The RNA helicase DDX6 promotes HIV-1 assembly in a co-opted cellular complex containing P body proteins and ABCE1.
Abstract
To produce progeny virus, human immunodeficiency virus type I (HIV-1) Gag assembles into capsids that package the viral genome and bud from the infected cell. During assembly of immature capsids, Gag traffics through a pathway of assembly intermediates (AIs) that contain the cellular adenosine triphosphatase ABCE1 (ATP-binding cassette protein E1). In this paper, we showed by coimmunoprecipitation and immunoelectron microscopy (IEM) that these Gag-containing AIs also contain endogenous processing body (PB)–related proteins, including AGO2 and the ribonucleic acid (RNA) helicase DDX6. Moreover, we found a similar complex containing ABCE1 and PB proteins in uninfected cells. Additionally, knockdown and rescue studies demonstrated that the RNA helicase DDX6 acts enzymatically to facilitate capsid assembly independent of RNA packaging. Using IEM, we localized the defect in DDX6-depleted cells to Gag multimerization at the plasma membrane. We also confirmed that DDX6 depletion reduces production of infectious HIV-1 from primary human T cells. Thus, we propose that assembling HIV-1 co-opts a preexisting host complex containing cellular facilitators such as DDX6, which the virus uses to catalyze capsid assembly.
Introduction
Capsid assembly is a key step in the HIV-1 life cycle and involves multimerization of ∼3,000 Gag polypeptides at the plasma membrane (PM) to form the immature capsid shell that surrounds and protects the viral genome. Gag is synthesized in the cytoplasm and then targets to the PM, where the spherical immature capsid assembles and undergoes budding and release (Demirov and Freed, 2004; Martin-Serrano and Neil, 2011). Several studies have advanced our understanding of these events. Specifically, a recent study suggested that genomic RNA (gRNA) first associates with a small number of Gag polypeptides in the cytoplasm (Kutluay and Bieniasz, 2010). This Gag–gRNA complex, which may also contain host RNAs, then targets to the site of assembly at the PM. Gag targeting requires exposure of the N-terminal myristate in Gag, which in turn is stabilized by Gag multimerization and by Gag binding to phosphatidylinositol-(4,5)-bisphosphate at the PM (Bieniasz, 2009; Chukkapalli and Ono, 2011). Additionally, recent light microscopy studies suggest that Gag stably anchors the gRNA to the PM (Jouvenet et al., 2009; Kemler et al., 2010), where Gag continues to multimerize, ultimately forming fully assembled immature capsids.
Despite these advances, the role of cellular proteins in facilitating events of capsid assembly remains unclear. A biochemical approach we previously established to help address this question showed that assembling HIV-1 Gag progresses through a stepwise, ATP-dependent pathway of discrete assembly intermediates (AIs), defined by their S values (∼10S, ∼80S, ∼150S, and ∼500S), culminating in production of fully assembled immature capsids (∼750S; Lingappa et al., 1997; Dooher et al., 2007). AIs contain HIV-1 Gag, GagPol, and Vif but form even when HIV-1 Gag is expressed in the absence of other HIV-1 gene products (Lingappa et al., 1997; Zimmerman et al., 2002; Dooher et al., 2007). Pulse-chase analyses established that Gag progresses sequentially through these AIs and into released virus (Lingappa et al., 1997; Dooher et al., 2007). The ordered progression of Gag through AIs was corroborated by the observation that assembly-defective Gag mutants are arrested at specific points in this assembly pathway (Lingappa et al., 1997; Singh et al., 2001; Dooher and Lingappa, 2004; Dooher et al., 2007; Klein et al., 2011). Progression of Gag through the assembly pathway was found to be ATP dependent even though Gag does not bind ATP (Lingappa et al., 1997), suggesting that cellular ATP-binding proteins facilitate capsid assembly. This led to the identification of ABCE1 (ATP-binding cassette protein E1), a cellular ATPase that associates with Gag in the ∼80S, ∼150S, and ∼500S AIs, termed high–molecular weight (HMW) AIs, and facilitates HIV-1 capsid formation (Zimmerman et al., 2002). Interestingly, ABCE1 was found to be essential for West Nile virus replication using an siRNA screen (Krishnan et al., 2008), suggesting that it may facilitate replication of several viruses. However, because ABCE1 is an essential host protein, elucidating its mechanism of action during HIV-1 assembly has been difficult.
Here, we asked whether HIV-1 HMW AIs contain proteins found in processing bodies (PBs). PBs are sites where nontranslating RNAs localize for decapping, 5′ to 3′ degradation, translational repression, and silencing in eukaryotic cells (Parker and Sheth, 2007). PB proteins (PBPs) were of interest to us because they are required for retrotransposition by two yeast retrotransposons (Griffith et al., 2003; Irwin et al., 2005; Checkley et al., 2010; Dutko et al., 2010) and facilitate replication of three positive-strand RNA viruses: brome mosaic, hepatitis C, and dengue (Díez et al., 2000; Ahlquist et al., 2003; Kushner et al., 2003; Noueiry et al., 2003; Ariumi et al., 2007; Randall et al., 2007; Beckham and Parker, 2008; Scheller et al., 2009; Ward et al., 2011). Moreover, PBPs are implicated in Ty3 assembly, as Ty3 GagPol, Ty3 mRNA, and viruslike particles (VLPs) accumulate during assembly in structures that contain PBPs (Beliakova-Bethell et al., 2006), including Dhh1, a homologue of the human DEAD-box RNA helicase DDX6 (also called RCK/p54), which is required for efficient Ty3 retrotransposition (Irwin et al., 2005). These PB-like Ty3 structures, termed retrosomes (Sandmeyer and Clemens, 2010), are altered by deletion of genes encoding yeast PBPs (Beliakova-Bethell et al., 2006) and by Ty3 Gag mutations (Larsen et al., 2007, 2008). It has been proposed that retrosomes sequester Ty3 assembly away from translation and RNA degradation and provide factors critical for packaging and assembly (Sandmeyer and Clemens, 2010). However, to date, assembling HIV-1 Gag has not been observed associated with PBs. Here, we demonstrate that HIV-1 forms AIs by co-opting a novel complex of PBPs and ABCE1 that existed in cells before infection and contains cellular facilitators that HIV-1 utilizes to catalyze immature capsid assembly.
Results
HMW AIs contain Gag, ABCE1, and PBPs
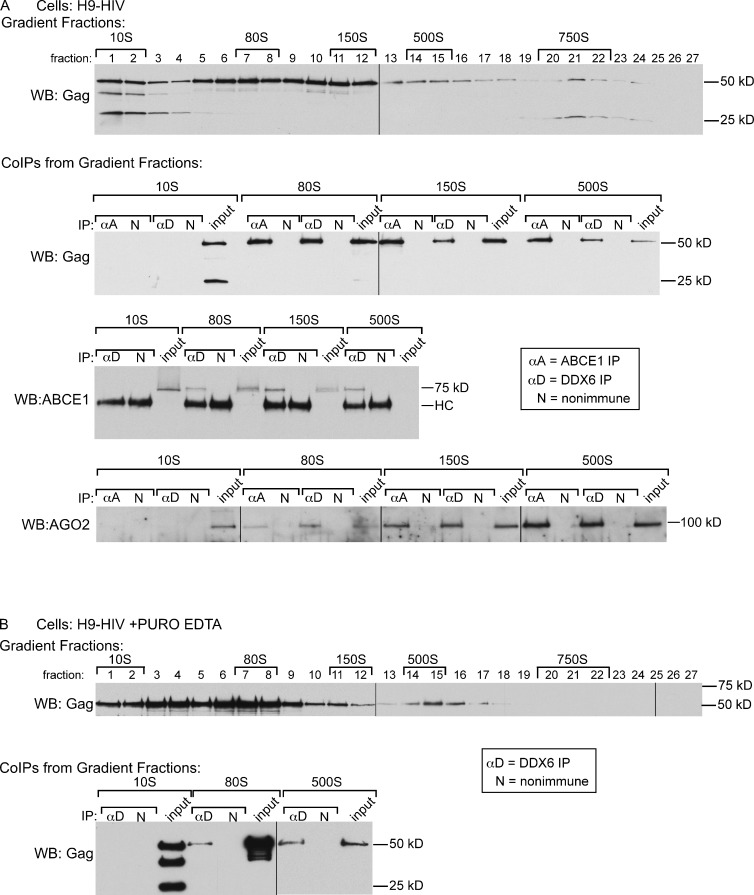
Immunoprecipitation (IP) with antibody to ABCE1 (α-ABCE1) can be used to isolate HMW AIs (∼80S, ∼150S, and ∼500S) from HIV-infected cells (Zimmerman et al., 2002). When AIs from a human H9 T cell line chronically infected with HIV-1 (H9-HIV) were subjected to IP with α-ABCE1, a large number of unidentified cellular proteins were coimmunoprecipitated along with HIV-1 capsid components (unpublished data). These data suggested that formation of AIs may result from recruitment of HIV-1 Gag into a normal cellular complex. RNA granules, which are sensitive to treatment with RNase A (Teixeira et al., 2005), constitute a class of cellular complexes that could be co-opted by HIV-1 Gag to form AIs. To assess their RNase sensitivity, AIs from lysates of COS-1 cells transfected with HIV-1 were separated by velocity sedimentation (VS), and fractions containing the isolated ∼80S AI were treated with buffer, SDS, or RNase A (Fig. 1). The integrity of the treated complexes was then determined by repeat VS followed by Western blot (WB) for Gag. Upon repeat VS, buffer treatment altered the migration of the ∼80S complex only slightly; in contrast, SDS treatment completely disrupted the integrity of the ∼80S AI, shifting Gag from the ∼80S to the ∼10S position, as expected. Like SDS treatment, RNase A treatment completely disrupted the ∼80S AI. Thus, the ∼80S AI is a ribonucleoprotein (RNP) complex, a category that includes ribosomes and cellular RNA granules. Previously, we found that, unlike ribosomes (Connolly and Gilmore, 1986), the ∼80S AI is not disrupted by treatment with puromycin and 10 mM EDTA (Dooher and Lingappa, 2004; also see top blot in Fig. 3 B); additionally, the experiment shown in Fig. 1 was performed in 10 mM EDTA to ensure disassembly of ribosomes. Together, these data suggested that the ∼80S AI is likely not a ribosome but might be related to an RNA granule.
Figure 1.
The 80S HIV-1 capsid AI is sensitive to RNase A treatment. Lysate of COS-1 cells expressing HIV-1 analyzed by VS followed by WB for Gag (starting gradient). The pooled ∼80S AI was treated with buffer, SDS, or RNase A and reanalyzed by VS followed by WB for Gag (posttreatment gradients). Brackets indicate approximate S values. Blots are representative of three independent experiments. Black lines indicate splice sites between lanes of gels.
Figure 3.
ABCE1 is associated with Gag, DDX6, and AGO2 in HIV-1 HMW AIs. (A) The top blot shows a lysate of H9-HIV cells analyzed by VS followed by WB for Gag, as in the top of Fig. 1. Pooled gradient fractions containing specific AIs were analyzed by IP, as in Fig. 2 B, followed by WB for Gag, ABCE1, or AGO2. Equivalent amounts of IP input are shown. The 500S input in the ABCE1 blot was seen on long exposures (not depicted). HC, IgG heavy chain detected by secondary antibody. Blots are representative of three independent experiments. (B) Same as for A, except H9-HIV cell lysate was treated with 1 mM puromycin (PURO) at 37°C and 10 mM EDTA to disassemble ribosomes before analysis by VS followed by gradient fraction coIP. Data are representative of two experiments. Black lines indicate splice sites between lanes of gels.
Many types of RNA granules have been described, each composed of overlapping sets of proteins (Anderson and Kedersha, 2006). We wondered whether HIV-1 AIs might contain PBPs, as PBPs play a role in assembly of Ty3 VLPs (Sandmeyer and Clemens, 2010), and retrotransposons and retroviruses share a common ancestor (Pélisson et al., 1997). Arguing against this possibility is the fact that complexes containing PBPs typically form large structures that are visible by light microscopy (Balagopal and Parker, 2009); in contrast, the ∼80S AI is smaller than typical RNA granules and likely falls below the threshold for detection by typical light microscopy. However, the small size of AIs would not rule out the presence of functional PBPs in AIs because PB disruption does not necessarily alter PB function (Balagopal and Parker, 2009), thus suggesting the existence within cells of functional PB complexes that are not readily detected by light microscopy.
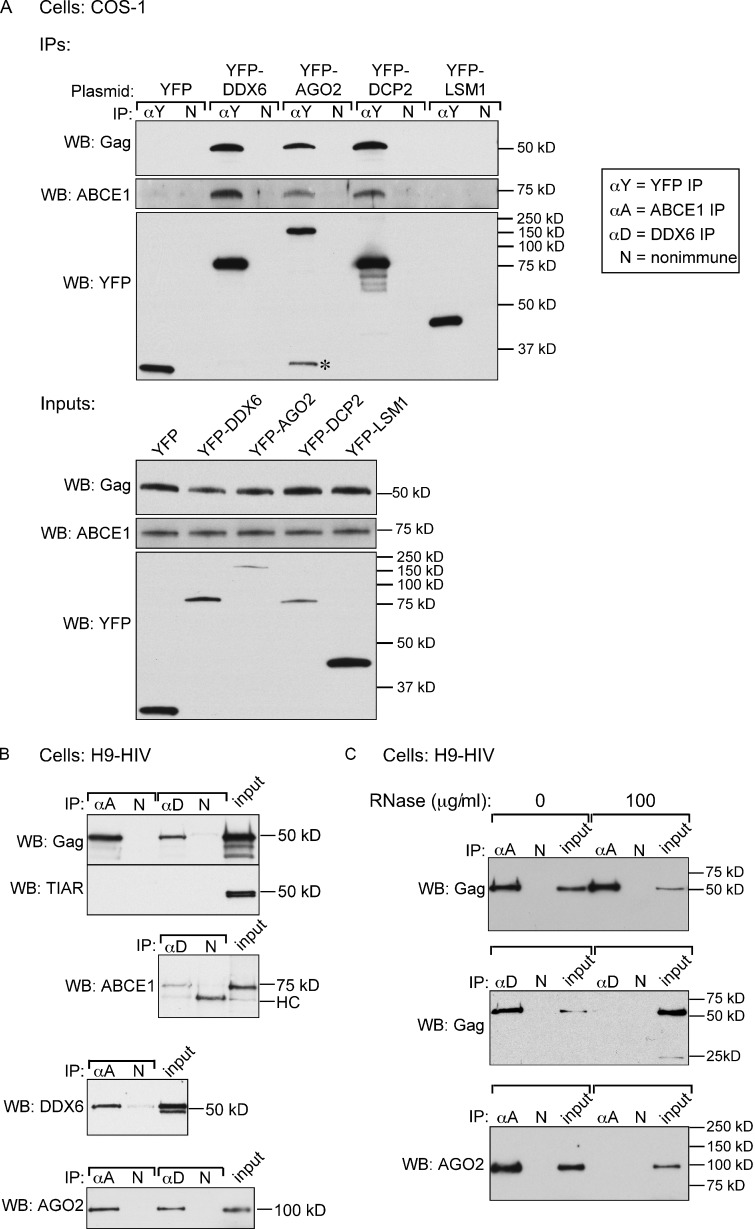
To determine whether PBPs are associated with HIV-1 Gag in AIs, we transfected COS-1 cells to express HIV-1 and one of four different YFP-tagged PBP constructs (DDX6, DCP2, AGO2, and LSM1). Cell lysates were immunoprecipitated with YFP antibody and analyzed by WB. YFP blots revealed that all the YFP-tagged proteins were expressed and immunoprecipitated by YFP antibody. Notably, YFP-tagged DDX6, DCP2, and AGO2 were associated with HIV-1 Gag and ABCE1 by coIP, whereas LSM1 was not (Fig. 2 A), either because the epitope tag blocks LSM1 incorporation, LSM1 is not present in these complexes, or the association of LSM is labile. Thus, at least three tagged PBPs were found associated with Gag.
Figure 2.
PBPs are associated with Gag and ABCE1 in cells expressing HIV-1, but the association is RNase A sensitive. (A) Lysates of COS-1 cells expressing HIV-1 and the indicated YFP-tagged plasmids analyzed by IP under native conditions with α-YFP (αY) or a nonimmune control antibody (N) followed by WB for Gag, ABCE1, or YFP. WB of equivalent aliquots of IP inputs is shown below IPs. The asterisk indicates a likely degradation product. (B) H9-HIV cell lysate analyzed by IP under native conditions with α-ABCE1, α-DDX6, or a matched nonimmune antibody, in parallel, followed by WB for Gag, TIAR, ABCE1, DDX6, or AGO2. (C) H9-HIV lysate was mock treated or RNase A treated and analyzed by IP, as in B, followed by WB for Gag or AGO2. Blots are representative of three independent experiments.
We also demonstrated that endogenous PBPs are associated with HIV-1 Gag and ABCE1. When H9-HIV cells were immunoprecipitated with α-ABCE1, endogenous DDX6 and AGO2 as well as HIV-1 Gag were detected by WB (Fig. 2 B). Similarly, antibody to DDX6 (α-DDX6) coimmunoprecipitated HIV-1 Gag, AGO2, and ABCE1 from H9-HIV cell lysates. TIAR, a stress granule protein, was not associated with ABCE1 or DDX6 by coIP (Fig. 2 B), suggesting that only a specific subset of RNA granule proteins is stably associated with HIV-1 Gag and ABCE1 in AIs. Additionally, we showed that the DDX6–Gag and AGO2–ABCE1 interactions in H9-HIV cells are sensitive to RNase A treatment (Fig. 2 C). In contrast, the ABCE1–Gag interaction, which was examined in parallel, was RNase A insensitive (Fig. 2 C), as previously shown (Lingappa et al., 2006). Together, these data indicated that Gag and ABCE1 are associated with PBPs in one or more RNase-sensitive complexes.
Next, we addressed whether endogenous DDX6 and AGO2 are each associated with HIV-1 Gag in separate complexes or whether DDX6, AGO2, and Gag are all together in one or more AIs. For this, we used VS to separate AIs present in H9-HIV lysates (Fig. 3 A) and then subjected fractions containing the isolated ∼10S, ∼80S, ∼150S, or ∼500S AIs to IP with α-ABCE1 or α-DDX6 followed by WB for Gag, ABCE1, or AGO2. As previously observed, α-ABCE1 failed to coimmunoprecipitate Gag from the ∼10S AI but did coimmunoprecipitate Gag from each of the HMW AIs (Zimmerman et al., 2002; Klein et al., 2011). Here, we observed the same pattern for DDX6 coIPs of Gag, AGO2, and ABCE1 and for α-ABCE1 coIPs of AGO2, with positive coIPs observed only for gradient fractions containing the HMW AIs. Moreover, although DDX6, AGO2, and ABCE1 were not associated with unassembled Gag in the ∼10S AI, these four proteins were present in the ∼10S fraction (Fig. 3 A and not depicted). Additionally, when H9-HIV lysates were treated with puromycin plus EDTA to dissociate ribosomes, Gag-containing complexes were still observed in the ∼80S and∼500S regions of the gradient, and DDX6 was associated with Gag in these complexes (Fig. 3 B), confirming that the HMW AIs do not represent ribosomes or polysomes. Together, these data indicated that ABCE1, DDX6, and AGO2 are likely associated with HIV-1 Gag in each of the HMW AIs.
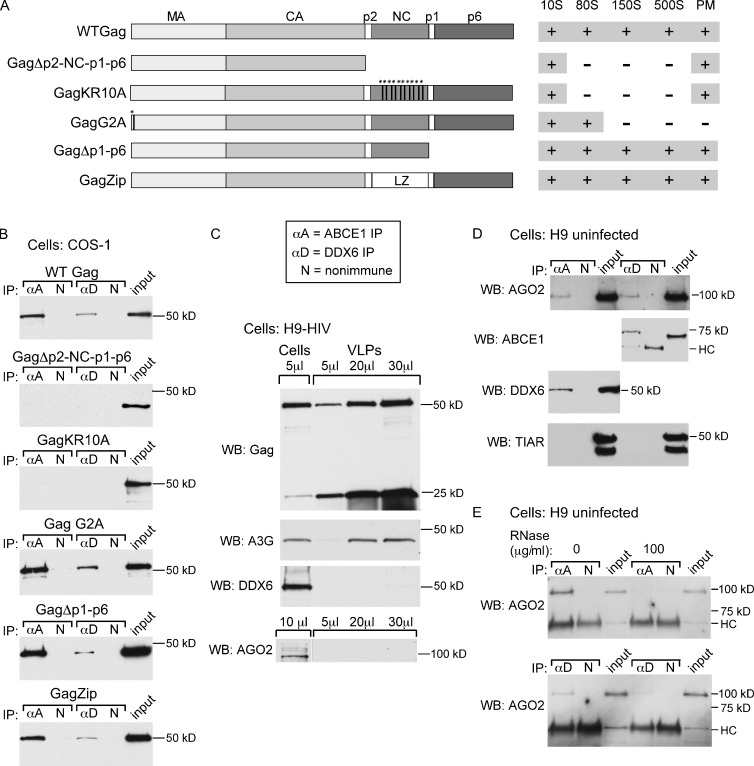
The gradient data also suggested that Gag first associates with DDX6, AGO2, and ABCE1 when it enters the ∼80S AI. If this is the case, then Gag mutants arrested at the ∼10S AI should not be associated with DDX6, unlike Gag mutants that progress into the ∼80S AI or beyond. Previously, it has been shown that a truncated Gag from which the nucleocapsid domain (NC), p6 domain, and spacer peptides p1 and p2 have been deleted (GagΔp2-NC-p1-p6) is assembly incompetent, fails to associate with ABCE1, and is arrested at the ∼10S AI (Lingappa et al., 1997; Dooher and Lingappa, 2004; Dooher et al., 2007; Klein et al., 2011). Similar results were obtained for GagKR10A, in which 10 basic residues in NC were substituted with alanines (Lingappa et al., 2006). In contrast, GagG2A, a Gag mutant that fails to be myristoylated, does not target to the PM (Göttlinger et al., 1989; Bryant and Ratner, 1990) and instead remains arrested in the ∼80S ABCE1-associated AI (Lingappa et al., 1997; Klein et al., 2011). For comparison, we also examined two assembly-competent mutants that associate with ABCE1 and progress through the entire assembly pathway (Lingappa et al., 1997; Klein et al., 2011): GagΔp1-p6, a C-terminally truncated Gag, and GagZip, in which the NC domain of Gag is replaced with a leucine zipper (LZ) domain from the GCN4 transcription factor (Fig. 4 A). We found that α-DDX6 did not coimmunoprecipitate GagΔp2-NC-p1-p6 or GagKR10A but did coimmunoprecipitate GagG2A, GagΔp1-p6, and GagZip (Fig. 4 B). This same subset of Gag constructs was also coimmunoprecipitated by α-ABCE1 (Fig. 4 B), consistent with previous findings (Lingappa et al., 1997; Zimmerman et al., 2002; Klein et al., 2011). Thus, it appears that Gag mutants that fail to form HMW AIs and fail to bind ABCE1 also fail to associate with endogenous DDX6; moreover, Gag first associates with DDX6 and ABCE1 when it enters the ∼80S AI. Additionally, these data suggested that DDX6 first associates with Gag in the cytoplasm, as DDX6 is associated with the GagG2A mutant, which is arrested in the cytoplasm (Ono and Freed, 1999). Notably, in this experiment, we used subgenomic plasmids encoding only HIV-1 Gag and HIV-1 Rev, a nuclear export factor required for Gag expression. Thus, these findings also demonstrated that the Gag–DDX6 association does not depend on HIV-1 proteins other than Gag and Rev; nor does it require other regions of the HIV-1 genome not encoded by these subgenomic constructs.
Figure 4.
PBPs are not associated with 10S-arrested Gag mutants or VLPs but are associated with ABCE1 in uninfected H9 T cells. (A) Diagram showing domains of Gag (matrix [MA], capsid [CA], nucleocapsid [NC], p6, p1, and p2), mutations encoded into Gag constructs, which AIs each construct forms, and whether they target to the PM. Asterisks indicate point mutations. (B) Lysates of COS-1 cells expressing the indicated Gag protein and HIV-1 Rev analyzed by IP, as in Fig. 2, followed by WB for Gag. (C) H9-HIV cell lysates and VLPs analyzed by WB for Gag, APOBEC3G (A3G), DDX6, and AGO2. Amounts loaded are indicated above the blot. (D) Lysates of uninfected parental H9 cells subjected to IP, as in B, followed by WB for AGO2, ABCE1, DDX6, or TIAR. HC, IgG heavy chain detected by secondary antibody. (E) Lysates of uninfected H9 cells were either mock treated or RNase A treated and analyzed by IP, as in B, followed by WB for AGO2. Blots are representative of two independent experiments.
Although DDX6 and AGO2 are associated with the ∼500S AI, the last intermediate formed before completion of capsid assembly, we were unable to detect significant amounts of DDX6 or AGO2 in VLPs released from H9-HIV cells (Fig. 4 C), in contrast to APOBEC3G, a cellular protein that associates with Gag and is found in VLPs (Sheehy et al., 2002). Thus, DDX6 and AGO2 appear to be largely released from immature capsids before virus budding occurs, as is the case for ABCE1 (Zimmerman et al., 2002; Dooher and Lingappa, 2004; Dooher et al., 2007).
We also examined whether ABCE1 is associated with endogenous PBPs in the uninfected H9 T cells used to generate H9-HIV (Fig. 4 D). Although α-ABCE1 did not coimmunoprecipitate the stress granule protein TIAR, it did coimmunoprecipitate DDX6 and AGO2; similarly, α-DDX6 failed to coimmunoprecipitate TIAR but did coimmunoprecipitate ABCE1 and AGO2. Moreover, although we did not test all the combinations of PBP interactions in H9 cells for RNase A sensitivity, those tested were RNase A sensitive (Fig. 4 E). Thus, ABCE1 forms an RNase-sensitive complex with DDX6 and AGO2 even in uninfected cells, suggesting that assembling HIV-1 Gag co-opts a normal cellular complex containing ABCE1 and a subset of PBPs.
Endogenous PBPs are colocalized with Gag at PM sites of assembly
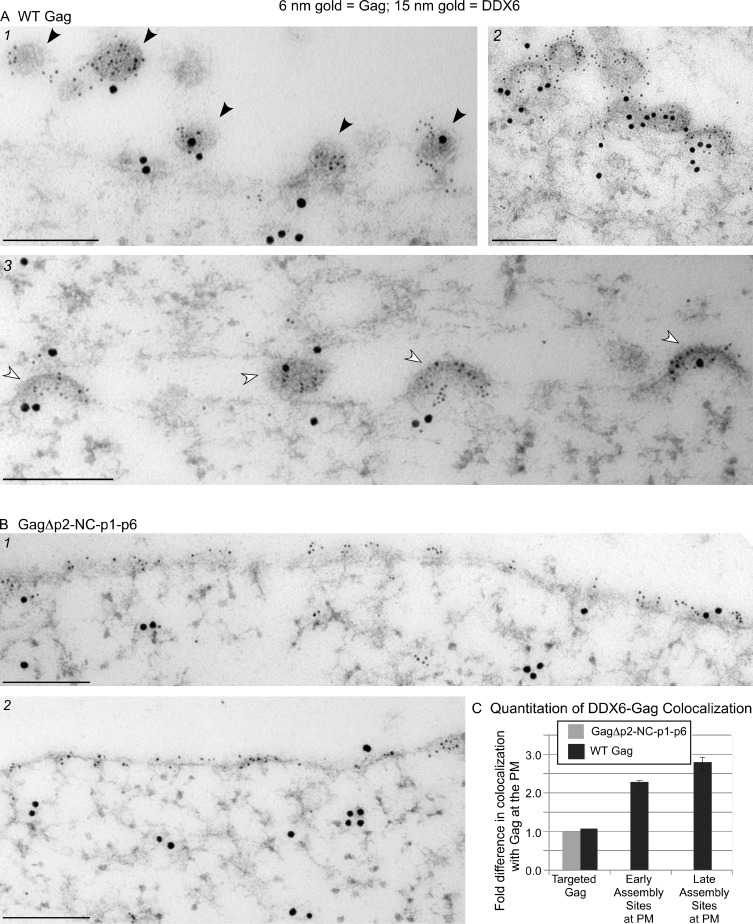
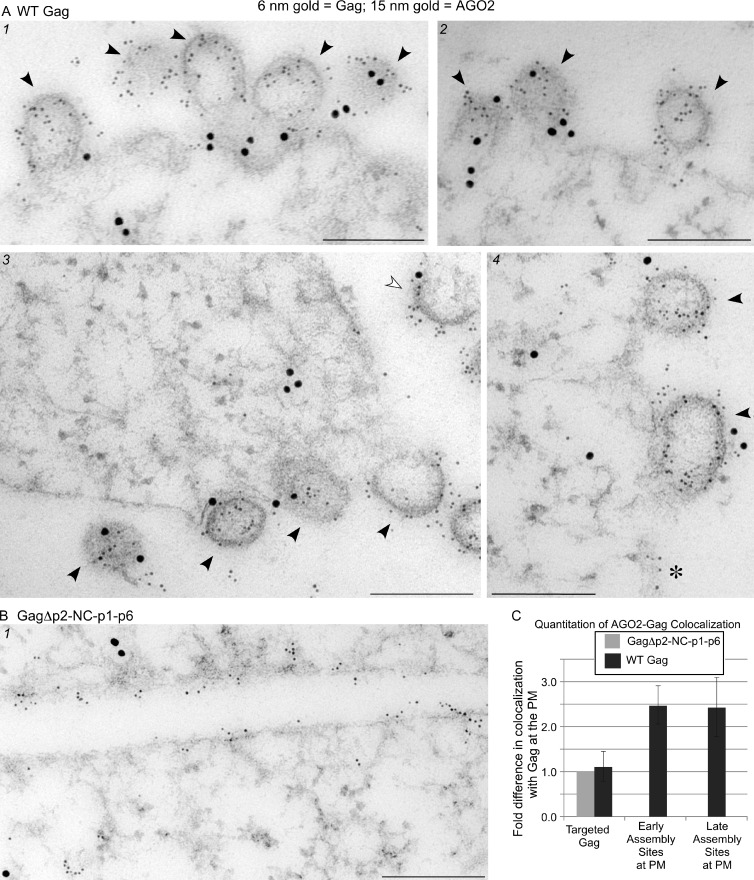
As coIPs showed that PBPs are associated with Gag in all of the HMW AIs, we predicted that DDX6 and AGO2 would be colocalized with HIV-1 Gag at PM sites of assembly. To test this, we used immunoelectron microscopy (IEM) instead of fluorescence microscopy because IEM would allow us to analyze endogenous DDX6 and AGO2, thereby avoiding any artifacts related to the overexpression of PB-GFP fusion proteins, to detect complexes that are too small to resolve by light microscopy, and to identify early and late stages of capsid assembly at the PM. Sections from cells expressing wild-type (WT) HIV-1 were labeled with antibodies against Gag (6-nm gold) and either DDX6 or AGO2 (15-nm gold), and colocalization of Gag-DDX6 and Gag-AGO2 at PM sites of assembly was examined. Although DDX6 and AGO2 are typically cytoplasmic, we controlled for the possibility that they might be associated with the PM at low levels in the absence of HIV-1 assembly by parallel labeling of cells expressing HIV-1 encoding the assembly-defective mutant GagΔp2-NC-p1-p6, which targets to the PM but fails to form HMW AIs (Dooher and Lingappa, 2004; Lingappa et al., 2006) or associate with ABCE1 (Zimmerman et al., 2002) or DDX6 (Fig. 4 B). When DDX6 and Gag were labeled in cells expressing WT Gag, extensive colabeling was observed at early and late sites of capsid assembly present at the PM (Fig. 5 A). In contrast, in cells expressing GagΔp2-NC-p1-p6, abundant Gag labeling was seen at the PM, but PM sites of assembly were absent, as expected, and Gag-DDX6 colabeling was noticeably less frequent than in cells expressing WT Gag (Fig. 5 B). Similar results were obtained when cells expressing WT Gag versus GagΔp2-NC-p1-p6 were double labeled for AGO2 and Gag (Fig. 6, A and B).
Figure 5.
Immunogold double labeling demonstrates that DDX6 is enriched at early and late sites of Gag assembly at the PM. (A and B) Infected COS-1 cells expressing WT Gag or GagΔp2-NC-p1-p6 were analyzed by immunogold double-label EM. Small gold particles represent HIV-1 Gag; large gold particles represent DDX6. Images show representative sites of labeling. Bars, 200 nm. (A) Infected cells expressing WT Gag. (1) Five late assembly sites (black arrowheads), four of which are double labeled. (2) Early and late assembly sites. (3) Four early assembly sites (white arrowheads), all of which are double labeled. (B) Infected cells expressing GagΔp2-NC-p1-p6, which targets to the PM but does not assemble. (C) Two independent labeling experiments were quantified to determine the number of Gag clusters at the PM that were colocalized with DDX6 in infected cells expressing WT Gag or the assembly-defective GagΔp2-NC-p1-p6. Colocalization in cells expressing GagΔp2-NC-p1-p6 was assumed to be random; this amount was therefore set to 1.0, and colocalization of DDX6 with WT Gag was expressed as fold increase above this negative control value. Targeted Gag is defined as nonassembling Gag at the PM, early assembly sites are those in which less than half of a virus bud has formed, and late assembly sites are those in which more than half of the virus bud has formed. Error bars indicate SD. Quantitation details are provided in Table 1.
Figure 6.
Immunogold double labeling demonstrates that AGO2 is enriched at early and late sites of Gag assembly at the PM. (A and B) COS-1 cells infected as in Fig. 5. Small gold particles represent HIV-1 Gag; large gold particles represent AGO2. Images show representative sites of labeling. Bars, 200 nm. (A) Infected cells expressing WT Gag. (1) Five late assembly sites (black arrowheads), four of which are double labeled. (2) Three late assembly sites (black arrowheads), two of which are double labeled. (3) Four late assembly sites (black arrowheads), three of which are double labeled, and one early assembly site that is also double labeled (white arrowhead). (4) Two late assembly sites (black arrowheads), both of which are double labeled, and a cluster of targeted Gag (asterisk) that is not double labeled. (B) Infected cells expressing GagΔp2-NC-p1-p6, which targets to the PM but does not assemble. (C) Quantitation of AGO2-Gag colocalization, as in Fig. 5 C. For definitions of targeted Gag and early and late assembly sites, see Fig. 5 legend. Error bars indicate SD. Details are provided in Table 2.
To quantify the association of Gag with DDX6 and AGO2, we categorized Gag clusters at the PM by their stage in assembly and counted how often a DDX6 or AGO2 label was present within a circular zone 170 nm in diameter surrounding a Gag cluster. In cells expressing assembly-defective GagΔp2-NC-p1-p6, Gag label at the PM was not associated with electron-dense structures representing multimerizing Gag or membrane deformation typical of budding (Figs. 5 B and 6 B). We called this category of nonassembling Gag at the PM targeted Gag. Targeted Gag was also found in cells expressing WT Gag (asterisk in image 4 of Fig. 6 A). However, in cells expressing WT Gag, two other categories of PM Gag labeling were frequently observed: early assembly sites, in which less than half of a virus bud had formed (e.g., white arrowheads in image 3 of Figs. 5 A and 6 A) and late assembly sites, in which more than half of the virus bud had formed (e.g., black arrowheads in Figs. 5 A [image 1] and 6 A [images 1–4]). In cells expressing WT Gag, DDX6 and AGO2 were 2.5–3 times more likely to be associated with Gag at both early and late assembly sites at the PM when compared with cells expressing GagΔp2-NC-p1-p6 (Figs. 5 C and 6 C and Tables 1 and 2). In contrast, targeted WT Gag was not associated with DDX6 and AGO2 when compared with the negative control cells. It is possible that targeted WT Gag serves as a source of Gag monomers that are added to AIs that contain DDX6 and AGO2 at the PM. Together, these data corroborated results of our coIP experiments indicating that DDX6 and AGO2 are specifically associated with HMW AIs during HIV-1 assembly.
Table 1.
Quantification of the colocalization of DDX6 with Gag in HIV-1–infected cells
| Experiment 1 | Experiment 2 | Experiment 1 and 2 combined | Fold colocalization | |||||
| WT Gag | GagΔp2-NC-p1-p6 | WT Gag | GagΔp2-NC-p1-p6 | WT Gag | GagΔp2-NC-p1-p6 | WT Gag | GagΔp2-NC-p1-p6 | |
| Membrane length (µm) | 111 | 150 | 155 | 166 | 266 | 316 | ||
| Total DDX6 label | 643 | 536 | 624 | 679 | 1,267 | 1,215 | ||
| Total targeted Gag clusters | 67 | 499 | 67 | 379 | 134 | 878 | ||
| Targeted Gag clusters with DDX6 | 22 | 165 | 19 | 108 | 41 | 273 | 1.00 | (1.00) |
| Gag clusters at early assembly sites | 58 | 0 | 56 | 0 | 114 | 0 | ||
| Gag clusters at early assembly sites with DDX6 | 41 | n/a | 36 | n/a | 77 | n/a | 2.20a | |
| Gag clusters at late assembly sites | 20 | 0 | 57 | 0 | 77 | 0 | ||
| Gag clusters at late assembly sites with DDX6 | 17 | n/a | 46 | n/a | 63 | n/a | 2.70a | |
Cells infected with the indicated proviruses were double immunogold labeled for Gag and DDX6. In two independent labeling experiments, the following parameters were quantified: the total length of membrane examined, the total number of independent DDX6 labeling events within 100 nm of the membrane, the total number of independent targeted, early, and late Gag clusters, and the total number of Gag clusters that were associated with DDX6 classified by stage of assembly (i.e., targeted Gag or Gag at early or late assembly sites). Fold colocalization is defined as the number of WT Gag clusters colocalized with DDX6 for each stage of assembly, normalized to Gag-DDX6 colocalization in the GagΔp2-NC-p1-p6 negative control (indicated by parentheses). Fold colocalization (bold) was calculated for each experiment. The fold colocalization averaged from the two experiments is shown and is also graphed with SD in Fig. 5.
P < 0.05.
Table 2.
Quantification of the colocalization of AGO2 with Gag in HIV-1 infected cells
| Experiment 1 | Experiment 2 | Experiment 1 and 2 combined | Fold colocalization | |||||
| WT Gag | GagΔp2-NC-p1-p6 | WT Gag | GagΔp2-NC-p1-p6 | WT Gag | GagΔp2-NC-p1-p6 total | WT Gag | GagΔp2-NC-p1-p6 total | |
| Membrane length (µm) | 178 | 174 | 158 | 168 | 336 | 342 | ||
| Total AGO2 label | 387 | 442 | 1,006 | 544 | 1,393 | 986 | ||
| Total targeted Gag clusters | 106 | 461 | 80 | 358 | 186 | 819 | ||
| Targeted Gag clusters with AGO2 | 19 | 95 | 27 | 90 | 46 | 185 | 1.11 | (1.00) |
| Gag clusters at early assembly sites | 27 | 0 | 55 | 0 | 82 | 0 | ||
| Gag clusters at early assembly sites with AGO2 | 12 | n/a | 38 | n/a | 50 | n/a | 2.45a | |
| Gag clusters at late assembly sites | 102 | 0 | 43 | 0 | 145 | 0 | ||
| Gag clusters at late assembly sites with AGO2 | 41 | n/a | 31 | n/a | 72 | n/a | 2.41b | |
Cells infected with the indicated proviruses were double immunogold labeled for Gag and AGO2 and analyzed as described for Table 1. Fold colocalization is defined as the number of WT Gag clusters colocalized with AGO2 for each stage of assembly, normalized to Gag-AGO2 colocalization in the GagΔp2-NC-p1-p6 negative control (indicated by parentheses). Fold colocalization (bold) with SD is graphed in Fig. 6.
P < 0.05.
P < 0.1.
DDX6 acts catalytically to facilitate capsid assembly independent of RNA packaging
Having established by biochemical and ultrastructural approaches that HMW AIs contain PBPs, we next asked whether PBPs facilitate events involved in HIV-1 capsid formation. We chose to examine DDX6 because RNA helicases are critical for RNA remodeling, during which one set of RNPs is removed from an RNA and replaced with a new set of RNPs, allowing the fate or function of the RNA to be reprogrammed (Linder and Jankowsky, 2011). In principle, a role could be envisioned for RNA helicases in at least two events critical for HIV-1 virus formation: capsid assembly and packaging. As cellular and/or viral RNAs may nucleate capsid assembly (Campbell and Rein, 1999), helicases could facilitate assembly by remodeling RNAs required for this process. Additionally or alternatively, helicases could facilitate RNA encapsidation by making genomic and/or cellular RNA available for encapsidation.
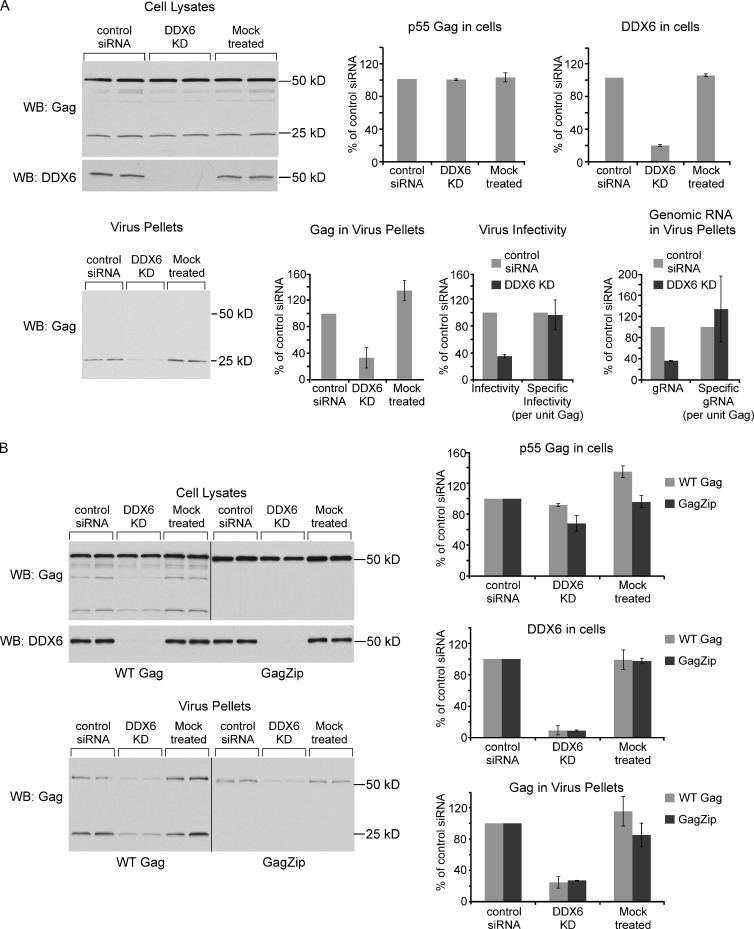
To test whether DDX6 plays a role in virus production, we depleted endogenous DDX6 from COS-1 cells using siRNA transfection. Unexpectedly, while choosing a control siRNA, we observed that transfection with some nontargeting siRNAs reduced intracellular steady-state Gag levels as well as VLP production by 2–100 fold when compared with mock-transfected cells (Fig. S1). For our experiments, we chose a control siRNA pool (control siRNA1 in Fig. S1) that had minimal effects on steady-state Gag levels and particle production relative to mock-transfected cells. When DDX6 in COS-1 cells was depleted to ∼20% of control levels, steady-state Gag levels were unaffected, but virus infectivity decreased to ∼35% of control (Fig. 7 A). The reduction in infectivity was a result of decreased VLP production, as both VLP production and total viral gRNA fell proportionately to ∼35% of control. The small amount of virus that was produced after DDX6 depletion may have resulted from the 20% of DDX6 that was not depleted; not surprisingly, the residual virus displayed infectivity per unit Gag similar to WT virus. The finding that DDX6 depletion reduces VLP production indicated that DDX6 facilitates formation of HIV-1 particles per se and does not act exclusively on gRNA packaging.
Figure 7.
DDX6 KD reduces virus production by cells expressing WT HIV-1 or the GagZip chimera. (A) COS-1 cells were either mock treated or transfected in duplicate with siRNA directed against DDX6 (DDX6 KD) or a control siRNA and then transfected with HIV-1. Cell lysates and virus pellets were analyzed by WB (blots and graphs shown; control siRNA set to 100%). Also graphed are infectivity of virus (determined by titering on TZM-Bl cells), specific infectivity (infectivity per unit Gag in virus), HIV-1 gRNA content of pelleted virus, and specific RNA content (gRNA per unit Gag in virus). The experiment was repeated three times; data are from one representative experiment. Graphs of protein levels and gRNA show means ± SD for duplicates within one experiment; infectivity graphs show means of two experiments ± SEM. (B) Same as for A, except that cells were also transfected with HIV-1 encoding GagZip in parallel with WT HIV-1. Note that GagZip is not cleaved, resulting in VLPs containing only p55. Black lines between panels indicate splice sites between lanes of gels. The experiment was repeated four times. Blots are from one representative experiment; graphs show means ± SD from two independent experiments.
To further test whether DDX6 acts on capsid assembly independent of RNA packaging, we expressed the aforementioned HIV-1 GagZip chimera in DDX6-depleted cells. The LZ domain, which replaces NC, promotes dimerization, as does NC; but, unlike NC, LZ does not bind RNA. For this reason, GagZip VLPs fail to encapsidate significant amounts of viral or cellular RNAs, as indicated by RiboGreen analysis and RT-PCR, although they very closely resemble WT VLPs in nearly all other respects (Zhang et al., 1998; Accola et al., 2000; Johnson et al., 2002; Zennou et al., 2004; Crist et al., 2009; Klein et al., 2011). We previously showed that although NC is required for ABCE1 binding by WT Gag, replacement of NC with LZ preserves ABCE1 binding, allowing GagZip to form HMW AIs that closely resemble WT HIV-1 HMW AIs (Klein et al., 2011). These data indicated that the NC-mediated dimerization, not NC-mediated RNA binding, is critical for ABCE1 binding and that the direct binding site for ABCE1 is not in NC but resides elsewhere in Gag and forms, or is exposed, upon Gag dimerization (Klein et al., 2011). Given that GagZip forms ABCE1-containing HMW AIs, we were not surprised to find that GagZip is also associated with DDX6 (Fig. 4 B). Importantly, in DDX6-depleted cells, whereas expression of GagZip was modestly reduced (to 68% of control), VLP production was reduced to a much greater extent (to 27% of control). These data demonstrate that DDX6 facilitates assembly of capsids that lack significant encapsidated gRNA or cellular mRNA, further confirming that DDX6 promotes Gag multimerization independent of gRNA or mRNA encapsidation.
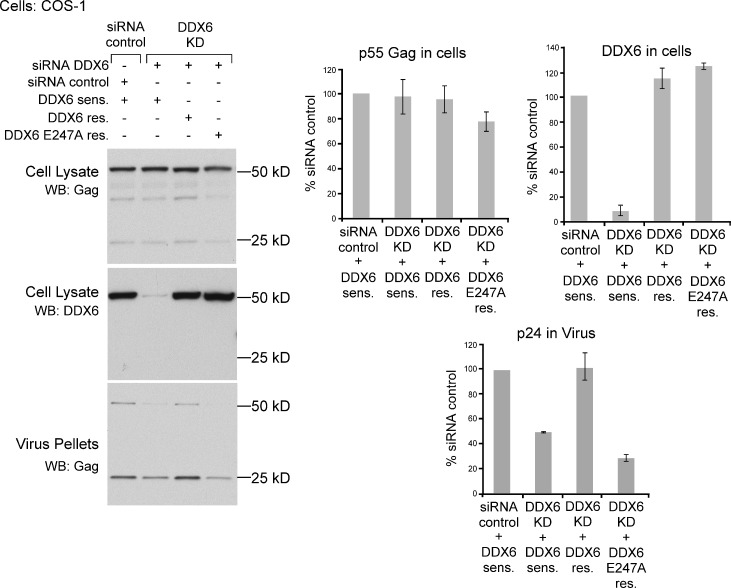
We also demonstrated that the inhibition in virus production in DDX6-depleted cells was not simply a result of indirect effects on cell proliferation. Cell counts performed on each day of the knockdown (KD) experiments revealed no reduction in the number of viable cells (Fig. S2). Additionally, expression of siRNA-resistant DDX6 completely rescued the defect in virus production caused by DDX6 depletion (Fig. 8), indicating that the effect of the KD on VLP production was a result of DDX6 depletion and was not a result of off-target effects of the DDX6-targeted siRNA. Notably, the 50% reduction in virus production in the KD cells in rescue experiments was less than we had observed in previous KD experiments (reduction to 32 and 27% of control in Fig. 7 [A and B], respectively). This may have been a result of the cotransfection of siRNA-sensitive DDX6 plasmid in the KD group on the day before HIV transfection in the rescue experiments. However, the incomplete KD phenotype was fortuitous, as it revealed that transfection of an siRNA-resistant DDX6 containing an E247A point mutation in the DEAD-box not only failed to rescue the defect in VLP production but appeared to display a dominant-negative effect, reducing VLP production from 50 to 28% of control (Fig. 8). As this mutation inactivates a motif required for ATP binding and hydrolysis (Linder, 2006), these data demonstrate that the role of DDX6 in facilitating HIV-1 capsid formation is dependent on its catalytic activity.
Figure 8.
The DDX6 KD phenotype is rescued by WT DDX6 but not by a DDX6 E247A DEAD-box mutant. COS-1 cells were transfected with siRNAs (control or DDX6 specific), as in Fig. 7, and subsequently transfected with plasmids encoding either siRNA-sensitive WT DDX6 (DDX6 sens.), siRNA-resistant WT DDX6 (DDX6 res.), or siRNA-resistant DDX6 E247A (DDX6 E247A res.). Finally, cells were transfected with HIV-1 provirus. WB was performed as in Fig. 7. The experiment was repeated three times. Blots are from one representative experiment; graphs show means ± SD from two independent experiments.
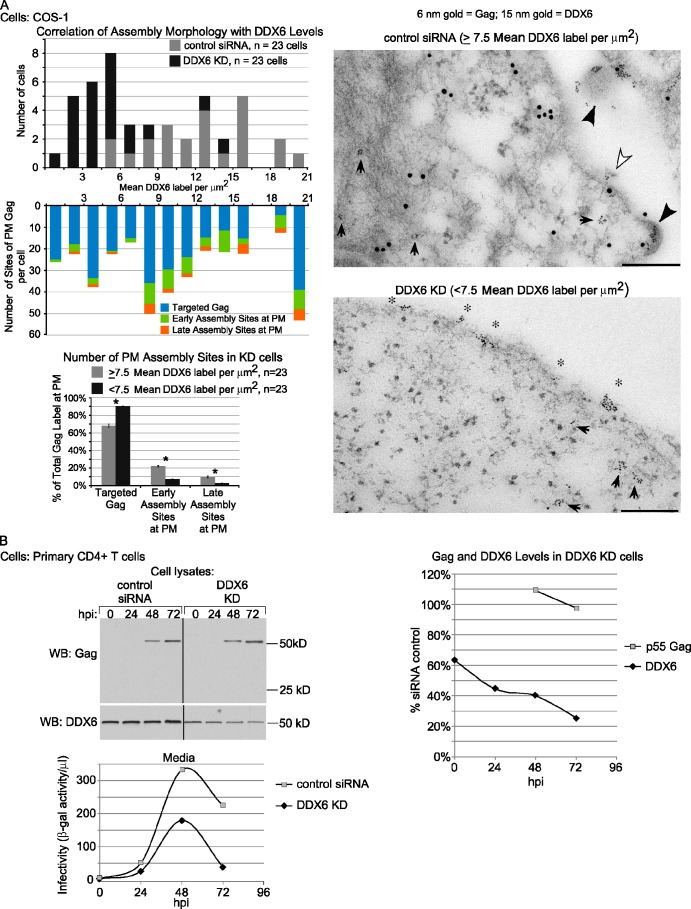
To further confirm that DDX6 facilitates immature capsid assembly, we examined DDX6-depleted cells using IEM with double labeling of Gag and DDX6 (Fig. 9 A). Quantitation of DDX6 labeling confirmed that siRNA treatment reduced levels of DDX6 in a large subpopulation of DDX6-depleted cells. We also quantified morphology at sites of Gag labeling at the PM to determine the number of assembly events that corresponded to targeted Gag (not associated with multimerization) versus early or late capsid assembly sites (Fig. 9 A). Notably, in cells containing ≥7.5 DDX6 labels/µm2 (which consisted mainly but not exclusively of control cells), 32% of Gag at the PM was in early and late AIs, and 68% was targeted Gag. In contrast, in cells containing <7.5 DDX6 labels/µm2 (which consisted mainly but not exclusively of DDX6-depleted cells), only 10% of Gag at the PM was in early and late AIs, and 90% was targeted Gag (Fig. 9 A). Importantly, we did not observe formation of capsids with abnormal morphology in DDX6-depleted cells. Although Gag label in the cytoplasm could not be reliably quantified as a result of difficulties in accurately detecting 6-nm gold particles against the high background found in the cytoplasm, our biochemical experiments demonstrated that steady-state Gag levels were comparable in KD and control cells. Thus, these data support the conclusion that DDX6 facilitates capsid formation at the PM.
Figure 9.
DDX6 KD reduces late assembly sites at the PM as well as infectious virus production in primary CD3+/CD4+ T cells. (A) COS-1 cells treated with control and DDX6-specific siRNA, as in Fig. 7, and analyzed by IEM with double labeling for Gag and DDX6, as in Fig. 5. Quantitation was performed on 23 randomly chosen cells from each group from two independent experiments. The top graph shows the number of cells containing the indicated amounts of DDX6 label. The middle graph shows the number of assembly sites per cell (categorized as targeted Gag or early or late assembly sites), stratified by the amount of DDX6 in cells. The bottom graph shows the number of early or late assembly sites or targeted Gag at the PM as a percentage of total PM sites of assembly in cells that had either ≥7.5 or <7.5 DDX6 labels/µm2 (mean DDX6 label) ±SEM. n, total number of cells examined taken from two independent experiments. *, P ≤ 0.01. Micrographs show an example of double labeling from each group. White and black arrowheads indicate early and late assembly sites, respectively. Small arrows intracellular Gag clusters. Asterisks indicate targeted Gag. Bars, 200 nm. (B) Activated primary CD3+/CD4+ T cells nucleofected with the indicated siRNAs and infected with replication-competent HIV-GFP. Cells from replicate wells were harvested at the indicated times and analyzed by WB for Gag and DDX6. Intracellular Gag and DDX6 levels were determined by quantifying WBs (graph on right). Media were collected at 24-h intervals and analyzed for infectivity in parallel (bottom graph). Data are from one experiment and are representative of three independent experiments. hpi, hours postinfection. Black lines indicate removal of splice sites between lanes of gels.
Finally, we demonstrated that depletion of DDX6 inhibits production of infectious HIV-1 from primary human T cells, a physiological relevant cell type (Fig. 9 B). Activated CD4+ T cells were transfected with siRNA and infected with replication-competent HIV-1 followed by media and cell lysate collections at 24-h intervals. Depletion reduced cellular DDX6 to ∼25% of control by 72 hours after infection. At the same time, media infectivity was reduced to 17% of control levels, whereas steady-state Gag levels in KD cells were essentially unchanged. Notably, as DDX6 levels dropped, a corresponding decrease in virus infectivity relative to control was observed. Trends were the same in two additional experiments performed similarly. The nearly sixfold reduction in infectious virus production from primary CD4+ T cells after DDX6 KD, with no change in steady-state Gag levels, supports the conclusion that DDX6 facilitates virus production.
Discussion
Here, we demonstrated that HIV-1 capsid AIs are formed when Gag co-opts a cellular complex that contains ABCE1 as well as PBPs (Figs. 2–4). We used multiple approaches to demonstrate that one of these AI-associated PBPs, DDX6, acts during assembly by promoting Gag multimerization. First, we showed that DDX6 depletion reduced VLP production by as much as approximately sixfold but with minimal effects on steady-state Gag levels; thus, nuclear export and translation were intact, and reduced virus production was not simply a result of a reduced concentration of intracellular Gag (Fig. 7 A). Second, our demonstration that DDX6 depletion also reduced production of GagZip VLPs, which resemble WT Gag VLPs but lack encapsidated gRNA or cellular mRNA (Fig. 7 B), established that DDX6 is required for capsid assembly independent of gRNA or cellular mRNA encapsidation. Third, using a quantitative IEM, we localized the defect in DDX6-depleted cells to Gag multimerization, which, in the case of HIV-1, occurs at the PM (Fig. 9 A). Lastly, we also showed that DDX6 acts catalytically to facilitate capsid formation (Fig. 8). From these diverse approaches, we conclude that in cells, HIV-1 capsid assembly is a host-catalyzed process.
Although it may seem surprising that an RNA helicase would promote capsid assembly, this conclusion is actually consistent with current thinking about the importance of RNA in promoting Gag multimerization. One obvious role for RNA helicases would be to remodel RNAs to make them available to bind to viral proteins during packaging. However, RNA is also critical for nucleating Gag multimerization, as shown by in vitro studies of recombinant Gag assembly. In these in vitro studies, recombinant nonmyristoylated, p6-deleted Gag is present at very high concentrations in the absence of other proteins but only forms capsidlike particles when single-stranded nucleic acid is added (Campbell and Rein, 1999). Interestingly, GagZip also undergoes multimerization upon addition of purified nucleic acid even though it lacks the RNA-binding properties of the NC domain, suggesting that other regions of Gag can bind to RNA (Crist et al., 2009). Importantly, RNAs in cells are covered with RNPs and would likely need to be remodeled before they could be used to nucleate assembly. Thus, we speculate that in cells, DDX6 may be the helicase that remodels RNA to make it available for binding by WT Gag or GagZip, thereby enabling nucleation of assembly. In the case of GagZip, gRNA or cellular mRNA made available by DDX6 may bind to GagZip in a manner that promotes capsid assembly but is not adequate for encapsidation, which could require additional RNA chaperoning activity provided by NC in the case of WT Gag (Rein et al., 2011). Alternatively, as data from one group suggest that GagZip VLPs contain fragments of 7SL RNA (Keene et al., 2010), 7SL RNA may be remodeled to allow nucleation of GagZip assembly followed by encapsidation.
Although we detected at least three PBPs in HIV-1 HMW AIs, other enzymes involved in RNA metabolism may also be present in these complexes. Studying the function of such enzymes is complicated by several issues. First, using siRNA KD approaches can be problematic, as indicated by our finding that some control siRNAs inhibit Gag expression most likely via indirect effects on RNA metabolism. Second, even small amounts of an enzyme remaining after KD could result in significant catalytic activity, thereby masking the full consequences of depletion. Third, such critical enzymes likely have functional homologues; thus, activity of other RNA helicases could also have contributed to residual VLP production after DDX6 depletion. Finally, such proteins may play multiple roles in RNA metabolism. For example, we could not address the effects on assembly of depleting DCP2 or AGO2 because these KDs severely reduced Gag expression in our hands (unpublished data). Thus, although it is possible that DCP2 or AGO2 also acts during HIV-1 assembly, their function in earlier steps of HIV-1 RNA metabolism makes their roles in assembly difficult to address. This problem of upstream effects preventing analysis of downstream functions also impacted our ability to study the role of DDX6 in packaging gRNA. Although our data argue that DDX6 facilitates capsid assembly per se, we cannot rule out a role for DDX6 in both assembly and packaging because defects in assembly inhibit VLP formation, and packaging can only be assayed if VLPs are formed. Importantly, although the small amount of residual virus formed after depletion did contain gRNA, that virus does not provide a reliable readout of packaging in the absence of DDX6 because it likely resulted from the activity of residual DDX6. Interestingly, a recent study found that DDX6 facilitates packaging of gRNA into human foamy virus (HFV), a spumaretrovirus distantly related to HIV-1 (Yu et al., 2011). Thus, it is possible that the role of DDX6 is different during assembly of HFV versus HIV-1, perhaps because these viruses assemble differently (Linial and Eastman, 2003). Alternatively, residual DDX6 activity in DDX6-depleted cells could be sufficiently functional for HFV assembly but not for HIV assembly or HFV packaging.
Our finding that HIV-1 Gag associates with ABCE1 and PBPs in the cytoplasm and that assembly is catalyzed by host enzymes differs from the currently accepted view in which Gag is thought to form oligomers associated with gRNA, with assembly driven solely by Gag–Gag, Gag–RNA, or Gag–phospholipid interactions (Rein et al., 2011). Our model of assembly occurring in complexes containing cellular enzymes (Fig. 10) is supported by the similarities between HIV-1 HMW AIs and Ty3 retrosomes. In both cases, WT Gag is associated with the DDX6 and DCP2 homologues (Figs. 2–4; Irwin et al., 2005; Beliakova-Bethell et al., 2006) in an NC-dependent manner (Fig. 4 B; Larsen et al., 2008). The association of Ty3 Gag with AGO2 cannot be assessed because Saccharomyces cerevisiae does not have an AGO2 homologue (Shabalina and Koonin, 2008). However, we also noted differences between retrosomes and HIV-1 HMW AIs, not surprisingly given the evolutionary distance between Ty3 and HIV-1 and their different life cycles. For example, HIV-1 capsids bud from the PM, whereas Ty3 capsids remain intracellular; consistent with this, HIV-1 AIs are found both in the cytoplasm and at the PM, whereas Ty3 retrosomes are only cytoplasmic. Additionally, Ty3 assembly occurs at a few larger centralized sites, whereas HIV-1 assembly occurs at numerous dispersed sites; this difference may explain why Ty3 retrosomes are easily visible using light microscopy, whereas some HIV-1 HMW AIs are likely below the threshold for detection by light microscopy. The general similarities suggest that formation of PBP-containing complexes that facilitate assembly and isolate it from host defenses may have been conserved during retroelement and retrovirus evolution, with variations evolving to accommodate life cycle differences. Consistent with this, in the aforementioned study, HFV Gag colocalized with DDX6 in perinuclear sites where HFV assembles (Yu et al., 2011). It remains to be determined whether other primate lentiviruses also form Gag-containing complexes associated with PBPs, although they have been shown to form HMW AIs containing Gag and ABCE1 (Dooher and Lingappa, 2004). It would also be interesting to determine whether Ty3 retrosomes contain ABCE1.
Figure 10.
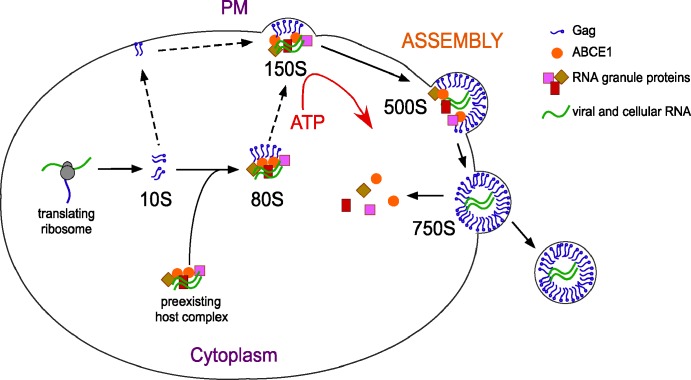
Model of the association of PBPs with Gag during immature HIV-1 capsid assembly. Newly synthesized Gag forms the ∼10S AI, a possible source of targeted Gag at the PM, which may be recruited for oligomerization at sites of assembly (dotted lines). Gag co-opts a complex of ABCE1 and PBPs, present in uninfected cells, to form the ∼80S AI. Analysis of G2A Gag mutants suggests that the ∼80S AI or the ∼150S AI targets Gag to the PM (dotted line). Gag continues to multimerize at the PM, forming the ∼500S AI, which also contains ABCE1 and PBPs. DDX6 facilitates multimerization of Gag at the PM. Upon formation of completed capsids (∼750S), PBPs and ABCE1 are released, and budding occurs.
Although HIV-1 HMW AIs contain PBPs, they appear to differ from typical PBs found in uninfected cells in multiple ways. First, they contain HIV-1 components, such as Gag, GagPol, and Vif (Zimmerman et al., 2002; Dooher et al., 2007). Second, they contain the ATPase ABCE1, which functions in translation termination and ribosome recycling in uninfected cells (e.g., Becker et al., 2012) and has not been reported in PBs. Third, PBs are typically thought to be visible using light microscopy, although, in fact, PB complexes of <30S and up to ∼1,000S have been reported in uninfected cells (Kanai et al., 2004; Höck et al., 2007). Whereas the ∼500S AI is likely visible using light microscopy, the ∼80S and ∼150S AIs most likely are not, as they have the same S value as a single ribosome. Notably, even when PBs are dispersed, they can remain functional (Balagopal and Parker, 2009), arguing that PB functionality is independent of size. Thus, functional PBP-containing structures could encompass a diverse range of sizes, with smaller PBP-containing complexes being less well studied than easily visible PBs. Importantly, the small size of some HIV HMW AIs may make them difficult to detect by methods that involve light microscopy; thus, our use of biochemical and ultrastructural approaches may have been advantageous. A fourth distinction is that although typical PBs are found in the cytoplasm or at the ER (Wilhelm et al., 2005; Squirrell et al., 2006), they have not been reported at the PM, unlike some HMW AIs. Interestingly, another group reported that during HIV-1 infection, the number of PBs per cell decreases from ∼28 to ∼7 (Milev et al., 2010). Therefore, when HIV-1 co-opts PB components to form HMW AIs, it may inhibit formation of typical PBs.
Notably, although our study shows that DDX6 is associated with Gag and facilitates virus production, DDX6 was not identified as a Gag interactor in a recent protein interaction screen (Jäger et al., 2012) nor as a facilitator of HIV-1 replication in large-scale siRNA screens (Brass et al., 2008; König et al., 2008; Zhou et al., 2008). Thus, apparently, such screens can miss important Gag interactors.
Although DDX6 has not been implicated previously in HIV-1 replication, other cellular RNA helicases are known to affect HIV-1 replication, including DDX3, RNA helicase A, DHX30, DDX1, RH116, DDX24, and MOV10 (Ranji and Boris-Lawrie, 2010). In the case of the PBP MOV10, three groups reported that overexpression reduces Gag expression and reverse transcription of HIV-1 and murine leukemia virus (Burdick et al., 2010; Furtak et al., 2010; Wang et al., 2010); interestingly, one study found that replication was inhibited by MOV10 depletion (Furtak et al., 2010), arguing that MOV10 may play a facilitating role like DDX6. Additionally, the RNA-binding protein Staufen, which is found mainly in stress granules (Thomas et al., 2009), associates with HIV-1 Gag and gRNA and influences virus production (Chatel-Chaix et al., 2004, 2007, 2008; Abrahamyan et al., 2010; Milev et al., 2010). Together with our findings, these data support a model in which RNA granule proteins, such as MOV10 and DDX6, modulate HIV-1 replication in different ways. This could occur via different cellular complexes; consistent with this, others have proposed that multiple classes of PBs exist (Decker and Parker, 2006).
At odds with our findings are two studies reporting that depletion of DDX6 increases viral gene expression and virus production by disrupting microRNA (miRNA) effector complexes that silence HIV-1 RNA (Chable-Bessia et al., 2009; Nathans et al., 2009). In contrast, we found that DDX6 KD reduced virus production, with minimal effects on steady-state Gag levels. Unlike our study, neither miRNA study showed a group that was not treated with siRNA in their KD figures. This omission could be problematic, given our finding that many control siRNAs cause large reductions in Gag expression and therefore on virus production, which would only be recognized if mock-treated cells are analyzed in parallel (Fig. S1). Thus, in the absence of a mock control, one cannot distinguish between a DDX6 KD that increases virus production and a control KD that decreases virus production. Alternatively, the discrepancy between our findings and those of the miRNA studies could be explained if DDX6 facilitates both miRNA inhibition and assembly, depending on its context. Related to this, a recent study found that AGO2 is associated with HIV-1 Gag and facilitates HIV-1 production (Bouttier et al., 2012). The conclusion of this study is consistent with our finding that AGO2 is present in HIV-1 AIs, but, on the surface, it is at odds with the aforementioned miRNA studies implicating AGO2 in miRNA-mediated inhibition of HIV-1 replication. A model in which PBPs could inhibit or promote HIV-1 replication, depending on the complex they are in, could reconcile such contradictions.
In conclusion, our findings suggest that HMW AIs are derived from novel cellular complexes that contain ABCE1 as well as PBPs and are present in uninfected cells (Fig. 10). Once co-opted by HIV-1, they become virus-specific HMW AIs, which function as assembly machines with cellular factors such as DDX6 and ABCE1 acting enzymatically to facilitate assembly. The general similarities to yeast Ty3 retrosomes suggest that this mechanism of assembly represents an ancient and highly conserved feature of retroelements that enables efficient, host-catalyzed capsid formation.
Materials and methods
Plasmids and cell lines
Proviruses were used in all experiments except Fig. 4 B, which expressed only HIV-1 Gag and Rev. The proviral plasmids, which were derived from an HIV-1 LAI strain provirus backbone obtained from M. Emerman (Fred Hutchinson Cancer Research Center, Seattle, WA), contain the entire HIV-1 genome, including the long terminal repeats, except for substitutions, deletions, and point mutations indicated below, and also contain the SV40 origin (Kimpton and Emerman, 1992). The replication-competent HIV-GFP plasmid used for infections, obtained from M. Emerman and M. Yamashita (Fred Hutchinson Cancer Research Center, Seattle, WA), encodes the entire genome except for nef, which was replaced by EGFP from Takara Bio Inc. (Yamashita and Emerman, 2004). For transfections, we used HIV-GFP ΔEnv, which was derived from HIV-GFP (Yamashita and Emerman, 2004), or HIV-1ΔEnvPro−, which encodes both a deletion in env and three point mutations that inactivate pro (D25K, G49W, and I50W). Gag mutants were generated in the HIV-1ΔEnv backbone, which encodes only the deletion of env and have been previously described (Dooher and Lingappa, 2004; Dooher et al., 2007; Klein et al., 2011). These include GagΔp2-NC-p1-p6 (previously termed HIV-1 pBruΔEnvGagTr or GagΔNCΔp6), which was generated by inserting a double stop codon after amino acid 363 in Gag, resulting in truncation after the capsid domain of Gag; GagG2A, in which the second amino acid of Gag is converted from glycine to alanine; and GagΔp1-p6, in which a double stop codon was inserted after amino acid 432 in Gag. GagZip has been previously described (Klein et al., 2011) and was constructed by introducing GCN4 zipper in place of NC using two-step PCR in the HIV-1ΔEnvPro− backbone. This led to loss of the ribosomal frameshift sequence in Gag and a failure to express Pol, as previously described (Accola et al., 2000). Thus, GagZip expressed from this construct remains unprocessed, resulting in VLPs containing only p55 Gag.
The YFP-AGO2, YFP-DDX6, YFP-DCP2, and YFP-LSM1 plasmids, which were generated in the EYFP-C1 vector (BD) and encode EYFP fused to the C terminus of the relevant PBPs, were obtained from T. Rana (University of Massachusetts Medical School, Worcester, MA) and have been previously described (Wichroski et al., 2006). To generate the pcDNA DDX6 siRNA–sensitive plasmid used in siRNA rescue experiments, DDX6 was PCR amplified from YFP-DDX6 and transferred to the pcDNA vector (Invitrogen). Then, site-directed mutagenesis was used to add sequence encoding MSTARTTENPVI to the N terminus to conform to the annotated DDX6 gene sequence (available from GenBank/EMBL/DDBJ under accession no. NM_004397.4). The pcDNA DDX6 siRNA–resistant plasmid (used in rescue experiments) was generated by introducing three silent mutations into the site targeted by the siRNA using the following site-directed mutagenesis primers: sense 5′-GAATTCCCATTTGCAAAAGCCATATGAGATTAACCTG-3′ and antisense 5′-CAGGTTAATCTCATATGGCTTTTGCAAATGGGAATTC-3′ (mutated residues are italicized). From this, the DEAD-box mutant pcDNA DDX6 E247A siRNA resistant was generated by site-directed mutagenesis with the following primers: sense 5′-CAGATGATAGTATTGGATGCGGCAGATAAGTTGCTG-3′ and antisense 5′-CAGCAACTTATCTGCCGCATCCAATACTATCATCTG-3′ (mutated residues are italicized).
The HIV-DDX6 siRNA–resistant plasmid used in the rescue experiments was made by amplification of an insert via two rounds of PCR. In the first round, two reactions were performed to yield two fragments with overlapping complementary ends as follows: using HIV-GFP as a template, a fragment was produced with sense primer no. 1 5′-GGCAGTCTAGCAGAAGAAG-3′ and antisense primer no. 2 5′-CGTGCTCATGGTTCTAGACTTATAGCAAAATCCTTTCCAAGC-3′, and, using pcDNA DDX6 siRNA resistant as a template, a fragment was produced with sense primer no. 3 5′-GGATTTTGCTATAAGTCTAGAACCATGAGCACGGCCAGAACAG-3′ and antisense primer no. 4 5′-NNNNCTCGAGTTAAGGTTTCTCATCTTCTACAGGCTC-3′. These fragments were gel purified and then combined into a final reaction with primer no. 1 and primer no. 4. The product from the final reaction was cloned into HIV-GFP using BamHI and XhoI to yield HIV-DDX6 siRNA resistant. The HIV-DDX6 E247A siRNA–resistant construct was produced by amplifying DDX6 E247A resistant from pcDNA DDX6 E247A resistant using primer no. 3 and primer no. 4 followed by cloning with XbaI–XhoI into HIV-GFP.
To express only Gag and Rev (Fig. 4 B), we cotransfected the subgenomic expression plasmids pSVGagRRE-R and pCMV Rev, which were obtained from D. Rekosh (State University of New York at Buffalo, Buffalo, NY; Smith et al., 1990), at a ratio of 8:1. pSVGagRRE-R encodes bp 679–2,483 of the BH10 strain of HIV-1 (containing the Gag-coding region) as well as the HIV-1 Rev response element. pCMV Rev encodes the rev gene from the same HIV-1 strain under the control of the cytomegalovirus promoter. The G2A and GagΔp1-p6 plasmids (previously called Gag p46) were generated in the pSVGagRRE-R backbone and encode the aforementioned mutations (Lingappa et al., 2006). The KR10A mutation was generated in the same backbone and encodes alanine substitutions of 10 basic residues in the NC domain of Gag at amino acids 391, 397, 403, 406, 409, 410, 411, 415, and 418 in Gag (Lingappa et al., 2006).
The H9-HIV cell line stably expresses the HIV-1 provirus–encoding deletions in env and vif, a frameshift in vpr, and substitution of nef with a puromycin resistance gene and was generated by infecting the parental H9 T cell line with the aforementioned provirus and selecting with puromycin (Dooher and Lingappa, 2004).
siRNA oligonucleotides
The siRNA oligonucleotide used for KD experiments was directed against human DDX6 (accession no. NM_004397.4). The region targeted by the siRNA is identical in the macaque sequence; therefore, the same siRNA was used for KD in COS-1 cells, which are derived from African green monkeys. The sequence for the DDX6-targeting siRNA is as follows (sense strand): 5′-GCAGAAACCCUAUGAGAUUUU-3′. As a control for the siRNA KD, a nontargeting siRNA was transfected in parallel. Both the KD and control siRNAs were synthesized by Thermo Fisher Scientific. The following control siRNAs were compared for effects on HIV-1 Gag expression in Fig. S1: control siRNA1 (ON-TARGETplus nontargeting pool; D-001810-10; Thermo Fisher Scientific), control siRNA2 (siGENOME nontargeting siRNA no. 1; D-001210-01; Thermo Fisher Scientific), control siRNA3 (Scramble II; D-001205; Thermo Fisher Scientific), control siRNA4 (RISC-free siRNA; D-001220-01; Thermo Fisher Scientific), and control siRNA5 (ON-TARGETplus nontargeting pool siRNA no. 1 with a Dy547 5′ sense strand modification). Control siRNA1 was used for siRNA KD experiments shown in Figs. 7, 8, and 9.
Antisera
Commercial DDX6 and AGO2 antibodies (A300-461 for DDX6 [Bethyl Laboratories, Inc.] and EIF2C2 M01 for AGO2 [Abnova]) were used for all experiments except those involving IEM (Figs. 5, 6, and 9) or antibody validation (Fig. S3). A commercial GFP antibody for YFP detection (632375; Takara Bio Inc.) and TIAR antibody (610352; BD) were used for WB. Because large quantities of antibodies are needed for IEM experiments, we generated peptide-specific antisera to DDX6 and AGO2, which were then affinity purified and validated by IP and WB (Fig. S3) and IEM using cells from which DDX6 and AGO2 were depleted using siRNA (Fig. 9 A and not depicted). For generation of peptide-specific antisera, the following peptides were chosen because of their high degree of antigenicity and surface probability using MacVector: 84RIKTSDVTSTKGNEFEDYC102 in DDX6 (accession no. BC065007) and 235CEVLDFKSIEEQQKPLTDSQR255 in AGO2 (accession no. NP_001158095). Rabbits were injected with the indicated peptide coupled to keyhole limpet hemocyanin (Lampire Biological Laboratories). Immune sera and preimmune sera were compared by WB and IP using the aforementioned commercial antibodies as positive controls. Selected lots with good reactivity were affinity purified against the peptide used to generate antiserum (SulfoLink column; Thermo Fisher Scientific). For IEM experiments, affinity-purified antibodies were desalted and concentrated to 1–2 mg/ml. ABCE1 and APOBEC3G antisera were generated as previously described and are directed against peptides containing the C-terminal 19 amino acids of ABCE1 and the C-terminal 29 amino acids of APOBEC3G (Zimmerman et al., 2002; Thielen et al., 2007). Gag was detected using a monoclonal antibody from the HIV-1 p24 Hybridoma (183-H12-5C) generated by B. Chesebro (National Institute of Allergy and Infectious Diseases, Rocky Mountain Laboratories, Hamilton, MT) and obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.
Transfections, cell harvests, IPs, and WB
COS-1 cells were transfected with the indicated plasmids (∼1.0–3.0 µg per well of a 6-well plate) using polyethylenimine (Polysciences, Inc.). Cells were harvested 38 h after transfection in one of the harvest buffers (0.35% Triton X-100, 10 mM NaCl, and 20 mM Hepes, pH 7.9, with either 1 mM MgAc and 0.2 mM EDTA or with 10 mM EDTA or 0.625% NP-40, 100 mM NaCl, 50 mM KAc, and 10 mM TrisAc, pH 7.4, with or without 10 mM EDTA), clarified by centrifugation for 10 min at 160 g using a rotor (GH3.8; Beckman Coulter) followed by centrifugation for 1–10 min at 18,000 g in a microcentrifuge at 4°C. H9-uninfected cells and H9-HIV cells were harvested following the same protocol as for transfected COS-1 cells. Where indicated, cell lysates were treated with RNase A (QIAGEN) at 100 µg/ml at 37°C for 10 min, incubated on ice for 30 min, and then cleared by centrifugation for 1–10 min at 18,000 g in a microcentrifuge at 4°C or were treated with puromycin (Invitrogen) at 1 mM final concentration at 37°C for 10 min. For IPs from total cell lysates, 1 µg of antibody was added to lysates for 30 min at 4°C followed by addition of protein G or protein A Tris–acrylamide beads (Thermo Fisher Scientific) and rotation for 1–4 h at 4°C. Antibodies used for IPs include the aforementioned commercial antibodies to DDX6 and AGO2, α-GFP (632460; Takara Bio Inc.) for YFP IPs, the affinity-purified, peptide-specific α-ABCE1 that we previously generated (Zimmerman et al., 2002), and the affinity-purified, peptide-specific α-AGO2 and α-DDX6 generated in this study (Fig. S3 only). IPs were washed twice with harvest buffer and once with harvest buffer lacking detergent. Lysates and IP eluates were analyzed by SDS-PAGE followed by WB with either a murine monoclonal antibody directed against HIV-1 Gag p24 (HIV-1 p24 Hybridoma; 183-H12-5C; obtained from B. Chesebro through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, and National Institute of Allergy and Infectious Diseases) or the aforementioned antibodies directed against ABCE1, DDX6, AGO2, TIAR, or anti-GFP (632375; Takara Bio Inc.). WBs were analyzed using ECL (Thermo Fisher Scientific) or Odyssey Imaging (LI-COR Biosciences).
Gradient analysis and RNase treatment
Cell lysates harvested as described in the previous section were subjected to VS for 45 min at 4°C in a rotor (MLS-50; Beckman Coulter) at 45,000 rpm (217,000 g) on step gradients (175-µl sample per gradient). The following sucrose solutions, made in the harvest buffers described in the previous section, were used for step gradients: 675 µl each of 10, 15, 20, 40, 50, 66, and 80% sucrose, which were similar to those previously described (Klein et al., 2011). Gradients were fractionated from top to bottom (175 µl per fraction), and 20-µl aliquots of each fraction were analyzed by SDS-PAGE followed by WB for Gag. For IPs, the following gradient fractions were pooled, diluted twofold, and subjected to IP, as described in the previous section: 1 and 2 (10S), 6 and 7 (80S), 10 and 11 (150S), 14 and 15 (500S), and 19 and 20 (750S). S values of AIs were estimated using the formula S = ΔI/ω2t, where S is the sedimentation coefficient of the particle in Svedberg units, ΔI is the time integral for sucrose at the separated zone minus the time integral for sucrose at the meniscus of the gradient, ω is rotor speed in radians/s, and t is time in seconds (McEwen, 1967). Values for I were determined for particles of a density of 1.3 g/cm3, according to tables published in McEwen (1967). Markers such as BSA (5S), macroglobulin (20S), hepatitis B virus capsids (100S), ribosomal subunits (40S and 60S), and polysomes (>100S) were used to calibrate the gradients and to confirm the calculated S values. However, it should be noted that the S-value assignments for each Gag-containing complex are approximate estimates that allow a general description of the complex and should not be interpreted as constituting a detailed biophysical analysis.
For RNase treatments of gradient fractions, fractions 6–9 representing the ∼80S fraction were pooled and subjected to diafiltration and concentration with a 4-ml, 50-kD molecular weight cutoff ultrafilter (Amicon; EMD Millipore) until the sucrose concentration was <5%. Concentrated 80S material was divided and treated with either 1% SDS and boiling for 5 min, 1× NP-40 buffer for 15 min at 37°C, or 50 µg/ml RNase A (QIAGEN) for 15 min at 37°C. Samples were then subjected to gradient analysis, and fractions were analyzed by WB for Gag. We have previously shown that AIs are insensitive to 1 µg/ml RNase A for 20 min at 22°C (Lingappa et al., 1997), but a 50-fold higher concentration of RNase A was used in the experiments described here.
siRNA depletion of DDX6
COS-1 cells were plated at a density of ∼100,000–140,000 cells per well of a 6-well plate (day 0). The next day (day 1) and 48 h later (day 2), cells were transfected using Lipofectamine (Invitrogen) without siRNA (mock), with control siRNA1 (100 pmol), or with DDX6-specific siRNA (100 pmol). 48 h later (day 4), cells were transfected using Lipofectamine with plasmids encoding HIV-1Δenv (1.5 µg) and VSV-G (0.375 µg; Fig. 7 A). Experiments in which infectivity was not assessed were not cotransfected with VSV-G on day 4 (Figs. 7 B and 8). 24 h later (day 5), media were collected for virus pelleting (using methods described under VLP preparation), and, where indicated, infectivity of released virus was measured by titering on TZM-bl cells. Cells were harvested in NP-40 buffer, as described above, and equivalent amounts of cell lysate were used for gel loading, as determined by total protein measurements (BCA protein assay reagent; Thermo Fisher Scientific). For EM analysis of cells depleted of DDX6 or treated with control siRNA1, the same KD protocol was followed, except cells were infected on day 4 and harvested for HPF 36 hours after infection.
gRNA quantification
Using the siRNA transfection protocol described in the Transfections, cell harvests, IPs, and WB section, COS-1 cells, initially seeded at ∼100,000 cells per well, were counted on days 2, 3, 4, and 5 of the KD protocol. Extracted RNA was DNase I treated (Ambion) followed by DNase I heat inactivation and subsequently subjected to reverse transcription using random primers and reverse transcription (SuperScript II; Life Technologies) according to the manufacturer’s protocol. Quantitative PCR was performed using iQ SYBR green Supermix (Bio-Rad Laboratories) with the following forward and reverse primers, respectively: 5′-CAAAACTCTTGCCTTATGGCCGGGTCCTCC-3′ and 5′-GACTATGTAGACCGGTTCTAT-3′. Serial dilutions of plasmid encoding our HIV-1 Δenv were subjected to quantitative PCR in parallel to generate a standard curve for quantification of RNA from viral pellets.
Cell counts
Following the siRNA transfection protocol in the COS-1 cells described in the Transfections, cell harvests, IPs, and WB section, cells were plated at ∼100,000 cells and counted on days 2, 3, 4, and 5 of the KD protocol. Media from cells were removed, cells were washed once with PBS, and cells were trypsinized (25200; Gibco). Trypsinized cells were resuspended in media, and twofold dilutions of cells were prepared in 0.4% Trypan blue solution (T8154; Sigma-Aldrich) to stain nonviable cells. For each well, two dilutions of cells were counted in duplicate for viable cells using a hemocytometer (VWR 15170–208). The mean cell counts were calculated and plotted as a log10 viable cell number over time (days) ±SD.
Rescue of siRNA depletion of DDX6
COS-1 cells were transfected with siRNA, as described in the previous section, with the following modifications. On day 3, cells transfected with control siRNA were transfected with 1 µg pcDNA DDX6 siRNA sensitive, and cells transfected with DDX6-specific siRNA were transfected with 1 µg pcDNA DDX6 sensitive (the same plasmid the control KD received), pcDNA DDX6 resistant (siRNA-resistant construct), or pcDNA DDX6 E247A resistant (siRNA-resistant mutant construct). On day 4, 1.5 µg HIV-GFP was transfected into wells that had received the siRNA-sensitive DDX6 construct. The remaining wells were transfected with an HIV construct that matched the DDX6 encoded by the pcDNA transfected on day 3 (either HIV-DDX6 siRNA resistant or HIV-DDX6 E247A siRNA resistant). Transfection of the rescue constructs as both pcDNA expression vectors and HIV expression vectors ensured that rescue constructs were expressed optimally.
CD4+ T cell isolation, culture, and siRNA nucleofection
Peripheral blood mononuclear cells were obtained from the Seattle Assay Control study. Following informed consent in accordance with procedures approved at the Fred Hutchinson Cancer Research Center, cells from healthy human donors were separated by leukapheresis and immediately frozen. Peripheral blood mononuclear cells from anonymous donors were thawed in serum-free media (AIM V; Invitrogen). CD4+/CD3+ T cells were isolated from thawed peripheral blood mononuclear cells by negative selection (EasySep Human CD4+ T/CD3+ cell enrichment kit; STEMCELL Technologies) and resuspended in AIM V supplemented with 2 mM Glutamax (Invitrogen) at a concentration of 5 × 106 cells/ml. Cells were allowed to rest overnight and then resuspended in activation media (AIM V media, 2 µg/ml PHA [50 U/ml IL-2; Thermo Fisher Scientific], 0.5 µl/ml mouse anti–human IFN-α [no. 21385-1; PBL InterferonSource], 0.125 µl/ml mouse anti–human IFN-α/β receptor [MAB411; EMD Millipore], and 2 mM Glutamax). After 72 h, activated cells were transfected with control siRNA1 or DDX6-specific siRNA by electroporation (500 pmol siRNA for 10 × 106 cells; Human CD4+ T cell nucleofector kit, VAPA-1002, program T-023; Lonza). After transfection, cells were resuspended in growth media (same as activation media but without PHA). At 72 h after transfection, cells were infected at a multiplicity of infection of two by incubating cells with virus for 5 h. After infection, cells were washed to remove remaining inoculum and resuspended in growth media. At 0, 24, 48, and 72 h after infection, cells and media were collected for analysis, and remaining cells were washed and resuspended in growth media.
VLP preparation
For VLP analysis, media were centrifuged (10 min at 900 g; GH3.8 rotor; Beckman Coulter) and passed through a 0.45-µm syringe filter to remove cells and then pelleted through a 30% sucrose cushion for 30 min at 4°C at 485,000 g using an SW60Ti rotor (Beckman Coulter). Supernatants were aspirated, pellets were harvested in NP-40 buffer, and aliquots were analyzed by SDS-PAGE and WB, as previously described.
For preparation of virus stocks, media were centrifuged (10 min at 900 g; GH3.8 rotor) and passed through a 0.45-µm syringe filter to remove cells and then pelleted using an SW40Ti rotor (Beckman Coulter) for either for 34 min at 4°C at 284,570 g or 90 min at 20°C at 50,000 g.
Virus infectivity measurements
COS-1 cells were transfected as previously described, and virus was harvested as described for VLP preparation. Infectivity was measured by titering stocks on TZM-bl cells (obtained from J.C. Kappes and X. Wu [University of Alabama, Birmingham, AL] and Tranzyme, Inc. through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, and National Institute of Allergy and Infectious Diseases). Infection of TZM-bl cells was facilitated by the addition of 20 µg/ml DEAE dextran and by spinoculation (2 h at 2,500 rpm in a centrifuge [Allegra 6; Beckman Coulter] using a GH3.8 rotor). Infectious units were determined by X-Gal staining and/or counting blue cells. For the primary T cell experiments, instead of X-Gal staining, a MUG (4-methylumbelliferone β-d-galactopyranoside) substrate was used, and the change in MUG fluorescence over time per volume of media was measured on a plate reader (FLx800; BioTek Instruments, Inc.) and used to compare infectivity between samples.
Infection of COS-1 cells for IEM
VSV-G–pseudotyped VLPs were generated by transfecting 293T cells with HIV-GFPΔenv and VSV-G (pMD2.G; obtained from D. Trono [Swiss Institutes of Technology, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland] via Addgene) at a ratio of 3:1 or 9:1 and harvesting virus by pelleting, as described for VLP preparation. Virus was titered on TZM-bl cells, as previously described, and cells were infected with virus at a multiplicity of infection of 10–50 using 20 µg/ml DEAE dextran and spinoculation, as described in the previous section. After incubation with virus for 12–15 h, cells were washed three times with media to remove virus inoculum. Infected cells were harvested at 36 h after infection in 3% PFA and 0.025% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, pelleted, and subjected to high-pressure freezing using the high-pressure freezer (EMPACT2; Leica) followed by freeze substitution. Samples were infiltrated overnight with embedding resin (LR White; London Resin Company Ltd) in ethanol, changed to straight LR White, embedded in gelatin capsules (Electron Microscopy Sciences), and cured overnight in a UV light cryochamber at 4°C. ∼50-nm sections were placed on grids, treated with 0.05 M glycine for 20 min at room temperature, rinsed in PBS, blocked for 45 min with 1–5% BSA (Electron Microscopy Sciences), and washed in PBS with 0.1% BSA-C (Electron Microscopy Sciences). The monoclonal antibody directed against HIV-1 Gag (described in the Transfections, cell harvests, IPs, and WB section) was used at a final concentration of 0.25 mg/ml diluted with 0.1% BSA-C in PBS and was detected with goat α–mouse IgG conjugated to 6-nm gold diluted 1:1 in 0.1% BSA-C in PBS (Electron Microscopy Sciences). Affinity-purified, peptide-specific rabbit α-DDX6 was prepared, as previously described, diluted with 0.1% BSA-C and 0.005% Tween 20 in PBS to ∼0.7 mg/ml, and detected with goat α–rabbit IgG conjugated to 15-nm gold diluted 1:1 with 0.1% BSA-C and 0.005% Tween 20 in PBS. Affinity-purified, peptide-specific rabbit α-AGO2 was prepared and used in the same manner as for α-DDX6, except that Tween was not added to the primary or secondary antibody incubations. For double immunolabeling, grids were subjected to the following series of 1-h incubations: α-DDX6 or α-AGO2 primary, gold-conjugated goat α–rabbit IgG secondary, α-Gag primary, and gold-conjugated goat α–mouse IgG. Between incubations, grids were rinsed three times with PBS and six times with 0.1% BSA-C in PBS. Sections were then fixed with 2% glutaraldehyde in PBS for 10 min at room temperature and rinsed in deionized water. Samples were poststained with uranyl acetate for 15 min and lead citrate for 5 min, and stained sections were examined using a transmission electron microscope (JEM-123; JEOL). WT Gag and GagΔp2-NC-p1-p6 sections were processed in parallel on the same day in each experiment, and the double-labeling experiment was performed twice. Single-labeling control experiments were performed to confirm specificity of primary antibody labeling and included comparisons to labeling with nonimmune rabbit IgG (Bethyl Laboratories, Inc.) and labeling in the absence of primary antibody. Pictures were taken using a 2k × 2k camera (UltraScan US10000; Gatan, Inc.) and DigitalMicrograph software (Gatan, Inc.), saved as TIFF files, and opened in Photoshop (Adobe). Contrast adjustments were made uniformly for the entire image in Photoshop.
To quantify colocalization, each grid was divided into nine nearly equivalent regions. If cells were present within a region, photographs were taken of Gag labeling at the membrane of up to two randomly selected cells at a magnification of 15k. Once all images were obtained, sites of assembly, referred to as regions of clustered Gag (RCGs), were marked. An RCG was defined as a 100-nm zone (which is approximately equivalent to the diameter of an immature VLP) plus an additional 35-nm zone on either side of the RCG. The additional 35 nm was included to account for the distance from the labeled antigen added by the primary and secondary antibody plus the gold bead. To be counted, RCGs had to contain at least three 6-nm gold particles. Sites where membrane deformation, dark staining, and Gag labeling were all present were marked first. Next, the remaining sites that met the aforementioned criteria for RCGs were marked, starting from one edge of the photograph and moving to the other. Clusters of Gag label that overlapped with an existing marked site <100 nm from the edge of the image field or further than 100 nm from the membrane were excluded from the analysis. For each RCG marked, the 100-nm inner diameter of the site was first centered on the Gag cluster and then aligned tangentially with the membrane such that the 100-nm zone was mostly within the cell. Colocalization was determined by examining each RCG for 15-nm gold DDX6 or AGO2 label. If 15-nm gold labeling occurred within the 170-nm RCG zone, it was scored as positive. The RCGs were classified as targeted Gag if there was no membrane deformation or electrondense staining at the membrane, an early PM site of assembly if there was electrondense staining with or without membrane deformation that formed a bud that was <50% complete, and a late PM site of assembly if membrane deformation formed a bud that was >50% complete. Percentage of colocalization was defined as the total number of RCGs that contained 15-nm gold labeling as a percentage of total RCGs within the same assembly category. Fold colocalization was defined as the ratio of percentage of colocalization of WT Gag over percentage of colocalization of GagΔp2-NC-p1-p6, which was calculated separately for targeted, early, and late assembly sites. Error bars represent SD from the two separate labeling experiments. Relevant quantification parameters are summarized in Table 1.
To quantify DDX6 labeling in cells depleted of DDX6 or cells treated with control siRNA1, two independent KD experiments were performed. Cells exhibiting Gag labeling were selected at random (13 cells from the first experiment and 10 cells from the second experiment). Once a cell was selected, the entire periphery of the cell was scanned for Gag labeling. If Gag labeling was present, the region was imaged. If after scanning the entire cell the total area imaged was less than ∼10 µm2, then additional images were taken from the cell at random to obtain at least ∼10 µm2 of area for analysis. For each cell, the total DDX6 label was determined and divided by the total area examined to obtain DDX6 label/µm2. Determination of targeted versus early versus late Gag at the PM is described in the legend for Fig. 5.
Densitometry
Where indicated with graphs or quantitation, bands on WBs were quantified using ImageJ software (National Institutes of Health) or Image Studio software (LI-COR Biosciences).
Online supplemental material
Fig. S1 shows that upon transfection, some but not all siRNA control oligonucleotides reduce steady-state Gag levels in cells. For KD experiments in this manuscript, we chose an siRNA control oligonucleotide that did not reduce Gag levels. Fig. S2 shows that DDX6 KD has no effect on cell viability using cell counts performed on successive days of a KD experiment. Fig. S3 uses commercial antibodies in IPs and WBs to validate the peptide antisera that we generated against DDX6 and AGO2 and used for IEM. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111012/DC1.
Supplementary Material
Acknowledgments
We thank M. Emerman, M. Yamashita, D. Rekosh, and T. Rana for reagents; A. Kell, K. Souza, and B. Vander Stoep Hunt for technical assistance; B. Schneider, S. MacFarlane, and the Fred Hutchinson Cancer Research Institute for assistance with transmission EM; A. Mouland, A. Groat-Carmona, and B. Robinson for comments on the manuscript; and the AIDS Research and Reference Reagent Program, Division of AIDS, and National Institute of Allergy and Infectious Diseases for reagents.
Funding sources included the National Institutes of Health R01AI48389 (to J.R. Lingappa) including an American Recovery and Reinvestment Act supplement, and National Science Foundation graduate fellowship DGE0203031 (to J.C. Reed). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. J.R. Lingappa is a cofounder of Prosetta Antiviral, Inc. and has donated two thirds of her founder’s shares to a nonprofit foundation.
Footnotes
Abbreviations used in this paper:
- AI
- assembly intermediate
- gRNA
- genomic RNA
- HFV
- human foamy virus
- HMW
- high molecular weight
- IEM
- immunoelectron microscopy
- IP
- immunoprecipitation
- KD
- knockdown
- LZ
- leucine zipper
- miRNA
- microRNA
- PB
- processing body
- PBP
- PB protein
- PM
- plasma membrane
- RCG
- region of clustered Gag
- RNP
- ribonucleoprotein
- VLP
- viruslike particle
- VS
- velocity sedimentation
- WB
- Western blot
- WT
- wild type
References
- Abrahamyan L.G., Chatel-Chaix L., Ajamian L., Milev M.P., Monette A., Clément J.F., Song R., Lehmann M., DesGroseillers L., Laughrea M., et al. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 123:369–383 10.1242/jcs.055897 [DOI] [PubMed] [Google Scholar]
- Accola M.A., Strack B., Göttlinger H.G. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402 10.1128/JVI.74.12.5395-5402.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 10.1128/JVI.77.15.8181-8186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Abe K., Dansako H., Ikeda M., Wakita T., Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81:13922–13926 10.1128/JVI.01517-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V., Parker R. 2009. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21:403–408 10.1016/j.ceb.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I., et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 482:501–506 10.1038/nature10829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C.J., Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 3:206–212 10.1016/j.chom.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N., Beckham C., Giddings T.H., Jr, Winey M., Parker R., Sandmeyer S. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 12:94–101 10.1261/rna.2264806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P.D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 5:550–558 10.1016/j.chom.2009.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouttier M., Saumet A., Peter M., Courgnaud V., Schmidt U., Cazevieille C., Bertrand E., Lecellier C.H. 2012. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res. 40:775–786 10.1093/nar/gkr762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319:921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Bryant M., Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA. 87:523–527 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick R., Smith J.L., Chaipan C., Friew Y., Chen J., Venkatachari N.J., Delviks-Frankenberry K.A., Hu W.S., Pathak V.K. 2010. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J. Virol. 84:10241–10253 10.1128/JVI.00585-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S., Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chable-Bessia C., Meziane O., Latreille D., Triboulet R., Zamborlini A., Wagschal A., Jacquet J.M., Reynes J., Levy Y., Saib A., et al. 2009. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 6:26 10.1186/1742-4690-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Clément J.F., Martel C., Bériault V., Gatignol A., DesGroseillers L., Mouland A.J. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 24:2637–2648 10.1128/MCB.24.7.2637-2648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Abrahamyan L., Fréchina C., Mouland A.J., DesGroseillers L. 2007. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J. Virol. 81:6216–6230 10.1128/JVI.00284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Boulay K., Mouland A.J., Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 5:41 10.1186/1742-4690-5-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley M.A., Nagashima K., Lockett S.J., Nyswaner K.M., Garfinkel D.J. 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30:382–398 10.1128/MCB.00251-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 410:512–524 10.1016/j.jmb.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T., Gilmore R. 1986. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J. Cell Biol. 103:2253–2261 10.1083/jcb.103.6.2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist R.M., Datta S.A., Stephen A.G., Soheilian F., Mirro J., Fisher R.J., Nagashima K., Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: Implications for retrovirus assembly. J. Virol. 83:2216–2225 10.1128/JVI.02031-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. 2006. CAR-1 and trailer hitch: Driving mRNP granule function at the ER? J. Cell Biol. 173:159–163 10.1083/jcb.200601153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov D.G., Freed E.O. 2004. Retrovirus budding. Virus Res. 106:87–102 10.1016/j.virusres.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Díez J., Ishikawa M., Kaido M., Ahlquist P. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA. 97:3913–3918 10.1073/pnas.080072997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooher J.E., Lingappa J.R. 2004. Conservation of a stepwise, energy-sensitive pathway involving HP68 for assembly of primate lentivirus capsids in cells. J. Virol. 78:1645–1656 10.1128/JVI.78.4.1645-1656.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooher J.E., Schneider B.L., Reed J.C., Lingappa J.R. 2007. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic. 8:195–211 10.1111/j.1600-0854.2006.00524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko J.A., Kenny A.E., Gamache E.R., Curcio M.J. 2010. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 84:5052–5066 10.1128/JVI.02477-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak V., Mulky A., Rawlings S.A., Kozhaya L., Lee K., Kewalramani V.N., Unutmaz D. 2010. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS ONE. 5:e9081 10.1371/journal.pone.0009081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlinger H.G., Sodroski J.G., Haseltine W.A. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 86:5781–5785 10.1073/pnas.86.15.5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J.L., Coleman L.E., Raymond A.S., Goodson S.G., Pittard W.S., Tsui C., Devine S.E. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics. 164:867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., Meister G. 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8:1052–1060 10.1038/sj.embor.7401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin B., Aye M., Baldi P., Beliakova-Bethell N., Cheng H., Dou Y., Liou W., Sandmeyer S. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15:641–654 10.1101/gr.3739005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., et al. 2012. Global landscape of HIV-human protein complexes. Nature. 481:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.C., Scobie H.M., Ma Y.M., Vogt V.M. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177–11185 10.1128/JVI.76.22.11177-11185.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Simon S.M., Bieniasz P.D. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA. 106:19114–19119 10.1073/pnas.0907364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. 2004. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 43:513–525 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Keene S.E., King S.R., Telesnitsky A. 2010. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J. Virol. 84:9070–9077 10.1128/JVI.00714-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I., Meehan A., Poeschla E.M. 2010. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 84:6352–6366 10.1128/JVI.00363-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpton J., Emerman M. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K.C., Reed J.C., Tanaka M., Nguyen V.T., Giri S., Lingappa J.R. 2011. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J. Virol. 85:7419–7435 10.1128/JVI.00288-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Zhou Y., Elleder D., Diamond T.L., Bonamy G.M., Irelan J.T., Chiang C.Y., Tu B.P., De Jesus P.D., Lilley C.E., et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 135:49–60 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan M.N., Ng A., Sukumaran B., Gilfoy F.D., Uchil P.D., Sultana H., Brass A.L., Adametz R., Tsui M., Qian F., et al. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature. 455:242–245 10.1038/nature07207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner D.B., Lindenbach B.D., Grdzelishvili V.Z., Noueiry A.O., Paul S.M., Ahlquist P. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA. 100:15764–15769 10.1073/pnas.2536857100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay S.B., Bieniasz P.D. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6:e1001200 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen L.S., Zhang M., Beliakova-Bethell N., Bilanchone V., Lamsa A., Nagashima K., Najdi R., Kosaka K., Kovacevic V., Cheng J., et al. 2007. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 81:6957–6972 10.1128/JVI.02207-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen L.S., Beliakova-Bethell N., Bilanchone V., Zhang M., Lamsa A., Dasilva R., Hatfield G.W., Nagashima K., Sandmeyer S. 2008. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 82:2501–2514 10.1128/JVI.01814-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. 2006. Dead-box proteins: A family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 34:4168–4180 10.1093/nar/gkl468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E. 2011. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12:505–516 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Lingappa J.R., Hill R.L., Wong M.L., Hegde R.S. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567–581 10.1083/jcb.136.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa J.R., Dooher J.E., Newman M.A., Kiser P.K., Klein K.C. 2006. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 281:3773–3784 10.1074/jbc.M507255200 [DOI] [PubMed] [Google Scholar]
- Linial M.L., Eastman S.W. 2003. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 277:89–110 10.1007/978-3-642-55701-9_4 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Neil S.J. 2011. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 9:519–531 10.1038/nrmicro2596 [DOI] [PubMed] [Google Scholar]
- McEwen C.R. 1967. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem. 20:114–149 10.1016/0003-2697(67)90271-0 [DOI] [PubMed] [Google Scholar]
- Milev M.P., Brown C.M., Mouland A.J. 2010. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 7:41 10.1186/1742-4690-7-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R., Chu C.Y., Serquina A.K., Lu C.C., Cao H., Rana T.M. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 34:696–709 10.1016/j.molcel.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A.O., Diez J., Falk S.P., Chen J., Ahlquist P. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094–4106 10.1128/MCB.23.12.4094-4106.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E.O. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: Role of the matrix amino terminus. J. Virol. 73:4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Pélisson A., Teysset L., Chalvet F., Kim A., Prud’homme N., Terzian C., Bucheton A. 1997. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica. 100:29–37 10.1023/A:1018336303298 [DOI] [PubMed] [Google Scholar]
- Randall G., Panis M., Cooper J.D., Tellinghuisen T.L., Sukhodolets K.E., Pfeffer S., Landthaler M., Landgraf P., Kan S., Lindenbach B.D., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA. 104:12884–12889 10.1073/pnas.0704894104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranji A., Boris-Lawrie K. 2010. RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biol. 7:775–787 10.4161/rna.7.6.14249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Datta S.A., Jones C.P., Musier-Forsyth K. 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 36:373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S.B., Clemens K.A. 2010. Function of a retrotransposon nucleocapsid protein. RNA Biol. 7:642–654 10.4161/rna.7.6.14117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N., Mina L.B., Galão R.P., Chari A., Giménez-Barcons M., Noueiry A., Fischer U., Meyerhans A., Díez J. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. USA. 106:13517–13522 10.1073/pnas.0906413106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S.A., Koonin E.V. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. (Amst.). 23:578–587 10.1016/j.tree.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 418:646–650 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Singh A.R., Hill R.L., Lingappa J.R. 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology. 279:257–270 10.1006/viro.2000.0706 [DOI] [PubMed] [Google Scholar]
- Smith A.J., Cho M.I., Hammarskjöld M.L., Rekosh D. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 64:2743–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell J.M., Eggers Z.T., Luedke N., Saari B., Grimson A., Lyons G.E., Anderson P., White J.G. 2006. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol. Biol. Cell. 17:336–344 10.1091/mbc.E05-09-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen B.K., Klein K.C., Walker L.W., Rieck M., Buckner J.H., Tomblingson G.W., Lingappa J.R. 2007. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 3:1320–1334 10.1371/journal.ppat.0030135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.G., Martinez Tosar L.J., Desbats M.A., Leishman C.C., Boccaccio G.L. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 122:563–573 10.1242/jcs.038208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Han Y., Dang Y., Fu W., Zhou T., Ptak R.G., Zheng Y.H. 2010. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 285:14346–14355 10.1074/jbc.M110.109314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.M., Bidet K., Yinglin A., Ler S.G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M.A. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 8:1173–1186 10.4161/rna.8.6.17836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski M.J., Robb G.B., Rana T.M. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41 10.1371/journal.ppat.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J.E., Buszczak M., Sayles S. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell. 9:675–685 10.1016/j.devcel.2005.09.015 [DOI] [PubMed] [Google Scholar]
- Yamashita M., Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670–5678 10.1128/JVI.78.11.5670-5678.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.F., Lujan P., Jackson D.L., Emerman M., Linial M.L. 2011. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 7:e1002303 10.1371/journal.ppat.1002303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V., Perez-Caballero D., Göttlinger H., Bieniasz P.D. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058–12061 10.1128/JVI.78.21.12058-12061.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qian H., Love Z., Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Xu M., Huang Q., Gates A.T., Zhang X.D., Castle J.C., Stec E., Ferrer M., Strulovici B., Hazuda D.J., Espeseth A.S. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 4:495–504 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Zimmerman C., Klein K.C., Kiser P.K., Singh A.R.S., Firestein B.L., Riba S.C., Lingappa J.R. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 415:88–92 10.1038/415088a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.