Abstract
Breast cancers are thought to be organized hierarchically with a small number of breast cancer stem cells (BCSCs) able to re-grow a tumor while their progeny lack this ability. Recently, several groups reported enrichment for BCSCs when breast cancers were subjected to classical anticancer treatment. However, the underlying mechanisms leading to this enrichment are incompletely understood. Using non-BCSCs sorted from patient samples, we found that ionizing radiation reprogrammed differentiated breast cancer cells into induced BCSCs (iBCSCs). iBCSCs showed increased mammosphere formation, increased tumorigenicity and expressed the same stemness-related genes as BCSCs from non-irradiated samples. Reprogramming occurred in a polyploid subpopulation of cells, coincided with re-expression of the transcription factors Oct4, Sox-2, Nanog, and Klf4, and could be partially prevented by Notch inhibition.
We conclude that radiation may induce a BCSC phenotype in differentiated breast cancer cells and that this mechanism contributes to increased BCSC numbers seen after classical anti-cancer treatment.
Keywords: Breast cancer stem cells, radiation, Notch, proteasome, dedifferentiation
Introduction
Recent clinical and preclinical data support the view that many solid cancers, including breast cancers, are organized hierarchically with a small number of cancer stem cells (CSCs) able to re-grow a tumor while their progeny lack this feature 1, 2. Clinically, CSCs have been associated with higher rates of recurrence and metastasis 3, 4. Importantly, CSCs in breast cancer and glioma have been found to be relatively resistant to radiation and chemotherapy compared to their non-tumorigenic progeny 5–7. Consistent with these reports, several groups reported enrichment for CSCs when solid cancers were subjected to classical anti-cancer treatments 5, 6, 8.
Using an in vitro system we quantified the number of breast CSCs (BCSCs) surviving after radiation treatment in patient samples as well as in several breast cancer lines. When we compared the absolute number of breast CSCs (BCSCs) that survived radiation treatment to the number of BCSCs expected to survive we found a profound enrichment in BCSCs after exposure to ionizing radiation, and such a drastic increase in numbers could not easily be explained by differences in radiation sensitivity and/or by active repopulation. Here we report that ionizing radiation induced a BCSC phenotype in previously non-tumorigenic cells. This transition was Notch-dependent and coincided with up-regulation of transcription factors used to generate induced pluripotent cells from differentiated normal cells.
Methods
Cell culture
Human SUM159PT breast cancer cell lines were purchased from Asterand (Asterand, Inc., MI). Human MCF-7 and T47D breast cancer cell lines were purchased from American Type Culture Collection (Manassas, VA). SUM159PT-ZsGreen-cODC, MCF-7-ZsGreen-cODC, T47D-ZsGreen-cODC, were obtained as described in Vlashi et al. 9. SUM159PT cells were cultured in log-growth phase in F12 Medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (Sigma Aldrich, St Louis, MO), penicillin (100 units/ml) and streptomycin (100 µg/ml) (both Invitrogen), and insulin (5µg/mL) and hydrocortisone (1 µg/ml). MCF-7 and T47D cells were cultured in log-growth phase in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, penicillin and streptomycin. All cells were grown in a humidified incubator at 37°C with 5% CO2.
Irradiation
Cells grown as monolayers were irradiated at room temperature using an experimental X-ray irradiator (Gulmay Medical Inc. Atlanta, GA) at a dose rate of 2.789 Gy/min for the time required to apply a prescribed dose. Corresponding controls were sham irradiated. Assessment of cell proliferation, the number of BCSCs, and sphere-forming assays were performed 5 days after radiation.
Flow cytometry
BCSCs were identified based on their low proteasome activity 9, 10 using the ZsGreen-cODC reporter system. Five days after radiation, cells were trypsinized and ZsGreen-cODC expression was assessed by flow cytometry (MACSQuant Analyzer, Miltenyi). Cells were defined as "ZsGreen-cODC positive" if the fluorescence in the FL-1H channel exceeded the fluorescence level of 99.9% of the empty vector-transfected control cells.
Aldefluor assay and separation of the ALDH1-negative population by FACS
Genestier et al. previously reported that breast cancer stem cells could be isolated based on their high ALDH1 activity 2. The ALDEFLUOR kit (StemCell Technologies, Durham, NC, USA) was used to isolate the population with no ALDH1 enzymatic activity. Cells obtained from breast cancer monolayer (SUM159PT and T47D) were suspended in ALDEFLUOR assay buffer containing ALDH1 substrate (BAAA, 1 μmol/l per 1×106 cells) and incubated during 40 minutes at 37°C. As negative control for each sample of cells an aliquot was treated with 50mmol/L diethylaminobenzaldehyde (DEAB), a specific ALDH1 inhibitor. The sorting gates were established using ALDEFLUOR-stained cells treated with DEAB as negative controls.
CD24/CD44 staining and separation of the CD24+/high/CD44− population by FACS
MCF-7 and T47D cells growing as monolayer cultures were stained for CD24 and CD44 expression as described previously 10. Briefly, cells were incubated with trypsin–EDTA, dissociated and passed through a 40µm sieve. Cells were pelleted by centrifugation at 500 × g for 5 minutes at 4°C, resuspended in 100 µL of monoclonal mouse anti-human CD24–fluorescein isothiocyanate (FITC) antibody (BD Pharmingen, San Jose, CA) and a monoclonal mouse anti-human CD44–phytoerythrin (PE) antibody (BD Pharmingen), and incubated for 20 minutes at 4°C. The sorting gates were established using cells stained with isotype controls (isotype control FITC-conjugated antibodies (BD pharmingen) and isotype control PE-conjugated antibodies (BD pharmingen), respectively).
Sphere forming capacity
After irradiation, cells were trypsinized and plated in mammosphere media (DMEM-F12, 0.4% BSA (Sigma), 10 ml/500ml B27 (Invitrogen) 5 µg/ml bovine insulin (Sigma), 4 µg/ml heparin (Sigma), 20 ng/ml fibroblast growth factor 2 (bFGF, Sigma) and 20 ng/ml epidermal growth factor (EGF, Sigma)) into 96-well ultra-low adhesion plates, ranging from 1 to 256 cells/well. Growth factors, EGF and bFGF, were added every 3 days, and the cells were allowed to form spheres for 20 days. The number of spheres formed per well was then counted and expressed as a percentage of the initial number of cells plated. Cells were also plated in mammosphere media into 100 mm suspension dishes at 10,000 cells/ml, and allowed to form spheres for 15 days, these cells were used for secondary sphere forming experiments. Three independent experiments were performed.
Quantitative Reverse Transcription-PCR
Total RNA was isolated using TRIZOL Reagent (Invitrogen). cDNA synthesis was carried out using the SuperScript Reverse Transcription III (Invitrogen). Quantitative PCR was performed in the My iQ thermal cycler (Bio-Rad, Hercules, CA) using the 2x iQ SYBR Green Supermix (Bio-Rad). Ct for each gene was determined after normalization to GAPDH or RPLP0 and ΔΔCt was calculated relative to the designated reference sample. Gene expression values were then set equal to 2−ΔΔCt as described by the manufacturer of the kit (Applied Biosystems). All PCR primers were synthesized by Invitrogen and designed for the human sequences of Oct4, Sox2, Nanog, Klf4, c-Myc. Primers for the customized stem cell gene expression array were synthesized by Real Time Primers LLC (Elkins Park, PA).
Two channel flow cytometry for OCT4/Sox2/Nanog/Klf4/c-Myc and DNA content
Cells were harvested at relevant time points, washed in cold TBS, and fixed overnight in cold (–20 °C) 70% ethanol. After two washes in TBS, cells were permeabilized with TBS/4% BSA/0.1% Triton X-100 for 10 min at RT. Samples were incubated with a rabbit polyclonal anti-Oct4 antibody (Cell Signaling), a monoclonal mouse anti-Sox2 (R&D Systems), a rabbit polyclonal anti-Nanog (Abcam), a monoclonal mouse anti-Klf4 (Abgen), monoclonal mouse anti-c-Myc (Abcam), or corresponding isotype control (Biolegend) in TBS/4% BSA/0.1% Triton X100 for 1 h at RT. Following three washes in TBS, cells were incubated with goat anti-rabbit Alexa Fluor 750-APC (Invitrogen) or goat anti-mouse PE-Cy7 (Sigma) antibodies in TBS/4%BSA/0.1% Triton X100, 1:200 for 1 h in the dark. DNA was counterstained with 10 µg/ml propidium iodide (PI) solution in PBS, containing 200 µg/ml RNAse (Sigma) and assessed by flow cytometry using a MACSQuant Analyzer (Miltenyi Biotec) and analyzed using the FlowJo Software (version 9.3.1).
Animals
Nude (nu/nu), 6–8-week-old female mice, originally obtained from The Jackson Laboratories (Bar Harbor, ME) were re-derived, bred and maintained in a pathogen-free environment in the American Association of Laboratory Animal Care-accredited Animal Facilities of Department of Radiation Oncology, University of California (Los Angeles, CA) in accordance to all local and national guidelines for the care of animals.
Tumor xenotransplantation
SUM159PT-ZsGreen-cODC negative cells derived from monolayer cultures and sorted by fluorescence activated cell sorting were plated in F12 media supplemented with 5% fetal bovine serum and penicillin (100 units/ml) and streptomycin (100 µg/ml) cocktail, Insulin (5µg/mL) and hydrocortisone (1 µg/ml). The following day, cells were irradiated with 0, 4, or 8Gy. Five days after irradiation, cells were injected subcutaneously into the thighs and shoulders of 6-week old female Nu/Nu mice (106, 105, 104, 103, or 102 cells per inoculum, n=8) within Matrigel (BD Biosciences). Tumor growth was assessed on a weekly basis, and the mice were sacrificed when the tumor size reached tumor diameters requiring euthanasia. The experiment was terminated after 13 weeks. Data was fitted using a sigmoidal regression model (Y=a*X^b/(c+X^b), Graphpad Prism 5.0). The number of cells needed to obtain tumors in 50% of the animals (TD50) was calculated for each radiation dose.
Human breast cancer primary specimens
Primary tumor specimens were obtained under a protocol approved by the University of California, Los Angeles Institutional Review Boards through the Translational Pathology Core Laboratory (TCPL) at UCLA (IRB# 02-02-057-22).
- Patient Sample 1: Invasive mammary carcinoma (90%), Lobular carcinoma in situ (10%), grade 2, TNM Stage: pT3, N0 (i-), Mx, ER3+, PR3+, HER/Neu1+, no amplification (FISH).
- Patient Sample 2: Invasive ductal carcinoma, grade 3, TNM Stage: pT2, N0, Mx, ER-, PR-, HER/Neu3+. Amplification (FISH).
- Patient Sample 3: Extensive ductal carcinoma in situ (90%), Invasive ductal carcinoma (10%), grade 1, TNM Stage: pT1b, N0 (i-)(sn), Mx, ER3+, PR2+, Her/neu1, no amplification (FISH).
The tumors specimen were digested and cells were expanded ex vivo for 2–3 weeks. ALDH-negative cells were isolated using fluorescence activated cell sorting.
Notch1-4 and Sox2/Nanog siRNA Transfection
SUM159PT cells were subjected to transfection with Notch1, Notch2, Notch3, Notch4, Sox2, or Nanog-specific siRNA (Sigma Aldrich). MISSION® siRNA universal negative control (Sigma Aldrich) was used as a transfection control. Briefly, siRNA (100ng total) and Lipofectamine (invitrogen) were diluted in OptiMEM I reduced serum media (Invitrogen), mixed and incubated for 20 min as described by the manufactor. Cells were rinsed with PBS 1X, twice, and incubated in 750 µL of OptiMEM I. siRNA/Lipofectamine mix was added on the top of the cells, and incubated at 37°C, 5% CO2 for 6h. After incubation, media was removed and 2 mL of serum containing F12 Medium were added. Twenty-four hours after transfection, cells were plated for a sphere forming capacity assay, or cells were sorted and treated as previously described.
Noscapine treatment and cell cycle analysis
In order to identify the contribution of polyploidy to CSCs generation, non-tumorigenic MCF-7, T47D, SUM159PT and 2 patient samples (2 and 3) were treated with Noscapine 11. Non-tumorigenic cells (CD24+/high/CD44−, ALDH1-negative, or ZsGreen-cODC-negative) were sorted and plated as monolayer cultures. Each following day, MCF-7 and T47D cells were treated with Noscapine (0, 10, or 25 µM). SUM159PT cells and patient samples 1 and 2 were treated with Noscapine at 0, 25, or 50 µM (Sigma Aldrich). At day 5, the presence of CD24−/low/CD44high, ALDH1-positive, or ZsGreen-cODC-positive cells was analyzed by flow cytometry. In parallel, cells were analyzed for polyploidy as described above.
Statistical methods
All results are expressed as mean values. A p-value of ≤ 0.05 in a paired two-sided Student's t-test was considered to indicate statistically significant differences. The test was applied to normalized data to compensate for the variance of measurements between biologically independent replicates of the same experiments.
Results
Radiation induces a BCSCs phenotype in previously non-tumorigenic cells
Consistent with a radioresistant phenotype for BCSCs, we and others previously reported that ionizing radiation increased the number of BCSCs in the overall breast cancer cell population 5, 6, 10. This has been ascribed to selective killing of non-tumorigenic cells and/or a switch from an asymmetric to symmetric type of cell division of BCSCs that gives rise to two identical daughter BCSCs and thus, leading to a relative and absolute increase in BCSCs.
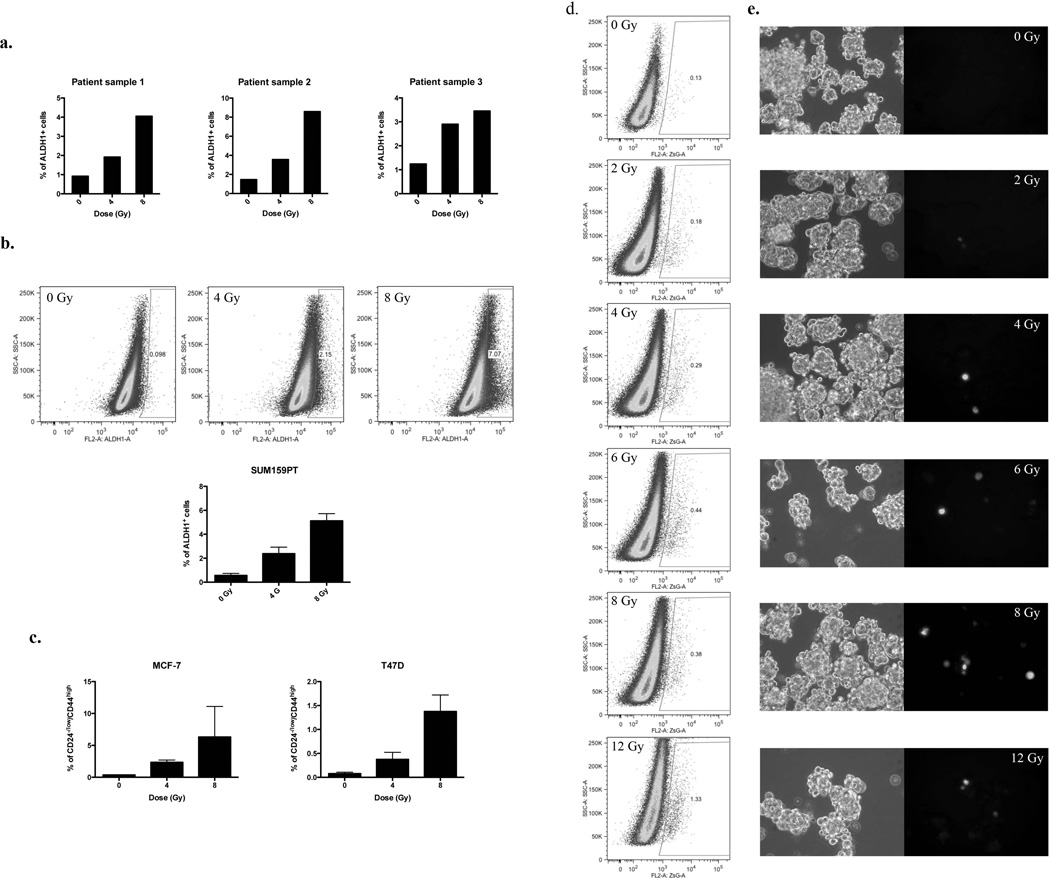
In the present study, we used single cell suspensions from fresh human breast specimen and stained them for ALDH1 activity, a recently described marker for BCSCs 2, 3. Using fluorescence activated cell sorting (FACS), we isolated non-BCSCs (ALDH1-negative cells) from these specimens after purging BCSCs (ALDH1-positive cells). Purified non-BCSCs (ALDH1-negative cells) were then plated and irradiated the following day with 0, 4, or 8 Gy. Five days after irradiation, we assessed the number of BCSCs arising within the non-BCSC population. We found that radiation led to a dose-dependent increase in the number of ALDH1-positive cells (Figure 1a; patient sample 1: 0Gy, 0.92%, 4Gy, 1.92%, 8Gy, 4.05%; Patient sample 2: 0Gy, 1.46%, 4Gy, 3.56%, 8Gy, 8.59%, Patient sample 3: 0Gy, 1.24%, 4Gy, 2.9%, 8Gy, 3.45%).
Figure 1. Radiation induces de novo generation of CSCs.
(a) Freshly isolated patient samples and (b) SUM159PT cells were stained for ALDH1 activity. ALDH1-negative cells were sorted, plated as monolayer cultures and irradiated with 0, 4, or 8Gy the following day. The presence of ALDH1-positive (ALDH1+) cells was analyzed 5 days after irradiation. Percentages of ALDH1-positive cells are show for 3 patient samples. Representative dot blots of SUM159PT and means of ALDH1-positive (ALDH1+) SUM159PT are shown (n=3). (c) MCF-7 and T47D were stained for CD24 and CD44, and purged by flow cytometry from CD24−/low/CD44high cells. Cells were then plated as monolayers and irradiated the following day with 0, 4, or 8Gy. Five days after treatment, cells were stained for CD24 and CD44, and the presence of CD24−/low/CD44high cells was analyzed by FACS. ZsGreen-cODC-negative cells from SUM159PT were sorted and plated as monolayers or mammospheres. The following day, cells were irradiated. Five days after treatment, the presence of ZsGreen-cODC-positive cells was analyzed by flow cytometry. (d) Representative dot blots and (e) pictures (phase contrast and green fluorescence) of SUM159PT-ZsGreen-cODC mammospheres 5 days after irradiation are shown. Means, s.e.m. and p-value for relative increases of ZsGreen-cODC-positive cell numbers are shown in supplementary table 1 and 2.
From previous experience with FACS-purified cell populations we were aware of the fact that in practice these purified cell population are not 100% pure 9. Indeed, a closer look at the FACS-purified populations of ALDH1-negative cells revealed that they always contained a very small population of contaminating ALDH1-positive cells. However, calculations based on the number of contaminating cells, the long doubling time of BCSCs (~72hours), which is not affected by irradiation 10, and the surviving fraction of BCSCs for each dose point (Supplementary figure 1), suggested that contaminating BCSCs present at the time of irradiation were unlikely to be the sole source of the absolute increase in BCSC numbers seen 5 days after irradiation. We therefore chose to test an alternative explanation, namely that non-tumorigenic breast cancer cells acquire a BCSC phenotype in response to ionizing radiation, thus contributing to the enrichment in BCSCs seen after radiation treatment.
To test this alternative hypothesis, we employed a panel of three established and widely used breast cancer cell lines (SUM159PT, MCF-7 and T47D). This panel allowed for the necessary number of experimental repeats in a large number of different assays. In addition to the Aldefluor test (Figure 1b) we used CD24 and CD44 to identify BCSCs and made use of a previously described imaging system for CSCs in breast cancer and glioma. This latter system is based on a fusion protein between the green fluorescent protein ZsGreen and the C-terminal degron of murine ornithine decarboxylase (cODC) and reports for 26S proteasome activity (Figure 1d, e), a protease activity that was found to be very low in CSCs 9. Cells with low proteasome activity accumulate the fluorescent fusion protein while in cells with high proteasome activity the cODC portion of the fusion protein directs ZsGreen to ubiquitin-independent degradation by the 26S proteasome. Importantly, in breast cancer, cells with intrinsically low proteasome activity overlapped with the CD24−/low/CD44high cell population 10 and with cells positive for ALDH1 (Supplementary figure 2a/b).
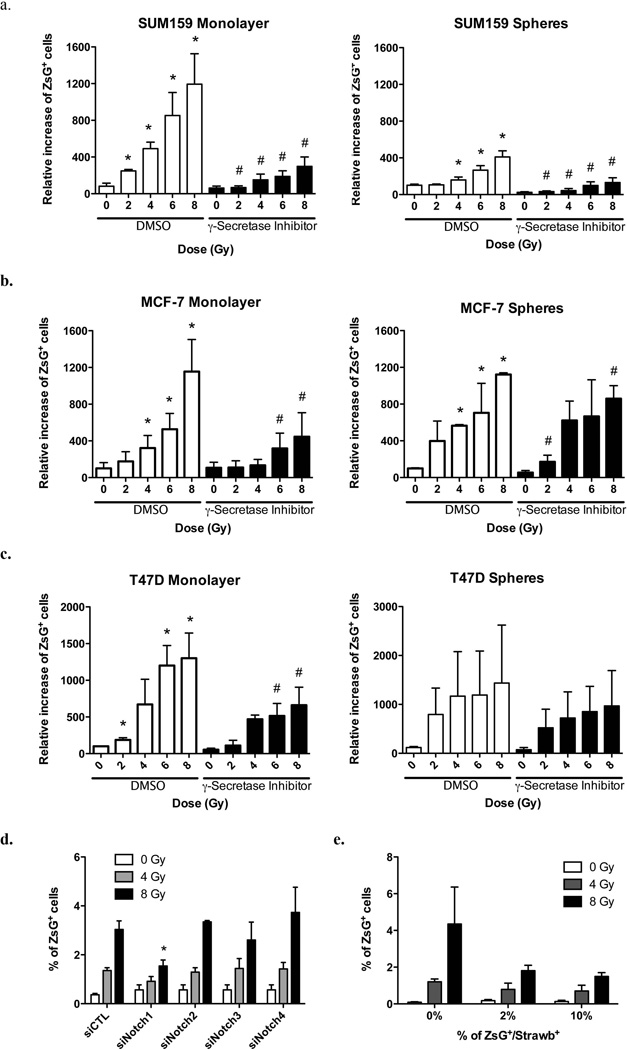
Next, we confirmed that radiation-induced increases in the number of BCSCs did not only occur in non-BCSCs from patient samples but also in non-BCSCs from established cells lines. Therefore, sorted ALDH1-negative cells from SUM159PT (Figure 1b) and T47D (Supplementary Figure 3a) cell lines were plated as monolayer cultures and irradiated the following day with 0, 4, or 8Gy. Like in patient-derived samples, we observed a significant dose-dependent increase in the number of ALDH1-positive cells, five days after irradiation (Figure 1b, SUM159PT: 0Gy: 0.3%; 4Gy, 1.76%, p = 0.008; 8Gy, 5.74%, p = 0.002; two-sided Student’s t-test). To confirm this observation we employed the CD24high/CD44low marker combination to isolate non-tumorigenic MCF-7 and T47D breast cancer cells. Again, five days after irradiation, we observed the induction of CD24−/low/CD44high from previously non-tumorigenic CD24high/CD44low cells (MCF-7: 0Gy: 0.37%: 4Gy: 2.35%, p=0.016; 8Gy: 6.33%, p=0.221. T47D: 0Gy: 0.08%: 4Gy: 0.38%, p=0.025; 8Gy: 1.38%, p=0.006). In parallel experiments we used SUM159PT, MCF-7, and T47D cells transfected with the ZsGreen-cODC reporter construct for proteasome activity. Using this third marker for BCSCs we also observed a dose-dependent, radiation-induced increase of ZsGreen-cODC-positive (low proteasome activity) cell numbers (Figure 1d/e, Figure 2, Supplementary Table 1 and 2). Importantly, cells positive for BCSC markers could not only be generated from differentiated cells in monolayer cultures but also from differentiating cells sorted from mammospheres (Figure 2, Supplementary figure 3b).
Figure 2. Radiation induces Notch-dependent de novo generation of CSCs.
(a) SUM159PT-, (b) MCF-7-, and (c) T47D-ZsGreen-cODC-negative cells were sorted and plated as monolayers or mammospheres. The following day, cells were then treated with 0, 2, 4, 6, 8, or 12 Gy (dose rate: 2.789 Gy/min). 1h before irradiation and every day after irradiation, cells were treated with a γ-secretase inhibitor (5µM). Five day after treatment, the presence of ZsG-cODC positive cells was analyzed by FACS. The mean for relative increases in ZsG-cODC positive cell numbers are shown (* and # indicates p<0.05, see supplementary table 1 and 2 for means, 95% CI and P value). (d) SUM159PT cells were transfected with Notch1, Notch2, Notch3, or Notch4-specific siRNA, and then sorted for ZsGreen-cODC-negative cells. Cells were plated as monolayers and irradiated with 0, 4, or 8 Gy the following day. Presence of ZsGreen-cODC-positive cells was analyzed 5 days after treatment. The mean percentage of ZsGreen-cODC-positive cell numbers are shown (*indicates p<0.05, n=4). (e) SUM159PT-ZsGreen-cODC cells were transfected with constitutively expressed Strawberry-Red vector. ZsGreen-cODC-positive/StrawberryRed-positive cells were isolated by follow cytometry and mixed with SUM159PT-ZsGreen-cODC-negative (Non-StrawberryRed transfected) cells at different concentration (0, 2, or 10%). Cells were plated and irradiated the following day. Five days after irradiation, the presence of ZsGreen-cODC-positive/StrawberryRed-negative cells was assessed by FACS. The mean percentages of ZsGreen-cODC-positive/StrawberryRed-negative cell numbers are shown (n=3).
Previous studies reported a role for the Notch signaling pathway in maintaining a stem cell phenotype in mammary epithelial cells 12. We decided to repeat the above radiation experiments in the presence of a γ-secretase inhibitor to block Notch signaling. Inhibition of Notch signaling attenuated the generation of cells positive for the BCSC marker (ZsGreen-cODC-positive) but did not completely abrogate the ability of non-tumorigenic cells (ZsGreen-cODC-negative) to give rise to cells with intrinsically low proteasome activity (Figure 2a-c, Supplementary Table 1 and 2). To further test the involvement of the Notch pathway in the induction of BCSCs, we inhibited the expression of Notch receptors by transfecting cells with targeting siRNA. SUM159PT-ZsGreen-cODC cells were transfected with specific siRNA targeting the Notch1, Notch2, Notch3, or Notch4 receptors using lipofectamine. Twenty-four hours after transfection, SUM159PT-ZsGreen-cODC-negative cells were isolated and plated as monolayer cultures. The following day, cells were irradiated with 0, 4, or 8 Gy. Five days after irradiation we observed a decrease in the number of BCSCs generated if compared to cells transfected with a scrambled siRNA control. Down-regulation of Notch2, 3 and 4 did not prevent induction of BCSCs (Figure 2d, 8Gy: siCTL: 3.04%, siNotch1: 1.54%, p=0.013). This indicated that signaling through the Notch1 receptor was either involved in inducing BCSCs or in maintaining the stem cell phenotype of newly generated BCSCs.
In order to study if non-tumorigenic cells could also generate BCSCs in the presence of pre-existing BCSCs, we reconstituted a mixed population of non-tumorigenic cells and StrawberryRed-labeled BCSCs at different dilutions (0, 2 and 10% BCSCs). After sorting, SUM159PT-ZsGreen-cODC-negative non-tumorigenic cells and SUM159PT-ZsGreen-cODC-positive/StrawberryRed-expressing BCSCs were mixed and plated as monolayer cultures. The following day, cells were irradiated with 0, 4, or 8Gy. When we analyzed the cells five days after irradiation by flow cytometry, we found that radiation-induced generation of BCSCs (ZsGreen-cODC-positive/StrawberryRed-negative) was decreased in the presence of pre-existing BCSCs (Figure 2e) suggesting a negative feedback loop of existing BCSC in the generation of induced BCSCs (iBCSCs).
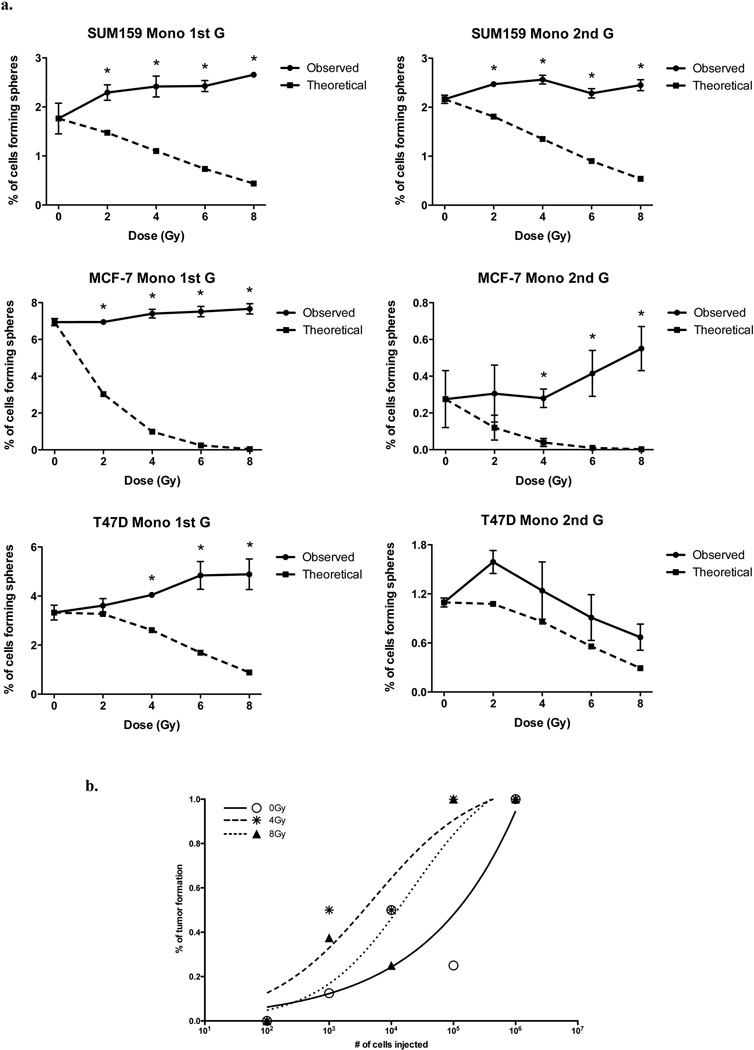
Using operational means, we next sought to test if the occurrence of ALDH1-positive cells and ZsGreen-cODC-positive cells with low proteasome activity reflected only an induction of BCSC marker expression or whether it was truly the induction of a BCSC phenotype. In order to assess the self-renewal capacity of iBCSC we employed a sphere-forming assay in which cells depleted from BCSCs with low proteasome activity were seeded at clonal densities into ultra-low adhesion plates in the absence of fetal calf serum to allow formation of mammospheres from single cells. In breast cancer, mammosphere formation is a measure of in vitro BCSC self-renewal capacity and correlates closely with tumorigenicity 13. In order to test if radiation induced a BCSC phenotype on the non-tumorigenic cells, we compared the sphere-forming capacity of irradiated samples and the non-irradiated control. Furthermore, the number of spheres formed for each radiation dose were compared with the hypothetical number of mammospheres expected. The expected number of mammospheres at each dose were calculated based on 1) the number of contaminating ZsGreen-cODC-positive cells after sorting, 2) a doubling time of 72 hours during the 5 days of culture, and 3) on their clonogenic surviving fraction at each dose point (Supplementary Figure 1).
In primary spheres, the self-renewal capacity of irradiated cells remained either the same or exceeded that of corresponding non-irradiated control cells. With the exception of T47D, this effect was also observed in secondary mammospheres. However, when the observed number of mammospheres was compared to the expected number of mammospheres at each dose point, mammosphere formation of irradiated cells significantly exceeded the numbers of mammospheres expected to be formed (Figure 3a and Supplementary figure 3b).
Figure 3. Radiation induces de novo generation of functional CSCs.
ZsGreen-cODC-negative cells from SUM159PT, MCF-7, and T47D were sorted and plated as monolayers or mammospheres (See Supplementary figure 3b). The following day, cells were irradiated. (a) Five days after irradiation, MCF-7, T47D, and SUM159PT cells were seeded at clonal densities to assess sphere-forming capacity. Means and s.e.m. are shown, *indicates p<0.05. Dark line graphs represent the mean of number of mammospheres observed (n=3), dashed lines represent the number of mammospheres expected to derive from contaminating BCSCs. Secondary sphere forming capacity was assessed 15 days after irradiation. (b) SUM159PT-ZsGreen-cODC-negative cells were sorted and plated as monolayer cultures. The following day, cells were irradiated with 0, 4, or 8 Gy. Five days after irradiation, cells were injected subcutaneously into nude mice. 13 weeks after injection, TC50 values were calculated.
Next, we sorted SUM159PT cells based on ZsGreen-cODC expression and irradiated non-BCSCs (ZsGreen-cODC-negative) with 0, 4, and 8 Gy. After 5 days, cells were injected into the thighs and shoulders of 6-week old female Nu/Nu mice in a limiting dilution assay (106, 105, 104, 103, or 102 cells per inoculum, n=8 per injection). 13 weeks after injection, the number of cells required to initiate tumor growth in 50% of the animals (TD50) was calculated. As expected, TD50 values of non-irradiated non-BCSCs were high (1.15×105 cells) consistent with a small number of contaminating BCSCs after FACS-sorting. However, TD50 values were reduced 32-fold after a single dose of 4Gy (3.6×103) and 9-fold (1.26×104) after a single dose of 8Gy suggesting a radiation-induced relative and absolute increase in BCSC numbers the (Figure 3b and Supplementary Table 3). Taken together, these data indicated that ionizing irradiation not only induced expression of BCSC markers in non-tumorigenic BSCSs but also led to the acquisition of cancer stem cell traits.
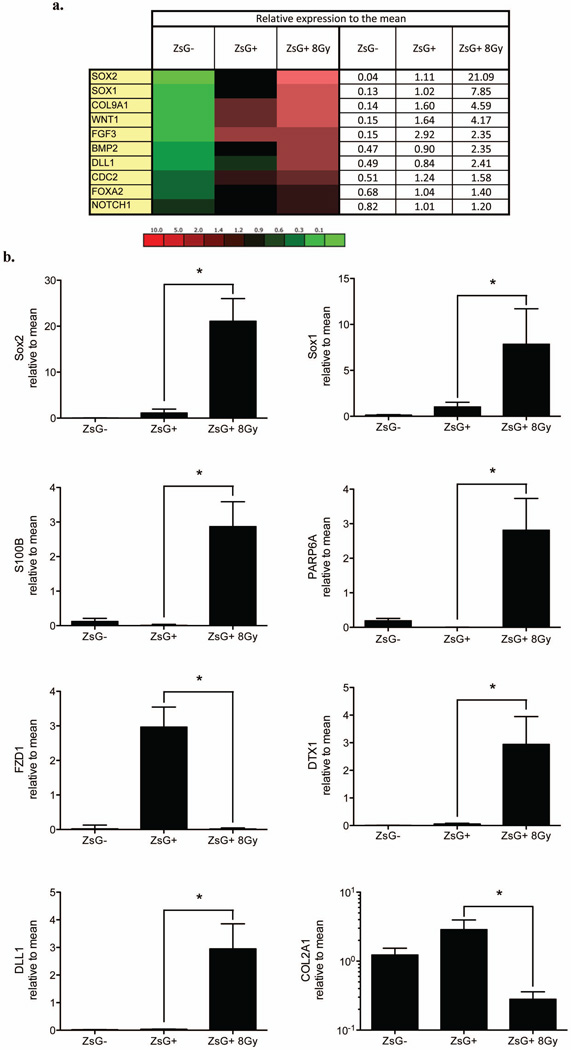
Finally, we decided to compare the expression profile of 86 genes associated with stem cell traits between non-irradiated ZsGreen-cODC-negative cells, non-irradiated ZsGreen-cODC-positive BCSCs and ZsGreen-cODC-positive iBCSCs induced by 8Gy. 10 genes were consistently and significantly up-regulated in ZsGreen-cODC-positive BCSCs and iBCSCs generated by 8Gy (Figure 4a and Supplementary Figure 4). Genes included key elements of the Notch, Wnt, Shh, and FGF signaling pathways as well as genes involved cell cycle regulation, cell adhesion, and cell-to-cell contact. The comparable gene expression profiles of ZsGreen-cODC-positive iBCSCs after a single radiation dose of 8Gy given to FACS-sorted ZsGreen-cODC-negative non-BCSC and preexisting ZsGreen-cODC-positive BCSCs, suggested that iBCSCs are driven by the same set of stem cell-related genes. There were however some differences in that expression of SRY (Sex determining region Y)-box 1 (Sox-1), SRY (Sex determining region Y)-box 2 (Sox-2), S100 calcium binding protein B (S100B), Par-6 partitioning defective 6 homolog alpha (PARP6A), Deltex homolog 1 (DTX1), and Delta-like 1 (DLL1) were significantly higher in iBSCSs, while expression of Frizzled homolog 1 (FZD1) and collagen type II alpha 1 (COL2A1) were significantly lower (Figure 6b). Overall, the expression profile of stemness-related genes in iBCSCs reassembled the expression profile found in BCSCs much closer than the expression profile found in the non-irradiated non-BCSC population they originated from.
Figure 4. Stem cell gene expression of BCSCs and iBSCS.
Expression of 86 stem cell related genes and 10 housekeeping genes was analyzed by semi-quantitative RT-PCR in ZsGreen-cODC-negative, -positive cells and iBCSCs (8 Gy). (a) Heat map of differentially expressed genes between ZsGreen-cODC-negative, ZsGreen-cODC-positive cells non-irradiated or iBCSCs 8Gy, and the mean expression (Mean of ZsGreen-cODC-negative cells, non-irradiated ZsGreen-cODC-positive cells and iBCSCs 8Gy cells) are shown (see also supplementary Figure 6). (b) Significant different expression between ZsGreen-cODC-positive non-irradiated BCSCs and iBCSCs 8Gy are shown (n=3), * indicates p<0.05.
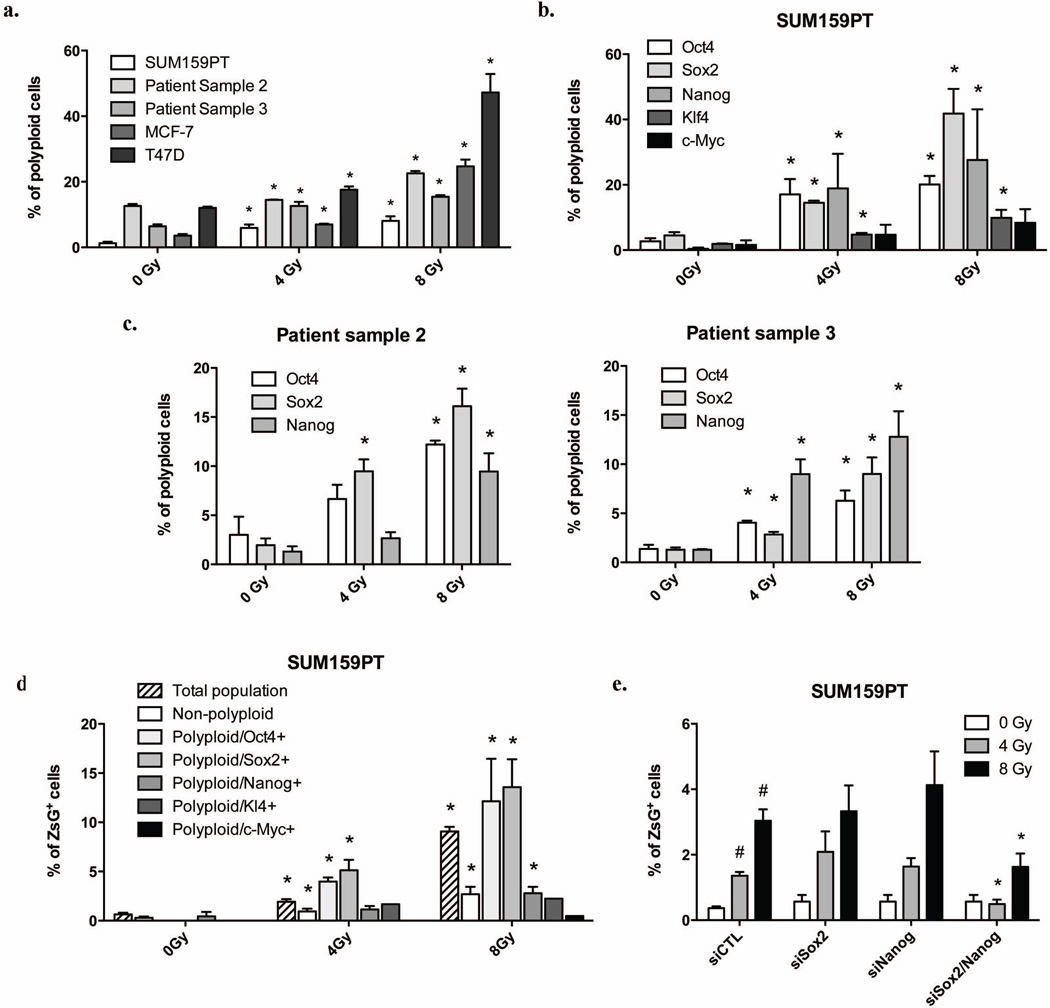
Figure 6. Irradiation-induced polyploid cells express Oct4, Sox2, Nanog, and Klf4 and are enriched for BCSCs.
Protein expression levels of ZsGreen-cODC, Oct4, Sox2, Nanog, Klf4 and c-Myc, and DNA content were analyzed by flow cytometry. (a) The means and s.e.m. of radiation-induced polyploid cells are shown. (b) The means and s.e.m. of the number of polyploid Oct4-, Sox2-, Nanog-, Klf4- and c-Myc-positive cells after irradiation are shown. (c) Expression of Oct4, Sox2 and Nanog in polyploid were analyzed in patient derived samples (For MCF-7 and T47D see Supplementary figure 5). (d) Distributions of SUM159PT BCSCs in the total population, the non-polyploid population, in polyploid Oct4-, Sox2-, Nanog-, Klf4-, or c-Myc-positive population are shown (see also Supplementary figure 6 for MCF-7 and T47D). Data are expressed as means and s.e.m., * indicates p<0.05. (e) SUM159PT-ZsGreen-cODC cells were transfected with Sox2 and/or Nanog-targeting siRNA, and ZsGreen-cODC-negative were sorted and irradiated. Means (± s.e.m.) of ZsGreen-cODC-positive cells found 5 days after irradiation are shown. * indicates p<0.05.
Radiation induces re-expression of Oct4, Sox-2, Nanog, and Klf4
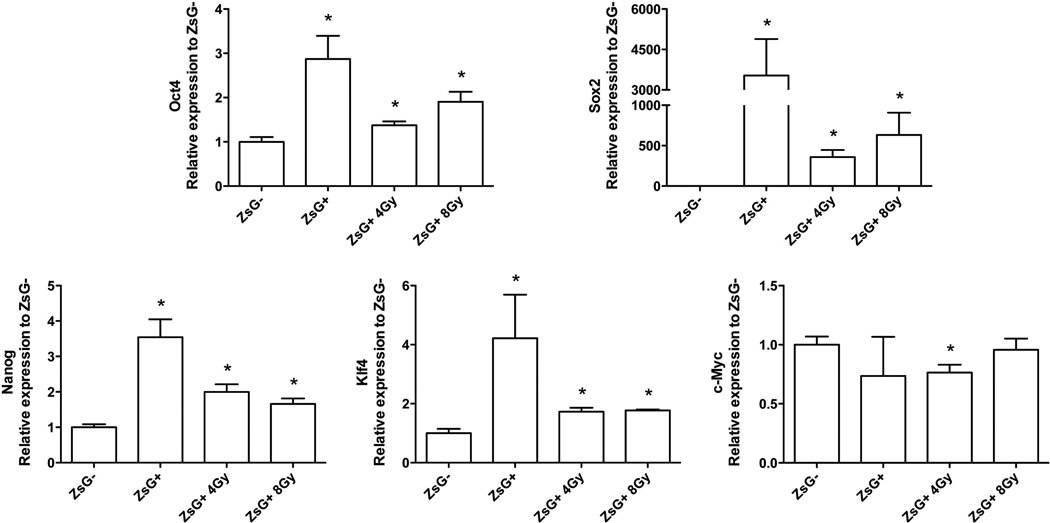
Acquisition of a stem cell phenotype has been described for non-malignant differentiated cells after overexpression of Oct4, Sox-2, Nanog, Klf4, and c-Myc. These transcription factors are now routinely used to generate induced pluripotent stem (iPS) cells from differentiated somatic cells 14, 15 and have also been shown to maintain the BCSCs phenotype 16. Interestingly, Oct4, Sox-2, Nanog, and Klf4 are known substrates of the 26S proteasome 17–20 and therefore expected to be stabilized in cells with low proteasome activity. To determine if these transcription factors were re-activated in non-BCSCs populations after irradiation, we analyzed their expression levels 5 days after 0, 4, or 8 Gy of radiation, the time point at which we observed absolute increases in BCSC numbers. As expected, we found a significant radiation dose-dependent increase of Oct4, Sox-2, Nanog, and Klf4 mRNA expression levels that matched the expression levels for these transcription factors in intrinsically occurring, non-irradiated BCSCs (Figure 5).
Figure 5. iBCSCs overexpress Oct4, Sox2, Nanog, and Klf4, but not c-Myc.
SUM159PT-ZsGreen-ODC cells were sorted into ZsGreen-cODC-positive and -negative cells. ZsGreen-cODC-negative cells were plated as monolayers, and irradiated the following day with 4 or 8 Gy. iBCSCs cells were sorted at day 5 post-irradiation. Expression of Oct4, Sox2, Nanog, Klf4, and c-Myc was analyzed by semi-quantitative RT-PCR. The means of transcription factor gene expression levels (n=4) are shown, * indicates p<0.05.
To test if re-expression of the above four transcription factors occurred randomly or in a specific subset of cells, we analyzed the expression of Oct4, Sox-2, Nanog, and Klf4 protein levels and correlated it to the DNA content of the cells. In irradiated cells we observed increased numbers of polyploid cells (SUM159PT: 0Gy: 0.74%, 4Gy: 4.63%, p<0.0001; 8Gy: 6.33%, p<0.0001; Patient sample 2: 0Gy: 12.6%, 4Gy: 14.4%, p<0.032; 8Gy: 22.5%, p<0.0003; Patient sample 3: 0Gy: 6.41%, 4Gy: 12.6%, p<0.012; 8Gy: 15.4%, p<0.0004; MCF-7: 0Gy: 3.60%, 4Gy: 6.96%, p<0.004; 8Gy: 24.7%, p<0.0006; T47D: 0Gy: 12.0%, 4Gy: 17.6%, p<0.007; 8Gy: 47.23%, p<0.003; Figure 6a and Supplementary figure 5a/b) in which Oct4, Sox-2, Nanog and Klf4 proteins were up-regulated (SUM159PT: Oct4: 0Gy, 2.71%, 4Gy: 17.02%, p=0.041; 8Gy: 20.1%, p=0.003; Sox2: 0Gy, 4.52%, 4Gy: 14.5%, p=0.0002; 8Gy: 48.98%, p<.0001; Nanog: 0Gy, 0.39%, 4Gy: 18.9%, p=0.223; 8Gy: 27.58%, p=0.222: Klf4: 0Gy, 1.89%, 4Gy: 4.74%, p=0.031; 8Gy: 9.86%, p=0.084; Figure 6b/c and supplementary figure 5c/d) and which were also highly enriched for ZsGreen-cODC-positive cells with low proteasome activity (Total population: 0Gy, 0.64%, 4Gy: 1.84%, p<0.0001; 8Gy: 9.09%, p<0.0001; Polyploid: 0Gy, 0.08%, 4Gy: 0.94%, p=0.01; 8Gy: 2.69%, p=0.005; Polyploid/Oct4+: 0Gy, 0%, 4Gy: 3.97%, p<0.0001; 8Gy: 12.14%, p=0.031; Polyploid/Sox2+: 0Gy, 0%, 4Gy: 3.84%, p=0.041; 8Gy: 10.19%, p=0.042; Polyploid/Nanog+: 0Gy, 0.45%, 4Gy: 1.14%, p=0.107; 8Gy: 2.79%, p=0.029; Figure 6d and Supplementary figure 5b). c-Myc levels were not increased (Figure 6b/d, 5d). Consistent with a previous report 21, inhibition of Notch activation attenuated the induction of polyploidy (4Gy: 4.87%; 4Gy + γ-secretase inhibitor: 2.72, p=0.03; 8Gy: 6.32%; 8Gy + γ-secretase inhibitor: 3.42, p=0.028; Supplementary figure 5b) and the induction of Oct4, Sox-2, Nanog, and Klf4 expressing polyploid cells (Supplementary Figure 5d). This again supported involvement of Notch signaling in radiation reprograming.
In order to identify the role of these transcription factors in the generation of iBCSCs, we used siRNA to target the expression of Sox2 and Nanog. Non-tumorigenic cells transfected with siRNA-Sox2 and siRNA-Nanog were sorted, plated, and irradiated with 0, 4, and 8Gy the following day. Down-regulation of either Sox2 or Nanog alone had no effect on the induction of iBCSCs. However, generation of iBCSCs was significantly reduced when Sox2 and Nanog were down-regulated simultaneously (0Gy: siRNA control: 0.37%, siRNA-Sox2/Nanog: 0.57%, p=0.505; 4Gy: siRNA control: 1.36%, siRNA-Sox2/Nanog: 0.49%, p=0.008; 8Gy: siRNA control: 3.04%, siRNA-Sox2/Nanog: 1.63%, p=0.047; Figure 6e).
Induction of polyploidy induced generation of iBCSCs
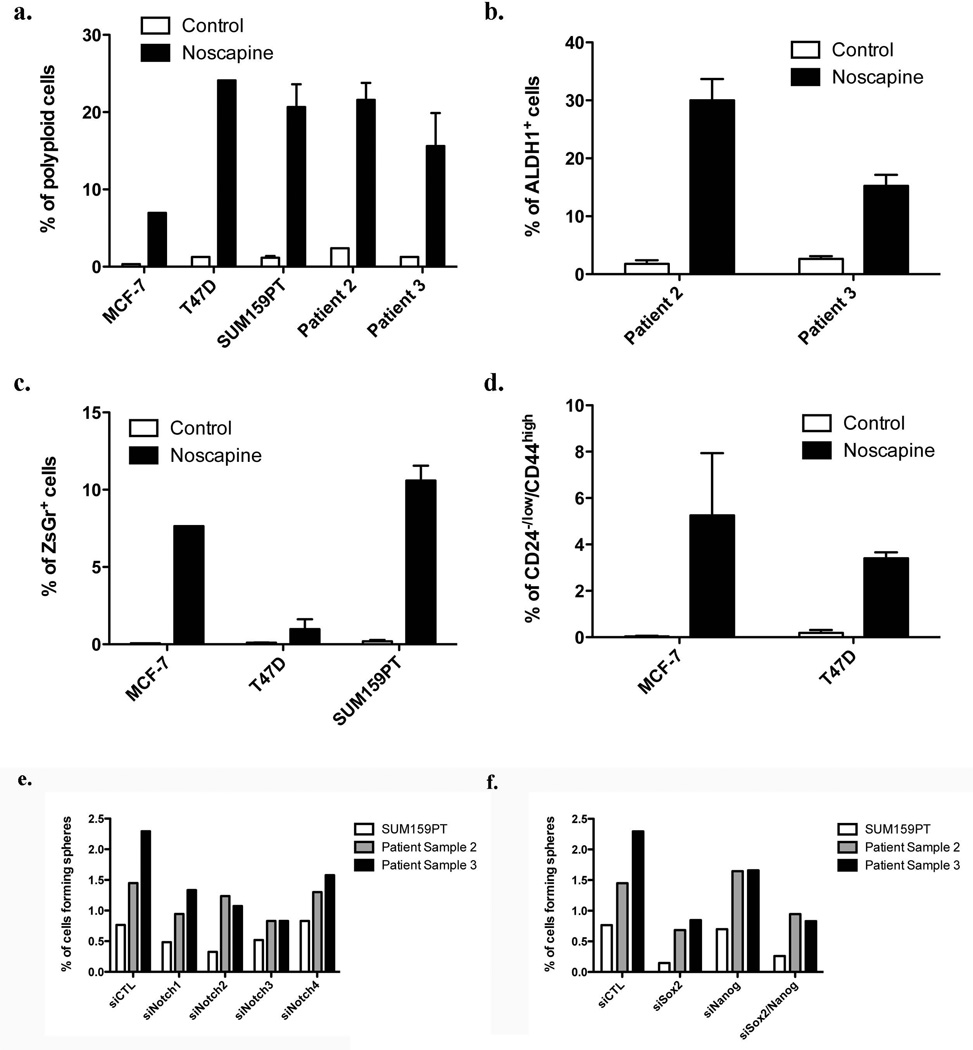
In differentiated cells, Sox2 and Oct4 are epigenetically silenced. The suppression of gene expression for both transcription factors is incomplete but still sufficient to maintain protein levels below a critical threshold. We hypothesized that in polyploid cells found after irradiation multiple copies of partially silenced Oct4 and Sox2 genes might be sufficient to drive gene expression beyond this threshold, thereby inducing a BCSC phenotype. To test this hypothesis we explored if the effect of radiation could be mimicked by pharmacological induction of polyploidy. Treatment of breast cancer cell lines and patient samples with noscapine caused a substantial increase in the number of polyploid cells on day 5 after addition of the drug. The observed increase was comparable to the amount of polyploidy found after irradiation (Figure 7a and Supplementary figure 7a/b). Next we tested if the number of BCSCs was also increased. In both patient samples noscapine treatment increased the number of ALDH1-positive cells significantly (Figure 7b). Increased numbers of BCSCs with low proteasome activity were also found when MCF-7, T47D or SUM159PT ZsGreen-cODC expressing cells were analyzed for ZsGreen-cODC-positive cells (Figure 7c and Supplementary figure 7c) and when MCF-7 and T47D cells were analyzed for the number of CD24low/−/CD44high cells (Figure 7d). These results suggested that increased gene dose in polyploid cells could indeed be one of the mechanisms leading to acquisition of a CSC phenotype in breast cancer in response to radiation treatment.
Figure 7. Polyploidy induces de novo generation of CSCs.
(a) Assessment of noscapine-induced polyploidy in MCF-7, T47D, SUM159PT cell lines and 2 patient derived samples, five days after drug treatment.
Non-tumorigenic cells ALDH1-negative patient-derived cells (b), MCF-7-, T47D-, and SUM159PT-ZsGreen-cODC-negative (c), and CD24+/CD44− MCF-7 and T47D cells (d) were treated with noscapine at 0, 25 or 50µM. The presence of iBCSCs was assessed after 5 days by flow cytometry.
SUM159PTcell and patient samples 2 and 3 were transfected with specific siRNA targeting Notch receptors (e), or Sox2 and Nanog (f). Scrambled sequences were used as control. Twenty-four hour after transfection, cells were plated for a sphere forming capacity assay. Percentages of cells able to form a sphere are shown for each condition.
Finally, we confirmed that BCSCs rely on Notch signaling using specific siRNAs targeting the Notch1, Notch2, Notch3, or Notch4 receptor. Cells were transfected with Lipofectamine, 24h after transfection cells were plated in sphere media for a sphere forming capacity assay. As expected, down-regulation of Notch receptor expression reduced the ability of the cells to form mammospheres (Figure 7e). Furthermore, in order to demonstrate that BCSCs rely on the transcription factors induced by radiation, we down-regulated Sox2 and Nanog using specific siRNAs. Cells were transfected with Lipofectamine, 24h after transfection cells were plated and tested for sphere formation. As expected, down-regulation of Sox2 and Nanog expression reduced self-renewal capacity of cells from established breast cancer cell lines and primary breast cancer samples and Sox2, which acts upstream of Nanog and has targets in addition to Nanog was more efficient in (Figure 7f).
Discussion
Treatment gaps in radiation therapy have long been known to worsen the outcome for patients suffering from epithelial cancers including cancers of the head and neck region and the breast 22, 23. The underlying mechanisms are incompletely understood but in general are attributed to accelerated repopulation, a phenomenon that refers to the increased growth rates of cancers during treatment gaps that far exceed their initial growth rates. It is thought that during accelerated repopulation CSCs switch from an asymmetric type of cell division, which leads to one daughter CSC and one differentiating cell, to a symmetric type of cell division which yields two identical daughter CSCs. Our data suggest that in addition to the classical view of accelerated repopulation in which CSCs switch from an asymmetric type of cell division that leads to one daughter CSCs and one differentiating cells to a symmetric type of cell division that yields in two daughter CSCs, differentiated cancer cells may also be able to acquire stem cell traits under certain conditions of tumor micro-environmental stress, including stress induced by ionizing radiation. Acquisition of stem cell traits by CD133-negative, non-tumorigenic glioma cells was previously reported under hypoxic conditions 24 and in response to nitric oxide-induced notch signaling 25, suggesting that cancer stem cell plasticity may be a common response to multiple stimuli including cancer therapies.
Our observation that ionizing radiation reactivated the same transcription factors in differentiated breast cancer cells that reprogram differentiated somatic cells into iPS cells is provoking. However, it is in line with recent reports that baseline levels of Sox2, Oct4, and Nanog expression can be detected in breast cancers 26, 27 and that ectopic overexpression of Oct4 in normal mammary epithelial cells induces a BCSCs phenotype 28. Our data further indicate that an increase number of gene copies of Oct4 and Sox2 in polyploid cells could be one possible mechanism behind radiation-induced reprogramming. This was supported by our data showing that down-regulation of Sox2 prevented mammosphere formation. Down-regulation of the Sox2 downstream target Nanog was less efficient, indicating that multiple genes downstream of Sox2 contribute to the acquisition of a cancer stem cells phenotype. Similar observations have been reported for lymphoma cells 29. Furthermore, this is in accordance with previous reports on Notch-dependent induction of polyploidy 21 and our data showing that inhibition of Notch-signaling partially prevented the occurrence of iBCSCs (Supplementary figure 5b), suggesting that targeting Notch-signaling might enhance local control after radiation therapy.
The CSC hypothesis was formulated more than a century ago 30. However, until recently prospective identification of CSCs was impossible. The discovery of marker combinations that identify CSCs have resulted in novel insights into the biology of cancer. Still, the CSC hypothesis has been challenged and some experimental data support a model of clonal evolution as an alternative organizational structure of tumors 31 in which every cancer cell may acquire stem cell traits at some point.
Our study unites the competing models of clonal evolution and hierarchical organization of cancers 32 as it suggests that undisturbed growing tumors indeed maintain a low number of CSCs. However, if challenged by various stressors including ionizing radiation, iCSCs are generated, which may together with the surviving CSCs repopulate a tumor. These findings have implications for the design of novel treatment protocols that target CSCs, including radiation therapy. The curability of a cancer may not only be dependent on the intrinsic radiosensitivity of CSCs but also on the radiosensitivity of induced CSCs and the rate at which they are generated. Controlling the radio-resistance of BCSCs and the generation of new iBCSCs during radiation treatment may ultimately improve curability and may allow for de-escalation of the total radiation doses currently given to breast cancer patients thereby reducing acute and long-term adverse effects.
Conclusion and summary
In summary, our study shows that ionizing radiation reactivated the expression of Oct4 and Sox2 and induced a CSC phenotype in previously non-tumorigenic breast cancer cells. The phenomenon was dependent on the induction of polyploidy and Notch signaling. We conclude that a detailed understanding of the underlying pathways could lead to novel combination therapies that will potentially enhance the efficacy of radiation treatment.
Supplementary Material
Acknowledgments
Grant Support
FP was supported by grants from the California Breast Cancer Research Program (15NB-0153) the Department of Defense (W81XWH-07-1-0065) and the National Cancer Institute (5RO1CA137110). CL was supported by an award from the Ward Family Foundation.
Footnotes
Authors contribution
Chann Lagadec: Conception and design, Collection and assembly of data, Data analysis and interpretation, Manuscript writing;
Erina Vlashi: Conception and design, Data analysis and interpretation;
Lorenza Della Donna: Conception and design;
Carmen Dekmezian: Collection and assembly of data;
Frank Pajonk: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript, Financial support;
References
- 1.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcato P, Dean CA, Pan D, et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells is Primarily Due to Isoform ALDH1A3 and Its Expression is Predictive of Metastasis. Stem Cells. 2010 doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 5.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 6.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li HZ, Yi TB, Wu ZY. Suspension culture combined with chemotherapeutic agents for sorting of breast cancer stem cells. BMC Cancer. 2008;8:135. doi: 10.1186/1471-2407-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao S. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Vlashi E, Kim K, Lagadec C, et al. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009;101:350–359. doi: 10.1093/jnci/djn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagadec C, Vlashi E, Della Donna L, et al. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010;12:R13. doi: 10.1186/bcr2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell ID, Carlton JB, Chan MY, et al. Noscapine-induced polyploidy in vitro. Mutagenesis. 1991;6:479–486. doi: 10.1093/mutage/6.6.479. [DOI] [PubMed] [Google Scholar]
- 12.Dontu G, Jackson KW, McNicholas E, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel M, Yang S. Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. 2010;6:367–380. doi: 10.1007/s12015-010-9123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao R, Daley GQ. From fibroblasts to iPS cells: induced pluripotency by defined factors. J Cell Biochem. 2008;105:949–955. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 16.Wu K, Jiao X, Li Z, et al. The cell-fate determination factor dachshund reprograms breast cancer stem cell function. J Biol Chem. 2010 doi: 10.1074/jbc.M110.148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ZY, Wang X, Zhou Y, et al. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 18.Baltus GA, Kowalski MP, Zhai H, et al. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–2184. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 19.Moretto-Zita M, Jin H, Shen Z, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Wang W, Li C, et al. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- 21.Baia GS, Stifani S, Kimura ET, et al. Notch activation is associated with tetraploidy and enhanced chromosomal instability in meningiomas. Neoplasia. 2008;10:604–612. doi: 10.1593/neo.08356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bese NS, Sut PA, Ober A. The effect of treatment interruptions in the postoperative irradiation of breast cancer. Oncology. 2005;69:214–223. doi: 10.1159/000087909. [DOI] [PubMed] [Google Scholar]
- 23.Withers HR, Maciejewski B, Taylor JM, et al. Accelerated repopulation in head and neck cancer. Front Radiat Ther Oncol. 1988;22:105–110. doi: 10.1159/000415101. [DOI] [PubMed] [Google Scholar]
- 24.Heddleston JM, Li Z, McLendon RE, et al. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charles N, Ozawa T, Squatrito M, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leis O, Eguiara A, Lopez-Arribillaga E, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2011 doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 27.Ezeh UI, Turek PJ, Reijo RA, et al. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 28.Beltran AS, Rivenbark AG, Richardson BT, et al. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res. 2011;13:R94. doi: 10.1186/bcr3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmina K, Jankevics E, Huna A, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010;316:2099–2112. doi: 10.1016/j.yexcr.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 31.Quintana E, Shackleton M, Sabel MS, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.