Abstract
Computer-aided drug design plays a vital role in drug discovery and development and has become an indispensable tool in the pharmaceutical industry. Computational medicinal chemists can take advantage of all kinds of software and resources in the computer-aided drug design field for the purposes of discovering and optimizing biologically active compounds. This article reviews software and other resources related to computer-aided drug design approaches, putting particular emphasis on structure-based drug design, ligand-based drug design, chemical databases and chemoinformatics tools.
Drug discovery and development is a very costly and time-consuming process in which every available discipline, including computer-aided drug design (CADD), is utilized in order to achieve the desired results. CADD provides valuable insights into experimental findings and mechanism of action, new suggestions for molecular structures to synthesize, and can help make cost-effective decisions before expensive synthesis is started. Numerous compounds that were discovered and/or optimized using CADD methods have reached the level of clinical studies or have even gained US FDA approval [1,2]. Many CADD techniques are used at various stages of a drug-discovery project, and one cannot designate a single ‘best’ computational drug-design technique in general. Hence, computational medicinal chemists should be aware of and willing to take advantage of all kinds of software and resources related to CADD during their routine work, although individually they may focus on, and subsequently become an expert in, the use of just one or a few specific techniques.
Ligands (be they inhibitors, activators, agonists, antagonists or substrate analogs) can be identified using conventional hit-identifying methods such as high-throughput screening (HTS) assays or employing various CADD techniques. Because of their respective strengths and weaknesses for drug discovery, HTS and CADD techniques are often seen as complementary to each other [3]. HTS has been used in combination with, or substituted by, CADD techniques, the latter being generally faster, more economical and easier to set up than HTS. In addition, by using CADD techniques, one can attempt to optimize ligands to imbue them with high-binding affinity and good selectivity, as well as acceptable pharmacokinetic properties, the latter not usually being within the scope of HTS.
Many of the techniques used in CADD are usually cheaper and faster than most of the experimental assaying methods, therefore large databases of compounds are often tested in silico before they – or, better, subsets of them – are submitted to in vitro testing. Nowadays, drug-design projects often start with hundreds of thousands or even millions of compounds, be they large corporate repositories, catalogs of commercially available screening samples or large virtual libraries. In such a scenario, one of the most valuable tools is so-called virtual screening (VS, also called in silico screening), which is the computational search for molecules with desired biological activities in large computer databases of small molecules that do not even have to physically exist [4].
Depending on the information obtainable at the beginning of the screening campaign about the target and/or existing ligands, VS can be divided into structure-based VS (SBVS) and ligand-based VS (LBVS). In the former, the 3D structure of a target is utilized; in the latter, established ligands of a known target are taken into account. Advances in parallel hardware and algorithms have enabled even large-scale VS runs to be completed in a reasonable time period. As the number of protein structures of interest to drug discovery has significantly increased, the distinction between ‘structure-based’ and ‘ligand-based’ drug-design methods has become blurred. The judicious use of conventional ligand-based methods, such as 3D pharmacophore searches, can greatly improve the efficiency and effectiveness of structure-based drug design (SBDD) [5]. Ligand-based search can act as the first stage in an SBVS workflow. In addition, to open more opportunities for hit identification/optimization for a target of interest, it is very common to employ many different design methods, including both SBVS and LBVS (see HIV-1 integrase as an example [6]).
Generally, molecular modeling techniques for drug design and discovery include not only VS methods, but also various other kinds of techniques summarized in Table 1. A large number of molecular modeling programs have been developed over the past three decades, implementing these techniques in both commercial and free software tools. Some of them are widely used in the pharmaceutical and biological industry as well as in academia and in government research laboratories. The extensive applications of these software tools and other resources, such as chemical databases, have made CADD a valuable asset in drug discovery and development.
Table 1.
Computer-aided techniques used in drug design and discovery.
| Technique | Roles in drug design and discovery |
|---|---|
| Docking | Predict binding mode and approximate binding energy of a compound to a target |
| Structure-based virtual screening | Identify active compounds for a specific target from a chemical library based on docking techniques |
| Pharmacophore modeling | Perceive and provide description of molecular features necessary for molecular recognition of a ligand by a biological macromolecule |
| Ligand-based virtual screening | Identify active compounds for a specific target from a chemical library based on pharmacophore modeling techniques |
| Homology modeling | Build a 3D structure for structure-based drug design for a target for which no crystal structure is available, based on related protein 3D structures |
| Molecular dynamics | Molecular mechanics-based simulation to understand the dynamic behavior of proteins or other biological macromolecules, to analyze the flexibility of the drug target for structure-based drug design and/or to calculate the binding affinity of a compound to a target |
| 2D quantitative structure–activity relationship | Finding a model that can be used to predict some property from the molecular structure of a compound |
| 3D quantitative structure–activity relationship | Technique used to quantitatively predict the interaction between a molecule and the active site of a target; 3D conformation-derived information is utilized in this technique |
| Quantum mechanics | An electron-orbital-based approach based on first principles to optimize structures of ligands and even protein–ligand complexes, improve the accuracy of docking and calculate, for example, free-binding energy |
| Absorption, distribution, metabolism, elimination, and toxicity prediction | Prediction of absorption, distribution, metabolism, elimination and toxicity of chemical substances in the human body to avoid costly later-stage failures in drug development |
The intention of this review is to present the readers with a broad overview of the software and resources commonly used in CADD. Given that it is an impossible task to provide all technical details of the background and applications of these software tools and resources, the reader is encouraged to go back to the referenced literature for additional information. Because of their importance in CADD, this review particularly focuses on SBDD, ligand-based drug design, chemical databases and chemoinformatics tools.
Comprehensive drug-design software packages
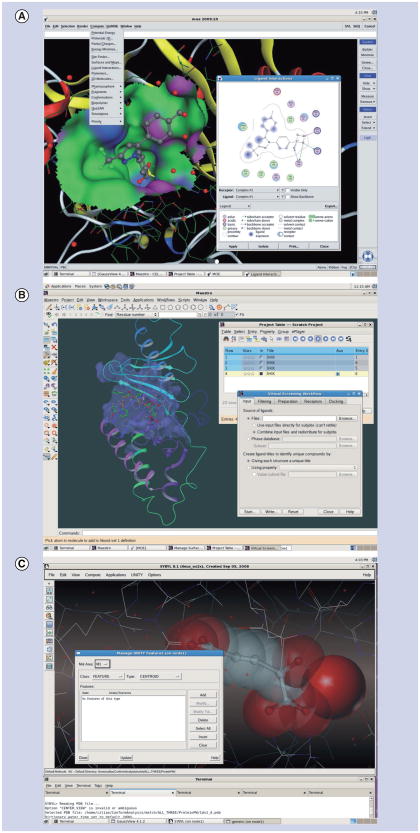
In 1979, a company named Tripos was established in St Louis, Missouri, USA. Tripos was the first company to deliver software for scientific computational drug discovery to the pharmaceutical industry. In the intervening three decades, numerous drug-design and simulation-software companies have come (and some gone). Most often they integrate different programs into comprehensive packages, although the individual programs of a package may require separate license keys to be purchased individually. Table 2 lists the most relevant, currently available drug-design packages and their included modules. Generally, a comprehensive drug-design package has a single, easy-to-use client interface (see Figure 1 for examples), from which the user can manipulate and build their models, manage jobs, and visualize and analyze results.
Table 2.
Commercial software packages for drug design.
| Name | Owned and distributed by | Modules | Ref. |
|---|---|---|---|
| Discovery Studio | Accelrys Inc. |
|
[241] |
| ICM | Molsoft LLC |
|
[242] |
| LeadIT | BioSolveIT GmbH |
|
[243] |
| MOE | Chemical Computing Group |
|
[244] |
| OpenEye† | OpenEye Scientific Software Inc. |
|
[245] |
| Schrödinger | Schrödinger Inc. |
|
[246] |
| SYBYL | Tripos Inc. |
|
[247] |
OpenEye software is free for academic users.
Figure 1. The graphical interfaces of three drug-design packages.
(A) MOE by Chemical Computing Group, (B) Maestro by Schrödinger, (C) SYBYL by Tripos.
Among these drug-design packages, Discovery Studio, MOE, the Schrödinger package and SYBYL are those with the most comprehensive tool set. Each of them supplies modules/programs for almost all kinds of CADD techniques listed in Table 1. Besides this, they also provide different assistant tools, workflows and scripting languages to help the users efficiently employing these packages or automate the drug-design procedures. Other packages are more specialized, that is, they focus on a few particular CADD techniques. The commercialization of these drug-design packages and their wide adoption by pharmaceutical industry as well as academia has, on the one hand, spurred the continued development of computational medicinal chemistry, and, on the other hand, supported the growth of these software packages themselves. Chemistry on the computer has become easier than before: designing and optimizing new drug candidates can be accomplished faster and more economically by efficiently employing one or more of these versatile drug-design packages.
It should be noted that some companies and organizations do not distribute their programs as packages although they have several programs related to drug design and modeling. These companies/organizations include Molecular Discovery [201], Cambridge Crystallographic Data Centre (CCDC) [202], SimBioSys Inc. [203], and MEDIT SA [204].
Programs for docking & SBVS
When the target protein’s structure is known, molecular docking is the preferred method to investigate how a ligand interacts with the protein. Molecular docking is an automated computer algorithm that determines how a compound may bind in the active site of a target and tries to predict how tightly it binds. This method attempts to mimic the process of bringing together a protein and a ligand to form a noncovalent complex, and to reveal the electrostatic and steric complementarity between the protein and ligand. Thus, an algorithm of a docking program faces two main tasks – the prediction of the correct poses of ligands at the active site of a protein and the correct ranking of these poses. Both tasks are of a challenging nature, and so far none of the reported docking programs are able to solve both of them perfectly. Prediction of possible binding modes in an active site is more straightforward and can be performed successfully by most programs. Because of its success at this task, docking is a well-established drug-design technology that is widely employed in SBDD. Nowadays, most docking programs available account for flexibility of ligands; however, handling of receptor flexibility remains a significant issue. Treatment of ligand flexibility can be divided into three basic categories: systematic methods (incremental construction and conformational search); random or stochastic methods (Monte Carlo, Genetic Algorithms and Tabu search); and simulation methods (molecular dynamics [MD] and energy minimization) [7]. Another crucial aspect is the scoring function applied during docking or SBVS to rank docking poses. Fundamentally, three classes of scoring functions are currently applied in docking programs: force field based, empirical and knowledge based. To date, more than 60 small-molecule docking programs and 30 scoring functions have been reported (see reviews [8–11]). Among the reported docking programs, AutoDock [12], DOCK [13], FlexX [14], FRED [15], Glide [16], GOLD [17], ICM [18] and Surflex-Dock [19] are perhaps the most popular docking tools (Table 3). Several benchmark studies have been published evaluating the performance of docking programs [20–25]. However, one cannot draw a simple conclusion from all these studies in that there would be a single docking program that outperforms all other programs in all aspects, for example, docking accuracy or hit enrichment. In addition, benchmarks evaluating different scoring functions have been reported [26,27].
Table 3.
The most used docking programs in structure-based drug design.
| Name | Developed by | Incorporated into software package | Free for academia | Drug-design applications | Ref. |
|---|---|---|---|---|---|
| AutoDock | Scripps Research Institute† | - | Yes | Aldose reductase inhibitors Rac1 Inhibitors Trypanothione reductase inhibitors |
[109] [110] [111] |
| DOCK | University of California, San Francisco‡ | - | Yes | STAT3 dimerization inhibitors Death-associated protein kinase inhibitors Inhibitors of osteoclast formation and bone resorption |
[112] [113] [114] |
| FlexX | BioSolveIT GmbH | LeadIT | No | Inhibitors of penicillin binding protein Inhibitors of ATP-phosphoribosyl transferase Human histamine H4 receptor ligands |
[115] [116] [117] |
| FRED | OpenEye Scientific Software | OpenEye | Yes | Proteasome inhibitors Heat-shock protein 90 inhibitors |
[29] [118] |
| Glide | Schrödinger, Inc. | Schrödinger | No | Inhibitors of dengue virus methyltransferase FGFR1 kinase inhibitors HIV-1 integrase inhibitors |
[119] [120] [121] |
| GOLD | Cambridge Crystallographic Data Centre | - | No | Topoisomerase I inhibitors MNK1 inhibitors Met tyrosine kinase inhibitors |
[122] [123] [124] |
| ICM | Molsoft LLC. | Molsoft | No | TNF-α inhibitors Aryl hydrocarbon receptor ligands GTP competitive inhibitors |
[125] [126] [127] |
| Surflex-Dock | Tripos Inc. | SYBYL | No | Glycogen synthase kinase inhibitors Proteasome inhibitors HIV-1 reverse transcriptase inhibitors |
[128] [29] [129] |
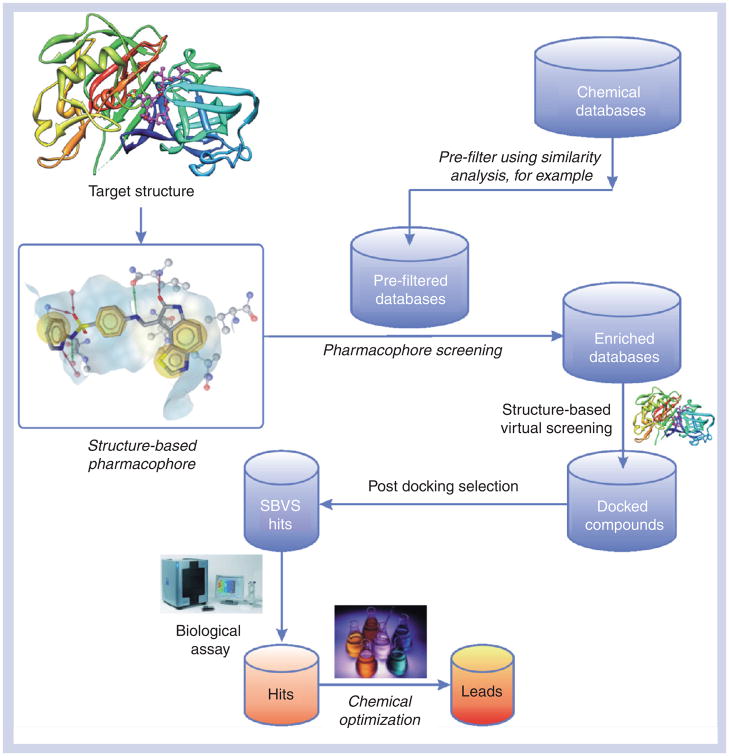
Figure 2 shows the schematic representation of a protocol commonly used in an SBVS campaign. The 3D structure of a target, which preferably is in complex with a ligand, is a prerequisite for docking or SBDD. The 3D structure may be a crystallographic x-ray structure or an NMR structure, often downloaded from the Protein Data Bank (PDB). However, experience has shown that, even though the experiment would seem to provide the ultimate answer to structural questions, some caution is warranted as possible ambiguities of some experimental structures can mislead the unwary medicinal chemists [28]. Hence, it is highly recommended to try to assess the validity and reliability of the chosen crystal structures before using them in drug-design projects.
Figure 2. Typical structure-based virtual screening-based drug-development protocol.
The italicized steps are not part of the SBVS in the narrow sense although they are often performed in a SBVS-based drug-discovery campaign.
SBVS: Structure-based virtual screening.
In order to reduce the sizes of the databases used in SBVS, they are prefiltered on the basis of calculated physicochemical descriptors, a pharmacophore model or simply by Lipinski’s Rule of Five. Although this step is not obligatory for SBVS, it is attractive for providing enrichment to speed up the identification of molecules binding the target receptor more quickly and to help ensure desired pharmacokinetic profiles of the identified binders. Once an appropriate set of molecules has been put together by the prefiltering steps listed above, they can be docked into the active site for further reduction of the number of candidates based on the fast (although not very accurate) scoring functions. To choose candidates for biological assays from the docking results, it is often helpful, if doable, to examine the docking poses visually and/or conduct further sophisticated computational studies such as MD simulations (see the section on MD simulations programs later for details).
In most cases, SBVS identifies hits with activity in the micromolar range, although nanomolar activities have, occasionally, been reported [4]. A prospective SBVS project can be regarded as successful if at least one new hit with a novel scaffold is yielded, especially if the efficiency of identifying these hits is significantly higher than HTS or traditional medicinal chemistry approaches would presumably have been. No guarantee, however, can usually be given for the success of such SBVS projects, since their outcome depends in an as yet unpredictable way on the combination of the investigated target, the chemical databases used and the applied search methods.
To improve the results of an SBVS experiment, different docking programs can be applied in combination. For example, in the identification of novel proteasome inhibitors, FRED, Surflex-DOCK and LigandFit were combined to screen the ChemBridge database [29]. In addition, several scoring functions can be employed simultaneously for predicting the binding affinity of a pose produced by a docking program [30].
Structure-based virtual screening methods can also be used in fragment-based drug discovery projects [31,32]. In this situation, libraries are screened that typically contain molecules with a molecular mass of less than 300 Da and with fewer than three hydrogen-bond donors and six hydrogen-bond acceptors. This helps with the design of small ligands that bind with high ligand efficiency and can be readily optimized to potent lead-like compounds. Sometimes, computational methods can also be applied to predict fragment binding: at first, a fragment library is docked into the binding site of interest; then the best orientations of some fragments are chosen and used as starting points for the attachment of substituents, with the aim of targeting new areas within the binding site where supplementary interactions may be made [33].
Many proteins are flexible targets, which are stabilized by ligand binding in one conformation out of an ensemble of conformers of similar energy in the unbound state. Taking into account the flexibility of the protein by docking programs is still an area of active development [34]. Currently, the algorithms accounting for receptor flexibility can be classified into two categories. The first one allows for protein conformational changes upon ligand binding. The best known of these is the induced-fit docking from Schrödinger (see [35] as an example), which is a protocol using a combination of the programs Prime and Glide. However, such methods cannot generally be used in SBVS mainly because of their unacceptably high computational demands (i.e., low speed) for screening large libraries. The second type of algorithms make use of multiple conformations of the target, in as much as a set of binding site conformations from different x-ray crystal structures [36], NMR ensembles [37] or extracted from MD or Monte Carlo simulations, are used [38–40].
Programs for 3D pharmacophore modeling & LBVS
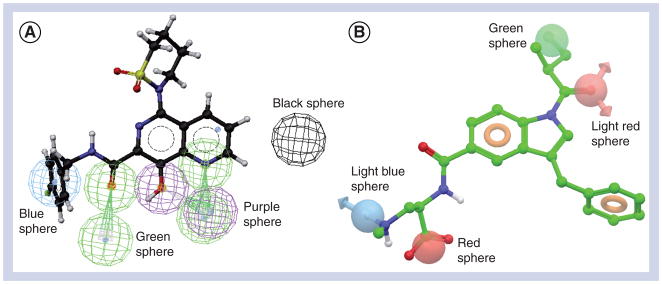
In the absence of a receptor structure, the identification or optimization of lead compounds can depend on pharmacophore modeling, which is typically performed by extracting common chemical features from 3D structures of a set of known ligands representative of essential ligand–macromolecule interactions. According to IUPAC, a pharmacophore is “an ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target and to trigger (or block) its biological response” [41]. The common chemical features that are usually used as types of the desired interactions are hydrogen-bond acceptors, hydrogen-bond donors, hydrophobic regions and positively or negatively charged groups (see examples in Figure 3). Exclusion volumes, inclusion regions or a combination of both can also be integrated into a pharmacophore. A pharmacophore is based on the concept of similarity between ligands (i.e., the pharmacophoric features have to be similar – not particularly the connectivity), and is used in LBVS to explore the diversity and complexity of molecular structures for the purpose of identifying novel structural hits. In medicinal chemistry, pharmacophores have found widespread use not only for hit-and-lead identification but also for subsequent lead optimization, and have been increasingly successful in drug discovery (see reviews [42–46]).
Figure 3. Examples of pharmacophores used in ligand-based virtual screening.
(A) A pharmacophore produced by Catalyst. Blue sphere: hydrophobic feature; green sphere: hydrogen-bond acceptor; purple sphere: hydrogen-bond donor; black sphere: excluded volume. (B) A pharmacophore produced by Phase. Light red sphere: hydrogen-bond acceptor; light blue sphere: hydrogen-bond donor; green sphere: hydrophobic feature; red sphere: negative feature; orange torus: aromatic ring.
Many programs, including Catalyst, DISCOtech, LigandScout [47,48], MOE (its pharmacophore module) and PHASE are widely used for pharmacophore elucidation and VS (Table 4). These programs differ mostly in the algorithms utilized for the handling of ligand flexibility and molecule alignment. None of these programs are free to academia; however, there is a ligand-based pharmacophore program called PharmaGist that can be accessed freely on the web [49,205].
Table 4.
Commonly used pharmacophore modeling programs.
| Name | Developed by | Incorporated into software package | Methods | Drug design applications | Ref. |
|---|---|---|---|---|---|
| Catalyst | Accelrys Inc. | Discovery Studio | Ligand based, includes the two methods HipHop and HypoGen for pharmacophore perception Produces conformers using pre-enumerating method by the Poling algorithm Uses feature-based method to align molecules |
Acetylcholinesterase inhibitors σ1 receptor ligands Tubulin inhibitors |
[50] [51] [130] |
| DISCOtech | Tripos Inc. | SYBYL | Ligand based Produces conformers using pre-enumerating method by Concord and Confort Uses Bron–Kerbosh clique- detection algorithm to align molecules |
Glycogen synthase kinase inhibitors SGLT2 inhibitors Ligands of AT2 |
[128] [131] [132] |
| LigandScout | Inte:Ligand† | Structure based Pharmacophoric feature points-based pattern- matching alignment algorithm | 11 β-HSD1 inhibitors Pim1 inhibitors HIV-1 transcriptase inhibitors |
[133] [134] [135] |
|
| MOE | Chemical Computing Group | MOE | Ligand based Produces conformers using pre-enumerating method by various methods ranging from molecular dynamics to stochastic methods and systematic search Uses property-based algorithm to align molecules |
Antitubercular agents Reversal agents Antimalarial agents |
[136] [137] [138] |
| PHASE | Schrödinger, Inc. | Schrödinger | Ligand based Produces conformers using pre-enumerating method by ConfGen Uses feature-based algorithm to align molecules |
Inhibitors of dengue virus methyltransferase Selective MDR1 agents γ-aminobutyric acid G1 receptor ρ1 antagonists |
[119] [139] [140] |
See [250].
Generally, ligand-based pharmacophore generation from a set of ligands involves two main steps: first, sampling of the conformational space for each ligand to take into account the conformational flexibility of the ligand, and second, alignment of the multiple ligands (in their various conformations) to determine the essential common chemical features needed to build a pharmacophore model. These two steps also pose the main challenges in ligand-based pharmacophore modeling. There are two types of pharmacophore models. The first type are the 3D quantitative structure–activity relationship (QSAR)-like models, which can be derived from a training set of ligands with biological activities typically spanning at least three orders of magnitude (see [50–52] as examples). With such models, the potencies of new compounds can be quantitatively predicted by evaluating how well each compound maps onto the model. The second type can be developed from a training set that includes only active ligands (see [53–55] as examples). The potencies of new compounds can be estimated qualitatively by whether they match the model. Representatives of these two types of methods are HypoGen and HipHop (both in Catalyst), respectively.
The performance and applicability of pharmacophore modeling primarily depends on two factors: the definition and placement of pharmacophoric features, and the alignment techniques used for overlaying the 3D pharmacophore model with a set of ligand molecules in a screened data set [45]. Ideally, the set of ligands has been derived from a number of different chemical series with limited conformational flexibility and not too many heteroatoms [42]. Since the application of pharmacophore matching is typically faster per compound than docking, large chemical structure databases can be subjected to pharmacophore searches for novel ligands. The hits obtained can exhibit novel and diverse chemotypes, enabling the medicinal chemist to pursue series with novel scaffolds. Lately, pharmacophore searching has also been used in industry to create small, focused sets for low-throughput, higher-quality assays to enhance the lead-identification process in parallel with HTS [56]. In such focused sets, the sources of compounds can be either in-house or purchased from compound vendors.
Before a chemical structure database can be screened with a 3D pharmacophore, it needs to be precomputed, that is, conformational sampling of every compound needs to be performed. Such corporate databases should at least contain conformational sampling of every compound in them. This allows rapid matching between the generated conformers as rigid bodies and the query. Before running the actual search on the full database, in order to assess the credibility of the used pharmacophore, it is recommended to use the derived pharmacophore performing on a small test database seeded with known actives and decoys. The list of compounds that matches the pharmacophore query should be evaluated for promiscuous matches, such as highly flexible, feature-rich molecules. Also, visually examining how much of the molecule falls within the pharmacophore and how much remains outside, can be used to rank the virtual hits for inclusion in the final set for screening [42].
A 3D pharmacophore can also be derived from a protein structure by observing the specific interactions between protein and ligand. In this case, shape and excluded volume information can be added to the pharmacophore. This has the advantage of finding hits that not only have the key binding elements but are also more likely to fit into the active site, which can reduce the false positive rate. Generally, database-searching methods based on 3D pharmacophores are much faster than structure-based methods, such as docking, which makes pharmacophore searching a more effective way to screen very large databases. Pharmacophore searching can, therefore, act as the first stage in an SBVS workflow.
Quantitative structure–activity relationship
Quantitative structure–activity relationship modeling has been used widely as a key computational tool for predicting physicochemical properties and rationalizing experimental binding data or inhibitory activity of chemical compounds. Typically, QSAR is performed in two diverse modes, referred to as 2D and 3D QSAR, which are quite different techniques for practical purposes. 2D QSAR is conceptually a way of finding a simple equation that can be used to predict some property from the molecular structure of a compound. It is a meaningful correlation (model) between a set of independent variables (chemical descriptors) calculated from chemical graphs, and a dependent variable such as binding affinity, log P, or the pKa value whose value one wishes to predict for the compound of interest [57]. There are many different algorithms for selecting 2D QSAR descriptors and building the model. Among them, the most used are regression-analysis algorithms, which automate the process of using correlation coefficients and cross-correlation coefficients to select chemical descriptors. Multivariate analysis algorithms, heuristic algorithms and genetic algorithms also are used. 2D QSAR in the narrow sense has the inherent advantage of being independent of the 3D conformation, while it has the weakness of being much less robust in terms of model interpretation. It has to be emphasized, however, that 2D QSAR models can be built from both 2D and 3D descriptors, the latter ones indeed requiring a (typically calculated) 3D conformation for each molecule both in the training and the test sets. An example of this type of QSAR program is BioEpisteme [206] of the Prous Institute for Biomedical Research.
Many comprehensive drug-design packages include their own 2D QSAR modules, with which the users can calculate different molecular descriptors and then build their 2D QSAR models. More standalone-type programs in the field include Codessa from Semichem [207] for building 2D QSAR models, which offers many algorithms for automatically selecting descriptors, and the structure–activity relationship (SAR) and QSAR programs PASS and GUSAR [208] with a large number of built-in (Q)SAR models. Software specifically generating molecular descriptors, but not necessarily QSAR models includes Dragon [209] and Mold2 [58]. 2D QSAR remains a valuable tool for predicting chemical properties of drug-like organic compounds, hence currently it is widely employed and an actively pursued methodology in the field of absorption, distribution, metabolism, elimination and toxicity (ADME/T) prediction.
Broadly speaking, 3D QSAR includes any QSAR approach based on 3D molecular structures. In this sense, QSAR built from molecular descriptors containing conformational coordinate-derived information could be classified as 3D QSAR (although, especially if mixed with 2D descriptors, can also be seen as a 2D QSAR technique, as mentioned previously). In a narrower sense, 3D QSAR is a technique that uses a 3D grid of points around the molecule, each point having properties associated with it that can vary in a field-like manner from point to point, such as steric interactions or electrostatic potential. The following discussion confines itself to this type of 3D QSAR. 3D QSAR is mainly used for predicting the binding affinity of a ligand to the active site of a specific target. It often requires 3D structures of the analyzed molecules, plus typically a molecular superposition step [59]. For building a 3D QSAR model, it is necessary to first select a training set, which ideally contains approximately 15 to 20 active compounds with preferably a wide of range of activity. The second step is to generate conformations and alignments of the training set compounds, which can be done manually or by algorithms. Most often, the most rigid molecules are aligned first, which provides a template with as little uncertainty as possible for further alignment of less rigid molecules. A dimensionality reduction step is then typically inserted to extract the features of the 3D interaction field that are most strongly determining the activity before the actual predictive model is built, often with a partial least squares (PLS) approach. Finally, a test set containing some active compounds (typically split off the original training set) is used to examine the robustness of the built 3D QSAR model.
There are several programs developed for 3D QSAR. The most well-known among them are comparative molecular field analysis (CoMFA) [60] and comparative molecular similarity indices analysis (CoMSIA) [61], both of which are integrated into SYBYL. References [62,63] describe their applications in drug discovery. The models built with the CoMFA or CoMSIA techniques are created to identify a correlation between the molecular fields and biological activity, which can be automatically achieved with a PLS algorithm. Another 3D QSAR program that has found application in drug design is molecular field analysis from Accelrys, which is similar in its approach to CoMFA [64,65].
In the early stage of drug design, if the active site of the target is unknown, 3D QSAR is useful to explain activities of existing compounds and to accurately predict the activities of analogs of those, whereas pharmacophore searches tend to be more valuable for quickly searching very large chemical databases and thus tend to be better for scaffold hopping to identify novel classes of active compounds. If the geometry of the active site is known, docking tends to replace 3D QSAR and will be the preferred prediction technique in many projects.
Recently, 3D QSAR modeling approaches have also been reported for use in VS [66]. For example, the QSAR modeling approaches of variable selection k-nearest neighbor and support vector machines using both MolconnZ and MOE chemical descriptors generated from 2D chemical graphs have been employed to identify histone deacetylase class 1 inhibitors by screening 9.5 million molecules compiled from the ZINC database, the World Drug Index database, the ASINEX Synergy libraries, and other commercial databases [67]. The same group also successfully employed similar QSAR and VS methods to discover geranylgeranyltransferase-I inhibitors [68].
Homology modeling
If the 3D model of a target protein is needed whose structure is not yet solved experimentally by x-ray crystallography or NMR, however the sequence of its amino acids is available and the experimental 3D structure(s) for one or more sufficiently similar proteins is known, homology modeling (also known as comparative modeling) is a useful approach to explain experimental facts, develop hypotheses, and/or carry out SBDD. Homology modeling attempts the construction of an atomic-resolution model of the target protein from its amino acid sequence using the experimental 3D structures of related homologous proteins as templates [69–71]. The concept is based on the experience that similar sequences lead to similar structures, that is, proteins descended from a common ancestor (a protein family) typically have similar sequences and similar 3D structures. Since experimental determination of protein structure through x-ray crystallography is still a difficult and costly process, homology modeling methods provide quick and easy ways to build models for further studies.
Typically, homology modeling of proteins includes the following four steps [69,72]: identification of one or more known experimental structures of a related protein that can serve as template, sequence alignment of target and template proteins, and model building for the target and refining/validation/evaluation of the models. Human intervention is typically needed to check for errors that may have been introduced during, for example, sequence alignment and refinement of models. Database search techniques using tools such as FASTA [210] and BLAST [211] are the simplest methods to identify templates for homology modeling. More advanced tools include PSI-BLAST [212] and FFAS [213].
The quality of a homology model is generally correlated with the quality of the template structure and the sequence alignment. Decreasing sequence identity between the target and the template will typically affect the quality of the homology model. If there are gaps in the alignment of structural regions between the target and template protein (these gaps are referred to as indels), homology modeling can become a quite error-prone process. Moreover, the quality of the model tends to decline if the resolution of the template protein is poor. The construction of less rigid regions, for example, loops, is generally also less accurate than the rest of the model. However, there is a general tendency that good accordance is obtained for the functional region of the protein as the active sites are usually highly conserved regions in the template structures [73]. How reasonable a homology model is, can be quantified, for example, by a Ramachandran plot, in which the distribution of backbone bond angles is shown. The quality of a homology model can also be examined by checking the inside and outside distribution of hydrophilic and lipophilic residues.
The most frequently used homology modeling programs and their application in drug design are listed in Table 5. Among them, SWISS-MODEL and Modeller are perhaps the most widely used, maybe because of their free availability. Several large-scale benchmarking experiments, most prominently Critical Assessment of Techniques for Protein Structure Prediction (CASP) [74], have been organized to assess the relative quality of various homology modeling methods. Biannually since 1994, CASP has invited research groups to blindly test their structure-prediction algorithms on a set of experimental solved, but not yet published, protein structures [214]. The results of each CASP round are released in a special annual issue of ‘Proteins: Structure, Function, and Bioinformatics,’ which the readers of this article are encouraged to read to obtain more information about how the CASP experiment was conducted, what kinds of homology modeling methods/programs were used, and which outperformed others.
Table 5.
Homology modeling programs used in drug design.
| Name | Developed by | Incorporated into software package | Free for academia | Drug design applications | Ref. |
|---|---|---|---|---|---|
| ICM | Molsoft LLC | Molsoft | No | Aryl hydrocarbon receptor ligands G-protein coupled receptor antagonists |
[126] [141] |
| Modeller | University of California, San Francisco† | Discovery Studio | Yes | Inhibitors of penicillin-binding protein Cdc25 phosphatase inhibitors G-protein coupled receptor antagonists |
[115] [142] [143] |
| MOE | Chemical Computing Group | MOE | No | Inhibitors of Jumonji domain-containing protein histone demethylases Inhibitors of human glutaminyl cyclase |
[144] [145] |
| Prime | Schrödinger, Inc. | Schrödinger | No | Janus kinase 3 inhibitors Inhibitors of the mammalian target of rapamycin kinase |
[146] [147] |
| SWISS-MODEL | Swiss Institute of Bioinformatics‡ | Yes | Inhibitors of osteoclast formation and bone resorption | [114] |
Models with more than 50% sequence identity are believed to be accurate enough for drug-design application. In this range, the root-mean-square deviation between the experimental structure and the model may be around 1 Å, which is equivalent to the typical resolution of structures solved by NMR. In the 25–50% identity range, errors can be more severe and are frequently located in the flexible loops. The homology model can be used for the assessment of druggability and mutagenesis experiments but should be applied with caution for drug design. Below 20–25% sequence identity, a model is usually not usable for drug design because serious errors can occur [69]. However, exceptions from this rule can be found, such as in G-protein coupled receptor modeling [75]. So far, homology modeling has been effectively employed to identify hits using VS, to suggest accurate binding modes and receptor–ligand interactions, to aid in mutagenesis experiments, to rationalize SAR data, and to optimize hit compounds [69]. Developing accurate enough homology models still remains a large challenge. However, a recent survey regarding VS surprisingly revealed that hits derived from docking into homology models had on average higher potency than hits identified by docking into experimental structures [4].
Chemical databases
The fact that the number of commercially and, even more so, publicly available databases of small-molecule compounds has increased considerably in recent years attests to the high relevance of such kinds of data collections for drug discovery and development. These databases may be just structure collections, such as of commercially available screening samples, or provide additional data such as measured bio-activity of the compounds and their protein targets, as well as targeted diseases. Quite a few of these databases (e.g., ChEMBL) attempt to link small-molecule data with information about their biological targets as well as available assay data.
Table 6 lists a selection of some of the better-known small-molecule databases relevant for drug discovery. Its focus is on publicly available databases but also references some commercial databases, which, for the most part, will be not discussed any further here.
Table 6.
Databases of interest for drug discovery.
| Database | Publisher | License type | Ref. |
|---|---|---|---|
| Open National Cancer Institute Database | National Cancer Institute | Publicly available | [253,254] |
| PubChem | National Center for Biotechnology Information | Publicly available | [216] |
| BindingDB | University of Maryland, USA | Publicly available | [255] |
| Relibase | Cambridge Crystallographic Data Centre | Freely accessible for academia, commercial version available | [256] |
| ChEMBLdb | European Bioinformatics Institute, Hinxton, UK | Publicly available | [257] |
| ChemSpider | Royal Society of Chemistry, UK | Publicly available | [258] |
| Human Metabolome Database | University of Alberta, Canada | Publicly available | [259] |
| DrugBank | University of Alberta, Canada | Publicly available | [260] |
| Therapeutic Target Database | National University of Singapore, Singapore | Publicly available | [261] |
| ZINC | University of California, San Francisco, USA | Publicly available | [262] |
| iResearch Library | ChemNavigator | Commercial | [263] |
| GVKBIO databases | GVK Biosciences Private Limited, India | Commercial | [264] |
| MDDR | Accelrys Inc. | Commercial | [265] |
| Wombat | Sunset Molecular Discovery | Commercial | [266] |
| World Drug Index | Thomson Reuters | Commercial | [267] |
All databases listed in Table 6 represent the outcome of substantial efforts of data-collection work by the corresponding groups or organizations. A comprehensive assessment of the quality of each database in a global sense, or for any particular entry, would require a similar size effort and is therefore an impossible task in the context of this review. To a good extent, we can only quote the providers of these databases as to what the specialty and value of the entries in them are. That said, there are chemoinformatics approaches that can be applied to check whether, for example, the correct structure is shown, whether stereochemistry is presented correctly and whether a reasonable tautomeric form is used [76]. Likewise, the values in any data fields should be spot-checked for plausibility and/or be reconfirmed through other sources. Finally, it should be regarded as good practice to carefully review search results obtained in any small-molecule database to the level needed.
Open National Cancer Institute Database
The Open National Cancer Institute (NCI) Database contains currently over 275,000 small-molecule structures, which represents the publicly available part of the over half-million structures collection assembled by the NCI in the course of a more than 50 years’ long effort of screening compounds against cancer and also AIDS [77]. This undertaking has been, and is still, managed by NCI’s Developmental Therapeutics Program, which made most of the open part of the database freely available on their website in the 1990s. Various companies are offering this database, or parts thereof, in the original or processed format, often in conjunction with their chemical database programs. A fully searchable version of the Open NCI Database, enhanced with additional experimental or calculated data, is freely accessible via a web-based interface that was implemented in its original form in 1998 and is still maintained on the web server of the NCI/ CADD Group [215]. While the pace of acquiring new compounds for testing by Developmental Therapeutics Program has slowed in the recent past and also has been partially superseded by other programs of the NIH (see PubChem [216]), the Open NCI Database can still be regarded as a very useful resource for researchers. It was one of the first large-scale small-molecule resources made freely available on the web.
PubChem
Arguably the highest profile of the more recently started database projects is PubChem, which has been implemented by the National Center for Biotechnology Information at the National Library of Medicine, NIH, as support for the NIH Roadmap (now called NIH Common Fund) initiative and launched publicly 2004. PubChem is an open public repository containing chemical structures and biological properties of molecules including small molecules and siRNA reagents. It comprises three interconnected databases: PubChem Substance, PubChem Compound and PubChem BioAssay [78]. PubChem Substance contains information about the original structure records submitted by more than 140 different database providers, such as chemical vendors, publishers or other government agencies. PubChem Compound is the index of unique chemical structures collected in PubChem Substance. PubChem BioAssay stores bioactivity screens of chemical substances described in PubChem Substance and acts as a repository of the small-molecule screening data generated by (historically) the Molecular Library Screening Center Network and (currently) the Molecular Library Probe Production Center Network under the NIH Molecular Libraries Program. It also includes biological property data contributed from other organizations. As of March 2011, PubChem has collected 85 million entries (also comprising mixtures, extracts, complexes and uncharacterized substances) in its substance database, which represents more than 32 million unique structure entries indexed in PubChem Compound. The subset of assays in PubChem BioAssay associated with Molecular Library Screening Center Network or Molecular Library Probe Production Center Network currently numbers more than 3400.
BindingDB
BindingDB contains experimentally determined enzyme kinetic data, measured or derived binding affinities of protein–ligand complexes and protein targets for small-molecule ligands [79]. Most of the data in BindingDB have been manually extracted from journals by curators, although some have been submitted by external authors and contributors directly. The database focuses on proteins that are drug targets or candidate drug targets. As of March 2011, the database contained more than 284,000 small molecules, approximately 5600 protein targets, a collection of approximately 649,000 binding datasets and measured results from 822 isothermal titration calorimetry experiments.
Relibase
Relibase was developed with the focus on providing a database and search system for the handling of protein–ligand complex data and the systematic investigation of protein–ligand interactions [80]. For the analysis of such interactions, 3D constraints can be specified allowing the search of desirable combinations of functional groups and their preferred interaction geometries. Relibase is available in a web-based version, which is free to use for academia. This version includes access to all experimental structures available in the PDB. Some important features of Relibase are standard text searching, 2D substructure searching, 3D protein–ligand interaction searching, ligand similarity searching, 3D visualization (using AstexViewer) and automatic superposition of related binding sites (allowing for, e.g., the comparison of ligand-binding modes, water positions and ligand-induced conformational changes). In addition, a commercial version of Relibase is offered as Relibase+, which provides a number of additional features including the ability to make proprietary (in-house) databases searchable in the same way as, and together with, the PDB version.
ChEMBL
ChEMBL is a database of bioactive drug-like small molecules [81]. The data in the current release (ChEMBL_09, as of March 2011) have been extracted from nearly 35,000 papers taken from 12 prominent medicinal chemistry journals that cover a significant fraction of global drug R&D published output. The current version contains more than 3 million activities of approximately 758,000 compounds, measured for approximately 8000 biological targets. Of those, more than half are protein targets and the others are cell lines or organisms. The mappings between targets and assay results include extensive compound sets against kinases and G-protein coupled receptors as well as approved drugs and clinical candidates. An important part of the curation work carried out for ChEMBL is the normalization of the bio-activities into a uniform set of end-points and units, and adding a set of varying confidence levels to the links between a molecular target and a published assay.
ChemSpider
ChemSpider, first released in 2007 and officially launched in 2008, is a freely accessible chemical compound database that was initially implemented by a group of volunteers. Since 2009, ChemSpider has been owned by the Royal Society of Chemistry (UK). It remains a resource offered free of charge. ChemSpider links together compound information across the web and provides free text and structure search access to currently approximately 25 million chemical structure entries (as of March 2011). Each structure entry in ChemSpider is associated with a list of predicted molecular properties as well as possibly available experimental data, spectra, links back to the almost 400 original data sources/databases, and reference resources such as other Royal Society of Chemistry databases, patent databases, PubMed, MeSH literature, pharmacological web-links (e.g., DailyMed and PillBox) or Google Scholar/Books.
Human Metabolome Database
The Human Metabolome Database (HMDB) [82] provides a detailed collection of information about small-molecule metabolites found in the human body. The data in HMDB was derived from literature or from experimental metabolite concentration data. It currently (March 2011, version 2.5) contains more than 7900 small-molecule metabolite entries that are associated with approximately 7200 protein (and DNA) sequences compiled from hundreds of mass spectra and NMR metabolomic analyses performed on urine, blood and cerebrospinal fluid samples. On the basis of this, HMDB is probably one of the most complete and comprehensively curated collections of human metabolite and metabolism data currently available. Each HMDB entry is organized into chemical, clinical and molecular biochemical data. In addition, links to other public databases are provided where available (e.g., to PubChem, KEGG [83], MetaCyc [217], ChEBI [84], PDB, Swiss-Prot [218] and GenBank [219]).
DrugBank
The DrugBank database (maintained by the same group as the HMDB) collates detailed drug data with target and mechanism of action information [85]. Approximately half of the information in the DrugBank data is dedicated to drug information; the other half is devoted to target sequences, pharmacological properties, pharmacogenomic data, food–drug interactions, drug–drug interactions and experimental ADME data. In its current version 3.0 (released January 2011), the database contains over 6800 drug entries including more than 1400 FDA-approved small-molecule drugs, 133 FDA-approved biotechnology (protein/ peptide) drugs, 83 nutraceuticals and over 5200 experimental drugs. In addition, more than 4400 nonredundant protein (i.e., drug target) sequences have been linked to the group of FDA-approved drug entries.
Therapeutic Target Database
The Therapeutic Target Database (TTD) provides information about drugs, targeted diseases and known and explored therapeutic protein and nucleic acid targets, as well as information about biochemical pathways [86]. TTD is conceptually similar to DrugBank but the mapping between compounds and targets is more focused on primary targets. Another difference is the classification of targets and compounds into marketed, clinical trial and research-phase compounds. The current version of the database contains more than 5100 drugs, including approximately 1500 approved drugs, approximately 1100 drugs in clinical trials and approximately 2300 experimental drugs. All drugs are linked to more than 1900 biological targets, of which 350 are marked as successful, 250 as in clinical trials, 43 as discontinued and approximately 1250 as research targets. The data in TTD have been collected by a comprehensive search of the literature, approved drug reports from the FDA, and latest reports from several pharmaceutical companies that describe clinical trial and other pipeline drugs.
ZINC database
The ZINC database, which has been especially prepared for VS, is a highly curated collection of commercially available chemical compounds gathered from more than 120 original vendor catalogs or compound collections [87]. The original compound databases have been filtered from duplicates, salt counter ions, compounds with atom types other than H, C, N, O, F, S, P, Cl, Br or I, molecules with a formula weight greater than 700, calculated log P greater than 6 or less than −4, number of hydrogen-bond donors greater than 6, number of hydrogen-bond acceptors greater than 11 and number of rotatable bonds greater than 15. In addition, ZINC aims to represent the biologically relevant form for each of its molecule entries, which it defines as the most relevant, correctly protonated forms or tautomers of the molecule between pH 5 and 9.5, the form with deprotonated carboxylic acids and tetrazoles and with generally protonated aliphatic amines (as the major normalized structural features). Also, for all molecules that are biologically relevant, 3D representation of the molecule is available (in case stereochemistry has not been fully specified for the original database structure, the enantiomer or a maximum of four diastereomers is generated). The current version of the database is ‘ZINC Eleven’ and contains approximately 20 million compounds. Besides the full database, several specific subsets (classified as e.g., ‘lead-like’, ‘drug-like’, ‘purchasable’, ‘fragment-like’) of the database can be downloaded from the ZINC website.
ChemNavigator iResearch Library
One of the largest small-molecule databases in existence is the iResearch Library (iRL) from ChemNavigator, a formerly small company in San Diego, CA (USA) that was acquired in 2009 by Sigma-Aldrich. The iRL is ChemNavigator’s continually updated compilation of commercially available screening compounds from more than 300 international chemistry suppliers. As of January 2011, the iRL had registered over 95 million chemical samples representing approximately 60 million unique chemical structures. The iRL is not per se freely publicly available. It is, however, included for searches (although not for bulk download) in several web-based services offered by the NCI/CADD Group, such as the Chemical Structure Lookup Service (CSLS; see below) and the Chemical Identifier Resolver (CIR) [220]. It can therefore be regarded as having an intermediate nature between public and commercial as a resource for computational medicinal chemistry and drug discovery. The database can be directly licensed from the company on DVD/ROM or accessed through an online iResearch System subscription. A license includes access to regular updates, sourcing information, and ChemNavigator’s optional chemistry procurement service.
Chemoinformatics tools
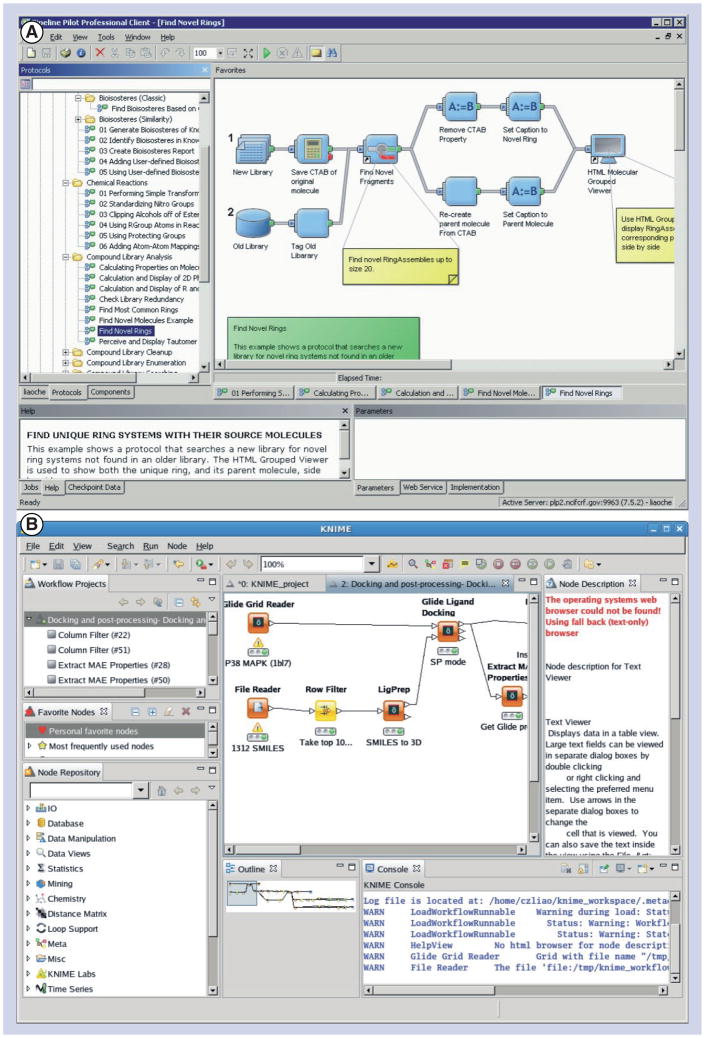
Chemoinformatics tools assist medicinal chemists in the acquisition, analysis and management of data and information relating to chemical compounds and their properties. In many research projects in drug development, a broad spectrum of programs is applied, which puts special emphasis on the management of data, as the interchange of information between different programs usually requires some effort and, quite often, also programming and/or scripting experience. In the past, such requirements were frequently regarded as barriers by medicinal chemists for using these programs themselves. However, with the advent of visual workflow/ data pipelining environments as implemented by Pipeline Pilot or Konstanz Information Miner (KNIME) (Figure 4), this problem has been mitigated to some extent. Since data-pipelining software packages enjoy high popularity not only with the ‘CADD professionals’ among the scientists engaged in drug development, but also with bench chemists, they will be described first.
Figure 4.
The interfaces of (A) Pipeline Pilot and (B) Information Miner.
Pipeline Pilot
Pipeline Pilot is a commercial scientific informatics platform providing a powerful data-pipelining engine based on configurable protocols. It provides a rapid application development environment to automate scientific data management, analysis and reporting processes. The student version of Pipeline Pilot (a light version that does not include all functionalities of the full version) is free to academia. Pipeline Pilot was developed by SciTegic, which became a subsidiary of Accelrys in 2004.
Pipeline Pilot was the first product that brought to the market the concept of ‘data pipelining,’ particularly in the fields of drug discovery and chemoinformatics. It provides the ability to graphically layout or build protocols and workflows, which can be reused, extended or rerun later also by other users. Hence, a Pipeline Pilot protocol represents a documentation of the process applied to a scientific problem by itself. Any functionality in Pipeline Pilot is organized into individual components that can be linked together to a protocol by a few mouse clicks. As part of a Pipeline Pilot license, different sets of component collections focused on topics such as chemistry, biology, life science modeling, materials modeling, reporting and visualization, analysis and statistics, imagining or database integration can be acquired from Accelrys. The Pipeline Pilot platform also exposes a web-services layer that allows a protocol to be integrated as part of a service-oriented architecture environment or other workflow frameworks. Pipeline Pilot provides the possibility to incorporate in-house solutions by writing one’s own, or modifying existing, components and protocols. The same mechanism allows an extensive list of third-party software providers to make their tools accessible as Pipeline Pilot components (e.g., Tripos, BioSolveIT and Molecular Networks).
Konstanz Information Miner
Konstanz Information Miner has been developed by the Institute for Bioinformatics and Information Mining at the University of Konstanz (Germany) [221]. Unlike Pipeline Pilot, KNIME is released under an open-source license; enterprise extensions and services for the deployment in a corporate environment are provided commercially by KNIME.com GmbH. KNIME was adopted early on by several pharmaceutical companies and a series of life-science software vendors that started offering their tools integrated into KNIME. However, the primary focus of KNIME lies on statistical data analysis and data mining, thus its application is not only restricted to the fields of life science and pharmaceutical research.
KNIME possesses various components for data integration (file I/O and database nodes supporting all common database management systems), data transformation (filter, converter, combiner), machine learning, data mining and data visualization. These components are organized as nodes that can be linked together by the modular data-pipelining concept utilized by KNIME to produce ‘data flows’ in KNIME terminology. The graphical user interface allows the user to visually create these data flows, to selectively execute some or all analysis steps and later to inspect the results, models and interactive views. Because of KNIME’s flexible application programming interface, custom nodes and types can be implemented quickly, extending KNIME to be able to read and process highly domain-specific data. In addition to the over 100 processing nodes incorporated into the basic package of the software, a series of third-party nodes are also available that provide access to methods available in packages such as the data-mining software Weka [222], the statistics package R [223], the open-source Chemistry Development Kit [88], BioSolveIT’s scientific software packages or Schrödinger’s suite of drug-design software.
CACTVS System
CACTVS, developed by Xemistry GmbH [224], is a universal multiplatform chemoinformatics toolkit for processing chemical information [89]. CACTVS is primarily a high-level chemistry-aware scripting environment that supports the rapid development of solutions for a broad range of information processing, exchange and reporting needs, such as those encountered in the pharmaceutical industry. CACTVS can be freely downloaded for evaluation, and is free for academic use.
CACTVS can be used to implement any type of structure, reaction or other chemistry object manipulation application either as web application, stand-alone software or as a batch tool. The CACTVS package also includes several standard applications for chemical data handling, for example, a visual molecular structure browser and a molecular structure editor. However, the strength of CACTVS lies in the possibility of implementating one’s own applications. For this, CACTVS provides a series of powerful algorithms or methods, for example, molecular properties calculation (including typical QSAR properties), structure and reaction depiction in many graphic formats (e.g., GIF, PNG, WMF, SVG and EPS), matching by SMARTS, recursive SMARTS, and macro SMARTS, full support for daylight-compatible SMIRKS transforms, Kekulé and tautomer set generators, manipulation of chemical structures and reactions (on the level of molecules, atoms, bonds, groups, rings, ring and pi systems), an extensive set of structure-identity hashcodes, I/O for dozens of chemistry exchange formats (e.g., SDF 2000/3000, ChemAxon, Tripos and Schrödinger) and table file formats such as Excel, tight integration of all PubChem databases for data lookups and direct access to other public online resources such as the NCI/ CADD CIR [220], Wikipedia or databases such as ChemSpider, ChemIDplus [225] and ChEBI [226]. As of the most recent version 3.386 (March 2011), CACTVS reads and writes native KNIME tables which allows dynamic linking between CACTVS and KNIME nodes via networked bi-directional table data exchange. A visual workflow/data pipelining environment for CACTVS is in development.
Open Babel
The Open Babel [227] project arose as a further development of the Babel chemistry file translation program and Babel’s successor OELib (released under the GNU General Public License by OpenEye Software). Open Babel is now a collaborative open-source effort of several academic groups and researchers in the fields of chemoinformatics and computational chemistry. Open Babel is designed as a toolset for the conversion of different chemical structure file formats and provides a data structure suitable for the representation of chemical structures and associated data. It is supplied as a C++ library including a command-line utility. The C++ library includes all of the file-translation code as well as a wide variety of utilities to help the development of other open-source scientific software in the fields of molecular modeling, chemistry, solid-state materials, biochemistry or related areas. Open Babel is used in a variety of open-source software packages (e.g., the 3D molecular editor Avogadro [228], the MySQL database extension MyChem for the handling of chemical structures [229] and the optical structure recognition package OSRA [90]) and provides bindings for a series of programming and scripting languages (e.g., Java, Perl and Python).
Auxiliary programs for drug design & discovery
Molecular dynamics simulations programs
Molecular dynamics simulations, in which atoms and molecules are allowed to interact for a period of time by approximations of known physics based on Newton’s equations of motion describing molecular mechanics (MM), are widely used computational techniques for the study of biological macromolecules [91,92]. MD is very useful for understanding the dynamic behavior of proteins or other biological macromolecules, from fast internal motions to slow conformational changes or even protein-folding processes. Owing to the enormous increase of computer power and improved algorithms, MD simulations of systems comprising 106 or more atoms and time periods on the order of microseconds or even milliseconds in explicit solvent environments, have become possible [93]. Commonly used MD simulations programs are Amber [94], CHARMM [95], Desmond [96], GROMACS [97], NAMD [98] (Table 7). A more comprehensive list of MD simulation programs can be found elsewhere [230].
Table 7.
Major molecular dynamics programs used in drug design.
| Name | Developed by | Free for academia | Drug design applications | Ref. |
|---|---|---|---|---|
| Amber | University of California, San Francisco, USA† | No | Human acetylcholinesterase inhibitors HIV-1 reverse transcriptase inhibitors |
[148] [149] |
| CHARMM | Harvard University, USA‡ | No | Glucose binding to insulin Flaviviral protease inhibitors |
[150] [39] |
| Desmond | D. E. Shaw Research§ | Yes | ||
| GROMACS | University of Groningen, The Netherlands¶ | Yes | Antiviral compounds for avian influenza neuraminidase | [151] |
| NAMD | University of Illinois, USA# | Yes |
Although SBVS against crystal or relaxed receptor structures is an established method for identifying potential inhibitors, the more-dynamic changes within a binding site cannot be readily taken into account by standard SBVS approaches. To accommodate full receptor flexibility, representative receptor ensembles derived from MD simulations can be used in docking studies [99]. The results from MD simulations can also be employed to refine docked complexes. Such simulations integrate flexibility of both the receptor and the ligand, thereby improving interactions and enhancing complementarity between the binding partners, and thus coming closer to the ideal of induced fit. Wrongly docked structures have a higher likelihood of generating unstable MD trajectories leading to the disruption of the complex, providing an additional filtering mechanism (albeit at a high computational cost) for false positives. MD simulations typically incorporate explicit solvent molecules, which is very important for understanding the role of the particular solvent and its effect on the stability of the ligand–protein complexes. While it is usually hard to reproduce correct compound binding affinities by docking studies, MD simulations can provide more reliable results for free-energy calculations using free-energy perturbation, thermodynamic integration, linear interaction energy methods or MM-Poisson–Boltzmann surface area methods. For more information about MD simulations, their use, and their accounting for the flexibility of docked complexes, see references [34,99].
However, notwithstanding the wide use of MD simulations in drug development, the setup of an MD simulation can be difficult. Another problem is that there are often no adequate parameters in the MD force fields parameter sets for nonstandard molecules, such as metalorganic compounds. In addition, MD simulations are still computationally expensive. All of these aspects have limited the use of MD simulations for high-throughput applications, that is, right now it is still impossible to apply MD simulations to the screening of entire chemical compound databases for the purpose of drug discovery, in contrast to what can be done with SBVS. Nevertheless, the combination of fast and inexpensive docking protocols with subsequent more costly MD techniques to subsets of the original screening database has become a feasible approach in rational drug design.
Quantum mechanics programs
In drug design, using classical MM approaches can have many pitfalls due to their possible inaccuracies based on all the approximations entering into MM. The most fundamental of them is that atoms and molecules are essentially described as balls and springs ruled by the laws of classical mechanics and not as nuclei held together by electron orbitals governed by the laws of quantum mechanics (QM), as they really are. Currently, with the increase of central processing unit performance and the improvement of algorithms and software, large-scale biological problems can be addressed using QM methods [100–102]. QM methods can be used to model unstable molecules such as radicals and, furthermore, estimate activation energies for chemical reactions, including those that are carried out by enzymes. The typical applications of QM in drug design include:
QM can be used to calculate energies and optimize structures of ligands and even protein–ligand complexes [103];
QM-derived atomic point charges have recently been shown to be important for the study of protein–ligand complexes, especially for docking studies attempting to obtain the correct binding mode of a ligand [104];
QM/MM methods are beginning to be employed for the calculation of free binding energies owing to their in-principle more accurate predictions. QM/MM approaches have shown promise for this; however, this technique still requires extensive sampling of ligand–receptor conformations through MD simulations and remains very time consuming [105];
The descriptors calculated from QM can also be used to build QSAR models. In this situation, 3D structures with all hydrogen atoms placed have to be utilized because of the need to have a complete description of all nuclei and electrons in the molecular species.
The most used QM programs in drug design are listed in Table 8. The more complete list of QM programs can be found elsewhere [231]. In spite of its age, Gaussian is generally perceived as the standard for density functional theory and ab initio calculations, certainly in terms of breadth of implemented capabilities and algorithms.
Table 8.
Quantum mechanics programs with frequent use in drug design.
| Name | Developed by | Free for academia | Ref. |
|---|---|---|---|
| Gamess | Iowa State University, USA | Yes | [273] |
| Gaussian | Gaussian Inc. | No | [274] |
| Ghemical | University of Kuopio, Finland | Yes | [275] |
| Jaguar | Schrödinger Inc. | No | |
| MOPAC | Stewart Computational Chemistry | Yes | [276] |
| NWChem | Environmental Molecular Sciences Laboratory | Yes | [277] |
| SPARTAN | Wavefunction, Inc. | No | [278] |
ADME/T prediction programs
Favorable ADME/T parameters are very important early requirements for drug candidates in order to reduce late-stage failure and minimize costs [106]. Numerous ADME/T properties are interdependent and therefore there is the need for optimizing them simultaneously during a drug-development project. The multiparameter ADME/T optimization is probably the least attractive stage but it may make the costly difference between success and failure. If fast and easy to use, in silico ADME/T prediction programs, capable of predicting potential ADME/T risks, can be of great benefit for medicinal chemists, and together with in vitro screens, guide syntheses and optimization strategies towards promising molecules only [107].
Many programs use built-in statistical models to calculate ADME/T endpoints. The idea behind these models, which are the core of the predicting programs, is QSAR, that is, to quantitatively define a structure–property relationship, which could be used to predict, in this case, ADME/T parameters. The quality of the models greatly depends on the right combination of statistical techniques, molecular descriptors, validation method, and most importantly on the quality and breadth of the experimental data used to derive them [108].
Many ADME/T prediction programs are available (Table 9). Some programs, such as ADMET Predictor, Sarchitect and ADME Suite can predict a broad spectrum of ADME/T parameters and can usually be used in batch mode, which makes them suitable for incorporation into pipelining data flow protocols, such as the ones built through KNIME and Pipeline Pilot. Some of them, for example, PASS, StarDrop and Leadscope, have auto-modeling capabilities in which the user can use their own experimental data to build and validate new predictive models in addition the models already available within the software.
Table 9.
Available ADME/T prediction programs.
| Program | Developed by | Free for academia | Prediction Spectrum | Ref. |
|---|---|---|---|---|
| ADMET Predictor |
Simulations Plus, Inc. | No† | ADME/T | [279] |
| StarDrop | Optibrium, Ltd | No | ADME/T | [280] |
| ADME Suite Tox Suite |
Advanced Chemistry Development, Inc. | No | ADME Toxicity | [281] |
| ADMEWORKS Predictor |
Fujitsu FQS | No | ADME/T | [282] |
| Sarchitect | Strand Life Sciences | No | ADME/T | [283] |
| QikProp | Schrödinger, Inc. | No | ADME/T | |
| TOPKAT | Accelrys, Inc. | No | Toxicity | |
| Leadscope | Leadscope, Inc. | No | Toxicity | [284] |
| Meteor Derek Nexus |
Lhasa, Ltd | No | Metabolism Toxicity |
[285] |
| PASS | Russian Academy of Medical Sciences | No | Toxicity | [286] |
| HazardExpert Pro MetabolExpert ToxAlert MEXAlert RetroMex |
CompuDrug, Ltd | No | Toxicity Metabolism Toxicity Metabolism Metabolism |
[287] |
| METAPC CASETOX |
Multicase, Inc. | No | Metabolism Toxicity |
[288] |
| VolSurf+ MetaSite |
Molecular Discovery, Ltd. | No | ADME Metabolism |
[289] |
| Bioclipse | Uppsala University, Sweden and European Bioinformatics Institute | Yes | Metabolism | [290] |
| MetaDrug | GeneGo, Inc. | No | Metabolism/Toxicity | [291] |
| TIMES | OASIS Lmc | No | Metabolism/Toxicity | [292] |
MedChem Designer of Simulations Plus is free to access. MedChem Designer can predict a few ADME/T properties.
Molecular visualization programs
Each of the drug-design packages mentioned in this review has a graphical user interface, through which the users can visualize and analyze their models and results, and can generate graphics for publications or reports. Even though the whole suite might only be available by purchasing a commercial license, some of the software vendors, for example, Accelrys, Molsoft and Schrödinger, have released the molecular visualization component of their suites for free download on the internet. For generating high-quality images or even animations for presentations and publications, five widely used programs, Chimera, Jmol, PyMOL, Swiss-PdbViewer (also known as DeepView), and VMD can be mentioned here (Table 10). Among them, Jmol is an open-source Java viewer for chemical structures in 3D. It is particularly useful for integrating figures into HTML pages.
Table 10.
Programs for molecular visualization.
| Name | Developed by | Free for academia | Ref. |
|---|---|---|---|
| Chimera | University of California, San Francisco, USA | Yes | [293] |
| Jmol | University of Notre Dame, USA | Yes | [294] |
| PyMOL | Schrödinger Inc. | No longer free for academia unless older versions are requested or a special request (teaching) is made | [295] |
| Swiss-PdbViewer (DeepView) | Swiss Institute of Bioinformatics | Yes | [296] |
| VMD | University of Illinois, USA | Yes | [297] |
The use of VMD and Chimera is not restricted to molecular visualization. Chimera is a highly extensible program for analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories and conformational ensembles. VMD can be used to animate and analyze the trajectory of an MD simulation. In particular, VMD can act as a graphical front-end for an external MD program by displaying and animating a molecule undergoing simulation on a remote computer.
Some useful web links
Click2Drug [232]: a directory of in silico drug-design tools. Helps find drug-design tools and links to their original web pages;
Protein Data Bank [233]: this archive contains information about experimentally determined structures of proteins, nucleic acids and complex assemblies. Users can perform simple and advanced searches based on annotations relating to sequence, structure and function. The PDB files can be downloaded for drug design, in particular SBVS. It should be noted that not every file deposited in PDB is good for drug design [28];
Ligand Expo [234] (formerly Ligand Depot): a sister site of the PDB and maintained by the same team at the Research Collaboratory for Structural Bioinformatics, provides chemical and structural information about small molecules within the structure entries of the PDB. Tools are provided to search the PDB chemical components dictionary of currently approximately 10,000 unique ligand structures, to identify structure entries containing particular small molecules, and to download the 3D structures of all the ligand instances in PDB entries (currently more than 360,000);
NCI CADD Group Chemoinformatics Tools and User Services [215]: this website provides access to several online databases and chemoinformatics resources, for example, the Enhanced NCI Database Browser, the CSLS and the CIR. The NCI Database Browser is a web service presenting and searching in the majority of Open NCI Database compounds (>250,000 structures). Different kinds of output features and links to other services for continued processing are offered. CSLS is a chemical database indexing service, currently providing access to almost 80 million structure records from more than 100 databases including databases such as the ChemNavigator iRL, PubChem, ChemSpider, ZINC and eMolecules. CIR allows the conversion of a given structure identifier (e.g., SMILES, chemical name, Standard InChI/InChIKey, NCI/CADD Identifier) into another representation or structure identifier. For the lookup of chemical names or hashed identifiers such as Standard InChIKeys, CIR currently connects to a database of approximately 120 million indexed chemical structures;
National Center for Biotechnology Information [235]: houses genome sequencing data in GenBank and an index of biomedical research articles in PubMed Central and PubMed, as well as other information relevant to biotechnology and drug design, such as the PubChem database. All these databases are available online through the Entrez search engine;
Virtual Computational Chemistry Laboratory [236]: numerous scientific programs, including molecular indices/property calculation and data analysis programs are provided on this website. This project’s overall objective is to develop multiplatform software allowing the computational chemist to perform a comprehensive series of molecular properties calculations and data analysis on the internet;
EPA’s SPARC Online Calculator [237]: the initial purpose of this website was to help environmental chemists predict data such as pKa values, hydrolysis, hydration, tautomer, kinetic and heat of formation of environmental chemicals. It is, however, also valuable to medicinal chemists to predict some physicochemical properties of small organic compounds;
Cambridge Crystallographic Data Centre [202]: originating in the Department of Chemistry at the University of Cambridge, UK, the CCDC is now a fully independent institution constituted as a nonprofit company. CCDC supports drug discovery through its industry-standard Cambridge Structural Database, containing more than half a million small-molecule crystal structures, and through knowledge-based tools to support receptor modeling, ligand design, docking, lead optimization and formulation studies;
ChemAxon [238]: provides chemical software-development platforms and desktop applications for the biotechnology and pharmaceutical industries. ChemAxon’s portfolio of software includes a set of chemoinformatics tools (e.g., MarvinSketch, MarvinView, MarvinSpace, MolConverter, JChem for Excel, JChem Base and JChem Cartridge) and a platform for the implementation of chemical communication web services (JChem Web Services). On the basis of these tools, ChemA xon has implemented Chemicalize [239] as a public web resource;
Molecular Networks GmbH [240]: provides a series of software tools for the chemical, biotechnology and pharmaceutical industry. Molecular Networks is well known for the development of the 3D-structure generator CORINA; however, the company’s suite of chemoinformatics applications covers many different areas in the areas of handling of chemical information, design of new chemical entities and prediction of physicochemical and biological properties of chemical compounds. Molecular Networks also has a strong academic background in the development of software for the prediction of chemical reactivity, computer-aided synthesis design and planning of organic reactions, synthesis-driven combinatorial library design, prediction of synthetic accessibility of compounds and prediction of enzyme-mediated chemical transformations.
Future perspective
After three decades of development, CADD has become a valuable component of drug discovery and development. To describe its typical use, at the beginning of a drug-discovery project, chemoinformatics tools are employed to choose compounds from available sources to be assayed. Some marginally active or better compounds may be found, and then chemical similarity searching techniques are used to find more compounds that should be assayed. If some compounds that are more active are discovered, computationally more expensive techniques are applied, such as docking and pharmacophore modeling, to identify more potent compounds or optimize more ADME/T favorable compounds. Techniques of CADD also provide other options for understanding chemical systems, which yield information that is not easy to obtain in laboratory analysis, and, furthermore, is typically (much) less costly than by experiment. After ups and downs of the perception of CADD in the field of drug development, and perhaps some over-hyping of its promises, especially in the initial phases of new trends in development, one can probably say that the discipline of computational medicinal chemistry has begun to mature and become a realistically assessed and routinely used component of modern drug discovery. The breadth of techniques and tools described in this article imply that, to become a successful computational medicinal chemist, it will be highly beneficial to master different kinds of CADD programs and utilize all computational resources that are valuable for drug design. In addition, having skills in one or more programming languages, such as Python, will help smooth routine drug-design work in a contemporary CADD setup.
While it would be desirable, one cannot bank on the fact that a quantum leap in precision of docking or pharmacophore search will occur in the next few years. Nevertheless, SBVS and LBVS are very likely to become routine in drug-discovery projects if they have not already done so. The use of more accurate methods, such as MD and QM, will continue to grow. Currently, sophisticated CADD tools are typically applied by modeling experts, but are increasingly spreading to the desktops of medicinal chemists as well.
Key Term
- High-throughput screening
Technology that allows for rapid testing of large molecular libraries against a particular target of interest in the search for biologically active compounds. If one or more compounds show promising activity, then, typically through several cycles of medicinal chemistry optimization, they are developed into a drug
- Fragment-based drug discovery
Method used for finding small chemical fragments that bind, though often weakly, to a biological target. The obtained fragments, which normally have better binding efficiency per atom than larger hit molecules but overall lower affinity, can be linked or combined to lead compounds with higher affinities
- Scaffold hopping
Identification of compounds with a different scaffold than existing active compounds but with similar or improved activity and other properties, typically based on presenting equivalent functionalities in a similar geometric manner but attached to a different core. Scaffold hopping can be achieved with the help of computational techniques or by traditional medicinal chemistry approaches
- Molecular mechanics
Method to calculate the properties of systems containing from a few atoms to a considerable number of atoms. The basis of molecular mechanics is the paradigm of classical physics, specifically Newton’s laws of motion, applied only to the nucleus without considering the electrons as individual components. The energy is a function of structural features such as angle bending, bond stretching, bond rotation (torsion), and non-bonding interactions. The set of these potential energy functions is the ‘force field’. Specific chemistries (atom types) are typically parameterized by large ‘parameter sets’, which are what truly defines the quantitative results obtained in molecular mechanics calculations
- QM/MM method
Combined QM and MM computational approach as a strategy to overcome the shortcomings of MM in MD simulations. The goals of using QM/MM are to improve the accuracy in specific parts of the system, such as when calculating the binding affinities between ligands and their targets, as well as to allow one to treat processes that are not usually within the scope of MM methods, such as bond breaking and formation. It combines the strength of both QM (accuracy) and MM (speed). Normally, a small portion of the macromolecular system (for example, the ligand or the ligand plus its interface with the protein) is treated by QM, while the remainder of the system is treated by MM
Footnotes
Financial & competing interests disclosure
The authors have no potential conflicts with the subject matter or materials discussed in this manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact reprints@future-science.com
Bibliography
- 1.Talele TT, Khedkar SA, Rigby AC. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr Top Med Chem. 2010;10(1):127–141. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- 2.Clark DE. What has computer-aided molecular design ever done for drug discovery? Expert Opin Drug Discov. 2006;1(2):103–110. doi: 10.1517/17460441.1.2.103. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira RS, Simeonov A, Jadhav A, et al. Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors. J Med Chem. 2010;53(13):4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ripphausen P, Nisius B, Peltason L, Bajorath J. Quo Vadis, virtual screening? A comprehensive survey of prospective applications. J Med Chem. 2010;53(24):8461–8467. doi: 10.1021/jm101020z. [DOI] [PubMed] [Google Scholar]
- 5.Peach ML, Nicklaus MC. Combining docking with pharmacophore filtering for improved virtual screening. J Cheminform. 2009;1(1):6. doi: 10.1186/1758-2946-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao C, Nicklaus MC. Computer tools in the discovery of HIV-1 integrase inhibitors. Future Med Chem. 2010;2(7):1123–1140. doi: 10.4155/fmc.10.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 8.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 9.Warren GL, Andrews CW, Capelli AM, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 10.Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast and highly accurate docking/ scoring methods: a long way to go. Br J Pharmacol. 2008;153(Suppl 1):S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer RT. Structure-based drug design: docking and scoring. Curr Protein Pept Sci. 2007;8(4):312–328. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- 12.Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 13.Lang PT, Brozell SR, Mukherjee S, et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15(6):1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein–ligand docking. Proteins. 1999;37(2):228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Mcgann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers. 2003;68(1):76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 16.Friesner RA, Banks JL, Murphy RB, et al. Glide: a new approach for rapid, accurate docking and scoring. 1 Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 17.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins. 2003;52(4):609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 18.Totrov M, Abagyan R. Flexible protein–ligand docking by global energy optimization in internal coordinates. Proteins Suppl. 1997;1:215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 19.Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des. 2007;21(5):281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 20.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., 3rd Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des. 2003;17(11):755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 21.Onodera K, Satou K, Hirota H. Evaluations of molecular docking programs for virtual screening. J Chem Inf Model. 2007;47(4):1609–1618. doi: 10.1021/ci7000378. [DOI] [PubMed] [Google Scholar]
- 22.Cross JB, Thompson DC, Rai BK, et al. Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J Chem Inf Model. 2009;49(6):1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Felts AK, Friesner RA, Levy RM. Comparative performance of several flexible docking programs and scoring functions: enrichment studies for a diverse set of pharmaceutically relevant targets. J Chem Inf Model. 2007;47(4):1599–1608. doi: 10.1021/ci7000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Li Y, Cheng T, Liu Z, Wang R. Evaluation of the performance of four molecular docking programs on a diverse set of protein–ligand complexes. J Comput Chem. 2010;31(11):2109–2125. doi: 10.1002/jcc.21498. [DOI] [PubMed] [Google Scholar]
- 25.Plewczynski D, Lazniewski M, Augustyniak R, Ginalski K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J Comput Chem. 2011;32(4):742–755. doi: 10.1002/jcc.21643. [DOI] [PubMed] [Google Scholar]
- 26.Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model. 2009;49(4):1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 27.Wang R, Lu Y, Fang X, Wang S. An extensive test of 14 scoring functions using the PDBbind refined set of 800 protein–ligand complexes. J Chem Inf Comput Sci. 2004;44(6):2114–2125. doi: 10.1021/ci049733j. [DOI] [PubMed] [Google Scholar]
- 28.Davis AM, St-Gallay SA, Kleywegt GJ. Limitations and lessons in the use of X-ray structural information in drug design. Drug Discov Today. 2008;13(19–20):831–841. doi: 10.1016/j.drudis.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basse N, Montes M, Marechal X, et al. Novel organic proteasome inhibitors identified by virtual and in vitro screening. J Med Chem. 2010;53(1):509–513. doi: 10.1021/jm9011092. [DOI] [PubMed] [Google Scholar]
- 30.Zhong S, Zhang Y, Xiu Z. Rescoring ligand docking poses. Curr Opin Drug Discov Devel. 2010;13(3):326–334. [PubMed] [Google Scholar]
- 31.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51(13):3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 32.Murray CW, Carr MG, Callaghan O, et al. Fragment-based drug discovery applied to Hsp90. Discovery of two lead series with high ligand efficiency. J Med Chem. 2010;53(16):5942–5955. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- 33.Vangrevelinghe E, Rudisser S. Computational approaches for fragment optimization. Curr Comput Aided Drug Des. 2007;3(1):69–83. [Google Scholar]
- 34.Cozzini P, Kellogg GE, Spyrakis F, et al. Target flexibility: an emerging consideration in drug discovery and design. J Med Chem. 2008;51(20):6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao C, Park JE, Bang JK, Nicklaus MC, Lee KS. Probing binding modes of small molecule inhibitors to the Polo-box domain of human Polo-like kinase 1. ACS Med Chem Lett. 2010;1(3):110–114. doi: 10.1021/ml100020e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barril X, Morley SD. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. J Med Chem. 2005;48(13):4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 37.Damm KL, Carlson HA. Exploring experimental sources of multiple protein conformations in structure-based drug design. J Am Chem Soc. 2007;129(26):8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- 38.Huang SY, Zou X. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins. 2007;66(2):399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 39.Ekonomiuk D, Su XC, Ozawa K, et al. Flaviviral protease inhibitors identified by fragment-based library docking into a structure generated by molecular dynamics. J Med Chem. 2009;52(15):4860–4868. doi: 10.1021/jm900448m. [DOI] [PubMed] [Google Scholar]
- 40.Amaro RE, Baron R, Mccammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des. 2008;22(9):693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wermuth G, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998) Pure Appl Chem. 1998;70(5):1129–1143. [Google Scholar]
- 42.Leach AR, Gillet VJ, Lewis RA, Taylor R. Three-dimensional pharmacophore methods in drug discovery. J Med Chem. 2010;53(2):539–558. doi: 10.1021/jm900817u. [DOI] [PubMed] [Google Scholar]
- 43.Yang SY. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today. 2010;15(11–12):444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 44.Gao Q, Yang L, Zhu Y. Pharmacophore based drug design approach as a practical process in drug discovery. Curr Comput Aided Drug Des. 2010;6(1):37–49. doi: 10.2174/157340910790980151. [DOI] [PubMed] [Google Scholar]
- 45.Wolber G, Seidel T, Bendix F, Langer T. Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov Today. 2008;13(1–2):23–29. doi: 10.1016/j.drudis.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Sun H. Pharmacophore-based virtual screening. Curr Med Chem. 2008;15(10):1018–1024. doi: 10.2174/092986708784049630. [DOI] [PubMed] [Google Scholar]
- 47.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 48.Wolber G, Dornhofer A, Langer T. Efficient overlay of small organic molecules using 3D pharmacophores. J Comp Aided Mol Des. 2006;20(12):773–788. doi: 10.1007/s10822-006-9078-7. [DOI] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36 (Web Server issue):W223–W228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhaery SS, Roy KK, Shakya N, et al. Novel carbamates as orally active acetylcholinesterase inhibitors found to improve scopolamine-induced cognition impairment: pharmacophore-based virtual screening, synthesis, and pharmacology. J Med Chem. 2010;53(17):6490–6505. doi: 10.1021/jm100573q. [DOI] [PubMed] [Google Scholar]
- 51.Zampieri D, Mamolo MG, Laurini E, et al. Synthesis, biological evaluation, and three-dimensional in silico pharmacophore model for sigma(1) receptor ligands based on a series of substituted benzo[d]oxazol-2(3H)-one derivatives. J Med Chem. 2009;52(17):5380–5393. doi: 10.1021/jm900366z. [DOI] [PubMed] [Google Scholar]
- 52.Onnis V, Kinsella GK, Carta G, et al. Virtual screening for the identification of novel nonsteroidal glucocorticoid modulators. J Med Chem. 2010;53(8):3065–3074. doi: 10.1021/jm901452y. [DOI] [PubMed] [Google Scholar]
- 53.Markt P, Feldmann C, Rollinger JM, et al. Discovery of novel CB2 receptor ligands by a pharmacophore-based virtual screening workflow. J Med Chem. 2009;52(2):369–378. doi: 10.1021/jm801044g. [DOI] [PubMed] [Google Scholar]
- 54.Neves MA, Dinis TC, Colombo G, Sá e Melo ML. Fast three dimensional pharmacophore virtual screening of new potent non-steroid aromatase inhibitors. J Med Chem. 2009;52(1):143–150. doi: 10.1021/jm800945c. [DOI] [PubMed] [Google Scholar]
- 55.Ismail MA, Barker S, Abou El-Ella DA, Abouzid KA, Toubar RA, Todd MH. Design and synthesis of new tetrazolyl- and carboxy-biphenylylmethyl-quinazolin-4-one derivatives as angiotensin II AT1 receptor antagonists. J Med Chem. 2006;49(5):1526–1535. doi: 10.1021/jm050232e. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, Duffy RA, Boykow GC, Chackalamannil S, Madison VS. Identification of novel cannabinoid CB1 receptor antagonists by using virtual screening with a pharmacophore model. J Med Chem. 2008;51(8):2439–2446. doi: 10.1021/jm701519h. [DOI] [PubMed] [Google Scholar]
- 57.Sprous DG, Palmer RK, Swanson JT, Lawless M. QSAR in the pharmaceutical research setting: QSAR models for broad, large problems. Curr Top Med Chem. 2010;10(6):619–637. doi: 10.2174/156802610791111506. [DOI] [PubMed] [Google Scholar]
- 58.Hong H, Xie Q, Ge W, et al. Mold2, molecular descriptors from 2D structures for chemoinformatics and toxicoinformatics. J Chem Inf Model. 2008;48(7):1337–1344. doi: 10.1021/ci800038f. [DOI] [PubMed] [Google Scholar]
- 59.Clark RD. Prospective ligand- and target-based 3D QSAR: state of the art 2008. Curr Top Med Chem. 2009;9(9):791–810. doi: 10.2174/156802609789207118. [DOI] [PubMed] [Google Scholar]
- 60.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1 Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc. 2002;110(18):5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 61.Klebe G, Abraham U, Mietzner T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem. 1994;37(24):4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 62.Salama I, Hocke C, Utz W, et al. Structure-selectivity investigations of D2-like receptor ligands by CoMFA and CoMSIA guiding the discovery of D3 selective PET radioligands. J Med Chem. 2007;50(3):489–500. doi: 10.1021/jm0611152. [DOI] [PubMed] [Google Scholar]
- 63.Sheng C, Zhang W, Ji H, et al. Structure-based optimization of azole antifungal agents by CoMFA, CoMSIA, and molecular docking. J Med Chem. 2006;49(8):2512–2525. doi: 10.1021/jm051211n. [DOI] [PubMed] [Google Scholar]
- 64.Patil R, Das S, Stanley A, Yadav L, Sudhakar A, Varma AK. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. Plos One. 2010;5(8):e12029. doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Zhao M, Cui G, Zhang X, Wang J, Peng S. Methyl (11aS)-1,2,3,5,11,11a-hexahydro-3,3-dimethyl-1-oxo-6H-imidazo-[3′,4′:1,2]p yridin[3,4-b]indol-2-substituted acetates: synthesis and three-dimensional quantitative structure–activity relationship investigation as a class of novel vasodilators. J Med Chem. 2008;51(15):4715–4723. doi: 10.1021/jm800249j. [DOI] [PubMed] [Google Scholar]
- 66.Tropsha A, Golbraikh A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr Pharm Des. 2007;13(34):3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- 67.Tang H, Wang XS, Huang XP, et al. Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J Chem Inf Model. 2009;49(2):461–476. doi: 10.1021/ci800366f. [DOI] [PubMed] [Google Scholar]
- 68.Peterson YK, Wang XS, Casey PJ, Tropsha A. Discovery of geranylgeranyltransferase-I inhibitors with novel scaffolds by the means of quantitative structure–activity relationship modeling, virtual screening, and experimental validation. J Med Chem. 2009;52(14):4210–4220. doi: 10.1021/jm8013772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cavasotto CN, Phatak SS. Homology modeling in drug discovery: current trends and applications. Drug Discov Today. 2009;14(13–14):676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 70.Kairys V, Gilson MK, Fernandes MX. Using protein homology models for structure-based studies: approaches to model refinement. ScientificWorldJournal. 2006;6:1542–1554. doi: 10.1100/tsw.2006.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kopp J, Schwede T. Automated protein structure homology modeling: a progress report. Pharmacogenomics. 2004;5(4):405–416. doi: 10.1517/14622416.5.4.405. [DOI] [PubMed] [Google Scholar]
- 72.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 73.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 74.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23(3):ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 75.Mobarec JC, Sanchez R, Filizola M. Modern homology modeling of G-protein coupled receptors: which structural template to use? J Med Chem. 2009;52(16):5207–5216. doi: 10.1021/jm9005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sitzmann M, Ihlenfeldt WD, Nicklaus MC. Tautomerism in large databases. J Comput Aided Mol Des. 2010;24(6–7):521–551. doi: 10.1007/s10822-010-9346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ihlenfeldt WD, Voigt JH, Bienfait B, Oellien F, Nicklaus MC. Enhanced CACTVS browser of the Open NCI Database. J Chem Inf Comput Sci. 2002;42(1):46–57. doi: 10.1021/ci010056s. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y, Bolton E, Dracheva S, et al. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38 (Database issue):D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35 (Database issue):D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hendlich M, Bergner A, Gunther J, Klebe G. Relibase: design and development of a database for comprehensive analysis of protein–ligand interactions. J Mol Biol. 2003;326(2):607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 81.Overington J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI) Interview by Wendy A Warr. J Comput Aided Mol Des. 2009;23(4):195–198. doi: 10.1007/s10822-009-9260-9. [DOI] [PubMed] [Google Scholar]
- 82.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37 (Database issue):D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32 (Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Matos P, Alcantara R, Dekker A, et al. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38(Database issue):D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30(1):412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent developments of the chemistry development kit (CDK) – an open-source java library for chemo- and bioinformatics. Curr Pharm Des. 2006;12(17):2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 89.Ihlenfeldt WD, Takahashi Y, Abe H, Sasaki S. Computation and management of chemical-properties in Cactvs – an extensible networked approach toward modularity and compatibility. J Chem Inf Comp Sci. 1994;34(1):109–116. [Google Scholar]
- 90.Filippov IV, Nicklaus MC. Optical structure recognition software to recover chemical information: OSRA, an open source solution. J Chem Inf Model. 2009;49(3):740–743. doi: 10.1021/ci800067r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schleif R. Modeling and studying proteins with molecular dynamics. Methods Enzymol. 2004;383:28–47. doi: 10.1016/S0076-6879(04)83002-7. [DOI] [PubMed] [Google Scholar]
- 92.Karplus M. Molecular dynamics simulations of biomolecules. Acc Chem Res. 2002;35(6):321–323. doi: 10.1021/ar020082r. [DOI] [PubMed] [Google Scholar]
- 93.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19(2):120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 94.Case DA, Cheatham TE, 3rd, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm – a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 96.Shaw DE. A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J Comput Chem. 2005;26(13):1318–1328. doi: 10.1002/jcc.20267. [DOI] [PubMed] [Google Scholar]
- 97.Christen M, Hunenberger PH, Bakowies D, et al. The GROMOS software for biomolecular simulation: GROMOS05. J Comput Chem. 2005;26(16):1719–1751. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 98.Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alonso H, Bliznyuk AA, Gready JE. Combining docking and molecular dynamic simulations in drug design. Med Res Rev. 2006;26(5):531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 100.Peters MB, Raha K, Merz KM., Jr Quantum mechanics in structure-based drug design. Curr Opin Drug Discov Devel. 2006;9(3):370–379. [PubMed] [Google Scholar]
- 101.Raha K, Peters MB, Wang B, et al. The role of quantum mechanics in structure-based drug design. Drug Discov Today. 2007;12(17–18):725–731. doi: 10.1016/j.drudis.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 102.Cavalli A, Carloni P, Recanatini M. Target-related applications of first principles quantum chemical methods in drug design. Chem Rev. 2006;106(9):3497–3519. doi: 10.1021/cr050579p. [DOI] [PubMed] [Google Scholar]
- 103.Liao C, Nicklaus MC. Tautomerism and magnesium chelation of HIV-1 integrase inhibitors: a theoretical study. ChemMedChem. 2010;5(7):1053–1066. doi: 10.1002/cmdc.201000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26(9):915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou T, Huang D, Caflisch A. Is quantum mechanics necessary for predicting binding free energy? J Med Chem. 2008;51(14):4280–4288. doi: 10.1021/jm800242q. [DOI] [PubMed] [Google Scholar]
- 106.Wang J, Hou T, Ralph AW. Chapter 5. Recent advances on in silico ADME modeling. Annu Rep Comput Chem. 2009;5:101–127. [Google Scholar]
- 107.Gleeson MP, Hersey A, Hannongbua S. In-silico ADME models: a general assessment of their utility in drug discovery applications. Curr Top Med Chem. 2011;11(4):358–381. doi: 10.2174/156802611794480927. [DOI] [PubMed] [Google Scholar]
- 108.Egan WJ. Chapter 29.Computational models for ADME. Annu Rep Med Chem. 2007;42:449–467. [Google Scholar]
- 109.Cosconati S, Marinelli L, La Motta C, et al. Pursuing aldose reductase inhibitors through in situ cross-docking and similarity-based virtual screening. J Med Chem. 2009;52(18):5578–5581. doi: 10.1021/jm901045w. [DOI] [PubMed] [Google Scholar]
- 110.Ferri N, Corsini A, Bottino P, Clerici F, Contini A. Virtual screening approach for the identification of new Rac1 inhibitors. J Med Chem. 2009;52(14):4087–4090. doi: 10.1021/jm8015987. [DOI] [PubMed] [Google Scholar]
- 111.Perez-Pineiro R, Burgos A, Jones DC, et al. Development of a novel virtual screening cascade protocol to identify potential trypanothione reductase inhibitors. J Med Chem. 2009;52(6):1670–1680. doi: 10.1021/jm801306g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matsuno K, Masuda Y, Uehara Y, et al. Identification of a new series of STAT3 inhibitors by virtual screening. Acs Med Chem Lett. 2010;1(8):371–375. doi: 10.1021/ml1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Okamoto M, Takayama K, Shimizu T, Ishida K, Takahashi O, Furuya T. Identification of death-associated protein kinases inhibitors using structure-based virtual screening. J Med Chem. 2009;52(22):7323–7327. doi: 10.1021/jm901191q. [DOI] [PubMed] [Google Scholar]
- 114.Ostrov DA, Magis AT, Wronski TJ, et al. Identification of enoxacin as an inhibitor of osteoclast formation and bone resorption by structure-based virtual screening. J Med Chem. 2009;52(16):5144–5151. doi: 10.1021/jm900277z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miguet L, Zervosen A, Gerards T, et al. Discovery of new inhibitors of resistant Streptococcus pneumoniae penicillin binding protein (PBP) 2x by structure-based virtual screening. J Med Chem. 2009;52(19):5926–5936. doi: 10.1021/jm900625q. [DOI] [PubMed] [Google Scholar]
- 116.Cho Y, Ioerger TR, Sacchettini JC. Discovery of novel nitrobenzothiazole inhibitors for Mycobacterium tuberculosis ATP phosphoribosyl transferase (HisG) through virtual screening. J Med Chem. 2008;51(19):5984–5992. doi: 10.1021/jm800328v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kiss R, Kiss B, Konczol A, et al. Discovery of novel human histamine H4 receptor ligands by large-scale structure-based virtual screening. J Med Chem. 2008;51(11):3145–3153. doi: 10.1021/jm7014777. [DOI] [PubMed] [Google Scholar]
- 118.Knox AJ, Price T, Pawlak M, et al. Integration of ligand and structure-based virtual screening for the identification of the first dual targeting agent for heat shock protein 90 (Hsp90) and tubulin. J Med Chem. 2009;52(8):2177–2180. doi: 10.1021/jm801569z. [DOI] [PubMed] [Google Scholar]
- 119.Podvinec M, Lim SP, Schmidt T, et al. Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J Med Chem. 2010;53(4):1483–1495. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]
- 120.Ravindranathan KP, Mandiyan V, Ekkati AR, Bae JH, Schlessinger J, Jorgensen WL. Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening. J Med Chem. 2010;53(4):1662–1672. doi: 10.1021/jm901386e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liao C, Karki RG, Marchand C, Pommier Y, Nicklaus MC. Virtual screening application of a model of full-length HIV-1 integrase complexed with viral DNA. Bioorg Med Chem Lett. 2007;17(19):5361–5365. doi: 10.1016/j.bmcl.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 122.Dong G, Sheng C, Wang S, Miao Z, Yao J, Zhang W. Selection of evodiamine as a novel topoisomerase I inhibitor by structure-based virtual screening and hit optimization of evodiamine derivatives as antitumor agents. J Med Chem. 2010;53(21):7521–7531. doi: 10.1021/jm100387d. [DOI] [PubMed] [Google Scholar]
- 123.Oyarzabal J, Zarich N, Albarran MI, et al. Discovery of mitogen-activated protein kinase-interacting kinase 1 inhibitors by a comprehensive fragment-oriented virtual screening approach. J Med Chem. 2010;53(18):6618–6628. doi: 10.1021/jm1005513. [DOI] [PubMed] [Google Scholar]
- 124.Peach ML, Tan N, Choyke SJ, et al. Directed discovery of agents targeting the Met tyrosine kinase domain by virtual screening. J Med Chem. 2009;52(4):943–951. doi: 10.1021/jm800791f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chan DS, Lee HM, Yang F, et al. Structure-based discovery of natural-product-like TNF-α inhibitors. Angew Chem Int Ed Engl. 2010;49(16):2860–2864. doi: 10.1002/anie.200907360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bisson WH, Koch DC, O’Donnell EF, et al. Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J Med Chem. 2009;52(18):5635–5641. doi: 10.1021/jm900199u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Odell LR, Howan D, Gordon CP, et al. The pthaladyns: GTP competitive inhibitors of dynamin I and II GTPase derived from virtual screening. J Med Chem. 2010;53(14):5267–5280. doi: 10.1021/jm100442u. [DOI] [PubMed] [Google Scholar]
- 128.Khanfar MA, Hill RA, Kaddoumi A, El Sayed KA. Discovery of novel GSK-3β inhibitors with potent in vitro and in vivo activities and excellent brain permeability using combined ligand- and structure-based virtual screening. J Med Chem. 2010;53(24):8534–8545. doi: 10.1021/jm100941j. [DOI] [PubMed] [Google Scholar]
- 129.Herschhorn A, Hizi A. Virtual screening, identification, and biochemical characterization of novel inhibitors of the reverse transcriptase of human immunodeficiency virus type-1. J Med Chem. 2008;51(18):5702–5713. doi: 10.1021/jm800473d. [DOI] [PubMed] [Google Scholar]
- 130.Chiang YK, Kuo CC, Wu YS, et al. Generation of ligand-based pharmacophore model and virtual screening for identification of novel tubulin inhibitors with potent anticancer activity. J Med Chem. 2009;52(14):4221–4233. doi: 10.1021/jm801649y. [DOI] [PubMed] [Google Scholar]
- 131.Wu JS, Peng YH, Wu JM, et al. Discovery of non-glycoside sodium-dependent glucose co-transporter 2 (SGLT2) inhibitors by ligand-based virtual screening. J Med Chem. 2010;53(24):8770–8774. doi: 10.1021/jm101080v. [DOI] [PubMed] [Google Scholar]
- 132.Georgsson J, Skold C, Plouffe B, et al. Angiotensin II pseudopeptides containing 1,3,5-trisubstituted benzene scaffolds with high AT2 receptor affinity. J Med Chem. 2005;48(21):6620–6631. doi: 10.1021/jm050280z. [DOI] [PubMed] [Google Scholar]
- 133.Yang H, Shen Y, Chen J, Jiang Q, Leng Y, Shen J. Structure-based virtual screening for identification of novel 11β-HSD1 inhibitors. Eur J Med Chem. 2009;44(3):1167–1171. doi: 10.1016/j.ejmech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 134.Olla S, Manetti F, Crespan E, et al. Indolyl-pyrrolone as a new scaffold for Pim1 inhibitors. Bioorg Med Chem Lett. 2009;19(5):1512–1516. doi: 10.1016/j.bmcl.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 135.Barreca ML, De Luca L, Iraci N, et al. Structure-based pharmacophore identification of new chemical scaffolds as non-nucleoside reverse transcriptase inhibitors. J Chem Inf Model. 2007;47(2):557–562. doi: 10.1021/ci600320q. [DOI] [PubMed] [Google Scholar]
- 136.Abdel-Aal WS, Hassan HY, Aboul-Fadl T, Youssef AF. Pharmacophoric model building for antitubercular activity of the individual Schiff bases of small combinatorial library. Eur J Med Chem. 2010;45(3):1098–1106. doi: 10.1016/j.ejmech.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 137.Lokhande TN, Viswanathan CL, Joshi A, Juvekar A. Design, synthesis and evaluation of naphthalene-2-carboxamides as reversal agents in MDR cancer. Bioorg Med Chem. 2006;14(17):6022–6026. doi: 10.1016/j.bmc.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 138.Joshi AA, Narkhede SS, Viswanathan CL. Design, synthesis and evaluation of 5-substituted amino-2,4-diamino-8-chloropyrimido-[4,5-b]quinolines as novel antimalarials. Bioorg Med Chem Lett. 2005;15(1):73–76. doi: 10.1016/j.bmcl.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 139.Hall MD, Salam NK, Hellawell JL, et al. Synthesis, activity, and pharmacophore development for isatin-β-thiosemicarbazones with selective activity toward multidrug-resistant cells. J Med Chem. 2009;52(10):3191–3204. doi: 10.1021/jm800861c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kumar RJ, Chebib M, Hibbs DE, et al. Novel γ-aminobutyric acid rho1 receptor antagonists; synthesis, pharmacological activity and structure–activity relationships. J Med Chem. 2008;51(13):3825–3840. doi: 10.1021/jm7015842. [DOI] [PubMed] [Google Scholar]
- 141.Cavasotto CN, Orry AJ, Murgolo NJ, et al. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J Med Chem. 2008;51(3):581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 142.Park H, Bahn YJ, Jung SK, et al. Discovery of novel Cdc25 phosphatase inhibitors with micromolar activity based on the structure-based virtual screening. J Med Chem. 2008;51(18):5533–5541. doi: 10.1021/jm701157g. [DOI] [PubMed] [Google Scholar]
- 143.Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the β2-adrenergic receptor. J Med Chem. 2008;51(10):2907–2914. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamada S, Suzuki T, Mino K, et al. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53(15):5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- 145.Buchholz M, Hamann A, Aust S, et al. Inhibitors for human glutaminyl cyclase by structure based design and bioisosteric replacement. J Med Chem. 2009;52(22):7069–7080. doi: 10.1021/jm900969p. [DOI] [PubMed] [Google Scholar]
- 146.Chen X, Wilson LJ, Malaviya R, Argentieri RL, Yang SM. Virtual screening to successfully identify novel janus kinase 3 inhibitors: a sequential focused screening approach. J Med Chem. 2008;51(21):7015–7019. doi: 10.1021/jm800662z. [DOI] [PubMed] [Google Scholar]
- 147.Nowak P, Cole DC, Brooijmans N, et al. Discovery of potent and selective inhibitors of the mammalian target of rapamycin (mTOR) kinase. J Med Chem. 2009;52(22):7081–7089. doi: 10.1021/jm9012642. [DOI] [PubMed] [Google Scholar]
- 148.Cavalli A, Bottegoni G, Raco C, De Vivo M, Recanatini M. A computational study of the binding of propidium to the peripheral anionic site of human acetylcholinesterase. J Med Chem. 2004;47(16):3991–3999. doi: 10.1021/jm040787u. [DOI] [PubMed] [Google Scholar]
- 149.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc. 2001;123(22):5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 150.Zoete V, Meuwly M, Karplus M. Investigation of glucose binding sites on insulin. Proteins. 2004;55(3):568–581. doi: 10.1002/prot.20071. [DOI] [PubMed] [Google Scholar]
- 151.Cheng LS, Amaro RE, Xu D, Li WW, Arzberger PW, Mccammon JA. Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J Med Chem. 2008;51(13):3878–3894. doi: 10.1021/jm8001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.Molecular Discovery. www.moldiscovery.com.
- 202.Cambridge Crystallographic Data Centre. www.ccdc.cam.ac.uk.
- 203.SimBioSys Inc. www.simbiosys.ca.
- 204.MEDIT SA. http://medit-pharma.com.
- 205.PharmaGist. http://bioinfo3d.cs.tau.ac.il/PharmaGist.
- 206.BioEpisteme. www.prousresearch.com/spage/technology/testpage/pageid-79/epage/BioEpisteme.aspx.
- 207.Semichem. Codessa. www.semichem.com/codessa/default.php.
- 208.Prediction of activity spectra for substances. http://195.178.207.233/PASS2008/en/index.html.
- 209.Talete. Dragon software. www.talete.mi.it.
- 210.University of Virginia. FASTA sequence comparison. http://fasta.bioch.virginia.edu.
- 211.The National Center for Biotechnology Information. Basic Local Alignment Search Tool. http://blast.ncbi.nlm.nih.gov.
- 212.European Bioinformatics Institute. PSI-BLAST. www.ebi.ac.uk/Tools/sss/psiblast.
- 213.Fold and Function Assignment. http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl.
- 214.Protein Structure Prediction Center. http://predictioncenter.org.
- 215.NCI/CADD Group. http://cactus.nci.nih.gov.
- 216.PubChem. http://pubchem.ncbi.nlm.nih.gov.
- 217.MetaCyc. http://metacyc.org.
- 218.Swiss-Prot. http://ca.expasy.org/sprot.
- 219.GenBank. www.ncbi.nlm.nih.gov/genbank.
- 220.NCI/CADD Chemical Identifier Resolver. http://cactus.nci.nih.gov/chemical/structure.
- 221.University of Konstanz. Institute for Bioinformatics and Information Mining. www.knime.org.
- 222.Weka. www.cs.waikato.ac.nz/ml/weka.
- 223.Statistics package R. www.r-project.org.
- 224.Xemistry GmbH. http://xemistry.com.
- 225.ChemIDplus. http://chem.sis.nlm.nih.gov/chemidplus.
- 226.ChEBI. www.ebi.ac.uk/chebi.
- 227.Open Babel. http://openbabel.org.
- 228.Avogadro. http://avogadro.openmolecules.net.
- 229.MySQL database extension. MyChem; http://mychem.sourceforge.net. [Google Scholar]
- 230.Wikipedia. MD simulation program list. http://en.wikipedia.org/wiki/Molecular_dynamics#Major_software_for_MD_simulations.
- 231.Wikipedia. QM program list. http://en.wikipedia.org/wiki/List_of_quantum_chemistry_and_solid_state_physics_software.
- 232.Click2Drug. www.click2drug.org.
- 233.Protein Data Bank. www.pdb.org.
- 234.Ligand Expo. http://ligand-expo.rcsb.org.
- 235.National Center for Biotechnology Information. www.ncbi.nlm.nih.gov.
- 236.Virtual Computational Chemistry Laboratory. www.vcclab.org.
- 237.EPA’s SPARC Online Calculator. http://ibmlc2.chem.uga.edu/sparc.
- 238.ChemAxon. www.chemaxon.com.
- 239.Chemicalize. www.chemicalize.org.
- 240.Molecular Networks GmbH. www.molecular-networks.com.
- 241.Accelrys Inc. http://accelrys.com.
- 242.Molsoft LLC. www.molsoft.com.
- 243.BioSolveIT GmbH. www.biosolveit.de.
- 244.Chemical Computing Group. www.chemcomp.com.
- 245.OpenEye Scientific Software Inc. www.eyesopen.com.
- 246.Schrödinger Inc. www.schrodinger.com.
- 247.Tripos Inc. www.tripos.com.
- 248.Scripps Research Institute. http://autodock.scripps.edu.
- 249.University of California, San Francisco. DOCK. http://dock.compbio.ucsf.edu.
- 250.Inte:Ligand. Ligand Scout. www.inteligand.com.
- 251.University of California, San Francisco. Modeller. www.salilab.org/modeller.
- 252.Swiss Institute of Bioinformatics. MODEL. http://swissmodel.expasy.org.
- 253.Enhanced NCI database browser. http://cactus.nci.nih.gov/ncidb2.
- 254.NCI discovery services. http://dtp.nci.nih.gov/webdata.html.
- 255.University of Maryland. BindingDB. www.bindingdb.org.
- 256.Cambridge Crystallographic Data Centre. Relibase. www.ccdc.cam.ac.uk/free_services/relibase_free.
- 257.European Bioinformatics Institute. ChEMBLdb. www.ebi.ac.uk/chembl.
- 258.Royal Society of Chemistry. ChemSpider. www.chemspider.com.
- 259.University of Alberta. Human Metabolome Database. www.hmdb.ca.
- 260.University of Alberta. DrugBank. www.drugbank.ca.
- 261.National University of Singapore. Therapeutic Target Database. http://xin.cz3.nus.edu.sg/group/ttd/ttd.asp/
- 262.University of California, San Francisco. ZINC. http://zinc.docking.org.
- 263.ChemNavigator. iResearch Library. www.chemnavigator.com.
- 264.GVK Biosciences Private Limited. GVKBIO databases. www.gvkbio.com.
- 265.Accelrys Inc. MDDR. http://accelrys.com/products/databases/bioactivity/mddr.html.
- 266.Sunset Molecular Discovery. Wombat. www.sunsetmolecular.com.
- 267.Thomson Reuters World Drug Index. http://thomsonreuters.com/products_services/science/science_products/a-z/world_drug_index/
- 268.University of California, San Francisco, USA. Amber. http://ambermd.org.
- 269.Harvard University. CHARMM. www.charmm.org.
- 270.DE Shaw Research. www.deshawresearch.com.
- 271.University of Groningen. GROMACS. www.gromacs.org.
- 272.University of Illinois. NAMD. www.ks.uiuc.edu/Research/namd.
- 273.Iowa State University. Gamess. www.msg.chem.iastate.edu/gamess.
- 274.Gaussian Inc. Gaussian. www.gaussian.com.
- 275.University of Kuopio. Ghemical. www.uku.fi/~thassine/projects/ghemical.
- 276.Stewart Computational Chemistry. MOPAC. http://openmopac.net.
- 277.Environmental Molecular Sciences Laboratory. www.emsl.pnl.gov.
- 278.Wavefunction, Inc. www.wavefun.com.
- 279.Simulations Plus, Inc. www.simulations-plus.com.
- 280.Optibrium, Ltd. StarDrop. www.optibrium.com.
- 281.Advanced Chemistry Development, Inc. www.acdlabs.com.
- 282.Fujitsu FQS. www.fqs.pl.
- 283.Strand Life Sciences. www.strandls.com.
- 284.Leadscope, Inc. www.leadscope.com.
- 285.Lhasa, Ltd. www.lhasalimited.org.
- 286.Russian Academy of Medical Sciences. PASS. http://pharmaexpert.ru/passonline.
- 287.CompuDrug, Ltd. www.compudrug.com.
- 288.Multicase, Inc. www.multicase.com.
- 289.Molecular Discovery, Ltd. www.moldiscovery.com.
- 290.Uppsala University. Sweden and European Bioinformatics Institute Bioclipse. www.bioclipse.net.
- 291.GeneGo, Inc. www.genego.com.
- 292.OASIS Lmc. http://oasis-lmc.org.
- 293.University of California, San Francisco. Chimera. www.cgl.ucsf.edu/chimera.
- 294.University of Notre Dame. Jmol. www.jmol.org.
- 295.Schrödinger Inc. PyMOL. www.pymol.org.
- 296.Swiss Institute of Bioinformatics. Swiss-PdbViewer. http://spdbv.vital-it.ch.
- 297.University of Illinois VMD. www.ks.uiuc.edu/Research/vmd.