional viral and/or host proteins) are known as the ‘pre-integration complex’ (PIC). After transport of the PIC into the nucleus, IN catalyzes the joining of the preprocessed 3′-ends to opposite strands of the host DNA. The integration process is finally completed by cleavage of the unpaired dinucleotides from the 5′ ends of the viral DNA and repair of the gaps between the viral and host DNA, presumably by host enzymes. For the integration reaction, divalent cations are required for catalytic activity of IN [3]. It is commonly assumed that Mg2+ is the physiologically relevant cofactor in vivo; however, Mn2+ has often been favored in vitro, arguably because it gives better inhibition results for many compounds [4].
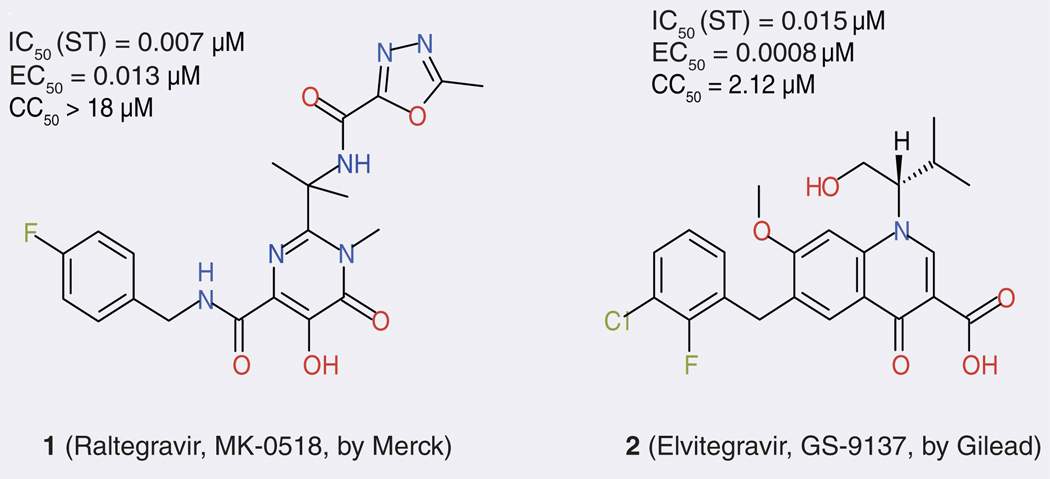
Currently, the treatment of HIV-1 infection often relies on a combination of several therapeutic agents, typically three to four, that target the viral enzymes reverse transcriptase (RT) and protease (PR), as well as viral entry. This is known as highly active antiretroviral therapy (HAART) [1]. Several factors (including the emergence of multidrug-resistant HIV strains, drug toxicity and patients’ ability to comply with the prescribed therapy) make it highly desirable to develop novel drugs that target other viral replication processes. IN is such a target, particularly because there is no known human counterpart of HIV-1 IN [5]. A wide variety of compounds have been reported as IN inhibitors during 20 years of IN inhibition-based anti-HIV drug development efforts [6–8]. However, only one such drug has made it to the market so far. In October 2007, the first drug based on IN inhibition, raltegravir (MK-0518, brand name Isentress; FIGURE 1) produced by Merck & Co, was approved by the US FDA [9–11]. However, to date, it is only used in individuals whose infection has proved resistant to other HAART drugs. As with any HAART medication, raltegravir is unlikely to show durability if used as monotherapy. Altogether, this signifies that HIV-1 IN remains a worth-while therapeutic target for continued drug development efforts, including application of computer tools in the form of computer-aided drug design (CADD) work.
Figure 1. Structures and antiviral data of raltegravir and elvitegravir.
ST: Strand transfer.
As a sign of this undiminished attractiveness of IN as a drug target, several other pharmaceutical companies continue to develop anti-HIV therapeutics based on IN inhibition. The compound that is currently the most advanced in the testing and approval process is elvitegravir or GS-9137 (FIGURE 1), discovered at Japan Tobacco and developed by Gilead Sciences. It is currently in Phase III clinical trial [12,13]. Several other compounds, including S-1360 (co-developed by Shionogi and GlaxoSmithKline), L-870,810 (developed by Merck), GS-9160 (developed by Gilead), GS-9224 (developed by Gilead), GSK-364735 (co-developed by Shionogi and GlaxoSmithKline) and BMS-707035 (developed by Bristol-Myers Squibb), have been in clinical trials, but have been halted for various reasons [9]. All these compounds selectively inhibit ST more potently than the 3′-P reaction and are therefore often called IN ST inhibitors (INSTIs). They are typically characterized by a region that is planar and well-suited to chelate two metal ions (presumably two Mg2+ as discussed earlier) and are assumed to be held in place by the catalytic triad Asp-64, Asp-116 and Glu-152 [14,15]. They represent the current major leads in the development of anti HIV-1 IN drugs and in this sense can be called ‘authentic HIV-1 IN inhibitors’ in contrast to the thousands of compounds that were found to possess activity in enzymatic assays, but little or no activity in cell-based assays, let alone in vivo. We present a more detailed account of these authentic IN inhibitors in an accompanying publication [16].
Overview on CADD & its application for developing IN inhibitors
Computer-aided drug design represents a significant tool to accelerate the discovery of new drugs and reduce costs by, among other tasks, helping identify leads to a specific target receptor with techniques including molecular mechanics and molecular dynamics (MD) simulations, quantum mechanical computations, homology modeling, docking, pharmacophore search and quantitative structure–activity relationship (QSAR) analyses [17]. Depending on the type and amount of structural information available and used, and the strategy employed, CADD approaches can be generally divided into structure-based drug design (SBDD), which is used, for example, when something about the macromolecular target structure is known; ligand-based drug design (LBDD), which is used, for example, when some inhibitors (or, if appropriate, activators) are known; or a combination of both [17–19].
Numerous 3D structures of human proteins and proteins of disease-causing organisms have been elucidated, which makes SBDD possible because such structures often include complexed ligands and thereby provide information regarding binding modes. Docking, an automated computer algorithm that determines how a compound may bind in the active site or an allosteric site of a protein, is probably the most commonly used tool in SBDD [20]. Compared with high-throughput screening, which is a very powerful tool for identifying hits, docking methods (virtual screening) can help limit the number of compounds, out of often many millions of screening samples, to a subset of molecules that is more likely to yield hits much faster and with less expense. Therefore, docking-based virtual screening techniques have gained popularity and delivered drugs to the market in recent years for all kinds of diseases, including cancer, AIDS, glaucoma and hypertension [21]. De novo design, another tool in SBDD, attempts to automate the process of SBDD to generate novel active molecules ‘from scratch’ [22]. Such programs attempt to design compounds to fit in a particular active site, or conform to a particular pharmacophore model, which is a worthwhile goal but an extremely complex task. Thus, they are used less commonly than docking in ‘real-life’ drug development projects.
Ligand-based drug design is dependent on knowledge about compounds with known biological effects but without structural information for the functional target. The methods used for LBDD include pharmacophore identification and QSAR. A pharmacophore is fundamentally a static representation of the spatial relationships between distinct chemical features of the compound(s) pivotal to biological activity. QSAR methods can be divided into four different classes based on the type of molecular descriptors used to correlate with activity: 1D-, 2D-, 3D- and 4D-QSAR [23], although mixtures, especially of 2D and 3D descriptors, are not unknown. 2D-QSAR is an equation for predicting some property from molecular descriptors and coefficients of those variables based on just the connectivity of the molecules [24]. 3D-QSAR refers to the application of force-field calculations, requiring 3D structures of studied molecules, often determined by molecule super-position [23]. 4D-QSAR analysis incorporates conformational and alignment freedom into the development of 3D-QSAR models for training sets of structure–activity data by performing molecular state ensemble averaging (the fourth dimension) [23]. The constructed pharmacophores and QSAR models can also be used to optimize already known leads or virtually screen chemical libraries to identify new leads [21].
For both RT and PR, plenty of structures of the full-length apoenzymes and complexes with substrates, products and inhibitors have been solved and successfully applied to drug discovery. However, the third enzyme encoded by the HIV-1 genome, IN, lacks well-resolved experimental structures of the full-length protein complexed with its substrate (viral DNA). This has severely hampered SBDD efforts for anti-HIV drugs based on this target. To alleviate this deficiency, various IN–DNA models have been computationally constructed and drug-discovery efforts have been based with some degree of success on some of them. Moreover, QSAR and pharmacophore studies, sometimes combined with docking methods and other computer techniques, have been employed to try to identify novel IN inhibitors. Theoretical studies have also been performed on how INSTIs chelate the metal cofactor to guide the design of novel INSTIs.
Pharmacophore search
Pharmacophore developed from known IN inhibitors
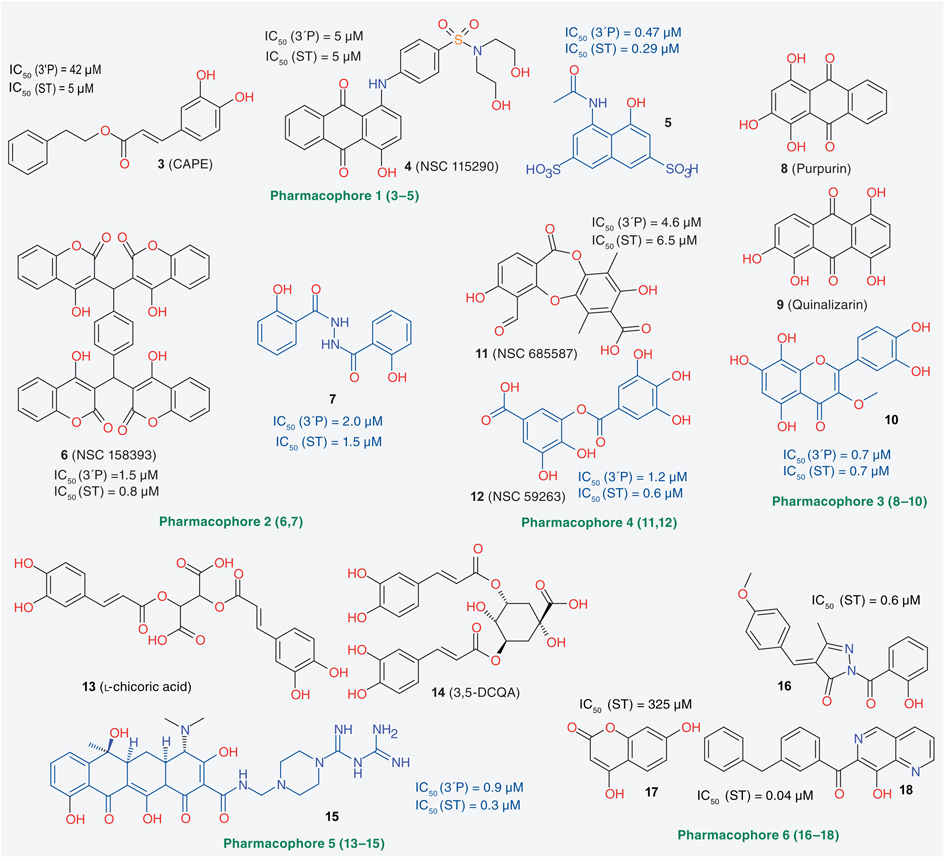
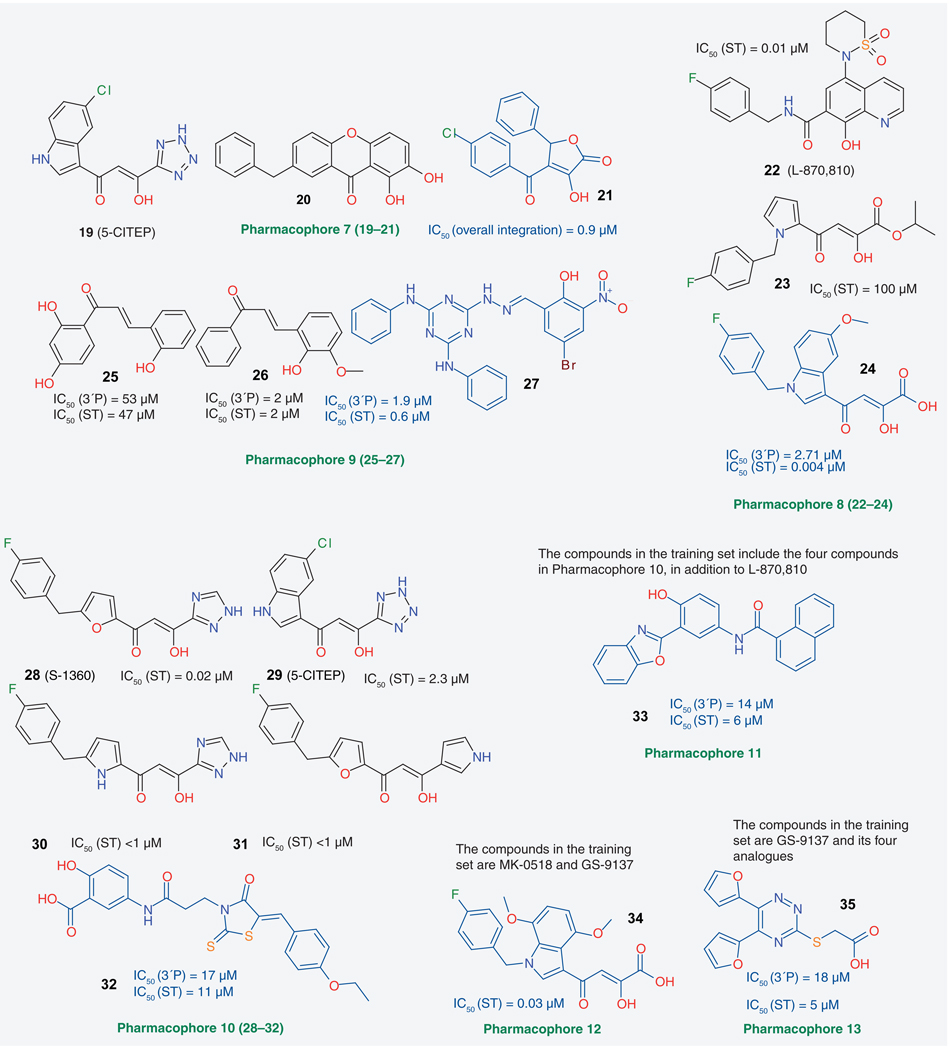
Pharmacophore search in 3D databases is a way of identifying compounds whose 3D structures, in some conformations, contain the pharmacophore used in the search query. The mentioned lack of an experimental structure of full-length IN complexed with a ligand and viral DNA and the discovery of a number of genuine IN inhibitors has led to a certain emphasis in the IN field on LBDD, in particular pharmacophore search. TABLE 1 provides an overview of published pharmacophore search studies [25–37]. A selection of compounds of these studies’ training sets (drawn in black) and of the most active hits (drawn in blue) are shown in FIGURE 2.
Table 1.
Reported pharmacophore studies to identify integrase inhibitors.
| ID | Type | Program | Method | Number of training set compounds |
Number of tested compounds |
Number of active compounds |
Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Three-point | Chem-X | 2 | 60 | 19 | [25] | |
| 2 | Three-point | Chem-X | 1 | 29 | 10 | [26] | |
| 3 | Three-point | Chem-X | 16 | 42 | 27 | [27] | |
| 4 | Four-point | Chem-X | 2 | 39 | 20 | [28] | |
| 5 | Four-point | Chem-X | 2 | 60 | 19 | [29] | |
| 6 | Four-feature | Catalyst | HypoGen | 26 | [30] | ||
| 7 | Four-feature | Catalyst | 2 | 10 | 8 | [31] | |
| 8† | Four-feature | Catalyst | HypoGen | 17 | [32] | ||
| 9 | Four-feature | Catalyst | HipHop | 4 | 52 | 11 | [33] |
| 10 | Four-feature | Catalyst | HipHop | 5 | 27 | 13 | [34] |
| 11 | Six-feature | Catalyst | 2 | 71 | 44 | [35] | |
| 12† | Seven-feature | Catalyst | HipHop | 2 | [36] | ||
| 13 | Four-feature | Catalyst | HipHop | 5 | 11 | 11 | [37] |
This model was not used directly by the authors in virtual screening, but prompted them to synthesize a series of new 1H-indole derivatives.
Figure 2. Training set compounds (in black) used to produce pharmacophore models and identified best integrase inhibitors (in blue) from each reported pharmacophore study.
Not all training set compounds for every study are shown here. 3′P: 3′-end processing; ST: Strand transfer.
Most of the early pharmacophore studies (1–5) used one or two compounds as training sets, and all the models produced were three-or four-point pharmacophores, consisting of admitted atom types (oxygen or nitrogen), dimensions between these atom types and element equivalences. The 3D NCI database and the Chem-X program were employed in both early 3D database building and search processes [38]. Most of the pharmacophore models published later (6–13) were based on compounds that had proven effective in human clinical trials. These compounds include S-1360, L-870,810, MK-0518 and GS-9137. Pharmacophore perception was typically performed employing the methods HypoGen or HipHop, embedded in the program Catalyst. Often, up to ten hypotheses were produced in a study, but only the top scoring hypothesis was used to search chemical databases. Some structurally novel IN inhibitors, most with micromolar level ST activities, were identified. Other IN inhibitors with higher activities (e.g., 1H-indole derivatives) were synthesized as a result of work with these pharmacophore models. Some compounds were in fact selected for biological testing based not only on the constructed pharmacophore model, but on their docking scores, typically obtained from docking into the IN active site based on PDB structures 1QS4 and 1BIS as starting structures [33–35,37].
The pharmacophore models 6, 8 and 12 are partly consistent with the putative two-metal chelation mechanism [14,15]. Kawasuji et al. devised a more elaborate 3D pharmacophore model when designing 2-hydroxy-3-heteroaryl acrylic acid derivatives as IN inhibitors [39,40]. This manually constructed model contains a hydrophobic feature, whose position was obtained by superimposing several INSTIs’ terminal phenyl rings, and a hydrophilic feature, which contains both an enolized hydroxyl group and two possible heteroatoms that provide a lone pair. In a paper on discordant resistance between mechanistically identical inhibitors of HIV-1 IN, Hazuda et al. modeled pharmacophores based on a diketo acid and L-870,810, which suggested two potential binding modes for some HIV-1 IN inhibitors [41].
Dynamic pharmacophore models
The so-called dynamic pharmacophore models are generated using snapshots from MD trajectories in order to incorporate the dynamic nature of the active site region into the pharmacophore model. This somewhat more recent method not only accounts for the inherent flexibility of the active site and aims to reduce the entropic penalties associated with binding of a ligand, but also overcomes the limitation of an incomplete crystal structure of the target protein. The multiunit search for interacting conformers (MUSIC) procedure simultaneously performs multiple, gas-phase minimizations for hundreds of probe molecules within the active site using the BOSS program [42]. The results for each snapshot of the protein are then over-laid to reveal conserved binding regions that are highly occupied over the course of the MD simulation, despite the inherent motion of the active site [43].
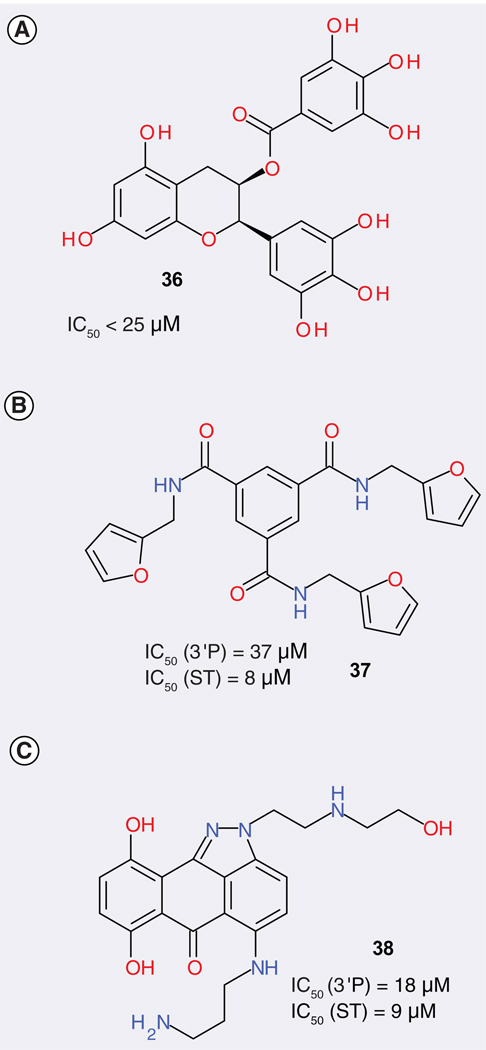
Three different groups have developed three dynamic pharmacophore models for IN and used them to screen chemical databases. Some micromolar level IN inhibitors were identified by this method [43–45]. For the first reported IN receptor-based pharmacophore model, the authors pursued compounds that bind to the essential residues Asp-64 and Asp-116 without associating with the metal dication, based on the assumption that inhibitors that bind to the essential residues may reduce the rapid rate of escape mutants known to be a serious complication in treating AIDS [43]. A crystal structure without Mg2+, but with the missing regions added by modeling, was used to perform MD simulations [46]. The protein conformations were collected at 50 ps intervals from the 500 ps MD trajectory. The overlay process displayed six conserved binding regions, which were used as hydrogen bond donors in the pharmacophore model. The average positions of Cγ of Asp-64, Cγ of Asp-116 and Cδ of Gln-62 were used as the centers for three excluded volumes. Six queries were systematically generated by removing one of the hydrogen bond donors and used to search a database. A set of 39 compounds were chosen for testing, among which four showed activities at 200 µM and two showed activities at 25 µM (one of which is illustrated in FIGURE 3A).
Figure 3. The best compounds identified by three different dynamic pharmacophore models.
3′P: 3′-end processing; ST: Strand transfer.
The second dynamic pharmacophore model was based on the crystal structure of 1QS4, which contains an inhibitor, 5-CITEP and one Mg2+ ion, for a 1-ns MD simulation [44]. The dynamic pharmacophore model, derived from the overlay of 11 static pharmacophore models, each of which was based on a representative snapshot, contains two hydrogen bond acceptors, five hydrogen bond donors, one hydrophobic area and six excluded volumes (defined by the key residues, Asp-64, Asp-116, Glu-152, Lys-156, Lys-159 and the Mg2+ ion, which was treated as a charged protein atom). This model was used to screen a library from which ultimately 23 compounds were chosen for testing. Among those, nine compounds effectively blocked the ST reaction at IC50s of less than 100 µM (the most effective is illustrated in FIGURE 3B). In subsequent work, the same authors extended the dynamic pharmacophore studies by considering more key residues in the active site and Mg2+ as an excluded volume or a charged feature [45]. Different types of functional fragments as probes were applied to map the complementary features of the IN binding. All ten of the snapshots from a MD simulation, together with their associated probe clusters were overlaid via five residues (Asp-64, Asp-116, Glu-152, Lys-156, Lys-159) to identify the consensus probe binding sites, which were used to locate the elements of the dynamic pharmacophore model. Two models were developed, both of which contained two hydrogen bond donors, two hydrogen bond acceptors, two features that could be both hydrogen bond donors and acceptors, two positively ionizable features and five excluded volumes. The difference between the models is that one treated Mg2+ as an excluded volume, whereas the other one treated it as a positively ionizable feature. A total of 22 compounds obtained from the database search were assayed. Two of them showed IC50 values of up to 10 µM for ST (one of which is illustrated in FIGURE 3C).
QSAR studies
More than 20 QSAR studies, including 2D-, 3D- and even 4D-QSAR analyses, have been reported over the past 15 years on a whole variety of IN inhibitors to elucidate the quantitative correlations between the chemical structures of IN inhibitors and their biological activities (TABLE 2). The fact that different structural classes of IN inhibitors emerged makes IN an ideal target for QSAR studies. The 2D-QSAR methods employed include electrotopological state (E-state) indices [47]; genetic function approximation (GFA) [48]; atom linear indices [49]; geometry, topology and atom-weights assembly (GETAWAY) [50]; probabilistic neural network (PNN) [51] or other methods based on numerical description of the molecular structure independent of the small molecule’s 3D conformation. The 3D-QSAR methods employed include comparative molecular field analysis (CoMFA) [52]; comparative molecular similarity indices analysis (CoMSIA) [53]; eigen value analysis (EVA) [54]; comparative molecular surface analysis (CoMSA) [55]; generating optimal linear partial least-squares (PLS) estimation (GRID/GOLPE) [56,57]; molecular field analysis (MFA; implemented in the Cerius2 program); molecular shape analysis (MSA; another method implemented in the Cerius2 program) and comparative residue interaction analysis (CoRIA) [58]. Among these methods, CoMFA, CoMSIA and GRID/GOLPE allow graphical representation of the 3D-QSAR models by means of PLS coefficients. Especially for CoMSIA, the contour plots provide designers with insights into how steric, electrostatic, hydrophobic and hydrogen-bonding interactions influence ligand activity. The 4D-QSAR study reported on IN inhibitors used 4D fingerprints [59] and classical 2D descriptors. The predictive ability of a QSAR model is customarily measured by a cross-validated r2 value and a predictive r2pred. Using these values as ranking criteria, we show the best QSAR models (TABLE 2) from QSAR studies of IN inhibitors (since almost every QSAR study comprises several QSAR models) [60–83]. Among these 24 studies, CoMFA and CoMSIA were used most often and, most of the time, CoMSIA demonstrated better predictive power and greater robustness than CoMFA.
Table 2.
Reported quantitative structure–activity relationship studies of integrase inhibitors.
| ID | Type | Method | Data set | Cross-validated r2 | r2pred | Drug design application? |
Ref. |
|---|---|---|---|---|---|---|---|
| 1 | 3D | CoMFA | Flavone analogs | 0.81 (3′-P) 0.77 (ST) |
NR | [60] | |
| 2 | 2D | Electrotopological state indices |
Flavone analogs | 0.51 (3′-P) 0.73 (ST) |
NR | [61] | |
| 3 | 3D | Eigen value analysis |
Salicylhydrazines, lichen acids, coumarins, quinones and thiazolothiazepines |
0.806(3′-P) 0.677 (ST) |
0.761 (3′-P) 0.591 (ST) |
NR | [62] |
| 4† | 3D | CoMFA and CoMSIA |
Cinnamoyl derivatives | 0.721 (3′-P)‡ 0.804 (3′-P)§ |
0.156 (3′-P)‡ 0.413 (3′-P)§ |
NR | [63] |
| 5 | 2D | Genetic function approximation |
Catechols and noncatechols | 0.728 (3′-P)¶ 0.796 (ST)¶ 0.616 (3′-P)# 0.544 (ST)# |
0.688 (3′-P)¶ 0.736 (ST)¶ 0.628 (3′-P)# 0.787 (ST)# |
NR | [64] |
| 6 | 3D | CoMSIA | Salicylpyrazolinones, dioxepinones, coumarins, quinones and benzoic hydrazides |
0.821 (3′-P) 0.759 (ST) |
0.608 (3′-P) 0.660 (ST) |
NR | [65] |
| 7 | 3D | CoMFA and CoMSIA |
Thiazolothiazepines | 0.508 (3′-P)‡ 0.675 (ST)‡ 0.603 (3′-P)§ 0.744 (ST)§ |
0.901 (3′-P)‡ 0.971 (ST)‡ 0.804 (3′-P)§ 0.970 (ST)§ |
NR | [66] |
| 8† | 2D | Genetic function approximation |
11 structurally different classes | 0.710 (3′-P) †† 0.742 (3′-P)‡‡ |
0.645 (3′-P) †† 0.822 (3′-P)‡‡ |
NR | [67] |
| 9† | 3D | GRID/GOLPE | Cinnamoyl derivatives and 2,6-bisbenzylidenecylohexane-1- one derivatives |
0.97 (3′-P) | 0.90 (3′-P) | NR | [68] |
| 10 | 3D | CoMFA | Salicylpyrazolinones, dioxepinones, coumarins, quinones and benzoic hydrazides |
0.830(3′-P) 0.746 (ST) |
0.608 (3′-P) 0.660 (ST) |
Yes | [69] |
| 11 | 3D | CoMFA and CoMSIA |
2-mercaptobenzenesulfonamides | 0.630 (3′-P)‡ 0.679 (ST)‡ 0.658 (3′-P)§ 0.719 (ST)§ |
0.625 (3′-P)‡ 0.671 (ST)‡ 0.639 (3′-P)§ 0.688 (ST)§ |
Yes | [70] |
| 12 | 3D | CoMSIA | β-ketoamide derivatives | 0.82 (ST) | 0.74 (ST) | NR | [71] |
| 13† | 2D | Atom linear indices |
Flavone derivatives | 0.721 (3′-P) | NR | [72] | |
| 14 | 3D | GRID/GOLPE | Diketohexenoic and diketobutanoic acids |
0.78 (EC50) | NR | [73] | |
| 15† | 3D | Molecular field analysis |
11 structurally different classes | 0.85 (3′-P)†† 0.91 (3′-P)‡‡ |
NR | [74] | |
| 16 | 3D | CoMFA and CoMSIA |
11 structurally different classes | 0.698 (3′-P)‡ 0.724 (3′-P)§ |
0.704 (3′-P)‡ 0.524 (3′-P)§ |
NR | [75] |
| 17 | 3D | CoMSA | Styrylquinolines | Yes | [76] | ||
| 18 | 2D | Probabilistic neural network |
Database of 2720 compounds | NR | [77] | ||
| 19 | 2D | GETAWAY | 11 structurally different classes | 0.669 (3′-P) | NR | [78] | |
| 20† | 3D | Molecular shape analysis |
Styrylquinoline derivatives | 0.516 (3′-P) | 0.611 (3′-P) | NR | [79] |
| 21† | 4D+2D | 4D fngerprints | 12 structurally different classes | 0.91 (3′-P) | 0.80 (3′-P) | NR | [80] |
| 22† | 3D | CoRIA | 13 structurally different classes | 0.37 (3′-P) | 0.63 (3′-P) | NR | [81] |
| 23 | 2D | TSAR | Phthalimide derivatives | 0.709 (ST) | 0.512 (ST) | NR | [82] |
| 24 | 2D | OPS | 4,5-dihydroxypyrimidine carboxamides |
0.58 (ST) | 0.87 (ST) | NR | [83] |
This QSAR model was built on biological data for only 3′-P.
For CoMFA.
For CoMSIA.
For catechol compounds.
For noncatechol compounds.
Two QSAR models were built, and this is for the first model.
For the second model.
3′P: 3′-end processing; CoMFA: Comparative molecular field analysis; CoMSIA: Comparative molecular similarity indices analysis; CoRIA: Comparative residue interaction analysis; GRID/GOLPE: Generating optimal linear partial least-squares estimation; NR: Not reported; OPS: Ordered predictors selection; QSAR: Quantitative structure–activity relationship; ST: Strand transfer; TSAR: Tools for structure–activity relationship.
Some QSAR studies used several different structural classes of IN inhibitors as datasets to try to explore different inhibitory mechanisms of structurally diverse IN inhibitors. In each of the QSAR studies 8 and 15, two QSAR models were derived using five and six structural classes of IN inhibitors, respectively (TABLE 2) . The authors first tried to use all these 11 structural classes of IN inhibitors, but did not obtain meaningful results. Descriptor-based cluster analysis was then employed, indicating that these 11 structural classes of IN inhibitors belonged to two clusters, which suggested that the known HIV-1 IN inhibitors may interact with IN at more than one binding site. QSAR study 21 used 12 structurally diverse classes of IN inhibitors as a dataset. These inhibitors were partitioned into five clusters, from which corresponding QSAR models (mostly composed of 4D fingerprint descriptors) were constructed. QSAR study 22 was performed on 13 different series of IN inhibitors. The major advantage of the CoRIA technique this study employed is that it can quantitatively extract crucial residues and identify the nature of the interactions between the ligand and receptor that modulate activity. This study suggested that Asp-64, Thr-66, Val-77, Asp-116, Glu-152 and Lys-159 are the key residues influencing the binding of ligands.
Although almost every QSAR study published for IN inhibitors stated something similar to the following: “this study was performed for the purpose of designing new chemotypes with enhanced potencies against HIV-1 IN”, few of the reported QSAR studies have actually been directly applied to drug design; for example, few led to the synthesis and assaying of new compounds based on the models. Of the 24 QSAR studies presented here, only studies 10, 11 and 17 report efforts towards this direction. The authors of QSAR study 10 utilized their CoMFA model to design four different structural classes of compounds (isoquinoline sulfonamides, furoyl pyrazolones, indole-2,3-diones and 2-phenyl-methanesulfonyl-benzothiazoles) as IN inhibitors using LeapFrog, a de novo drug design program. Some compounds thus designed showed weak activities toward both 3′-P and ST [69]. QSAR study 11, combined with docking results, was used to guide the rational design of new inhibitors: 12 new analogs of 2-mercaptobenzenesulfonamides were synthesized and used as a test set for validation of the models. The compound that showed the best activity is given in FIGURE 4. QSAR study 17 used the constructed 3D-QSAR model for the analysis of molecular diversity in a virtual combinatorial library of styrylquinolines with the aim of designing new synthetic drugs. This effort finally led to the design of 11 compounds; however, these did not show improved IC50 (3′-P) [76].
Figure 4. An IN inhibitor identified by QSAR study 11.
3′P: 3′-end processing; ST: Strand transfer.
De novo design of IN inhibitors
The power of de novo design is that it can, in principle, produce completely novel compounds. By the same token, however, it implies more uncertainty and is probably therefore used less often than other drug design methods. Presumably because of this, and because the structure of the IN active site is still uncertain, only two papers have been published to date that report de novo design of IN inhibitors. The first paper, as just discussed in the section on ‘QSAR studies’, used the LeapFrog program to design IN inhibitors [69]. A pharmacophore model derived from the CoMFA model was used as input to LeapFrog. Inhibitors were designed using the molecular evolution process and their binding energies were calculated. Compounds showing improved binding energies were selected for synthesis.
The authors of the second de novo study constructed a very large virtual diversity space containing more than 1013 chemical compounds, which were built from about 400 combinatorial libraries [84]. This diversity space was used as a vast source of compounds by a de novo drug design program to generate IN inhibitors. Briefly, a control library with a styrylquinoline scaffold was first included in the diversity space, based on the β-diketone motif from known IN inhibitors, in order to determine whether the de novo design program would generate and present candidate ligands from the library. Three styrylquinoline structures from the best-scoring ligand candidates were selected for synthesis and exhibited activity in the micromolar range. However, the structures of these compounds were not disclosed by the authors.
HIV-I IN–DNA models
Over the past 15 years, quite a number of experimental structures of IN have been solved. However, these structures only contain one or two of the three domains and do not provide important answers necessary for structure-based drug design. The antiviral activity of at least large classes of IN inhibitors is due exclusively to inhibition of ST, implying that IN at this stage is bound to viral DNA in a complex (PIC) that can and should be inhibited to prevent integration into host DNA [16,85]. This implies that not a single one of the experimental structures comes even close to representing the true macromolecular complex needed to achieve maximum antiviral efficiency. The more experimental structures became available, the less clear it seems to become how full-length IN in its physiologically relevant multimeric state is complexed with the 3′-processed viral DNA and how the host DNA and the metal ions are assembled to form the integration-competent PIC.
The availability of these experimental fragmental structures prompted some modelers to try to computationally assemble the full-length enzyme by various modeling techniques. There were two desired outcomes: first, to elucidate the true structure of IN and, second, to provide a 3D structure as a template for the design of IN inhibitors by SBDD methods. Despite all the partial structures available, it became apparent that it is no trivial task to assemble them into the full-length enzyme in a straightforward way, especially when one wants to add the viral DNA, host DNA and/or the divalent metal ions. Therefore, a variety of more or less sophisticated methods (e.g., MD) was used in the construction of such IN–DNA models.
Reported IN–DNA models
So far, to our knowledge, 13 IN–DNA models have been reported in the literature (TABLE 3) [86–109]. They can be divided into two main categories: first, models to better understand the enzyme–DNA binding in a general manner, typically constructed as a multimer, most often a tetramer (1–10); and, second, ternary IN–Mg– DNA complexes that focus on the active site to try to elucidate the binding modes of INSTIs constructed by docking methods, based solely on the residues of the CAT domain plus DNA and Mg2+ (11–13). Only a few applications of models of the first category for docking experiments to elucidate possible binding modes of IN inhibitors have been reported.
Table 3.
Integrase–DNA models reported in the literature.
| ID | Multimer modeled |
Viral DNA | Host DNA | Metal ions | Experimental structures used | Software tools used |
Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Octamer | Yes (16 bp) | Yes (26 bp) | No | 1ITG [87], 1IHV [88] and 1ASV [89] | Insight II, GRASP |
[86] |
| 2 | Dimer | Yes (18 bp) | No | No | 1EX4 [90] | MIDAS (docking) |
[90] |
| 3 | Tetramer | Yes (20 bp) | Yes (26 bp) | No Mg2+, contains Zn2+ | Unclear | Unclear | [91] |
| 4 | Tetramer | Yes (14 bp) | No | No Mg2+, contains Zn2+ | 1K6Y [92], 1EX4 [90] | Unclear | [92] |
| 5 | Tetramer | Yes (13 bp) | Yes (17 bp) | No | 1WJA [94], 1BIS [95] and 1IHV [88] | Insight II | [93] |
| 6 | Dimer | Yes (27 bp) | No | Two Mg2+ | 1QS4 [98] and 1K6Y [92] | CHARMM, ESCHER (docking), Insight II, NAMD |
[96,97] |
| 7† | Tetramer | Yes (20 bp) | Yes (25 bp) | Two Mg2+, contains Zn2+ | 1EX4 [90], 1K6Y [92] and 1VSH [100] | Sybyl/ Biopolymer, CHARMM, Insight II, GRASP |
[99] |
| 8 | Tetramer | Yes (22 bp) | Yes (26 bp) | One Mg2+ contains Zn2+ |
1WJA [94], 1BL3 [102], 1IHV [88] and 1MUH [103] |
Insight II, GRID, Dali |
[101] |
| 9 | Tetramer | Yes (27 bp) | No | Two Mg2+ contains Zn2+ |
1QS4 [98], 1BIS [95], 1VSH [100], 1EX4 [90], 1K6Y [92] and 1WJD [94] |
GROMACS, DOT, APBS, Sybyl/ Biopolymer |
[104] |
| 10 | Tetramer | Yes (20 bp) | No | One Mg2+ contains Zn2+ |
1EX4 [90], 1K6Y [92] and 1K61 [85] | AMMP | [105] |
| 11 | Monomer | Adenine only‡ | No | Two Mg2+ | 1BL3 [102] and 1VSH [100] | Swiss PDB, Cn3D, Gold | [106] |
| 12 | Monomer | Yes (16 bp) | No | Two Mg2+ | 1EX4 [90] | Sybyl | [107] |
| 13 | Monomer | Yes (22 bp) | No | Two Mg2+ | 1WKN [96,97] 1ZA9§ [101] |
Macromodel, Gold, Induced- Fit Docking |
[108] [109] |
The authors of [99] published three models. The data here are for ‘Model II’.
An adenine, but not a DNA molecule, was inserted by superimposition with the indole fragment of 5CITEP.
These two structures (1KWN and 1ZA9) are not from experiments, but come from the two integrase–DNA models 6 and 8.
The general models combined two or more of the one- or two-domain experimental structures to build a full-length protein. The unresolved residues were typically either modeled de novo or simply omitted, especially if located at the termini. One of the largest challenges in this context is how to connect and align the domains in 3D space. Furthermore, it must be decided whether to model a dimer, tetramer or octamer. The choices made here have a substantial effect on the positioning of the viral DNA models placed in the next step. To help with these decisions, all modelers took additional experimental evidence into account that provided indication of relative spatial arrangements and outright interactions between protein regions and/or DNA regions. The other challenge is how to deal with the viral and host DNA. When binding to a protein, poly nucleotides typically do not adopt canonical B-DNA or A-DNA conformation [110], yet authors had to shy away from imposing any specific conformational changes due to lack of structural data.
Model 1 was the first reported IN–DNA model. It is an octamer of so-called ‘protomers’ consisting of the HIV-1 core and C-terminal domains, combining photo-cross-linking results pinpointing IN features that are in close proximity to specific viral and target DNA sites [86]. Model 2 was constructed by docking a model of the 18-bp viral DNA end onto the IN dimer (1EX4) along a contiguous strip of positive charge extending outward from the active site [90]. Model 3 is a well-packed tetramer displaying twofold symmetrical DNA contacts and overall geometry, which was based on an extensive set of cross-linking experiments [91]. Model 4 was built based on the authors’ crystal structure 1K6Y and other biochemical experimental finding. No further computational refinement was performed on this model because no major steric clashes were observed [92]. Model 5 is a tetramer containing both viral and host DNA. Apart from exploiting the usual experimental results as constraints for the three-step model building, such as cross-linking and protein footprinting data, the authors used a comparison of computed and measured hydrodynamic properties as additional guidance in the relative positioning of the three IN domains to each other [93]. Model 6 represents the full-length HIV-1 IN dimer, in which a 27-bp model of the viral U5 LTR was placed by means of an automated docking procedure [96,97]. Model 7 was constructed by exploiting structural information from several different protein data bank (PDB) structures and adding missing residues via modeling techniques to complete the 288-residue monomer sequence, the initial dimer models. After MD simulation, two of the dimeric IN–DNA complexes were combined in a centrally symmetrical fashion to form a tetramer. Finally, a model of host DNA was added in such a way that modeled insertion points of the two 3′-ends of viral DNA were spaced about five host nucleotides apart [99]. Model 8 is a model of the integration complex based upon experimental evidences, including a comparison with the homologous Tn5 transposase (Tnp), which contains bound DNA (1MUH) and an analysis of DNA binding sites [101]. The host DNA was modeled as noticeably bent, with the major groove close to the 3′-OH group of the U5 viral DNA. Model 9 was created by employing homology modeling and MD simulation. Placement of 27-bp models of viral U5 long terminal repeats (LTRs) was performed in four variants of the protein structure differing in type and number of metal ions added. A model of viral DNA was docked into the tetrameric enzyme models [104]. Model 10 was obtained after MD simulation of the tetramer and subsequent addition of the viral DNA models by means of a docking approach [105]. More information about models 1–10 is about to be published elsewhere [111].
In 2007, an electron microscopical single-particle reconstruction of a tetramer of HIV IN bound to DNA three-way junction substrates designed to resemble integration intermediates was reported [112]. It is attractive to assume that this computationally reconstructed 3D map may reflect a true intermediate in the integration reaction. It is striking, however, that none of the models 1–10 presented a close match to the electron-microscopical 3D map.
Models 11–13 focused on the CAT domain. Model 11 is not an IN–Mg–DNA complex in the strict sense because it uses an adenine to mark the terminal portion of the 3′-processed viral DNA [106]. The author positions this model, produced by structural superimpositions of HIV-1 IN CAT (1BL3) with Rous sarcoma virus IN CAT (1VSH), as a possible surrogate for an IN–Mg–DNA complex. Model 12, which places even more emphasis on the active site, was constructed using the viral DNA from the crystal structure of DNA-bound Tnp, and was intended to help identify a binding mode for inhibitors. The predicted conformation of the crystallographically ‘missing loop’ (Gly-140–Gly-149) was used to build a 3D structure to model ST. The final model was energetically refined and used for docking studies [107]. Model 13 was built based on the superposition of models 6 and 8. The core domain of model 6 and viral DNA of model 8 were combined in this model. High-energy contacts between the parts of this new complex were alleviated by energy-minimization while freezing the two Mg2+ ions and 3′-OH of the viral DNA adenosine [108].
Drug design application of the IN–DNA models
Use of IN–DNA models in the context of drug design has been published for models 5, 7 and 12–13. Most of these studies employed docking methods to try to answer the following questions: which residues make up the docking site? What are the possible interactions of inhibitors with metal ions and viral DNA? What can these models teach us about observed drug resistance mutations? Direct applications to drug design have been reported for models 7 and 13 [108,113].
Model 5 was used as docking target in an effort to gain more insight into the mechanism of action of the reported bifunctional quinolonyl diketo acid derivatives [114]. Model 7 was applied in drug discovery by classic methods; for example, virtual screening methods including docking and filtering by predicted ADME/Tox properties. First, 50 different INSTIs were used to produce 30 different pharmacophore models, which were employed to screen a database of 13.5 million purchasable samples. The obtained 234,894 hits were filtered to 167,479 compounds by deleting duplicate molecules, molecules violating Lipinski’s rule of five and inorganic compounds. All these compounds were docked into the active site of model 7. After visual analysis of approximately 1500 compounds from these docking results and consideration of the calculated ADME/Tox properties of these compounds, 88 samples were purchased and assayed. Several micromolar level inhibitors of the ST reaction catalyzed by wild-type HIV-1 IN were obtained [113].
Even though the lack of a well-defined binding pocket makes it difficult to model a plausible binding mode using the IN CAT, models 12 and 13 were used in attempts at obtaining a picture of the binding by docking some INSTIs into the active site.
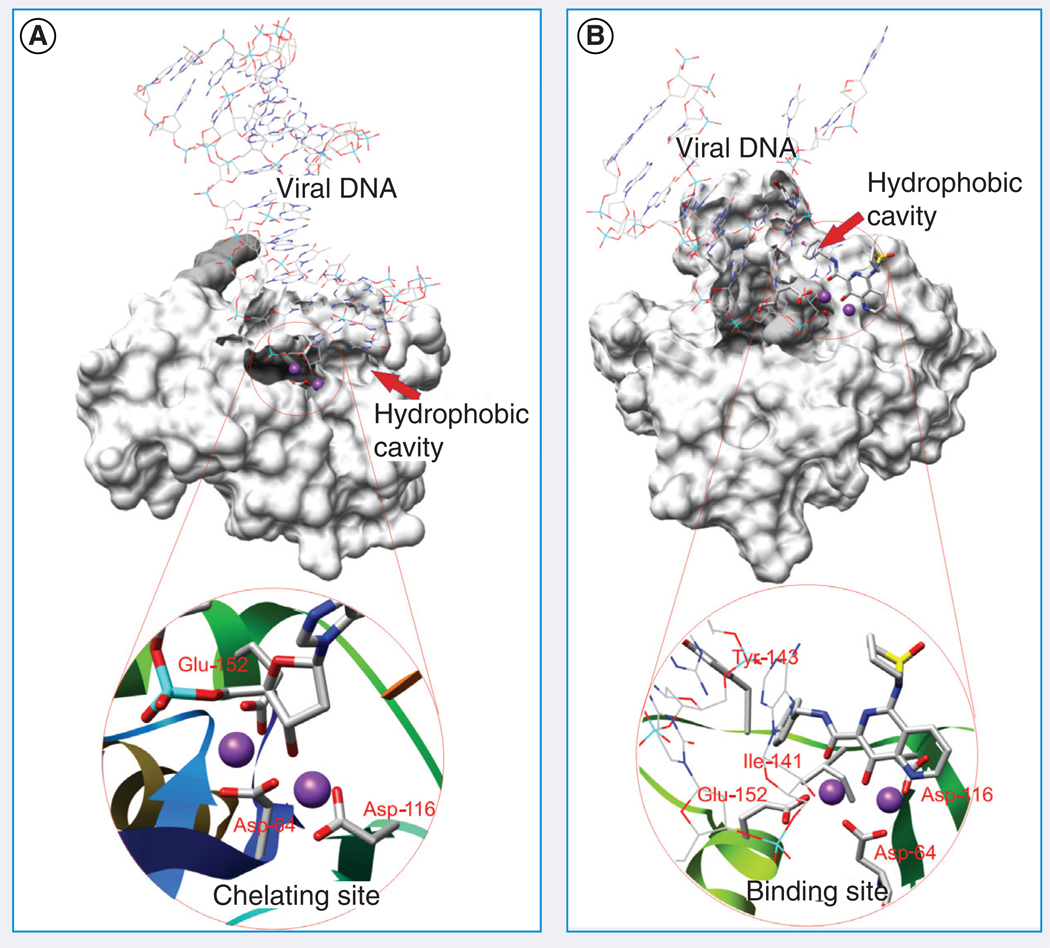
The active site model of model 12 contains two Mg2+ ions and one hydrophobic cavity, which comprises residues of Gln-62–Asp-64, His-114–Asn-117 and Asp-139–Glu-152 (FIGURE 5A) [107]. Several INSTIs were manually docked into the active site, the results of which demonstrated that the common binding mode includes four key elements: first, the binding site exists only upon 3′-processing of viral DNA, after removal of a GT dinucleotide; second, the hydrophobic tail of typical INSTIs binds in the hydrophobic pocket formed mostly by the flexible active-site loop; third, the polar inhibitor moiety forms a critical chelating interaction with only one Mg2+ and lastly, INSTIs have a polar interaction with an interesting triad motif consisting of Cys-65, His-67 and Glu-92.
Figure 5. Models 12 (A) and 13 (B), each of which contains a chelating site and a hydrophobic cavity.
Model 13 contains an integrase strand tramsfer inhibitor, L-870,810, which was docked using the induced-fit docking approach.
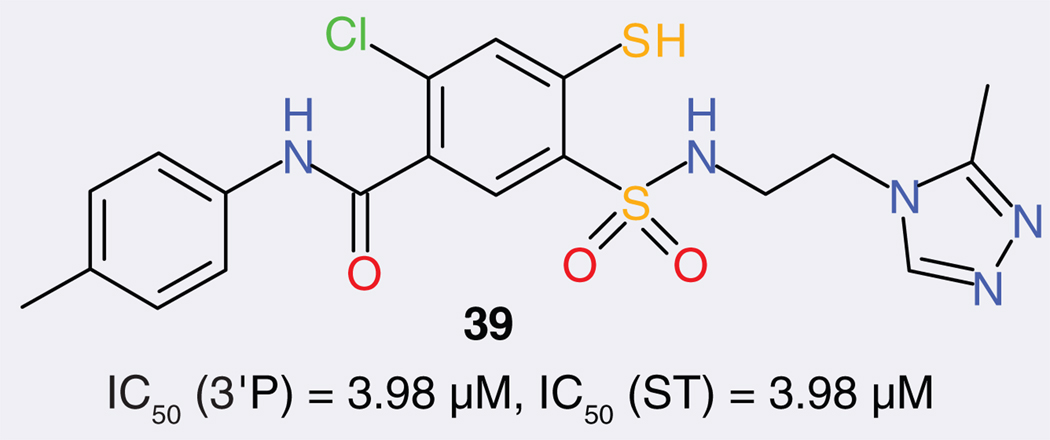
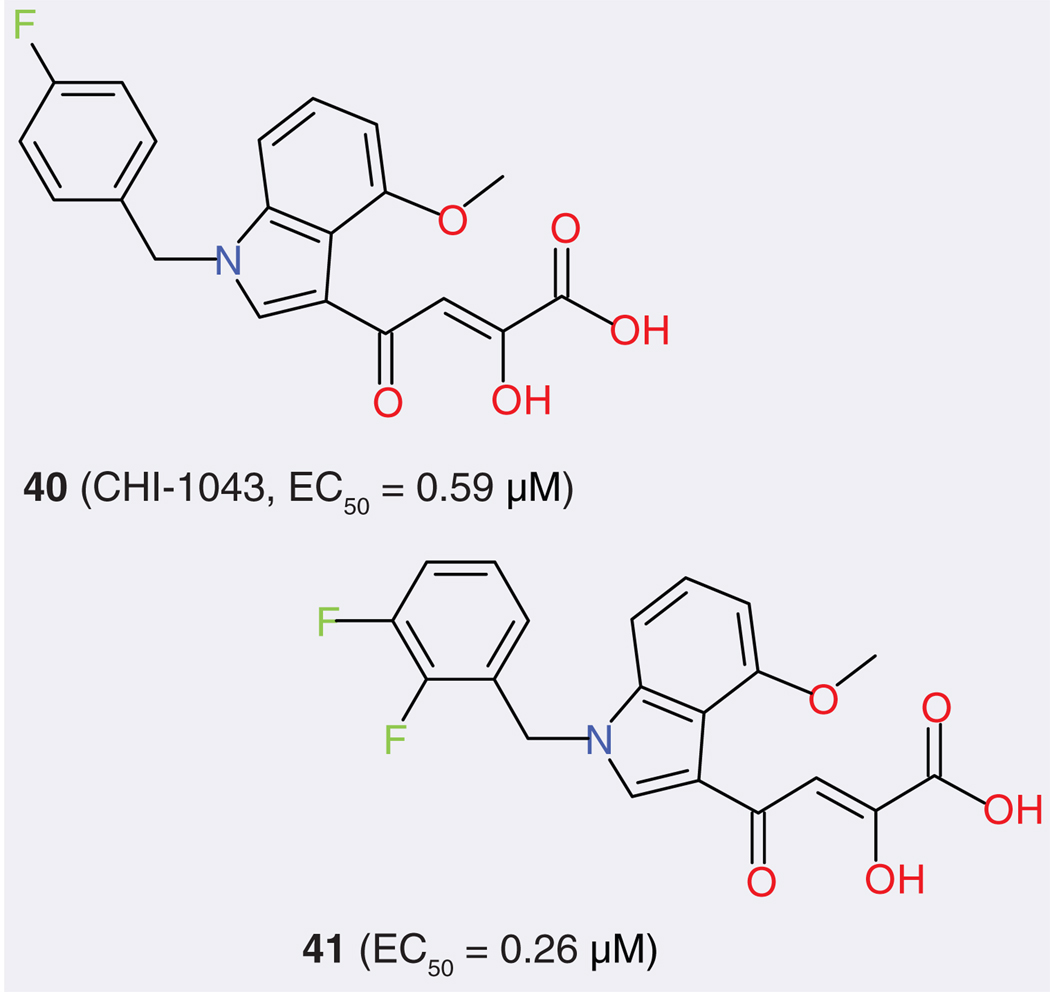
The active site of model 13 has two Mg2+ ions and a hydrophobic cavity with a volume of 46 Å3 defined by nonpolar residues Leu-68, Ile-73, Val-75, Leu-158 and Ile-162. One INSTI, CHI-1043 (FIGURE 6), was docked into the active site. The result showed that the diketo acid moiety is able to interact with the two Mg2+ ions, in agreement with the proposed two-metal chelation mechanism of action for INSTIs. The 4-fluorophenyl ring of the inhibitor occupies the hydrophobic cavity. These results prompted the authors to design and synthesize derivatives (e.g., compound 41), which demonstrated potent enzymatic inhibition at a nanomolar concentration, high antiviral activity and low toxicity [108]. This model was further used to dock six well-known INSTIs with the induced-fit docking approach allowing for receptor flexibility [115]. In addition, different docking modes were obtained, with quite a bit of change observed compared with the rigid docking: for example, the p -fluorobenzyl group made strong hydrophobic interactions with the Ile-141 side chain rather than with the hydrophobic cavity just mentioned (FIGURE 5B).
Figure 6. Structures of CHI-1043 (40) and an optimized compound (41) based on it.
Theoretical study on HIV-1 IN inhibitors
Like many organic molecules, some INSTIs can have multiple tautomers. One notable example are the keto-enol acid derivates, a fundamental innovative type of INSTIs discovered independently by Merck and Shionogi [16,116]. As a result of this tautomerism, they are often referred to as ‘diketo acids’. Detailed geometric and energetic elucidation of these compounds’ tautomerism and how the various possible tautomers could chelate two Mg2+ was seen as an important aid in the design of new chelating moieties, which was achieved in a computational study at a high level of theory [15].
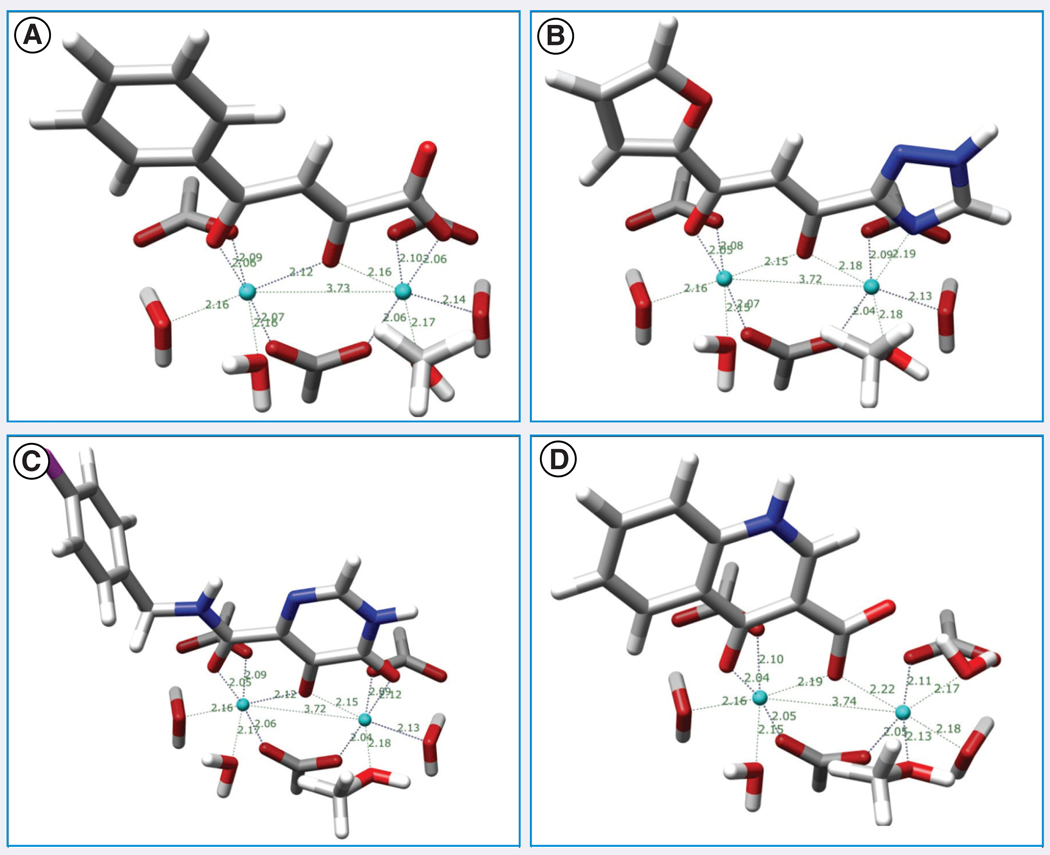
Density functional theory (DFT) at the B3LYP/6–311++ G(d,p) level using the Gaussian 03 suite of programs [117] was employed to calculate the tautomerism and corresponding transition states in both vacuum and aqueous solution of diketo acid, α,γ -diketotriazole, dihydroxypyrimidine carboxamide and 4-quinolone-3-carboxylic acid, each of which is the main portion and putative chelating moiety of four well-known INSTIs: L-708,906, S-1360, MK-0518 and GS-9137, respectively [15].
As aforementioned, the chelation mode of INSTIs with two magnesium ions is a process deemed important for their inhibitory activity. An assembly of three formic acids, which take the places of the residues of Asp-64, Asp-116 and Glu-152 of the IN, four water molecules, and two magnesium ions was modeled to partly mimic the binding site of IN. These components were arranged using the coordinates of Tn5 Tnp [118]. Among other conclusions, these DFT calculations showed that first, generally, the chelating moieties’ global energy minimum conformation had to be planar to form stable complexes; second, in aqueous solution, species with deprotonated enolized or the phenolic hydroxyl groups form some of the most stable complexes and, third, replacement of one water molecule by a methanol to mimic the terminal 3′-OH of viral DNA in the binding site of IN after 3′-P did not significantly affect the inhibitors’ capability to form good chelating complexes. FIGURE 7 shows optimized geometries of the lowest-energy complexes in aqueous solution.
Figure 7. Optimized geometries of complexes of (A) α,γ-diketo acid, (B) α,γ-diketotriazole, (C) dihydroxypyrimidine carboxamide and (D) 4-quinolone-3-carboxylic acid.
Note that all enolized or phenolic hydroxyl groups are deprotonated.
The calculation results were generally consistent with the experimental data, such as with the planar conformation of bound inhibitor 5-CITEP (compound 19) found in the crystal structure [98], suggesting that the keto-enol form is indeed the biologically active tautomer.
Patents involving IN inhibitors discovered by computer tools
As of now, there are dozens, if not hundreds, of patents of IN inhibitors and their preparation methods. However, like most patents claiming pharmaceutical compounds, very few of them mention the computer tools that were used in the discovery of these IN inhibitors. To our knowledge, only one patent document, US 20090088420, patented by the University of Southern California, describes computer tool applications. Pharmacophore models, combined with an in silico protocol and in vitro assays, to be used in IN inhibitors’ design and discovery, are claimed by this patent. Eight classified pharmacophore models, each of which contains four or five features produced by the Catalyst program and compounds with 49 formulas are described in the summary of the invention. Ten compounds, including six α,γ-diketo acids, S-1360 and 5-CITEP were used to develop the pharmacophore models. Compounds obtained by using the highest ranked pharmacophore models to search a chemical database containing approximately 5 million compounds were virtually screened by docking into subunit B of the core domain of an x-ray structure of IN (PDB ID: 1BIS). A Mg2+ ion was placed in the active site. FIGURE 8 shows three compounds with good ST inhibitory activities disclosed in this invention.
Figure 8. Three compounds disclosed in patent US 20090088420.
3′P: 3′-end processing; ST: Strand transfer.
Future perspective
Several factors, including undiminished growth in computing speed, high-performance cluster computing in most research centers, development of highly sophisticated algorithms, availability of large chemical databases and access to a growing body of experimentally determined structures of targets, have made CADD more important and valuable than ever in the lead identification and optimization process. This has been no different in the area of development of IN inhibitors, where CADD has been playing an important role.
In spite of the availability of 25 approved anti-AIDS drugs, there is continued need for the development of new agents for the treatment of AIDS for a whole number of reasons; the main one being a need for better resistance profiles [119]. The approval and coming to market of raltegravir has shown that IN is a worthwhile target. It is expected that in the near future, the ranks of INSTIs in clinical trials will grow, and hopefully at least some of them will progress to FDA approval. The continued lack of a full-length experimental 3D structure of HIV-1 IN complexed with an authentic ligand, viral DNA and possibly even host DNA (or at least shorter models thereof) still limits the role that SBDD can play in the discovery of structurally novel INSTIs with high antiviral activity and low toxicity. New hope has been given through the very recent publication of several crystal structures of full-length IN of the prototype foamy virus (PFV) in complex with its cognate DNA [120]. This may finally provide a structural basis for retroviral DNA integration and thus its inhibition, particularly because two of these crystal structures additionally contain raltegravir and elvitegravir, respectively, bound in the presence of viral DNA and two Mg2+ ions. The authors express the hope that, based on these new findings, the HIV-1 intasome may be modeled and that this will aid in the development of antiretroviral drugs. The verdict is still out regarding how useful these structures will truly be for anti-HIV drug development given the low sequence similarity of PFV IN compared with HIV-1 IN and the marginal crystallographice resolution (~3Å) of these structures. Therefore, LBDD should not yet be discounted and, thus, conducting pharmacophore searches to find moieties that can chelate two magnesium ions simultaneously and then embed these moieties into synthesizable scaffolds appears to remain a promising way to design and discover novel INSTIs.
Supplementary Material
KEY TERMS
- Integrase
Enzyme produced by a retrovirus (an RNA virus, such as HIV-1, which replicates in a host cell) that enables the viral DNA material to be integrated into the DNA of the infected cell. It is also produced for the same purpose by viruses containing double-stranded DNA.
- Computer-aided drug design
Uses computational methods to help in the process of drug design. CADD approaches aim to discover, enhance or analyze drugs and related biologically active molecules. Its most fundamental goal is to predict whether a given molecule will bind to a target. In a broader sense, modern CADD also assists in the subsequent steps of optimizing a ligand’s other properties, such as bioavailability, metabolic fate and half-life, absence of side effects and toxicities before it can become an approved drug.
- AIDS
Disease of the human immune system caused by the human immunodeficiency virus. This disease causes a gradual decrease of the effectiveness of the immune system and makes individuals progressively susceptible to opportunistic infections and tumors. In 2008, an estimated 33.4 million people lived with this disease and an estimated 2.0 million died from it worldwide.
Footnotes
Financial & competing interests disclosure
The authors have no potential conflicts with the subject matter or materials discussed in this manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Supporting information
Several different identifiers, including standard InChIKey, FICuS [121], SMILES and standard InChI of the compounds presented in this review have been calculated. This material is available free of charge via the internet at www.future-science.com
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Anthony NJ. HIV-1 integrase: a target for new AIDS chemotherapeutics. Curr. Top. Med. Chem. 2004;4(9):979–990. doi: 10.2174/1568026043388448. [DOI] [PubMed] [Google Scholar]
- 2.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249(4976):1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 3.Asante-Appiah E, Skalka AM. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus. Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 4.Grobler JA, Stillmock K, Hu B, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl Acad. Sci. USA. 2002;99(10):6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaFemina RL, Schneider CL, Robbins HL, et al. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 1992;66(12):7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egbertson MS. HIV integrase inhibitors: from diketoacids to heterocyclic templates: a history of HIV integrase medicinal chemistry at Merck West Point and Merck Rome (IRBM) Curr. Top. Med. Chem. 2007;7(13):1251–1272. doi: 10.2174/156802607781212248. [DOI] [PubMed] [Google Scholar]
- 7.Pace P, Rowley M. Integrase inhibitors for the treatment of HIV infection. Curr. Opin. Drug Discov. Devel. 2008;11(4):471–479. [PubMed] [Google Scholar]
- 8. Marchand C, Maddali K, Metifot M, Pommier Y. HIV-1 IN inhibitors: 2010 update and perspectives. Curr. Top. Med. Chem. 2009;9(11):1016–1037. doi: 10.2174/156802609789630910.. ▪ Up-to-date review covering approximately 25 years of development of HIV-1 integrase inhibitors.
- 9.Al-Mawsawi LQ, Al-Safi RI, Neamati N. Anti-infectives: clinical progress of HIV-1 integrase inhibitors. Expert Opin. Emerg. Drugs. 2008;13(2):213–225. doi: 10.1517/14728214.13.2.213. [DOI] [PubMed] [Google Scholar]
- 10.Anker M, Corales RB. Raltegravir (MK-0518), a novel integrase inhibitor for the treatment of HIV infection. Expert Opin. Investig. Drugs. 2008;17(1):97–103. doi: 10.1517/13543784.17.1.97. [DOI] [PubMed] [Google Scholar]
- 11.Summa V, Petrocchi A, Bonelli F, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008;51(18):5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Motomura T, Aramaki H, et al. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J. Med. Chem. 2006;49(5):1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 13.Shimura K, Kodama E, Sakagami Y, et al. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137) J. Virol. 2008;82(2):764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johns BA, Svolto AC. Advances in two-metal chelation inhibitors of HIV integrase. Expert Opin. Ther. Pat. 2008;18(11):1225–1237.. ▪▪ Comprehensive review of integrase inhibitors, which match a two-metal binding pharmacophore.
- 15.Liao C, Nicklaus MC. Tautomerism and magnesium chelation of HIV-1 integrase inhibitors: a theoretical study. ChemMedChem. 2010;5:1053–1066. doi: 10.1002/cmdc.201000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao C, Marchand C, Burke TR, Jr, et al. Authentic HIV-1 integrase inhibitors. Future Med. Chem. 2010;2(7):1107–1122. doi: 10.4155/fmc.10.199..▪ Comprehensive review of patents involving authentic integrase inhibitors.
- 17. Young DC. Computational Drug Design: A Guide for Computational and Medicinal Chemists. NY, USA: John Wiley & Sons, Inc.; 2009. . ▪▪ Wonderful book centered around the way computational techniques are utilized in the drug design process.
- 18.Skjevik ÅA, Teigen K, Martinez A. Overview of computational methods employed in early-stage drug discovery. Future Med. Chem. 2009;1(1):49–63. doi: 10.4155/fmc.09.7. [DOI] [PubMed] [Google Scholar]
- 19.Tanrikulu Y, Schneider G. Pseudoreceptor models in drug design: bridging ligand- and receptor-based virtual screening. Nat. Rev. Drug Discov. 2008;7(8):667–677. doi: 10.1038/nrd2615. [DOI] [PubMed] [Google Scholar]
- 20.Tuccinardi T. Docking-based virtual screening: recent developments. Comb. Chem. High. Throughput Screen. 2009;12(3):303–314. doi: 10.2174/138620709787581666. [DOI] [PubMed] [Google Scholar]
- 21. Clark DE. What has computer-aided molecular design ever done for drug discovery? Expert Opin. Drug Discov. 2006;1(2):103–110. doi: 10.1517/17460441.1.2.103.. ▪ Relates several successful stories about computer-aided drug design.
- 22.Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 2005;4(8):649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 23.Potemkin V, Grishina M. Principles for 3D/4D QSAR classification of drugs. Drug Discov. Today. 2008;13(21–22):952–959. doi: 10.1016/j.drudis.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Debnath AK. Quantitative structure-activity relationship (QSAR) paradigm-Hansch era to new millennium. Mini. Rev. Med. Chem. 2001;1(2):187–195. doi: 10.2174/1389557013407061. [DOI] [PubMed] [Google Scholar]
- 25.Nicklaus MC, Neamati N, Hong H, et al. HIV-1 integrase pharmacophore: discovery of inhibitors through three-dimensional database searching. J. Med. Chem. 1997;40(6):920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- 26.Hong H, Neamati N, Wang S, et al. Discovery of HIV-1 integrase inhibitors by pharmacophore searching. J. Med. Chem. 1997;40(6):930–936. doi: 10.1021/jm960754h. [DOI] [PubMed] [Google Scholar]
- 27.Neamati N, Hong H, Mazumder A, et al. Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J. Med. Chem. 1997;40(6):942–951. doi: 10.1021/jm960759e. [DOI] [PubMed] [Google Scholar]
- 28.Neamati N, Hong H, Sunder S, et al. Potent inhibitors of human immunodeficiency virus type 1 integrase: identification of a novel four-point pharmacophore and tetracyclines as novel inhibitors. Mol. Pharmacol. 1997;52(6):1041–1055. doi: 10.1124/mol.52.6.1041. [DOI] [PubMed] [Google Scholar]
- 29.Hong H, Neamati N, Winslow HE, et al. Identification of HIV-1 integrase inhibitors based on a four-point pharmacophore. Antivir. Chem. Chemother. 1998;9(6):461–472. doi: 10.1177/095632029800900602. [DOI] [PubMed] [Google Scholar]
- 30.Mustata GI, Brigo A, Briggs JM. HIV-1 integrase pharmacophore model derived from diverse classes of inhibitors. Bioorg. Med. Chem. Lett. 2004;14(6):1447–1454. doi: 10.1016/j.bmcl.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 31.Barreca ML, Rao A, De Luca L, et al. Efficient 3D database screening for novel HIV-1 IN inhibitors. J. Chem. Inf. Comput. Sci. 2004;44(4):1450–1455. doi: 10.1021/ci034296e. [DOI] [PubMed] [Google Scholar]
- 32.Barreca ML, Ferro S, Rao A, et al. Pharmacophore-based design of HIV-1 integrase strand-transfer inhibitors. J. Med. Chem. 2005;48(22):7084–7088. doi: 10.1021/jm050549e. [DOI] [PubMed] [Google Scholar]
- 33.Dayam R, Sanchez T, Clement O, et al. β-diketo acid pharmacophore hypothesis. 1. Discovery of a novel class of HIV-1 integrase inhibitors. J. Med. Chem. 2005;48(1):111–120. doi: 10.1021/jm0496077. [DOI] [PubMed] [Google Scholar]
- 34.Dayam R, Sanchez T, Neamati N. Diketo acid pharmacophore. 2. Discovery of structurally diverse inhibitors of HIV-1 integrase. J. Med. Chem. 2005;48(25):8009–8015. doi: 10.1021/jm050837a. [DOI] [PubMed] [Google Scholar]
- 35.Deng J, Sanchez T, Al-Mawsawi LQ, et al. Discovery of structurally diverse HIV-1 integrase inhibitors based on a chalcone pharmacophore. Bioorg. Med. Chem. 2007;15(14):4985–5002. doi: 10.1016/j.bmc.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 36.De Luca L, Barreca ML, Ferro S, et al. A refined pharmacophore model for HIV-1 integrase inhibitors: optimization of potency in the 1H-benzylindole series. Bioorg. Med. Chem. Lett. 2008;18(9):2891–2895. doi: 10.1016/j.bmcl.2008.03.089. [DOI] [PubMed] [Google Scholar]
- 37.Dayam R, Al-Mawsawi LQ, Zawahir Z, et al. Quinolone 3-carboxylic acid pharmacophore: design of second generation HIV-1 integrase inhibitors. J. Med. Chem. 2008;51(5):1136–1144. doi: 10.1021/jm070609b. [DOI] [PubMed] [Google Scholar]
- 38.Milne GW, Nicklaus MC, Driscoll JS, et al. National Cancer Institute drug information system 3D database. J. Chem. Inf. Comput. Sci. 1994;34(5):1219–1224. doi: 10.1021/ci00021a032. [DOI] [PubMed] [Google Scholar]
- 39.Kawasuji T, Yoshinaga T, Sato A, et al. A platform for designing HIV integrase inhibitors. Part 1: 2-hydroxy-3-heteroaryl acrylic acid derivatives as novel HIV integrase inhibitor and modeling of hydrophilic and hydrophobic pharmacophores. Bioorg. Med. Chem. 2006;14(24):8430–8445. doi: 10.1016/j.bmc.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 40.Kawasuji T, Fuji M, Yoshinaga T, et al. A platform for designing HIV integrase inhibitors. Part 2: a two-metal binding model as a potential mechanism of HIV integrase inhibitors. Bioorg. Med. Chem. 2006;14(24):8420–8429. doi: 10.1016/j.bmc.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 41.Hazuda DJ, Anthony NJ, Gomez RP, et al. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl Acad. Sci. 2004;101(31):11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen WL, Tirado-Rives J. Molecular modeling of organic and biomolecular systems using BOSS and MCPRO. J. Comput. Chem. 2005;26(16):1689–1700. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- 43.Carlson HA, Masukawa KM, Rubins K, et al. Developing a dynamic pharmacophore model for HIV-1 integrase. J. Med. Chem. 2000;43(11):2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- 44.Deng J, Lee KW, Sanchez T, et al. Dynamic receptor-based pharmacophore model development and its application in designing novel HIV-1 integrase inhibitors. J. Med. Chem. 2005;48(5):1496–1505. doi: 10.1021/jm049410e. [DOI] [PubMed] [Google Scholar]
- 45.Deng J, Sanchez T, Neamati N, Briggs JM. Dynamic pharmacophore model optimization: identification of novel HIV-1 integrase inhibitors. J. Med. Chem. 2006;49(5):1684–1692. doi: 10.1021/jm0510629. [DOI] [PubMed] [Google Scholar]
- 46.Lins RD, Briggs JM, Straatsma TP, et al. Molecular dynamics studies on the HIV-1 integrase catalytic domain. Biophys. J. 1999;76(6):2999–3011. doi: 10.1016/s0006-3495(99)77453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kier LB, Hall LH. An electrotopological-state index for atoms in molecules. Pharm. Res. 1990;7(8):801–807. doi: 10.1023/a:1015952613760. [DOI] [PubMed] [Google Scholar]
- 48.Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J. Chem. Inf. Comput. Sci. 2002;34(4):854–866. [Google Scholar]
- 49.Ponce YM. Total and local quadratic indices of the molecular pseudograph’s atom adjacency matrix: applications to the prediction of physical properties of organic compounds. Molecules. 2003;8(9):687–726. doi: 10.3390/91201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consonni V, Todeschini R, Pavan M. Structure/response correlations and similarity/diversity analysis by GETAWAY descriptors. 1. Theory of the novel 3D molecular descriptors. J. Chem. Inf. Comput. Sci. 2002;42(3):682–692. doi: 10.1021/ci015504a. [DOI] [PubMed] [Google Scholar]
- 51.Specht DF. Probabilistic neural networks. Neural Networks. 1990;3(1):109–118. doi: 10.1109/72.80210. [DOI] [PubMed] [Google Scholar]
- 52.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 2002;110(18):5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 53.Klebe G, Abraham U, Mietzner T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J. Med. Chem. 1994;37(24):4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 54.Ferguson AM, Heritage T, Jonathon P, et al. EVA: a new theoretically based molecular descriptor for use in QSAR/QSPR analysis. J. Comput. Aided Mol. Des. 1997;11(2):143–152. doi: 10.1023/a:1008026308790. [DOI] [PubMed] [Google Scholar]
- 55.Polanski J, Walczak B. The comparative molecular surface analysis (COMSA): a novel tool for molecular design. Comput. Chem. 2000;24(5):615–625. doi: 10.1016/s0097-8485(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 56.Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28(7):849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 57.Baroni M, Costantino G, Cruciani G, et al. Generating optimal linear PLS estimations (GOLPE): an advanced chemometric tool for handling 3D-QSAR problems. Quant. Struct. Act. Relat. 1993;12(1):9–20. [Google Scholar]
- 58.Datar PA, Khedkar SA, Malde AK, Coutinho EC. Comparative residue interaction analysis (CoRIA): a 3D-QSAR approach to explore the binding contributions of active site residues with ligands. J. Comput. Aided Mol. Des. 2006;20(6):343–360. doi: 10.1007/s10822-006-9051-5. [DOI] [PubMed] [Google Scholar]
- 59.Duca JS, Hopfinger AJ. Estimation of molecular similarity based on 4D-QSAR analysis: formalism and validation. J. Chem. Inf. Comput. Sci. 2001;41(5):1367–1387. doi: 10.1021/ci0100090. [DOI] [PubMed] [Google Scholar]
- 60.Raghavan K, Buolamwini JK, Fesen MR, et al. Three-dimensional quantitative structure-activity relationship (QSAR) of HIV integrase inhibitors: a comparative molecular field analysis (CoMFA) study. J Med. Chem. 1995;38(6):890–897. doi: 10.1021/jm00006a006. [DOI] [PubMed] [Google Scholar]
- 61.Buolamwini JK, Raghavan K, Fesen MR, et al. Application of the electrotopological state index to QSAR analysis of flavone derivatives as HIV-1 integrase inhibitors. Pharm. Res. 1996;13(12):1892–1895. doi: 10.1023/a:1016005813432. [DOI] [PubMed] [Google Scholar]
- 62.Makhija MT, Kulkarni VM. Eigen value analysis of HIV-1 integrase inhibitors. J. Chem. Inf. Comput. Sci. 2001;41(6):1569–1577. doi: 10.1021/ci0001334. [DOI] [PubMed] [Google Scholar]
- 63.Buolamwini JK, Assefa H. CoMFA and CoMSIA 3D QSAR and docking studies on conformationally-restrained cinnamoyl HIV-1 integrase inhibitors: exploration of a binding mode at the active site. J. Med. Chem. 2002;45(4):841–852. doi: 10.1021/jm010399h. [DOI] [PubMed] [Google Scholar]
- 64.Makhija MT, Kulkarni VM. QSAR of HIV-1 integrase inhibitors by genetic function approximation method. Bioorg. Med. Chem. 2002;10(5):1483–1497. doi: 10.1016/s0968-0896(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 65.Makhija MT, Kulkarni VM. Molecular electrostatic potentials as input for the alignment of HIV-1 integrase inhibitors in 3D QSAR. J. Comput. Aided Mol. Des. 2001;15(11):961–978. doi: 10.1023/a:1014888730876. [DOI] [PubMed] [Google Scholar]
- 66.Makhija MT, Kulkarni VM. 3D-QSAR and molecular modeling of HIV-1 integrase inhibitors. J. Comput. Aided Mol. Des. 2002;16(3):181–200. doi: 10.1023/a:1020137802155. [DOI] [PubMed] [Google Scholar]
- 67.Yuan H, Parrill AL. QSAR studies of HIV-1 integrase inhibition. Bioorg. Med. Chem. 2002;10(12):4169–4183. doi: 10.1016/s0968-0896(02)00332-2. [DOI] [PubMed] [Google Scholar]
- 68.Costi R, Santo RD, Artico M, et al. 2,6-bis(3,4,5-trihydroxybenzylydene) derivatives of cyclohexanone: novel potent HIV-1 integrase inhibitors that prevent HIV-1 multiplication in cell-based assays. Bioorg. Med. Chem. 2004;12(1):199–215. doi: 10.1016/j.bmc.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Makhija MT, Kasliwal RT, Kulkarni VM, Neamati N. De novo design and synthesis of HIV-1 integrase inhibitors. Bioorg. Med. Chem. 2004;12(9):2317–2333. doi: 10.1016/j.bmc.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Kuo CL, Assefa H, Kamath S, et al. Application of CoMFA and CoMSIA 3D-QSAR and docking studies in optimization of mercaptobenzene-sulfonamides as HIV-1 integrase inhibitors. J. Med. Chem. 2004;47(2):385–399. doi: 10.1021/jm030378i. [DOI] [PubMed] [Google Scholar]
- 71.Daeyaert FFD, Vinkers HM, Jonge MRD, et al. Ligand–based computation of HIV-1 integrase inhibition strength within a series of β-ketoamide derivatives. internet electron. J. Mol. Des. 2004;3(9):528–543. [Google Scholar]
- 72.Marrero-Ponce Y. Linear indices of the “molecular pseudograph’s atom adjacency matrix”: definition, significance-interpretation, and application to QSAR analysis of flavone derivatives as HIV-1 integrase inhibitors. J. Chem. Inf. Comput. Sci. 2004;44(6):2010–2026. doi: 10.1021/ci049950k. [DOI] [PubMed] [Google Scholar]
- 73.Di Santo R, Costi R, Artico M, et al. Design, synthesis and biological evaluation of heteroaryl diketohexenoic and diketobutanoic acids as HIV-1 integrase inhibitors endowed with antiretroviral activity. Farmaco. 2005;60(5):409–417. doi: 10.1016/j.farmac.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 74.Yuan H, Parrill A. Cluster analysis and three-dimensional QSAR studies of HIV-1 integrase inhibitors. J. Mol. Graph. Model. 2005;23(4):317–328. doi: 10.1016/j.jmgm.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 75.Nunthaboot N, Tonmunphean S, Parasuk V, et al. Three-dimensional quantitative structure: activity relationship studies on diverse structural classes of HIV-1 integrase inhibitors using CoMFA and CoMSIA. Eur. J. Med. Chem. 2006;41(12):1359–1372. doi: 10.1016/j.ejmech.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 76.Niedbala H, Polanski J, Gieleciak R, et al. Comparative molecular surface analysis (CoMSA) for virtual combinatorial library screening of styrylquinoline HIV-1 blocking agents. Comb. Chem. High. Throughput Screen. 2006;9(10):753–770. doi: 10.2174/138620706779026042. [DOI] [PubMed] [Google Scholar]
- 77.Vilar S, Santana L, Uriarte E. Probabilistic neural network model for the in silico evaluation of anti-HIV activity and mechanism of action. J. Med. Chem. 2006;49(3):1118–1124. doi: 10.1021/jm050932j. [DOI] [PubMed] [Google Scholar]
- 78.Saiz-Urra L, Gonzalez MP, Fall Y, Gomez G. Quantitative structure–activity relationship studies of HIV-1 integrase inhibition. 1. GETAWAY descriptors. Eur. J. Med. Chem. 2007;42(1):64–70. doi: 10.1016/j.ejmech.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 79.Leonard JT, Roy K. Exploring molecular shape analysis of styrylquinoline derivatives as HIV-1 integrase inhibitors. Eur. J. Med. Chem. 2008;43(1):81–92. doi: 10.1016/j.ejmech.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 80.Iyer M, Hopfinger AJ. Treating chemical diversity in QSAR analysis: modeling diverse HIV-1 integrase inhibitors using 4D fingerprints. J. Chem. Inf. Model. 2007;47(5):1945–1960. doi: 10.1021/ci700153g. [DOI] [PubMed] [Google Scholar]
- 81.Dhaked DK, Verma J, Saran A, Coutinho EC. Exploring the binding of HIV-1 integrase inhibitors by comparative residue interaction analysis (CoRIA) J. Mol. Model. 2009;15(3):233–245. doi: 10.1007/s00894-008-0399-4. [DOI] [PubMed] [Google Scholar]
- 82.Dessalew N. Investigation of the structural requirement for inhibiting HIV integrase: QSAR study. Acta. Pharm. 2009;59(1):31–43. doi: 10.2478/v10007-009-0006-y. [DOI] [PubMed] [Google Scholar]
- 83.de Melo EB, Ferreira MM. Multivariate QSAR study of 4,5-dihydroxypyrimidine carboxamides as HIV-1 integrase inhibitors. Eur. J. Med. Chem. 2009;44(9):3577–3583. doi: 10.1016/j.ejmech.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Nikitin S, Zaitseva N, Demina O, et al. A very large diversity space of synthetically accessible compounds for use with drug design programs. J. Comput. Aided Mol. Des. 2005;19(1):47–63. doi: 10.1007/s10822-005-0097-6. [DOI] [PubMed] [Google Scholar]
- 85.Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 2005;4(3):236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 86.Heuer TS, Brown PO. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37(19):6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 87.Dyda F, Hickman AB, Jenkins TM, et al. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266(5193):1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 88.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, et al. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34(31):9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 89.Bujacz G, Jaskolski M, Alexandratos J, et al. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 1995;253(2):333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 90.Chen JC, Krucinski J, Miercke LJ, et al. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl Acad. Sci. USA. 2000;97(15):8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 2001;20(13):3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20(24):7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Podtelezhnikov AA, Gao K, Bushman FD, McCammon JA. Modeling HIV-1 integrase complexes based on their hydrodynamic properties. Biopolymers. 2003;68(1):110–120. doi: 10.1002/bip.10217. [DOI] [PubMed] [Google Scholar]
- 94.Cai M, Zheng R, Caffrey M, et al. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997;4(7):567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 95.Goldgur Y, Dyda F, Hickman AB, et al. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA. 1998;95(16):9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Luca L, Pedretti A, Vistoli G, et al. Analysis of the full-length integrase-DNA complex by a modified approach for DNA docking. Biochem. Biophys. Res. Commun. 2003;310(4):1083–1088. doi: 10.1016/j.bbrc.2003.09.120. [DOI] [PubMed] [Google Scholar]
- 97.De Luca L, Vistoli G, Pedretti A, et al. Molecular dynamics studies of the full-length integrase-DNA complex. Biochem. Biophys. Res. Commun. 2005;336(4):1010–1016. doi: 10.1016/j.bbrc.2005.08.211. [DOI] [PubMed] [Google Scholar]
- 98.Goldgur Y, Craigie R, Cohen GH, et al. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl Acad. Sci. USA. 1999;96(23):13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karki RG, Tang Y, Burke TR, Jr, Nicklaus MC. Model of full-length HIV-1 integrase complexed with viral DNA as template for anti-HIV drug design. J. Comput. Aided Mol. Des. 2004;18(12):739–760. doi: 10.1007/s10822-005-0365-5. [DOI] [PubMed] [Google Scholar]
- 100.Bujacz G, Alexandratos J, Wlodawer A, et al. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 1997;272(29):18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 101.Wielens J, Crosby IT, Chalmers DK. A three-dimensional model of the human immunodeficiency virus type 1 integration complex. J. Comput. Aided Mol. Des. 2005;19(5):301–317. doi: 10.1007/s10822-005-5256-2. [DOI] [PubMed] [Google Scholar]
- 102.Maignan S, Guilloteau JP, Zhou-Liu Q, et al. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 1998;282(2):359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 103.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289(5476):77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 104.Wang LD, Liu CL, Chen WZ, Wang CX. Constructing HIV-1 integrase tetramer and exploring influences of metal ions on forming integrase-DNA complex. Biochem. Biophys. Res. Commun. 2005;337(1):313–319. doi: 10.1016/j.bbrc.2005.08.274. [DOI] [PubMed] [Google Scholar]
- 105.Chen A, Weber IT, Harrison RW, Leis J. Identification of amino acids in HIV-1 and avian sarcoma virus integrase subsites required for specific recognition of the long terminal repeat Ends. J. Biol. Chem. 2006;281(7):4173–4182. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Savarino A. In silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology. 2007;4:21. doi: 10.1186/1742-4690-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen X, Tsiang M, Yu F, et al. Modeling, analysis, and validation of a novel HIV integrase structure provide insights into the binding modes of potent integrase inhibitors. J. Mol. Biol. 2008;380(3):504–519. doi: 10.1016/j.jmb.2008.04.054. [DOI] [PubMed] [Google Scholar]
- 108.Ferro S, De Luca L, Barreca ML, et al. Docking studies on a new human immunodeficiency virus integrase-Mg-DNA complex: phenyl ring exploration and synthesis of 1H-benzylindole derivatives through fluorine substitutions. J. Med. Chem. 2009;52(2):569–573. doi: 10.1021/jm8009266. [DOI] [PubMed] [Google Scholar]
- 109.Barreca ML, Iraci N, De Luca L, Chimirri A. Induced-fit docking approach provides insight into the binding mode and mechanism of action of HIV-1 integrase inhibitors. ChemMedChem. 2009;4(9):1446–1456. doi: 10.1002/cmdc.200900166. [DOI] [PubMed] [Google Scholar]
- 110.Aishima J, Gitti RK, Noah JE, et al. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30(23):5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao C, Nicklaus MC. HIV-1 Integrase: Mechanism and Inhibitor Design. John Wiley & Sons, Inc.; 2010. HIV-1 integrase–DNA models. (In Press). [Google Scholar]
- 112.Ren G, Gao K, Bushman FD, Yeager M. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J. Mol. Biol. 2007;366(1):286–294. doi: 10.1016/j.jmb.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liao C, Karki RG, Marchand C, Pommier Y, Nicklaus MC. Virtual screening application of a model of full-length HIV-1 integrase complexed with viral DNA. Bioorg. Med. Chem. Lett. 2007;17(19):5361–5365. doi: 10.1016/j.bmcl.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 114.Di Santo R, Costi R, Roux A, et al. Novel bifunctional quinolonyl diketo acid derivatives as HIV-1 integrase inhibitors: design, synthesis, biological activities, and mechanism of action. J. Med. Chem. 2006;49(6):1939–1945. doi: 10.1021/jm0511583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barreca ML, Iraci N, De Luca L, Chimirri A. Induced-fit docking approach provides insight into the binding mode and mechanism of action of HIV-1 integrase inhibitors. ChemMedChem. 2009;4(9):1446–1456. doi: 10.1002/cmdc.200900166. [DOI] [PubMed] [Google Scholar]
- 116.Hazuda DJ, Felock P, Witmer M, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287(5453):646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 117.Frisch MJ, Trucks GW, Schlegel HB, et al. Gaussian 03, Revision E.01. Wallingford, CT, USA: Gaussian, Inc.; 2004. [Google Scholar]
- 118.Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 2004;14(1):50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 119. Flexner C. HIV drug development: the next 25 years. Nat. Rev. Drug Discov. 2007;6(12):959–966. doi: 10.1038/nrd2336.. ▪ Reviews current treatment options, novel mechanisms that can be exploited for existing drug targets and the potential of novel targets for the development of anti-AIDS drugs.
- 120. Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464(7286):232–236. doi: 10.1038/nature08784.. ▪▪ Important paper presenting crystal structures of full-length integrase of the prototype foamy virus in complex with its cognate DNA plus authentic HIV-1 integrase inhibitors (raltegravir and elvitegravir), which may provide a new structural basis for understanding retroviral DNA integration. The authors claim that their models provide plausible binding modes for authentic inhibitors of HIV-1 integrase, thereby helping advance anti-HIV drug development.
- 121.Sitzmann M, Filippov IV, Nicklaus MC. Internet resources integrating many small-molecule databases. SAR QSAR Environ. Res. 2008;19(1–2):1–9. doi: 10.1080/10629360701843540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.