The transition from a meiotic-like spindle formation characterized by lack of centrioles to a typical mitotic spindle occurs gradually in embryos during the preimplantation stage.
Abstract
The transition from meiosis to mitosis, classically defined by fertilization, is a fundamental process in development. However, its mechanism remains largely unexplored. In this paper, we report a surprising gradual transition from meiosis to mitosis over the first eight divisions of the mouse embryo. The first cleavages still largely share the mechanism of spindle formation with meiosis, during which the spindle is self-assembled from randomly distributed microtubule-organizing centers (MTOCs) without centrioles, because of the concerted activity of dynein and kinesin-5. During preimplantation development, the number of cellular MTOCs progressively decreased, the spindle pole gradually became more focused, and spindle length progressively scaled down with cell size. The typical mitotic spindle with centrin-, odf2-, kinesin-12–, and CP110-positive centrosomes was established only in the blastocyst. Overall, the transition from meiosis to mitosis progresses gradually throughout the preimplantation stage in the mouse embryo, thus providing a unique system to study the mechanism of centrosome biogenesis in vivo.
Introduction
The transition from meiosis to mitosis is a fundamental process in animal development. Although it has been widely assumed that fertilization by the sperm triggers the immediate start of mitotic divisions, very little is known about how the transition from meiosis to mitosis is achieved. One of the most remarkable features of this transition is the shift from acentrosomal to centrosomal spindle formation. The centrosome was originally identified as the structure in the cytoplasm from which spindle poles form (Boveri, 1887, 1888; van Beneden and Neyt, 1889) and has been defined by structure using electron microscopy (de Harven and Bernhard, 1956; Bessis et al., 1958) but not by function (Lüders and Stearns, 2007). Centrosomes contain two centrioles surrounded by pericentriolar material (Urbani and Stearns, 1999; Bettencourt-Dias and Glover, 2007). At early interphase, each cell has one pair of centrioles, which duplicate to give rise to two pairs to be segregated equally during cell division.
When sperm and oocyte fuse to generate a zygote, a single pair of centrioles is provided by the sperm in most organisms, whereas oocyte centrioles degenerate before fertilization (Szollosi et al., 1972; Schatten et al., 1986a,b). This mechanism has been suggested to serve as a safeguard against parthenogenetic development and centrosome overduplication (Simerly et al., 1995). However, in rodents, sperm centrioles also degenerate during spermiogenesis, becoming unidentifiable by electron microscopy (Woolley and Fawcett, 1973; Schatten, 1994; Manandhar et al., 1998). Nonetheless, centrioles are identified by electron microscopy in the blastocyst (i.e., 64-cell stage; Gueth-Hallonet et al., 1993). These findings suggest that the first few cell cycles in early mouse development may exhibit centriole-independent mitotic cell divisions. Furthermore, contrary to the classical view, the centriole may be generated de novo under physiological conditions (Strnad and Gönczy, 2008; Loncarek and Khodjakov, 2009).
Recent studies have begun to shed light on de novo centrosome formation under experimental conditions. Drosophila melanogaster embryos mutated for DSAS4 lose centrioles but still develop to term (Basto et al., 2006), and cultured cells in which centrioles were destroyed regenerated a centriole (Khodjakov et al., 2002; La Terra et al., 2005). Although the centriole appears dispensable under certain experimental conditions, it is essential for Drosophila embryogenesis because mutations in SAK/PLK4, a protein necessary for centriole duplication, lead to early embryonic lethality (Rodrigues-Martins et al., 2008).
The centrosome plays a major role in spindle assembly in most animal cells, acting as a scaffold to initiate microtubule polymerization by stabilizing microtubule minus ends. Spindles can also assemble in the absence of centrosomes (Hyman, 2000). Acentrosomal spindle assembly is particularly important in oocytes of many species, including mouse and human. In mouse oocytes, the spindle is assembled by microtubule-organizing centers (MTOCs) together with the plus end–directed motor kinesin-5 and possibly the minus end–directed motor dynein (Schuh and Ellenberg, 2007). Cytoplasmic MTOCs are recruited to the surface of the germinal vesicle (nucleus of the oocyte), in turn leading to stochastic self-organization of the barrel-shaped spindle. However, the mechanisms underlying mitotic spindle formation in the absence of centrioles in the early mouse embryo and how the transition proceeds from multipolar meiotic to the bipolar mitotic spindle assembly driven by centrosomes remain unknown.
Our present study uses a combination of quantitative live-embryo imaging, fixed-cell analysis, embryo micromanipulation, and small-molecule perturbation to address the transition from meiosis to mitosis in the mouse embryo. Although this transition is classically defined sharply by the time point of sperm fertilization, we found that spindle morphology and characteristic features of cell division change only very gradually toward centrosomal divisions over the first eight embryonic cleavages during the preimplantation stage from the zygote to the blastocyst. Our findings set the stage for exploring the molecular mechanisms by which the centrosome and mitotic cell division are established de novo in early embryonic development.
Results
Randomly distributed MTOCs form a multipolar spindle that progressively clusters into a barrel-shaped spindle in the mouse zygote
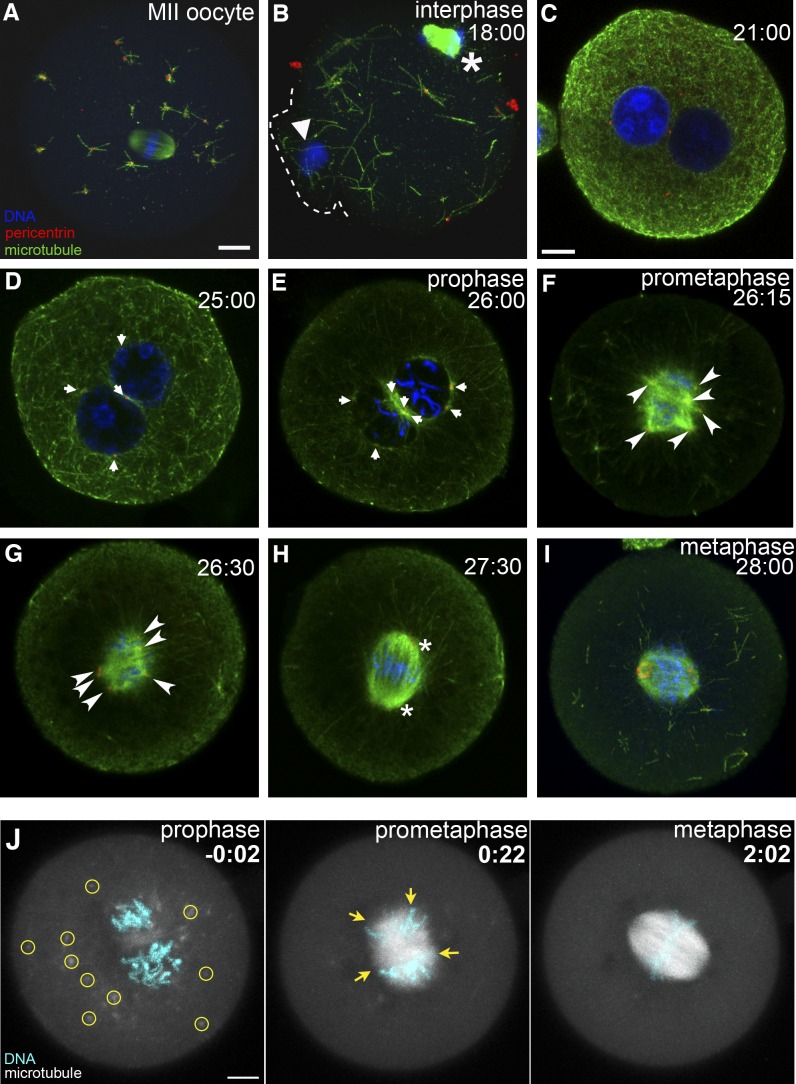
To investigate how the transition from meiosis to mitosis proceeds during oocyte to embryo development, we systematically analyzed MTOC organization, spindle assembly, and morphology in mouse oocytes, zygotes, and embryos throughout the preimplantation stage. First, we examined how microtubules are organized at the transition from oocyte to zygote by immunohistochemical analysis. In the oocyte at second meiotic metaphase (MII oocyte), MTOCs, defined herein by localization of pericentrin, were distributed throughout the cytoplasm and clustered at the spindle poles (Fig. 1 A). The microtubules nucleated at cytoplasmic MTOCs were relatively short, and a space-filling microtubule network was missing, consistent with the findings of Kubiak (1991). Similar analysis of the zygote upon fertilization revealed the random distribution of cytoplasmic MTOCs in early interphase (P > 0.9 for >40 MTOCs, each from 3 zygotes; Kolmogorov–Smirnov test; Fig. 1 B and Fig. S1, A and B). We detected no polarized distribution of MTOCs or the dense microtubule network in midzygotic interphase (Fig. 1 C).
Figure 1.
Stochastic MTOC assembly leads to formation of the multipolar spindle followed by progressive clustering into a barrel-shaped spindle. (A–I) Immunofluorescence staining of the mouse MII oocyte (A) and zygotes (B–I) fixed at consecutive stages of development: early interphase (18 h after hCG), in which the fertilization cone (dotted line) forms in response to sperm entry (B); mid (C)- and late (D) interphase (21 and 25 h after hCG, respectively); prophase (E); early (F), mid (G)-, and late (H) prometaphase; and metaphase (I; 28 h after hCG). Single-section images (C–H) or z-projected images of confocal sections (A, B, and I) show microtubules, pericentrin, and DNA. In B, arrowhead marks male chromatin delivered by the sperm; asterisk marks the second meiotic spindle. Note the absence of MTOC enhancement in the fertilization cone. Arrows in D and E mark MTOCs on the pronuclear surface. Upon NEBD, a multipolar spindle forms with no major axis (early prometaphase; F). Arrowheads in F and G mark the multipoles. A few major axes become visible in midprometaphase (G), consolidating into a single major axis with minor additional axes in late prometaphase (asterisks; H) and eventually forming a barrel-shaped spindle with pericentrin localized on two ring-shaped poles (metaphase; I). (J) Live imaging of mouse zygotes during the first division at prophase (left), prometaphase (middle), and metaphase (right). Z-projected images of confocal sections (3 µm thick) show microtubules (EGFP-MAP4; gray) and DNA (H2B-mRFP1). Circles and arrows mark MTOCs and the multipoles, respectively. Bars, 10 µm. Time is given in hours and minutes after NEBD.
At late zygotic interphase, the cytoplasmic microtubule network disappeared, whereas microtubules emanating from MTOCs became increasingly prominent (Fig. 1 D), suggesting that MTOCs become more actively engaged in microtubule polymerization. The mean size of MTOCs also increased from 0.54 ± 0.14 µm (n = 28) at 16.5 h after injection of human chorionic gonadotropin (hCG) to 0.67 ± 0.25 µm (n = 29) at 25 h after hCG (Welch two-sample t test, P < 0.05; see Materials and methods for definition of MTOC size; Fig. S1 C). At prophase, dense microtubule fibers surrounded all MTOCs and pronuclei (Fig. 1 E). Upon nuclear envelope breakdown (NEBD), microtubule fibers increased massively toward the center of the cell, leading to formation of multiarray fibers at prometaphase (Fig. 1 F). No enrichment of microtubules was detected in the vicinity of chromatin (Video 1). Thus, the stochastic multipolar spindle forms (Fig. 1 G), and its progressive organization (Fig. 1 H) leads to the barrel-shaped spindle at metaphase (Fig. 1 I), similar to that in oocytes (Schuh and Ellenberg, 2007).
To obtain a dynamic view of the spindle formation, we performed live 3D imaging of the mouse zygote microinjected with mRNA encoding EGFP-MAP4 and H2B-mRFP1 to visualize microtubules and DNA, respectively (Fig. 1 J and Video 2). Recording conditions were tested by transferring the embryos to foster mothers after image acquisition, in which 43% (n = 54; 74% in control, n = 54) of the embryos developed to full term. Live-embryo imaging revealed a progressive increase in number and activity of MTOCs around pronuclei (Fig. 1 J, −0:02 at NEBD) until, upon NEBD, microtubule polymerization increased massively in the region formerly occupied by the pronuclei (Fig. 1 J, 0:22). However, unlike the meiotic oocyte (Schuh and Ellenberg, 2007), no microtubule ball formed, and chromosomes remained aggregated as two clumps in the cell center. Most of the cytoplasmic asters that were not incorporated into the microtubule clusters disappeared in prometaphase, whereas MTOCs (based on pericentrin immunostaining) remained detectable (Fig. 1, F–I), suggesting that those peripheral cytoplasmic MTOCs lost their microtubule-polymerizing activity (Kubiak, 1991). A transient multipolar spindle formed, and the direction of multipoles appeared stochastic. Orientation of the major spindle axis, and possibly of the eventual cleavage plane, was not obvious at this stage in most cases. Progressive clustering of the multipoles, accompanied by fluctuation in the direction of the spindle, led to focusing and establishment of a bipolar spindle (Fig. 1 J, 2:02). Together, the findings indicate that the zygote inherits the random distribution of MTOCs from the oocyte, with no impact from the sperm, and that the stochastic assembly of MTOCs leads to formation of a multipolar spindle that progressively clusters to establish spindle bipolarity.
Dynein is essential for MTOC maturation and, together with kinesin-5, required for bipolar spindle establishment
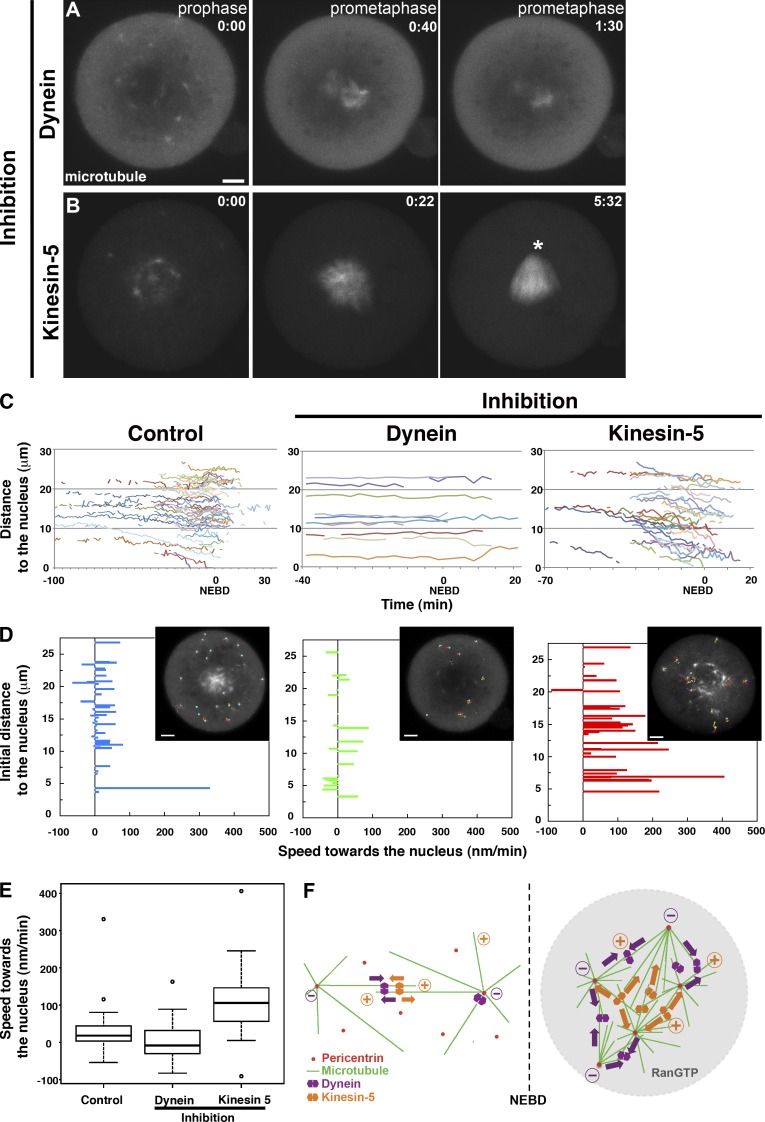
To examine the mechanism of the microtubule reorganization observed in the mouse zygote, we first investigated the potential involvement of cellular motor proteins in MTOC maturation, which refers herein to the change in the ability of MTOCs to polymerize microtubules and organize the microtubule network. Live imaging of embryos after inhibition of dynein activity by microinjecting the mouse zygote with P150-CC1 protein, a dominant-negative form of the P150Glued subunit that blocks dynein–dynactin interaction (Zhang et al., 2009), revealed that MTOC maturation was significantly reduced (Fig. 2 A, 0:00; and Video 3). Quantification of MTOC maturation based on the intensity of the microtubule signal centered around MTOCs indicated those in dynein-inhibited embryos were on average 2.7-fold brighter (n = 30 MTOCs in 6 embryos) than the cytoplasmic background, whereas those in control embryos were 3.6-fold brighter (n = 30 MTOCs in 6 embryos; P < 0.05, Welch two-sample t test; Fig. S2 A). Live imaging also revealed a decrease in the mean number of mature MTOCs at NEBD, from 30.2 (n = 7 embryos) to 18.8 (n = 8; P < 0.05, Welch two-sample t test). On the other hand, inhibition of kinesin-5 activity with monastrol did not alter MTOC maturation in the zygote (Fig. 2 B, 0:00; and Video 4). Thus, the maturation of MTOCs is dependent on dynein activity, possibly because of dynein-mediated recruitment of MTOC components (Quintyne et al., 1999).
Figure 2.
Dynein is essential for maturation of MTOCs and spindle, whereas kinesin-5 is required for spindle bipolarization. (A and B) Live imaging of mouse zygotes during the first division under inhibition of dynein by P150-CC1 (A) or of kinesin-5 by monastrol (B) at prophase (left), early (middle), and late prometaphase (right), respectively. Z-projected images of confocal sections (3 µm thick) show microtubules (EGFP-MAP4; gray). Time indicates hours and minutes after NEBD. Note that the monopolar spindle is formed in the kinesin-5–inhibited zygote (asterisk in B; right). (C) Distance of each cytoplasmic MTOC from the nucleus measured by tracking MTOCs in control, dynein-, and kinesin-5–inhibited zygotes (n = 4, 1, and 3 embryos, respectively) and plotted against time after NEBD. (D) Speed of each MTOC movement toward the nucleus in relation to the initial distance of the MTOC from the nucleus in control, dynein-, and kinesin-5–inhibited embryos. Insets show representative tracks in zygotes. (E) Whisker box plot of speed of MTOC movement toward the nucleus in control (n = 48 tracks derived from four zygotes), dynein-inhibited (n = 38 tracks derived from three zygotes), and kinesin-5–inhibited (n = 32 tracks derived from three zygotes) embryos. MTOC movement is significantly slower in dynein-inhibited embryos and faster in kinesin-5–inhibited embryos than in controls (P < 0.05, Welch two-sample t test). The lines near the middle of the boxes represent the median (50th percentile). The bottom and top of the boxes are the 25th and 75th percentile, respectively. The whiskers extend to the most extreme data point, which is no more than 1.5 times the percentile range of the box. The dots represent the extreme data point extending out of the 1.5 times percentile range of the box. (F) Summary of the potential mechanism leading to acentrosomal spindle assembly in the mouse zygote before (left) and after (right) nuclear envelope breakdown (NEBD). Bars, 10 µm.
Next, we analyzed the impact of motor protein inhibition on spindle assembly. After NEBD, microtubule polymerization in the center of the cell increased but to a substantially lesser extent in dynein-inhibited embryos (2.8 ± 2.0–fold brighter than cytoplasmic background at 20 min after NEBD; n = 6 embryos) than in controls (6.4 ± 1.7–fold; n = 6; P < 0.05, Welch two-sample t test). Two multipolar spindles formed around each pronucleus (Fig. 2 A, 0:40) but failed to fuse and faded away (Fig. 2 A, 1:30; and Video 3). Inhibition of kinesin-5 in zygotes initially produced a multipolar spindle (Fig. 2 B, 0:22; and Video 4), which finally focused into a monopolar spindle (Fig. 2 B, 5:32), and failure in cell division. Thus, the mouse zygote requires dynein for MTOC maturation and spindle assembly and kinesin-5 for separation of the two spindle poles.
Acentrosomal spindle assembly reportedly relies on RanGTP in promoting microtubule nucleation and stabilization as well as in activating motor proteins in proximity of the chromatin (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). Live imaging of mouse embryos after microinjection of the zygotes with RanT24N, a dominant-negative form of Ran, revealed no differences from control embryos until after NEBD, when the microinjected zygotes failed to exhibit the massive increase in microtubule polymerization and in which spindle formation and anaphase entry was delayed (5 h on average after NEBD [n = 6] compared with 2 h in controls [n = 10]; Fig. S2 B) or failed (38%, n = 5/13). Thus, RanGTP facilitates spindle assembly upon NEBD in the mouse zygote, consistent with the mechanism in mouse oocytes (Schuh and Ellenberg, 2007).
MTOC accumulation on the pronuclear membrane depends on dynein and microtubules
Because the spindle is assembled primarily by the MTOCs present on the pronuclear membrane at prophase, we investigated MTOC accumulation, in particular whether MTOCs are preferentially assembled on the pronuclear membrane. The number of MTOCs increased progressively during interphase on the pronuclear membrane (P < 0.05, Welch two sample t test), whereas the number in the cytoplasm remained relatively unchanged (Fig. S2 C). This specific increase of MTOCs on the pronuclear membrane was suppressed in the presence of nocodazole (Fig. S2, C and D), whereas inhibition of RanGTP (Fig. S2 B), dynein (not depicted), actin polymerization (latrunculin B and cytochalasin B; not depicted), or Polo-like kinase 1 (BI2536; not depicted; Lane and Nigg, 1996; Lénárt et al., 2007) had no impact on the ratio of nuclear to cytoplasmic MTOCs, suggesting that this preferential accumulation of MTOCs is dependent on polymerized microtubules. This microtubule dependency might also reflect the collection of MTOCs on the membrane of pronuclei as they form, expand in size, and move from the periphery to the center of the zygote, given that pronuclei movement is also dependent on microtubules (Schatten et al., 1986b).
Requirement of polymerized microtubules prompted us to test the involvement of motor proteins in the potential recruitment of MTOCs to the pronuclear membrane. Tracking of MTOC movement in live images revealed that MTOCs close to the pronuclear membrane (∼5 µm in distance) contribute to the pool on the surface of the pronuclei (n = 4; Fig. 2, C and D; and Video 5), distinct from the wide recruitment of MTOCs from the cytoplasm in oocytes (Schuh and Ellenberg, 2007). Inhibition of dynein activity with P150-CC1 blocked the movement of essentially all MTOCs, demonstrating that MTOC recruitment in the vicinity of nuclei is dependent on dynein in the mouse zygote (Fig. 2, A and C–E; and Video 6). In contrast, inhibition of kinesin-5 by monastrol enhanced the recruitment of distal MTOCs toward the pronuclear membrane (Fig. 2, B–E; and Video 7), although most of them did not reach the nucleus in time to participate in spindle assembly, suggesting that kinesin-5 is already active during zygotic prophase and may counteract attracting forces of dynein. Overall, although the first division of the mouse embryo partly shares the mechanism of spindle assembly with the meiotic division, some features and mechanisms are clearly distinct from those in meiosis (Fig. 2 F), suggesting that the transition from meiosis to mitosis starts already during the first division.
Progressive transition from acentrosomal to centrosomal spindle formation during mouse preimplantation development
Although our findings thus far demonstrated that the meiosis to mitosis transition starts during the first division in the mouse embryo, the mechanism of spindle assembly in the zygote has many similarities to meiosis and is very distinct from a typical mitotic division with centrosomal spindles. Thus, the transition from multipolar self-assembly of MTOCs to bipolar centrosomal spindle assembly is likely established during preimplantation development before the 64-cell blastocyst stage (Gueth-Hallonet et al., 1993). However, the characteristics and mechanism of this transition, e.g., whether the transition is abrupt or gradual during subsequent embryonic cleavages, remain unclear. Therefore, we analyzed spindle assembly throughout the preimplantation stage until late blastocyst.
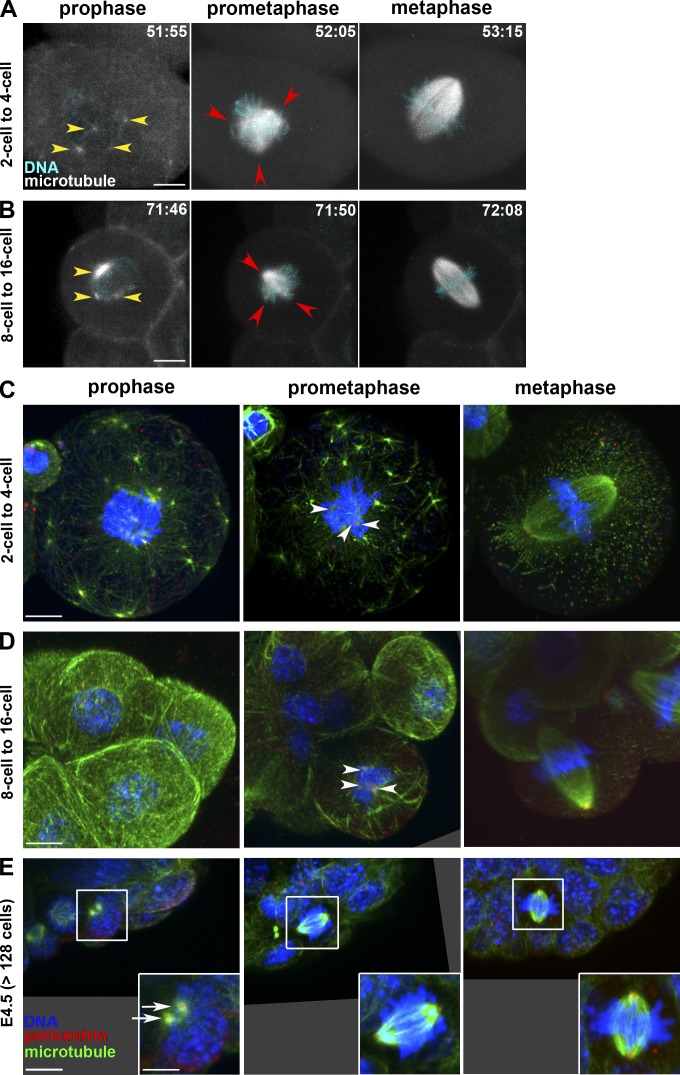
Live imaging of microtubule dynamics (Fig. 3, A and B; and Videos 8 and 9) and immunostaining for pericentrin (MTOC) and microtubules (Fig. 3, C–E) revealed several MTOCs and microtubule asters in the cytoplasm and around the nucleus at prophase not only in the first but also in the second (two cell to four cell) and third (four cell to eight cell) divisions (Fig. 3, A and C; and Fig. S3). After NEBD, microtubule clusters formed a multipolar spindle in prometaphase and assembled into a barrel-shaped bipolar spindle in metaphase (Fig. 3, A and C; and Video 8). From the eight-cell until the 32-cell blastocyst stage, multiple asters were present around the nucleus in prophase, whereas fewer were seen in the cytoplasm (Fig. 3, B and D). Prometaphase exhibited a transient multipolar spindle, with MTOCs along the axis but not yet exclusively localized at the pole (Fig. 3, B and D; and Video 9), which rapidly elongates to form the metaphase spindle reminiscent of the typical mitotic spindle, with pericentrin focused as dots at two poles (Fig. 3, B and D).
Figure 3.
Progressive transition from acentrosomal to centrosomal spindle formation during mouse preimplantation development. (A and B) Live imaging of mouse embryos during the second division (A) and the fourth division (B) at prophase, prometaphase, and metaphase. Note the small microtubule asters (presumably MTOCs; yellow arrowheads) in two-cell and eight-cell stage embryos before NEBD. Red arrowheads mark multipoles of the spindle at prometaphase. Z-projected images of 3-µm confocal sections showing microtubules (EGFP-MAP4; gray) and DNA (H2B-mRFP1). Time is shown in hours and minutes after hCG. (C–E) Immunostaining of embryos fixed during the second division (showing one of the two cells; C), the fourth division (D), and at 113 h after hCG (E4.5; E) at prophase, prometaphase, and metaphase and stained for DNA, microtubules, and pericentrin. In prometaphase at the two-cell and eight-cell stages, several small MTOCs are visible (arrowheads), whereas only two bright MTOCs (arrows) before and after NEBD are seen at E4.5. All pictures are projected images of 0.38-µm stacks. Insets in E represent a zoom of the boxes. Bars: (A–E, main images) 10 µm; (E, insets) 5 µm.
In the embryonic day 4.5 (E4.5) blastocyst (128-cell stage or later), interphase cells had one or two bright MTOC dots, particularly at the basal (blastocyst cavity) side next to an invagination of the nucleus in trophectoderm cells, in agreement with an electron microscopic study (Gueth-Hallonet et al., 1993). At prophase, two bright MTOCs next to the nucleus were recognizable, which became separated to form the spindle poles at prometaphase, with no other MTOC detectable along the spindle axis (Fig. 3 E). At metaphase, the spindle was focused on two bright pericentrin dots (Fig. 3 E), which are most likely functional centrosomes, as indicated by colocalization of one of the centrosomal components, centrin (Fig. 4 A). These divisions can thus be defined as typical mitosis, consistent with reappearance of the centriole at the 64-cell stage (Gueth-Hallonet et al., 1993).
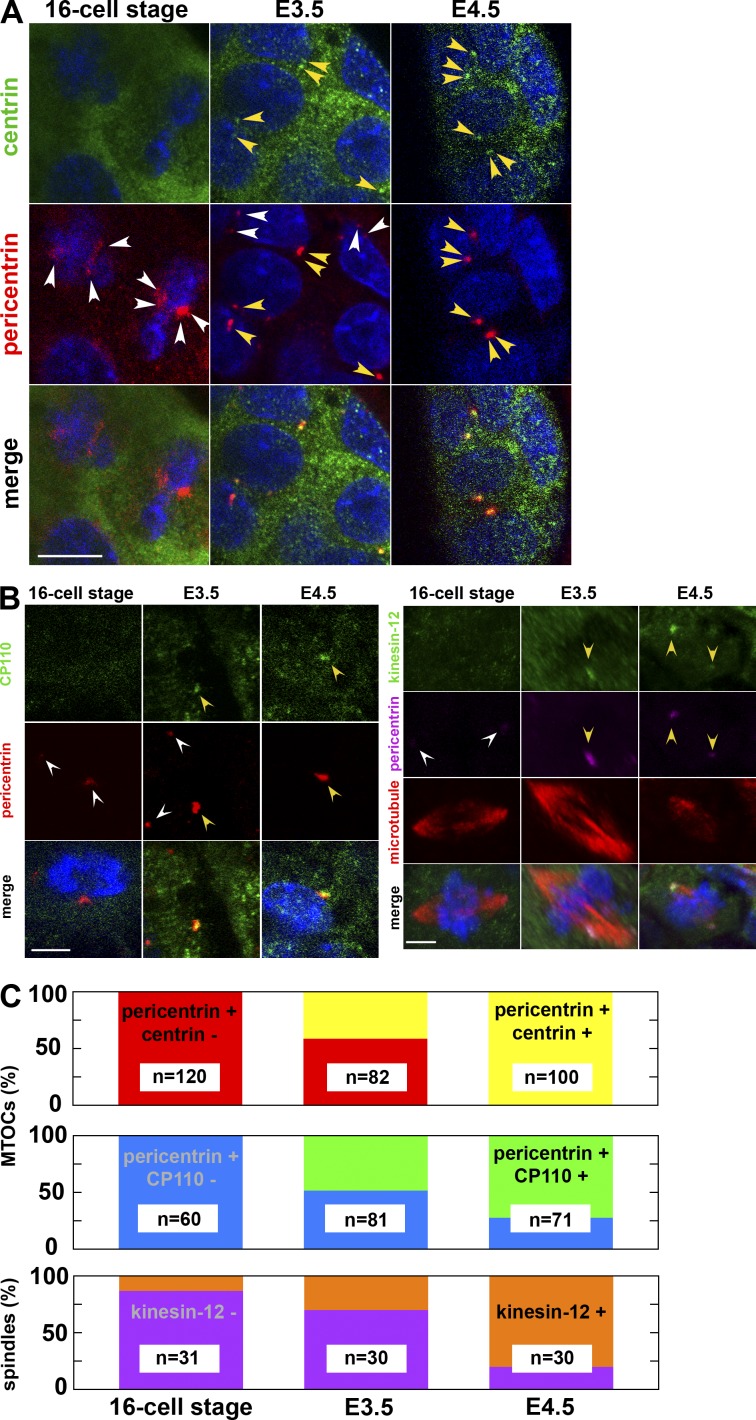
Figure 4.
Emergence of the centrin- and CP110-positive centrosome and the kinesin-12–positive spindle in the E3.5 blastocyst. (A) Immunostaining of embryos fixed at the 16-cell stage, E3.5 (between 32- and 64-cell stages), and E4.5 (with >128 cells) and stained for DNA (blue), centrin, and pericentrin. At the 16-cell stage, all pericentrin-positive MTOCs are negative for centrin. At E3.5, some MTOCs are positive for pericentrin and negative for centrin (white arrowheads), whereas others are positive for both (yellow arrowheads). At E4.5, one or two dots positive both for pericentrin and centrin are visible in each cell (yellow arrowheads). Note that the laser intensity for centrin detection was enhanced in 16-cell stage and E3.5 embryos, resulting in the enhanced signal in the cytoplasm. Because single-section images of confocal microscopy are shown, the centrin and pericentrin signal of other cells are out of focus. Bar, 10 µm. (B, left) Immunostaining of mouse embryos fixed at the 16-cell stage, E3.5, and E4.5 and stained for DNA (blue), CP110, and pericentrin. At the 16-cell stage, MTOCs are negative for CP110. At E3.5, only some MTOCs are positive for CP110 (yellow arrowheads; white arrowheads mark those negative for CP110), whereas at E4.5, all MTOCs are positive. (right) Immunostaining of mouse embryos fixed at the 16-cell stage, E3.5, and E4.5 and stained for DNA (blue), kinesin-12, pericentrin, and microtubules. At the 16-cell stage, the spindle is negative for kinesin-12 (white arrowheads). At E3.5, only some spindles are positive for kinesin-12 (yellow arrowheads), whereas at E4.5, most of the spindles are positive (yellow arrowheads). Z-projected sections of confocal images. Bars, 5 µm. (C) The fraction of centrin-positive (yellow) and -negative (red) MTOCs, CP110-positive (green) and -negative (blue) MTOCs, and of kinesin-12–positive (orange) and –negative (violet) spindles at the 16-cell, E3.5, and E4.5 stages.
Unlike the E4.5 embryo, in which all examined MTOCs (n = 100; Fig. 4 C) showed centrin–pericentrin colocalization, we detected this colocalization in only 41% of MTOCs (n = 34 of 82) in the E3.5 embryo (32- to 64-cell blastocyst stage; Fig. 4 A, yellow arrowheads). Remarkably, centrin-positive MTOCs (0.82 µm mean diameter of 16 MTOCs from 5 E4.5 blastocysts) were significantly larger than centrin-negative MTOCs (0.54 µm of 13 centrin-negative MTOCs from 5 E3.5 blastocysts; P < 0.05, Welch two-sample t test), suggesting that the centrin-positive MTOCs accumulate pericentrosomal materials as expected for centrosomes. Note that we found cells with centrin-positive MTOCs in both inner cell mass and trophectoderm within the blastocyst, in a manner similar to the lineage-independent shift from kinesin-5–dependent to –independent metaphase spindle function (FitzHarris, 2009). In agreement with the gradual emergence of centrin-positive centrosomes, other centrosomal components, odf2 (Kunimoto et al., 2012) and CP110 (Schmidt et al., 2009), also start expression in the E3.5 blastocyst (Fig. 4, B and C; and Fig. S4). It is particularly interesting to note that expression of kinesin-12, a motor protein involved in formation and maintenance of mitotic spindles (Tanenbaum et al., 2009; Liu et al., 2010), at E3.5 (Fig. 4, B and C) coincides with the disappearance of the kinesin-5 dependency for the spindle maintenance (FitzHarris, 2009).
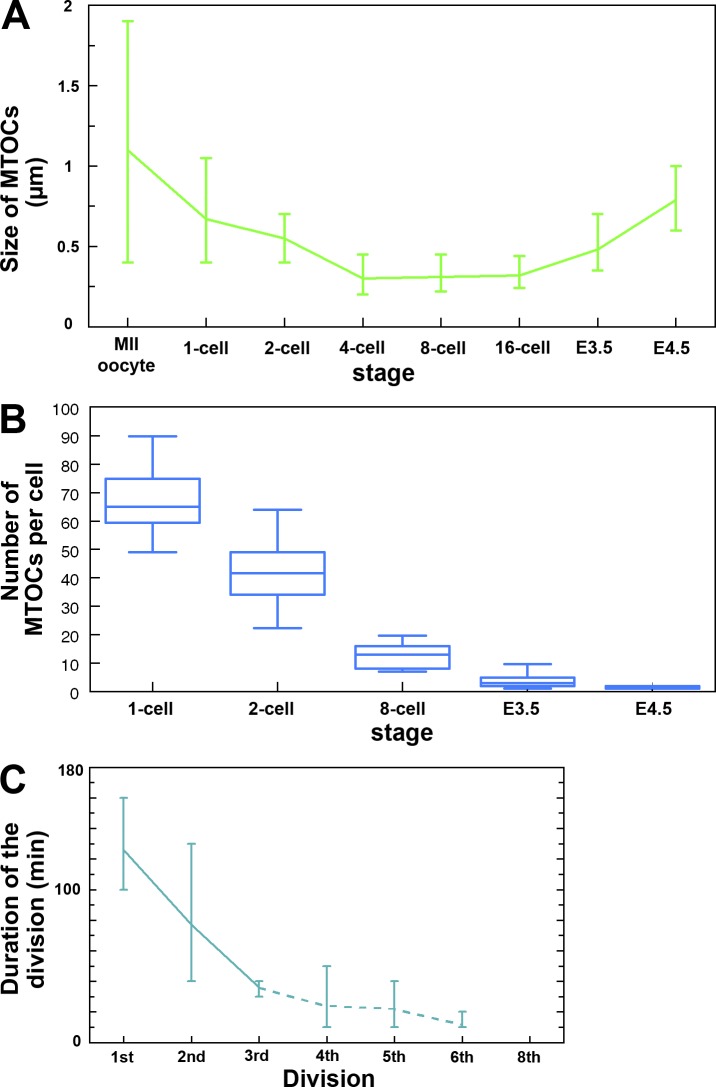
Next, we examined the size and number of MTOCs during the oocyte to embryo transition. After oocytes became fertilized, the mean size of MTOCs progressively decreased (Fig. 5 A). Only at E3.5 when centrin started colocalization at MTOCs, the size of MTOCs increased gradually to a size close to that for typical centrosomes (∼1.0 µm; Piehl et al., 2004; Decker et al., 2011). Although the number of MTOCs per cell progressively decreased during the preimplantation stage (Fig. 5 B), the total number per embryo increased, i.e., 68 on average in the zygote (n = 19), 82 in the two-cell (n = 16), and 102 in the eight-cell embryos (n = 13), pointing to de novo MTOC formation during the preimplantation stage.
Figure 5.
Progressive change in MTOCs and the time required for division. (A) Progressive change in diameter of MTOCs shortly before NEBD in the embryos at consecutive developmental stages. (B) Whisker box plot of the number of MTOCs per cell at one-cell, two-cell, eight-cell, E3.5, and E4.5 stages, shortly before NEBD. The lines near the middle of the boxes represent the median (50th percentile). The bottom and top of the boxes are the 25th and 75th percentile, respectively. The end of the bottom whisker is the 5th percentile, and the end of the top whisker is the 95 percentile. (C) Duration of cell division (from NEBD to the beginning of anaphase) at consecutive stages of preimplantation development. Note that from the eight-cell stage on, the duration of cell division (∼20 min or less) becomes only twice as long as the time interval of recording (every 10 min); thus, the data cannot be as precise (indicated by a broken line) as those for the earlier stages. In A and C, the vertical bars indicate the range of values.
Finally, we examined the change in the time required for division. Live imaging at 10-min intervals of embryos injected with mRNA encoding EGFP-MAP4 and H2B-mRFP1 revealed progressive decreases in the time between NEBD and the beginning of anaphase, with a mean of 126 min during the first division to 10 min at the 32- to 64-cell transition (Fig. 5 C). This progressive decrease in division time duration, possibly because of quicker spindle assembly from the smaller number of MTOCs, is not proportional to that of cell cycle duration (not depicted; Kurotaki et al., 2007) but correlates with the decrease in cell size (compare with Fig. 6 B). The features of spindle assembly and of MTOCs suggest a gradual transition rather than a sudden change from multiple MTOCs to functional centriole-containing centrosomes during mouse preimplantation development.
Figure 6.
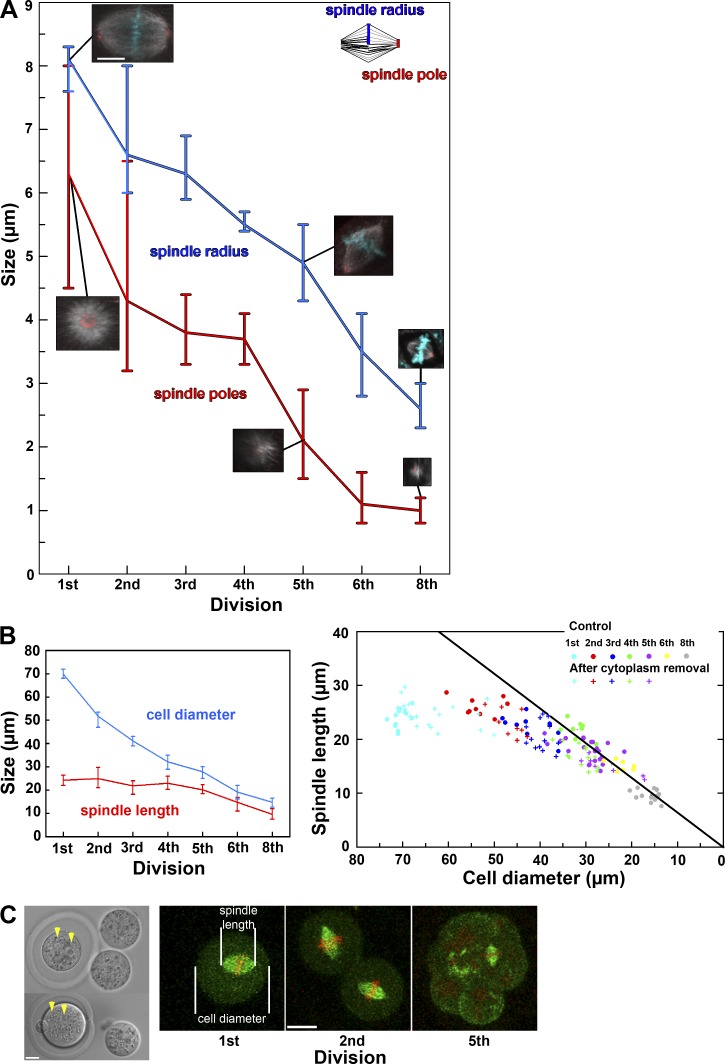
Gradual change in spindle characteristics and establishment of the spindle length regulation during mouse preimplantation development. (A) Progressive change in radius and diameter of the spindle in the embryos at consecutive developmental stages. Insets show a representative image of the spindle and its pole at each stage. Bar, 10 µm. (B, left) Change in the mean size of the spindle and of the cell in the embryos at consecutive developmental stages. (right) Cell diameter and spindle length are plotted as colored circles for individual embryos at different developmental stages, illustrating that their ratio (slope of the black line) remains constant from the fourth to eighth division. Those for experimentally micromanipulated embryos are shown as colored crosses. (C, left) Experimentally micromanipulated zygotes in which two thirds (top) or half (bottom) of the cytoplasm was removed. Note that two pronuclei are visible (yellow arrowheads) after cytoplasm removal. Metaphase spindle and measurement of its size and cell size by live imaging of the micromanipulated embryos during the subsequent divisions. All pictures are projected images of 4.5-µm stacks. Bars, 20 µm. In A and B, the vertical bars indicate the range of values.
Gradual change in spindle characteristics and establishment of spindle length regulation
In view of the gradual change in the manner of spindle assembly throughout the preimplantation stage, we examined in more detail the change in spindle characteristics. Unlike the circular deposition of MTOCs at the spindle poles in oocytes and embryos until the eight-cell stage, the pericentrin signal at eight-cell to 16-cell divisions was localized as dots at two poles, and the spindle poles became increasingly focused in the divisions from the 16-cell stage (Fig. 6 A and Fig. S5). Accordingly, the radius of the spindle and the size of the spindle poles (Fig. 6 A, see inset for definition) progressively decreased during the preimplantation stage (Fig. 6 A).
Spindle length was similar (mean of 24 µm) in the first three divisions but decreased from 22 µm at the eight-cell stage to 16 µm at the 32- to 64-cell stage (Fig. 6 B), suggesting an association between change in spindle size after the eight-cell stage and the meiosis to mitosis shift. Because cell size also progressively decreased and the slope of its decrease paralleled that for the spindle length after the fourth division (Fig. 6 B), we tested for a possible correlation between spindle size and cell size. Indeed, plotting these two parameters for each cell derived from embryos at the first to eighth division (Fig. 6 B, right) revealed a remarkably constant cell size to spindle size ratio of ∼1.6 from the fourth division onward (mean of 1.5, 1.7, 1.5, and 1.6 for the fourth, fifth, sixth, and eighth division, respectively), consistent with the reported scaling effect of cell size on spindle length (Wühr et al., 2008; Hara and Kimura, 2009). Spindle length was relatively constant at earlier divisions (mean of 22.8, 26.3, and 22.8 µm for the first, second, and third division, respectively), suggesting an upper limit to the mitotic spindle length in the mouse embryo, in agreement with that for Xenopus laevis embryos (Wühr et al., 2008).
To test whether spindle size regulation depends on the developmental stage or, instead, on cell size, we examined cell and spindle size during division of live embryos that were depleted of various amounts of cytoplasm (≤65%) by micromanipulation (see Materials and methods for detailed methods; Fig. 6 C). Live imaging of the embryos injected with mRNA for EGFP-MAP4 and H2B-mRFP1 demonstrated that the relationship between cell size and spindle length was conserved between control and manipulated embryos, regardless of the developmental stage, resulting in overlapping plots (Fig. 6 B, right). This result clearly suggests that the metaphase spindle length is not correlated to cell size during the first divisions. Once the cell size approaches <1.6-fold of the upper limit of the spindle length, the spindle size scales to cell size. The scaling effect thus becomes operational at around the eight-cell stage between third and fourth embryonic divisions and is independent of developmental cues. Overall, the progressive change in the morphology, characteristics, and molecular components of MTOCs and the spindle in the early mouse embryo supports the notion that the transition from meiosis to mitosis and establishment of the mitotic spindle properties proceed gradually throughout the preimplantation stage.
Discussion
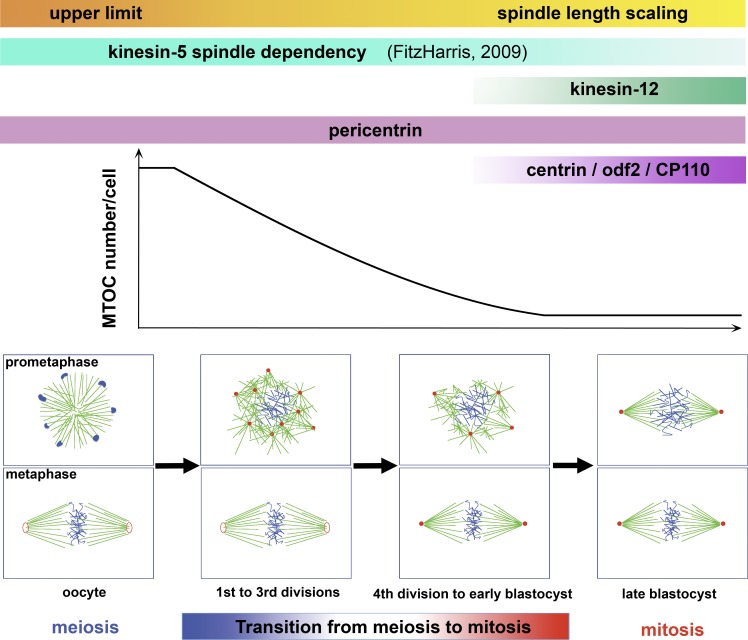
Our study demonstrates that during the meiosis to mitosis transition, changes in MTOC organization, spindle assembly, and characteristics and in cell division time do not occur abruptly upon fertilization but progress gradually instead throughout the first eight divisions of the preimplantation mouse embryo. The transition can be subdivided into three phases (Fig. 7): (1) the first three embryonic divisions, when the mechanism of acentrosomal spindle formation is largely shared with meiotic division; (2) the divisions from eight-cell stage until the blastocyst, in which multiple MTOCs or potentially some centrosomes are focused into a sharp bipolar spindle; and (3) the divisions after blastocyst, in which two centrosomes assemble a typical mitotic spindle. The first three divisions in the mouse embryo partially share the mechanism of meiotic spindle formation (summarized in Fig. 2 F, after NEBD). Given the comparable number and distribution pattern of MTOCs from MII oocyte to zygotes, presumably zygotes inherit MTOCs from oocytes but not from sperm. Nonetheless, the first three divisions also exhibit a change toward mitosis with respect to MTOC maturation and spindle assembly (Fig. 2 F, before NEBD). In the zygote, MTOCs are recruited for spindle assembly only from the vicinity of the pronuclei, whereas recruitment in the oocyte is from throughout the cytoplasm (Schuh and Ellenberg, 2007), possibly caused by the lack of the counteracting kinesin-5 force in oocytes. This shift toward mitosis could also account for the lack of microtubule ball formation upon NEBD in zygotes.
Figure 7.
Gradual transition from meiotic to mitotic spindle assembly throughout the preimplantation stage in the mouse embryo. Summary of the progressive transition from meiosis to mitosis throughout mouse preimplantation development. See Discussion for details.
The second phase, from eight-cell to 64-cell stage, is the time of transition from meiosis-like divisions to mitotic divisions. The number of MTOCs per cell progressively decreases during the preimplantation stage, whereas the total amount of MTOC material per embryo, as estimated by multiplying the mean MTOC number with the volume (Fig. 5, A and B), is 3.2, 2.2, 0.5, and 1.8 (arbitrary units) in the zygote, two-cell, eight-cell, and E3.5 stage embryos, respectively, compared with 20.2 in the E4.5 blastocyst. Thus, it is plausible that until blastocyst, noncentriolar MTOCs are generated by splitting the limited amount of materials inherited from the oocyte and available in the embryo in a manner similar to the centrosomal material reported for early Caenorhabditis elegans embryos (Greenan et al., 2010; Decker et al., 2011). When MTOC materials available in a cell become too few and weak, the centrosome generation might be activated. The precise evaluation of this model in the mouse embryo awaits further studies.
The regulation of spindle length according to cell size becomes active as soon as the ratio approaches 1.6, around the fourth division, similar to that observed in HeLa cells (1.4; Goshima and Scholey, 2010). Although the apparent plateau in spindle length during earlier divisions suggests an upper limit in metaphase spindle length, in agreement with Wühr et al. (2008), we cannot exclude the possible scaling correlation between the anaphase spindle length and cell size (Hara and Kimura, 2009) because of the difficulty in defining the spindle length in anaphase in which the chromosomes move toward the daughter cell poles beyond the eventual nuclear position in the early mouse embryo (e.g., Video 9). Overall, the transition is gradual during this period, not sharp or step wise, and not synchronous in timing, as indicated by the asynchronous emergence of cells with centrin-positive MTOCs (centrosomes) in E3.5 blastocysts.
In the third phase, i.e., after the 64-cell stage, the centriole is present (Gueth-Hallonet et al., 1993), and the spindle is clearly focused with a well-defined axis at prometaphase. All of these features define cell division later than the blastocyst stage as typical mitosis.
Mouse preimplantation development is not only viewed as a unique phase in terms of developmental mechanisms (O’Farrell et al., 2004; Motosugi et al., 2005) but also provides a unique system to study the transition from meiotic to mitotic cell division (this study; Kubiak et al., 2008; FitzHarris, 2009). This transition could possibly apply to other organisms: In human zygotes, centrioles are introduced by the sperm (Simerly et al., 1995) and are detectable by electron microscopy in one of the spindle poles (Sathananthan et al., 1991). The essential role of the centriole for spindle assembly in early human embryos, however, remains to be shown, as parthenogenetically activated embryos can develop up to the blastocyst stage (Paffoni et al., 2007; de Fried et al., 2008).
Conceivably, in mouse development, the molecular components necessary for reestablishment of the centriole and centrosome are produced during the preimplantation stage, given the lack of evidence for centriole propagation. It will be of particular interest to determine molecules operating during this progressive transition and the stage they become active. The trigger for de novo centriole formation in the blastocyst remains to be investigated but might reflect an exhausted supply of some MTOC components, transcriptional activation of key centriolar components during preimplantation development, or reaching threshold levels in some of the progressively changing cellular features. Because the mouse preimplantation embryo exhibits de novo centrosome formation under physiological conditions, it is an attractive system to investigate the mechanism of centrosome biogenesis and propagation. Future studies promise to identify the components essential for centrosome generation and propagation.
Materials and methods
Ethics statement
The handling of laboratory mice necessary to pursue all the proposed experiments (killing for collection of oocytes, fertilized eggs, and embryos, injections of exogenous gonadotropins for collection of the embryos, and embryo transfer to anesthetized foster mothers) was performed in the animal facility of the institute according to the permission from S. Aschhoff (European Molecular Biology Laboratory, Heidelberg, Germany; Animal Research Committee number TH11 00 11).
The European Molecular Biology Laboratory animal facility is operating according to international animal welfare rules (Federation for Laboratory Animal Science Associations guidelines and recommendations). Requirements of formal control of the German national authorities and funding organizations are satisfied and controlled by the Institutional Animal Care and Use Committee.
Embryo culture
B6C3F1 female mice were injected with 0.1 ml (5 IU) of pregnant mare serum gonadotropin followed 46–48 h later by injection with 0.1 ml (5 IU) hCG before mating. Embryos were cultured in 10-µl drops of KCl- and NaCl-enriched simplex optimized medium (KSOM; EMD Millipore; Zenith Biotech) covered with mineral oil (Acros Organics; Sigma-Aldrich) in a 5% CO2 atmosphere at 37°C. For live imaging, embryos were cultured in similar 5-µl drops prepared in MatTek 35-mm glass-bottom dishes in a 5% CO2 chamber (PeCon) at 37°C on the microscope stage. Depending on the experiments, medium was supplemented with 0.3 µM nocodazole (Sigma-Aldrich) or 100 µM monastrol (Sigma-Aldrich), for which control samples were prepared with equivalent amounts of DMSO. Note that under treatment with 0.3 µM nocodazole, the zygote retains part of the microtubule matrix (Fig. 3 B), whereas the oocyte loses essentially all microtubules (consistent with the results in Schuh and Ellenberg, 2007; see Table S1 for a summary comparing our experimental conditions to those in Schuh and Ellenberg, 2007), caused possibly by the reported difference in the dynamics of microtubule polymerization between oocytes and early embryos (Kubiak, 1991).
Immunofluorescence analysis
For embryo recovery, particular attention was paid to minimizing the time required for the recovery (∼1 h after sacrifice of the mother until fixation). After removal of the zona pellucida with pronase (Sigma-Aldrich), embryos were fixed for 30 min at 37°C with 100 mM Hepes, 50 mM EGTA, 10 mM MgSO4, 2% formaldehyde, and 0.2% Triton X-100 and permeabilized for 2 h at room temperature in PBS supplemented with 0.2% Triton X-100, based on the method used by Schuh and Ellenberg (2007; Table S1). For imaging specifically microtubules (as in Fig. 1 and Fig. 3), embryos were fixed in the fixation solution supplemented with 0.05% glutaraldehyde and washed for 1.5 h in PBS supplemented with 0.01 mg/ml NaBH4. For imaging odf2, CP110, and kinesin-12, embryos were fixed in 100% cold methanol (−20°C) for 10 min, rehydrated in 50% methanol-PBS (4°C) for 5 min, and washed in PBS supplemented with 0.1% Triton X-100. Embryos were then incubated in PBS with 3% BSA and 0.1% Triton X-100 with, depending on the experiments, rat antityrosinated α-tubulin (YL 1/2, MCA77G; 1:200,000; AbD Serotec), mouse antipericentrin (611814; 1:200; BD), rabbit antipericentrin (PRB-432C; 1:200; Covance), rabbit anti–centrin-1 (ab11257; 1:200,000; Abcam), mouse anti–γ-tubulin (6657; 1:40,000; Sigma-Aldrich), rabbit anti-odf2 (ab43840; 1:1,000; Abcam), rabbit anti-CP110 (1:1,000; gift from E. Nigg, Biozentrum, University of Basel, Basel, Switzerland; Schmidt et al., 2009), or rabbit anti–kinesin-12 (1:200; gift from P. Baas, Drexel University College of Medicine, Philadelphia, PA; Liu et al., 2010) primary antibodies followed by Alexa Fluor 488–, 546–, 555–, 633–, or 647–conjugated secondary antibodies (1:200; Molecular Probes). DNA was stained using Hoechst 33342 (1:2,000; Molecular Probes). Immunostained images were acquired using a confocal microscope (LSM 780; Carl Zeiss) equipped with a 40× water immersion C-Apochromat, 1.2 NA objective (Carl Zeiss) at room temperature (20°C) in PBS containing 1% BSA. The microscope (LSM 780) was controlled using the ZEN 2010 software (Carl Zeiss). The pinhole was open to the 1-µm thickness of the stack, and when the z stack was acquired, the interval used between stacks varied from 0.4 (optimal) to 0.5 µm. Excitation of Hoechst 33342 was performed using a 405-nm diode laser. Excitation of Alexa Fluor 488 was performed using a 488-nm argon laser, excitation of Alexa Fluor 546 and 555 was performed using a 561-nm diode-pumped solid-state laser, and excitation of Alexa Fluor 633 and 647 was performed using a 633-nm helium/neon laser.
Microinjection and micromanipulation
mRNA was synthesized using an mRNA message machine kit (mMESSAGE mMACHINE; Ambion). Plasmids for P150-CC1 and His-GFP were a gift from X. Zhu (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China). Proteins were expressed in Escherichia coli strain BL21(DE3) and purified on a nickel column using His tags accordingly (Zhang et al., 2009) with the help of M. Pfeiffer (Max-Planck Institut, Münster, Germany). Proteins were dialyzed in PBS and concentrated to 24 mg/ml using a centrifugal filter (Amicon Ultra; EMD Millipore). The RanT24N protein was a generous gift of R. Walczak and I. Mattaj (European Molecular Biology Laboratory, Heidelberg, Germany). Microinjections in the zygote or in both blastomeres of the two-cell embryo for blastocyst live imaging were performed at 32°C in 10-µl flushing-holding medium (EMD Millipore) drops covered with mineral oil (Sigma-Aldrich) on a microscope (Axio Observer.Z1; Carl Zeiss) using micromanipulators (Narishige). mRNA encoding H2B-mRFP1 (final concentration of 0.01 µg/µl) and 0.8 µg/µl EGFP-MAP4 were mixed together in RNase-free water (Ambion) and microinjected using an air microinjector (FemtoJet; Eppendorf). The protein P150-CC1 (24 mg/ml) in PBS or RanT24N (270 µM) in PBS, 0.15 M NaCl, 1.5 mM MgCl2, and 10% glycerol, each mixed with dextran–Texas red (molecular weight of 70,000; 1:10; Molecular Probes) to compare the injected amount between embryos, was microinjected using an oil microinjector (Narishige) and a Piezo microinjector controller (PMAS-CT150; PrimeTech, Ltd.). The respective control was injected with 24 mg/ml GFP-His protein in PBS mixed with dextran–Texas red (1:10) or with 280 µM BSA (Sigma-Aldrich) in PBS and 10% glycerol mixed with dextran–Texas red (1:10). Activity of P150-CC1 was confirmed in MII oocytes, in which it disassembled the spindle upon microinjection. mRNA injections were performed ≥3 h before imaging. To generate the embryo with reduced cell size, the zona pellucida was cut with a needle (Tsunoda et al., 1986), and a fraction of the cytoplasm was removed in KSOM containing Hepes (Zenith Biotech) and 5 µg/ml cytochalasin B (Sigma-Aldrich) using a pipette 25–30 µm in diameter. The amount of removed cytoplasm can be calculated from the reduced cell size: in which r = radius and V = volume of the cell. This was confirmed by measurement of the radius of the zygote after cytoplasmic removal, i.e., when one half or two thirds of the cytoplasm was removed, the radius of the micromanipulated zygote was ∼28–32 or 25–28 µm, respectively, in comparison to the radius of the control zygote, 35–40 µm.
Live imaging of embryos
Time-lapse images were acquired using a confocal microscope (LSM 780) equipped with a 40× water immersion C-Apochromat, 1.2 NA objective at 37°C in KSOM medium. The microscope (LSM 780) was controlled using the ZEN 2010 software. Several embryos were imaged simultaneously, and sometimes in different groups, using the multipoint acquisition function in ZEN 2010 software. The pinhole was open to the 3.5-µm thickness of the stack, and when the z stack was acquired, the interval used between stacks varied from 1.7 (optimal) to 3.5 µm. Excitation of EGFP and mRFP1 was performed using a 488-nm argon laser and a 561-nm diode-pumped solid-state laser, respectively. To verify the live-imaging conditions, embryos imaged under a typical imaging condition (five stacks every 2 µm at 5-min intervals at 0.5% laser intensity for both lasers overnight during the first division for a period of 10 h, i.e., 120 cycles) were transferred into a foster mother, with imaged embryos on one side and the control on the other side. Nine embryo transfers for a total of 54 imaged embryos and 54 control embryos were performed, giving rise to 23 and 40 live pups, respectively.
Image analysis and statistics
All image analyses were performed using Imaris (Bitplane). MTOCs were tracked automatically using the tracking function and verified manually. Maturation of MTOC was assessed by measuring the mean intensity value of a 1-µm sphere centered around the MTOC as compared with the background intensity of a 5-µm sphere in the cytoplasm.
The size of the MTOC was defined by the diameter of the circle at which the intensity of the immunostained pericentrin signal becomes half the maximal intensity (Fig. S1 C; Jaensch et al., 2010). Statistical analyses were conducted using R and SlideWrite Plus (Advanced Graphics Software, Inc.). Distribution of MTOCs in the zygote was evaluated by the two-sample Kolmogorov–Smirnov test applied on the observed value of K3(r) against the theoretical value for complete spatial randomness. K3(r) is the image of r, radius of the point, by the K function of a 3D point pattern calculated using the Spatstat package for R (Baddeley and Turner, 2005). A random noise pattern was generated on a cube using ImageJ (National Institutes of Health), which resulted in P = 1 (Schneider et al., 2012). An artificially biased and nonrandom pattern (Fig. S1), generated by choosing the 10 most prominent MTOCs and 25 additional dots in a restricted volume in the embryo, gave P = 0.006723 (<0.05).
Online supplemental material
Fig. S1 shows the measurement of distribution and size of MTOCs in the mouse zygote. Fig. S2 shows that dynein is essential for MTOC maturation, whereas RanGTP facilitates spindle assembly. Fig. S3 shows the presence of multiple MTOCs at prophase during four- to eight-cell division. Fig. S4 shows the progressive emergence of odf2, a centrosome marker. Fig. S5 shows the progressive focusing of spindle poles at 16- to 64-cell stage mouse embryos. Table S1 shows the comparison of the experimental conditions in this study to those used in Schuh and Ellenberg (2007). Video 1 shows a multipolar spindle at prometaphase during the first division of the mouse zygote. Video 2 shows a time lapse of the mouse zygote during the first division. Video 3 shows a time lapse of the dynein-inhibited mouse zygote during the first cleavage. Video 4 shows a time lapse of the kinesin-5–inhibited mouse zygote during the first cleavage. Video 5 shows the tracking of MTOCs during the first division. Video 6 shows the tracking of MTOCs in the dynein-inhibited mouse zygote during the first cleavage. Video 7 shows the tracking of MTOCs in the kinesin-5–inhibited mouse zygote during the first cleavage. Video 8 shows a time lapse of the mouse embryo during the second division. Video 9 shows a time lapse of the mouse embryo during the fourth division. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201202135/DC1.
Supplementary Material
Acknowledgments
The authors thank Xueliang Zhu for p150-CC1 and His-GFP constructs, Martin Pfeiffer for p150-CC1 and His-GFP protein purification, Rudolf Walczak and Iain Mattaj for RanT24N protein, Erich Nigg for the CP110 antibody, Peter Baas for the kinesin-12 antibody, Anton Pigge for help with mouse operations, and Kota Miura for help with statistical analysis. We also thank staff in the Hiiragi, Ellenberg, and Schuh laboratories for helpful and stimulating discussions, Zbigniew Polanski and Jacek Kubiak for critical reading and comments, and Christian Helker for his continuous encouragement.
Work in the laboratory of T. Hiiragi is supported by Max Planck Society, European Research Council under the European Community’s Seventh Framework Program, Stem Cell Network North Rhine Westphalia, German Research Foundation (Deutsche Forschungsgemeinschaft), and World Premier International Research Center Initiative, Ministry of Education, Culture, Sports, Science and Technology, Japan. M. Schuh’s and J. Ellenberg’s contribution to the research leading to these results has received funding from the European Community’s Seventh Framework Program (FP7/2007-2013-241548) within the MitoSys consortium.
Footnotes
Abbreviations used in this paper:
- hCG
- human chorionic gonadotropin
- KSOM
- KCl- and NaCl-enriched simplex optimized medium
- MTOC
- microtubule-organizing center
- NEBD
- nuclear envelope breakdown
References
- Baddeley A., Turner R. 2005. Spatstat: an R package for analyzing spatial point patterns. J. Stat. Softw. 12:1–42 [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bessis M., Breton-Gorius J., Thiery J.P. 1958. Centriole, corps de Golgi et aster des leucocytes. Etude au microscope electronique. Rev. Hematol. 13:363–386 [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D.M. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Boveri T. 1887. Ueber die Befruchtung. der Eier von Ascaris megalocephala. Sitz.-Ber. Ges. Morph. Phys. Mçnchen. 3:71–80 [Google Scholar]
- Boveri T. 1888. Zellen-Studien II. Die Befruchtung und Teilung des Eies von Ascaris megalocephala. Jena. Z. Naturwissen. 22:685–882 [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Decker M., Jaensch S., Pozniakovsky A., Zinke A., O’Connell K.F., Zachariae W., Myers E., Hyman A.A. 2011. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21:1259–1267 10.1016/j.cub.2011.06.002 [DOI] [PubMed] [Google Scholar]
- de Fried E.P., Ross P., Zang G., Divita A., Cunniff K., Denaday F., Salamone D., Kiessling A., Cibelli J. 2008. Human parthenogenetic blastocysts derived from noninseminated cryopreserved human oocytes. Fertil. Steril. 89:943–947 10.1016/j.fertnstert.2007.04.045 [DOI] [PubMed] [Google Scholar]
- de Harven E., Bernhard W. 1956. Etude au microscope electronique de l’ultrastructure du centriole chez les vertebres. Z. Zellforsch. Mikrosk. Anat. 45:378–398 10.1007/BF01106086 [DOI] [PubMed] [Google Scholar]
- FitzHarris G. 2009. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development. 136:2111–2119 10.1242/dev.035089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Scholey J.M. 2010. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26:21–57 10.1146/annurev-cellbio-100109-104006 [DOI] [PubMed] [Google Scholar]
- Greenan G., Brangwynne C.P., Jaensch S., Gharakhani J., Jülicher F., Hyman A.A. 2010. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol. 20:353–358 10.1016/j.cub.2009.12.050 [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Antony C., Aghion J., Santa-Maria A., Lajoie-Mazenc I., Wright M., Maro B. 1993. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J. Cell Sci. 105:157–166 [DOI] [PubMed] [Google Scholar]
- Hara Y., Kimura A. 2009. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 19:1549–1554 10.1016/j.cub.2009.07.050 [DOI] [PubMed] [Google Scholar]
- Hyman A.A. 2000. Centrosomes: Sic transit gloria centri. Curr. Biol. 10:R276–R278 10.1016/S0960-9822(00)00406-1 [DOI] [PubMed] [Google Scholar]
- Jaensch S., Decker M., Hyman A.A., Myers E.W. 2010. Automated tracking and analysis of centrosomes in early Caenorhabditis elegans embryos. Bioinformatics. 26:i13–i20 10.1093/bioinformatics/btq190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R.T., Dasso M. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484 10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L., Sluder G., Cassels G., Sibon O., Wang C.-L. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158:1171–1181 10.1083/jcb.200205102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak J.Z. 1991. Cell cycle-dependent behavior of microtubules in hybrids of mouse oocytes and blastomeres. Int. J. Dev. Biol. 35:421–427 [PubMed] [Google Scholar]
- Kubiak J.Z., Ciemerych M.A., Hupalowska A., Sikora-Polaczek M., Polanski Z. 2008. On the transition from the meiotic to mitotic cell cycle during early mouse development. Int. J. Dev. Biol. 52:201–217 10.1387/ijdb.072337jk [DOI] [PubMed] [Google Scholar]
- Kunimoto K., Yamazaki Y., Nishida T., Shinohara K., Ishikawa H., Hasegawa T., Okanoue T., Hamada H., Noda T., Tamura A., et al. 2012. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 148:189–200 10.1016/j.cell.2011.10.052 [DOI] [PubMed] [Google Scholar]
- Kurotaki Y., Hatta K., Nakao K., Nabeshima Y.-I., Fujimori T. 2007. Blastocyst axis is specified independently of early cell lineage but aligns with the ZP shape. Science. 316:719–723 10.1126/science.1138591 [DOI] [PubMed] [Google Scholar]
- Lane H.A., Nigg E.A. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713 10.1083/jcb.135.6.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., Khodjakov A. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722 10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.-M. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Liu M., Nadar V.C., Kozielski F., Kozlowska M., Yu W., Baas P.W. 2010. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J. Neurosci. 30:14896–14906 10.1523/JNEUROSCI.3739-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Khodjakov A. 2009. Ab ovo or de novo? Mechanisms of centriole duplication. Mol. Cells. 27:135–142 10.1007/s10059-009-0017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8:161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Manandhar G., Sutovsky P., Joshi H.C., Stearns T., Schatten G. 1998. Centrosome reduction during mouse spermiogenesis. Dev. Biol. 203:424–434 10.1006/dbio.1998.8947 [DOI] [PubMed] [Google Scholar]
- Motosugi N., Bauer T., Polanski Z., Solter D., Hiiragi T. 2005. Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev. 19:1081–1092 10.1101/gad.1304805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H., Stumpff J., Su T.T. 2004. Embryonic cleavage cycles: how is a mouse like a fly? Curr. Biol. 14:R35–R45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 284:1356–1358 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- Paffoni A., Brevini T.A.L., Somigliana E., Restelli L., Gandolfi F., Ragni G. 2007. In vitro development of human oocytes after parthenogenetic activation or intracytoplasmic sperm injection. Fertil. Steril. 87:77–82 10.1016/j.fertnstert.2006.05.063 [DOI] [PubMed] [Google Scholar]
- Piehl M., Tulu U.S., Wadsworth P., Cassimeris L. 2004. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA. 101:1584–1588 10.1073/pnas.0308205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Gill S.R., Eckley D.M., Crego C.L., Compton D.A., Schroer T.A. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321–334 10.1083/jcb.147.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., Bettencourt-Dias M. 2008. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 7:11–16 10.4161/cc.7.1.5226 [DOI] [PubMed] [Google Scholar]
- Sathananthan A.H., Kola I., Osborne J., Trounson A., Ng S.C., Bongso A., Ratnam S.S. 1991. Centrioles in the beginning of human development. Proc. Natl. Acad. Sci. USA. 88:4806–4810 10.1073/pnas.88.11.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G. 1994. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 165:299–335 10.1006/dbio.1994.1256 [DOI] [PubMed] [Google Scholar]
- Schatten G., Schatten H., Spector I., Cline C., Paweletz N., Simerly C., Petzelt C. 1986a. Latrunculin inhibits the microfilament-mediated processes during fertilization, cleavage and early development in sea urchins and mice. Exp. Cell Res. 166:191–208 10.1016/0014-4827(86)90519-7 [DOI] [PubMed] [Google Scholar]
- Schatten H., Schatten G., Mazia D., Balczon R., Simerly C. 1986b. Behavior of centrosomes during fertilization and cell division in mouse oocytes and in sea urchin eggs. Proc. Natl. Acad. Sci. USA. 83:105–109 10.1073/pnas.83.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.-D., Nigg E.A. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 130:484–498 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Simerly C., Wu G.J., Zoran S., Ord T., Rawlins R., Jones J., Navara C., Gerrity M., Rinehart J., Binor Z., et al. 1995. The paternal inheritance of the centrosome, the cell’s microtubule-organizing center, in humans, and the implications for infertility. Nat. Med. 1:47–52 10.1038/nm0195-47 [DOI] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. 2008. Mechanisms of procentriole formation. Trends Cell Biol. 18:389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., Donahue R.P. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11:521–541 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Janssen A., Geers E.F., Alvarez-Fernández M., Medema R.H. 2009. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19:1703–1711 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Tsunoda Y., Yasui T., Nakamura K., Uchida T., Sugie T. 1986. Effect of cutting the zona pellucida on the pronuclear transplantation in the mouse. J. Exp. Zool. 240:119–125 10.1002/jez.1402400115 [DOI] [PubMed] [Google Scholar]
- Urbani L., Stearns T. 1999. The centrosome. Curr. Biol. 9:R315–R317 10.1016/S0960-9822(99)80201-2 [DOI] [PubMed] [Google Scholar]
- van Beneden E., Neyt A. 1889. Nouvelles recherches sur la fécondation et la division mitosique chez l’Ascaride mégalocéphale. Bull. Acad. R. Med. Belg. 3:215–295 [Google Scholar]
- Wilde A., Zheng Y. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Woolley D.M., Fawcett D.W. 1973. The degeneration and disappearance of the centrioles during the development of the rat spermatozoon. Anat. Rec. 177:289–301 10.1002/ar.1091770209 [DOI] [PubMed] [Google Scholar]
- Wühr M., Chen Y., Dumont S., Groen A.C., Needleman D.J., Salic A., Mitchison T.J. 2008. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 18:1256–1261 10.1016/j.cub.2008.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang F., Cao J., Shen Y., Huang Q., Bao L., Zhu X. 2009. Nudel promotes axonal lysosome clearance and endo-lysosome formation via dynein-mediated transport. Traffic. 10:1337–1349 10.1111/j.1600-0854.2009.00945.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.