Abstract
The cancer stem cell (CSC) concept, which arose more than a decade ago, proposed that tumor growth is sustained by a subpopulation of highly malignant cancerous cells. These cells, termed CSCs, comprise the top of the tumor cell hierarchy and have been isolated from many leukemias and solid tumors. Recent work has discovered that this hierarchy is embedded within a genetically heterogeneous tumor, in which various related but distinct subclones compete within the tumor mass. Thus, genetically distinct CSCs exist on top of each subclone, revealing a highly complex cellular composition of tumors. The CSC concept has therefore evolved to better model the complex and highly dynamic processes of tumorigenesis, tumor relapse, and metastasis.
Introduction
Cancer remains one of the leading causes of mortality worldwide (Jemal et al., 2011). In the United States, 5-yr survival rates only increased from 50% in 1974–1976 to 68% in 1999–2006, underlining how much more progress is needed to understand and successfully treat this disease (American Cancer Society, 2011). Over the past decade, the concept of the cancer stem cell (CSC) has emerged after identification and characterization of CSC-enriched populations in several distinct cancer entities (Table 1; Lapidot et al., 1994; Reya et al., 2001; Trumpp and Wiestler, 2008). Although the concept remains controversial (Kelly et al., 2007; Quintana et al., 2010; Magee et al., 2012), new observations from clinical studies and basic research have led to a more comprehensive CSC model of tumorigenesis, tumor recurrence, and metastasis formation. The aim of this review is to illustrate the current dynamic view of CSCs to foster the development of better therapeutic approaches to target this highly complex and deadly disease.
Table 1.
Identification of human primary tumor CSC biomarkers using in vivo assays
| Cancer | Animal | Type of injection | Treatment of recipient mice | Injection with Matrigel | Percentage of CSC-enriched population in tumor | Biomarkers | Minimal number of biomarker + cells to obtain a tumor | Reference |
| ALL (B-ALL) | NOD/SCID/IL2rγc−/− newborns | Intravenous | Sublethal irradiation | No | 82.50% | CD34+/CD19+ | 2–6 × 104 | Kong et al., 2008 |
| AML | NOD/SCID | Intravenous | Sublethal irradiation | No | 0.75% | CD34+/CD38− | 2 × 105 | Bonnet and Dick, 1997 |
| AML | NOD/SCID, NOD/SCID/β2m−/− and NOD/SCID/IL2rγc−/− | Intravenous and intrabone | IVIG of CD122 pretreatment and sublethal irradiation | No | 0.076% (*) | CD34+/CD38− (*) or CD34+/CD38+ (**) (in samples with lowest CD34+/CD38− fraction) | 7.5 × 103 (*) or 106 (**) | Taussig et al., 2008 |
| AML | NOD/SCID | Intrafemoral | IVIG of CD122 pretreatment and sublethal irradiation | No | 0.06–0.00009% of bulk | NA | NA | Eppert et al., 2011 |
| Bladder | Rag2γcDKO | Intradermal | NA | Yes | 3–36.3% | CD44 | 100 | Chan et al., 2009 |
| Breast | NOD/SCID | Mammary fat pad | VP-16, estrogen pellets | No | 11–35% | ESA+/CD44high/CD24low-neg | 200 | Al Hajj et al., 2003 |
| Breast | NOD/SCID | Humanized mammary fat pad | Estrogen pellets | Yes | 3–10% | ALDH-1+ | 500 | Ginestier et al., 2007 |
| Brain | NOD/SCID | Intracranial | NA | No | 6–29% | CD133+ | 100 | Singh et al., 2004 |
| Colorectal | NOD/SCID | Renal capsule | Sublethal irradiation | Yes | 1.8–24.5% | CD133+ | 100 | O’Brien et al., 2007 |
| Colorectal | NOD/SCID | Subcutaneous | NA | No | 2.60% | ESAhigh/CD44+ | 200 | Dalerba et al., 2007 |
| Colorectal | NOD/SCID | Subcutaneous | NA | Yes | 3.50% | ALDH-1+ | 25 serially passaged | Huang et al., 2009 |
| Head and neck squamous cell carcinoma | NOD/SCID and Rag2γcDKO | Subcutaneous | NA | Yes | 10–12% | CD44+ | 5000 | Prince et al., 2007 |
| Liver | SCID | Intrahepatic | NA | No | 2.50% | CD45−/CD90+ | 103 | Yang et al., 2008 |
| Lung | SCID and NUDE | Subcutaneous, after in vitro expansion | NA | Yes | 0.4–1.5% | CD133+ | 104 | Eramo et al., 2008 |
| Lung | NOD/SCID/IL2rγc−/− | Subcutaneous | NA | Yes | Median 15% | lin-/CD166+ | ≤500 | Zhang et al., 2012 |
| Melanoma | NOD/SCID | Subcutaneous | NA | No | 1.6-20.4% | ABCB5+ | 105 | Schatton et al., 2008 |
| Melanoma | Rag2γcDKO | Intradermal | NA | Yes | 2.5–41% | CD271+ | 100 | Boiko et al., 2010 |
| Melanoma | NOD/SCID/IL2rγc−/− | Subcutaneous | NA | Yes | NA | NA | 1 (in 28% of cases) | Quintana et al., 2010 |
| Melanoma | NUDE, (NOD/SCID, NOD/SCID/IL2rγc−/−) | Subcutaneous | NA | Yes | 8–11% | CD271+ | 1000 | Civenni et al., 2011 |
| Pancreatic | NOD/SCID | Subcutaneous and intrapancreatic | NA | Yes | 0.2-0.8% | ESA+/CD44+/CD24+ | 100 | Li et al., 2007 |
| Pancreatic | NUDE | Intrapancreatic | NA | No | 3.6 cells per high-power field | ESA+/CD133+ | 500 | Hermann et al., 2007 |
Studies reporting the existence of enriched human CSC populations are listed. In the first five columns, the main parameters influencing the efficiency of tumor engraftment are listed. From left to right: the tumor entity, the type of immunocompromised mouse strain used, the route of transplantation of human tumor cells, preconditioning of the recipient mice, treatment of mice during the assay, and whether the tumor cells were mixed with Matrigel upon transplantation. In the next four columns, the main results of these studies are summarized. From left to right: the frequency of the identified CSC-enriched population observed in the given tumor entity, the biomarkers identified for this CSC-enriched population, and the minimal number of tumor cells expressing these biomarkers able to give rise to a human tumor as well as the reference of the corresponding study. *, results for the CD34+/CD38− population. **, results for the CD34+/CD38+ population.
The classical concept of CSCs
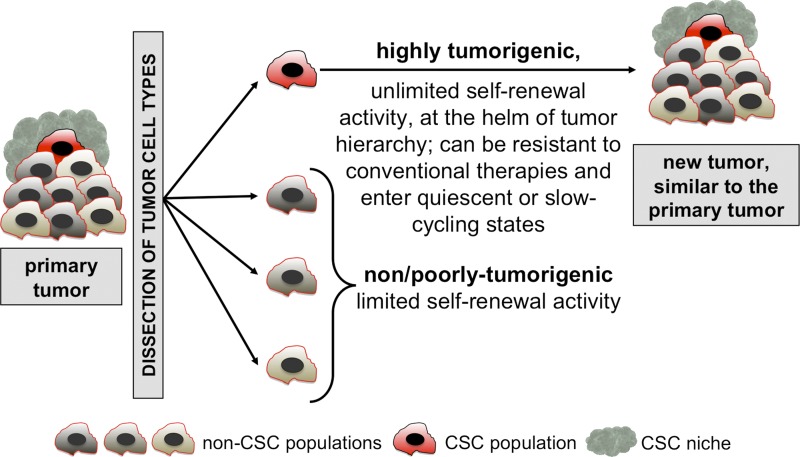
Adult regenerating tissues (such as the skin, the gastrointestinal mucosa, or the hematopoietic system) are hierarchically organized (Murphy et al., 2005; Fuchs and Nowak, 2008; van der Flier and Clevers, 2009; Seita and Weissman, 2010). At the top of the cellular organization, normal adult stem cells maintain tissues during homeostasis and facilitate their regeneration, for example in response to infection or to cell loss due to injury. These physiological stem cells are defined by their functional properties: they have the life-long capacity to self-renew (the ability to give rise to a new stem cell after cell division), are multipotent, and can reversibly enter quiescent or even dormant states and resist cytotoxic drugs (Fuchs and Nowak, 2008; Wilson et al., 2008; van der Flier and Clevers, 2009; Seita and Weissman, 2010). Similar to regenerative tissues, many tumors follow a hierarchical organization, and like physiological stem cells, CSCs are defined by a series of functional traits (Fig. 1; Reya et al., 2001; Dick, 2008; Clevers, 2011; Nguyen et al., 2012).
Figure 1.
The classical “cancer stem cell” (CSC) concept. Tumors are heterogeneous and hierarchically organized entities. Upon dissociation and transplantation into an immunocompromised animal, human CSCs can be functionally distinguished from non/poorly tumorigenic cell populations by their ability to reinitiate and grow a similar heterogeneous tumor in vivo.
Universal CSC functional traits
At the helm of tumor hierarchy.
First, CSCs can generate all cell types present in a tumor. Located at the top of the tumor hierarchy, CSCs can self-renew and also generate non-CSC progeny, which form the tumor bulk (differentiated progeny). Hierarchical organization of tumors, governed by CSCs, have been reported for many tumor types including germ cell cancers (Illmensee and Mintz, 1976), leukemia (Lapidot et al., 1994; Bonnet and Dick, 1997), breast cancer (Al-Hajj et al., 2003; Ginestier et al., 2007), brain cancer (Singh et al., 2004), colon cancer (Dalerba et al., 2007; O’Brien et al., 2007; Huang et al., 2009), pancreatic cancer (Hermann et al., 2007; Li et al., 2007), melanoma (Schatton et al., 2008; Boiko et al., 2010), and several others (Table 1).
Unlimited self-renewal potential.
In striking contrast with their differentiated progeny, CSCs can undergo unlimited self-renewing divisions. Typically, the presence of human CSCs within a cell population is experimentally addressed by serial transplantation of tumor cells into immunocompromised mice or rats (Table 1). Although considered state-of-the-art, this assay has limitations and only imperfectly recapitulates the in vivo situation found in patients. Indeed, the immunocompromised mouse models lack an adaptive immune system (neither mouse nor human) and express cytokines/chemokines and other environmental components of mouse origin, such as the tumor vasculature. Furthermore, the detection and enumeration of functional CSCs by these methods remains highly assay dependent, as several different immunocompromised mouse strains and many methods of tumor dissociation and implantation exist (Quintana et al., 2008; Ishizawa et al., 2010; Civenni et al., 2011). Nevertheless, human CSCs cannot be simply reduced to technical artifacts because of their detection in xenografts. Indeed, mouse CSCs have also been reported in syngenic mouse models of leukemia (Deshpande et al., 2006; Krivtsov et al., 2006; Yilmaz et al., 2006), breast cancer (Cho et al., 2008; Vaillant et al., 2008; Zhang et al., 2008), and skin cancer (Malanchi et al., 2008), providing strong evidence that CSCs govern many tumor types.
Other CSC functional traits
Quiescent or slow-cycling states.
Although cellular quiescence does not seem to be a universal feature, some CSCs have been reported to shuttle between quiescent, slow-cycling, and active states, similar to the behavior of many adult stem cell types (Wilson et al., 2008; Essers et al., 2009; Fuchs, 2009; Tian et al., 2011). For example, the presence of quiescent leukemic stem cells (LSCs) has been reported in a mouse model for acute myeloid leukemia (AML; Ishikawa et al., 2007; Saito et al., 2010). Moreover, using clonal tracking techniques, delayed-contributing CSC clones were identified both in AML and in colon carcinoma, which suggests the existence of long-term quiescent/dormant pools of human CSCs (Hope et al., 2004; Dieter et al., 2011). In line with these findings, a study showed a strong correlation between the number of slow-cycling breast cancer cells, as measured by retention of the membrane dye PKH26 in mammosphere cultures, and the frequency of CSCs, assessed by tumorigenicity assay in mice (Pece et al., 2010). Of major importance, quiescence or slow cycling states might render CSCs less likely to be responsive to conventional therapies, which mainly target cycling cells. Moreover, quiescent normal stem cells can reenter the cell cycle after injury to repair a tissue (Wilson et al., 2008; Essers et al., 2009; Essers and Trumpp, 2010; Saito et al., 2010; Seita and Weissman, 2010). In agreement with this stem cell feature, it has been hypothesized that potential reactivation of quiescent CSCs might induce tumor relapse, which sometimes occurs decades after completion of therapy (Aguirre-Ghiso, 2007; Pantel et al., 2009).
Increased resistance to conventional therapies.
Similar to physiological stem cells, some CSCs were reported to exhibit remarkable resistance to conventional therapies. For instance, breast CSCs were found to accumulate in women with locally advanced tumors after cytotoxic chemotherapy had eliminated the bulk of the tumor cells (Li et al., 2008). Similarly, in chronic myeloid leukemia (CML), BCR-ABL–driven LSCs are resistant to tyrosine kinase inhibitors (TKIs) such as Imatinib, whereas these compounds eliminate the rest of the leukemic cells, often even achieving a complete molecular response (undetectable levels of BCR-ABL mRNA by RT-PCR; Goldman et al., 2009; Oravecz-Wilson et al., 2009; Goldman, 2010; Perrotti et al., 2010). Accordingly, during STOP trials, in which TKI treatments are discontinued, tumor relapse was observed in the majority of patients. This was most likely caused by new tumor cell production by resistant CML-LSCs, as the relapsed “non-LSCs” leukemic cells remained sensitive to the initially used TKI (Barnes and Melo, 2006; Druker et al., 2006; Ross et al., 2011). Another case of resistant CSCs was reported for myelodysplasia carrying a 5q deletion. Although the majority of tumor cells were efficiently targeted by treatment with the immunomodulator Lenalidomide, leading to a complete clinical and cytogenic remission, most patients relapsed because of the outgrowth of remaining resistant CSCs (Tehranchi et al., 2010).
CSC resistance might first of all be caused by increased drug efflux capacities, mediated by expression of multidrug resistance (MDR) transporters (Dean et al., 2005). Indeed, cancer cells named “side population,” because of their ability to efflux the fluorescent dye Hoechst, have been found to be highly enriched for both normal cells and CSCs (Wulf et al., 2001; Ho et al., 2007; Lou and Dean, 2007; Moshaver et al., 2008). Second, aldehyde dehydrogenase 1 (ALDH-1), a cytosolic enzyme involved in the catalysis of aldehyde oxidation, was reported to be specifically active in several CSCs (Pearce et al., 2005; Ginestier et al., 2007; Huang et al., 2009; Ran et al., 2012). In a retrospective study, ALDH-1 activity was significantly higher in breast cancer metastatic cells, which developed resistance to cyclophosphamide, compared with sensitive cells (Sládek et al., 2002; Douville et al., 2009). This suggests that ALDH-1 might also play a role in cytotoxic drug resistance. Third, genotoxic treatments like ionizing radiation might be evaded by CSCs because of increased DNA damage check point response and DNA repair capacities, as observed in CD133-expressing glioblastoma CSC-enriched populations compared with CD133-negative populations (Bao et al., 2006). Last, CSCs might counterbalance the radiation-induced reactive oxygen species (ROS) production by increased expression of free radical scavengers, as reported for mouse and human breast CSCs (Diehn et al., 2009). This ability might selectively protect CSCs from ROS-mediated DNA damage and hence explain their resistance to irradiation treatments. Importantly, in vitro studies of drug-sensitive tumor cell lines suggest that cancer cells might transiently and reversibly acquire drug resistance, indicating that drug resistance might not always be a stable trait (Sharma et al., 2010).
The CSC niche
The tumor microenvironment is composed of diverse immune cells and stromal cells, as well as extracellular components (Bissell and Hines, 2011; Shiao et al., 2011; Hanahan and Coussens, 2012). CSC functional traits might be sustained by this microenvironment, termed “niche” (Cabarcas et al., 2011). For instance, vascular endothelial cells maintained self-renewal and promoted tumorigenicity of glioma CSCs in a mouse xenograft model (Calabrese et al., 2007). Hypoxia, notably via the hypoxia-inducible factor 2-α (HIF2α), increased glioma CSC self-renewal and tumorigenic capacities (Li et al., 2009b). Moreover, inflammatory molecules such as interleukin 6, secreted by infiltrating immune cells in the tumor, enhanced the proliferation of colitis-associated CSCs (Grivennikov et al., 2009).
The CSC niche might not only regulate CSC traits but might also directly provide CSC features to non-CSCs. For instance, tumor-associated myofibroblasts enhanced self-renewal of colorectal CSCs via hepatocyte growth factor (HGF) production but also strongly enhanced the in vivo tumorigenicity of non-CSCs through their secreted factors (Vermeulen et al., 2010).
In leukemia (Colmone et al., 2008) and in prostate carcinoma (Shiozawa et al., 2011), cancer cells could hijack existing physiological stem cell niches. Although physiological stem cell niches are known to play important roles for quiescence maintenance and resistance to stress-inducing treatments (Ehninger and Trumpp, 2011; Park et al., 2012), the influence of the tumor niche on CSC function at the cellular and molecular level still remains to be elucidated.
Origin of CSCs
Importantly, the CSC concept has to be separated from the “cell of origin” question: as outlined in the previous sections, CSCs are defined by a series of functional tumor-propagating traits. In contrast, the tumor cell of origin defines the cell type, from which the disease is derived, meaning the cell type first hit by an oncogenic mutation. However, this cell of origin does not necessarily immediately acquire a CSC phenotype (Visvader, 2011). Having said this, CSCs might indeed originate from stem cells, as they already are capable of almost limitless self-renewal. For instance, embryonic stem cells can form teratomas when transplanted subcutaneously in recipient mice (Sell and Pierce, 1994).
Nevertheless, CSCs can also originate from more differentiated progenitors that acquire stemness traits by accumulation of genetic or epigenetic abnormalities. For instance, progenitors derived from stem cells that already carry initiating genetic mutations acquire further mutations during differentiation that will finally lead to transformation. This would mean that the cell of origin for the first mutation is a stem cell, but that the CSC that drives the tumorigenic clone would be a more differentiated progenitor. Such a scenario has been reported for CML. Although the BCR-ABL fusion protein is the first event and is present in hematopoietic stem cell (HSC)-like CML cells (suggesting that the “cell of origin” of the disease is a normal HSC), advanced-stage LSCs during blast crisis were found to be in a state similar to granulocyte/macrophage progenitors (GMPs; Jamieson et al., 2004). Similarly, in AML, the AML-ETO fusion protein is present in HSCs (cell of origin of the disease), but the functional LSCs were detected in a Thy1− progenitor cell state by an in vitro colony forming assay (Miyamoto et al., 2000). However, in acute promyelocytic leukemia (APL), the MLL-AF9 fusion protein was not detected in HSCs (Turhan et al., 1995), but when introduced in mouse GMPs, it could induce leukemia, indicating that both the cell of origin and the LSCs were found in progenitors rather than in HSCs (Krivtsov et al., 2006).
Recent work on induced pluripotent stem cells (iPS) has demonstrated that, contrary to all expectations, the acquisition of self-renewal and pluripotency starting from any cell type can be achieved by activation of as few as four transcription factors (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). Importantly, this reprogramming process requires the transient repression of p53 and INK4a, two of the most frequently mutated tumor suppressor loci in human cancers (Li et al., 2009a; Utikal et al., 2009; Goh et al., 2011; Romagosa et al., 2011). Similarly, loss of the same tumor suppressors causes an activation of a self-renewal program in normally non–self-renewing hematopoietic progenitors; consequently, these cells start to behave as HSCs in vivo despite maintaining a progenitor phenotype. Thus, these cells do not de-differentiate, but rather acquire self-renewal potential after loss of tumor suppressors (Akala et al., 2008). These data raise the possibility that the accumulation of mutations in certain oncogenes and tumor suppressors may lead to a process that may be described as a partial reprogramming, with the acquisition of self-renewal activity paralleling the development of CSC activity. Indeed, in vitro reprogramming of human skin fibroblasts by stable expression of hTERT, H-RasV12, and SV40 LT and ST antigens led to the generation of cells with CSC properties, able to form hierarchically organized tumors in mice (Scaffidi and Misteli, 2011). Similarly, experimentally induced expression of the depolarization-inducing transcription coactivator TAZ (Cordenonsi et al., 2011), as well as experimentally induced expression of the epithelial-to-mesenchymal transition (EMT)-inducing transcription factors TWIST or SNAIL (Mani et al., 2008), was reported to provide CSC-like properties to non-CSC cells.
Identification of CSC biomarkers
The CSC concept proposes that one of the major parameters to evaluate therapy efficacy is the quantification of remnant CSCs within minimal residual disease, and not gross measurement of tumor regression, as is typically used in clinical trials (Blagosklonny, 2006). Therefore, one big aim in the field is to identify reliable and specific CSC biomarkers for each tumor type.
Lapidot et al. (1994) first demonstrated that only a small subset of human AML cells display tumorigenic properties. Subsequently, Bonnet and Dick (1997) identified LSCs in AML as CD34+/CD38− leukemic cells, closely resembling normal HSCs (Bonnet and Dick, 1997), by limiting dilution transplantations in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice. However, in the meantime, examination of a large set of AML patients revealed that functional CSC clones reside within several distinct immunophenotypically defined cellular compartments, showing significant interpatient heterogeneity (Eppert et al., 2011; Gibbs et al., 2012).
For solid tumors, a first leap forward was achieved in 2003, when Michael Clarke and colleagues reported the identification of breast CSCs within the ESA+/CD44+/CD24low-neg population of mammary pleural effusion and tumor samples. As few as a hundred ESA+/CD44+/CD24low-neg cells were able to reinitiate the original tumor in NOD/SCID mice, whereas ten thousand cells with an alternate phenotype were not (Al-Hajj et al., 2003).
After these two landmark publications, CSCs were identified in many more solid and hematopoietic human tumors as summarized in Table 1. Importantly, several studies indicate the existence of CSC-enriched populations displaying different, sometimes nonoverlapping sets of markers for the same tumor type. For example, only 1% of breast cancer cells simultaneously express both reported CSC phenotypes ESA+/CD44+/CD24low-neg and ALDH-1+ (Ginestier et al., 2007). This discrepancy might be due to several different factors. First, differences in methods could be responsible for these differences (Quintana et al., 2008; Civenni et al., 2011). Second, several CSC clones may coexist within primary tumors (Anderson et al., 2011; Eppert et al., 2011), and the research groups may have detected different CSC populations. Third, functional CSC clones might reside within several immunophenotypically defined cellular compartments (Eppert et al., 2011; Gibbs et al., 2012). Last but not least, cancer entities are rarely uniform. For instance, during the last few years, various genome-wide gene expression profiling efforts combined with biomarker and clinical approaches have led to the subclassification of breast cancers into increasing numbers of molecular subtypes (Perou et al., 2000; Herschkowitz et al., 2007; Curtis et al., 2012). The mutational patterns vary between the different subtypes, and this genetic heterogeneity is likely paralleled by a heterogeneous CSC complexity. Different CSC phenotypes might thus be associated with different cancer subtypes.
Therapeutic targeting of CSCs
Recent work on AML shows that the signature of functionally validated LSCs is a good predictor for poor patient survival independent of any biomarker (Eppert et al., 2011). These data strongly suggest that therapeutic targeting of CSCs should be relevant for patients. However, many technical hurdles still need to be overcome.
Targeting CSC biomarkers.
CD44, a transmembrane glycoprotein involved in cell–cell and cell–matrix adhesion, has been identified as being expressed by many tumor CSCs (Table 1; Zöller, 2011). A CD44 antibody restricted human AML LSC proliferation in xenograft studies, probably by inhibiting LSC–niche interactions (Jin et al., 2006; Krause et al., 2006). However, universal targeting of CD44, which is also expressed by many adult stem cells, might be deleterious for patients. Undesirable effects might be evaded by targeting different isoforms of CD44 specifically expressed by tumor cells, such as CD44v6 (Heider et al., 2004; Orian-Rousseau, 2010). However, severe skin toxicity has been reported in a phase I clinical trial of bevatuzumab (an anti-CD44v6 antibody) in the case of head and neck squamous cell carcinoma (Riechelmann et al., 2008).
More recently, successful eradication of AML LSC was described by targeting of CD123 (interleukin-3 [IL-3] receptor α chain) using a blocking antibody in xenografted mice (Jin et al., 2009). However, because CD123 is also expressed by normal HSCs, there is a risk of severe side effects (Taussig et al., 2005). Targeting ABC transporters expressed by CSCs has also been explored (Lou and Dean, 2007) but was put on hold after realizing that these transporters play pivotal roles in blood–brain barrier maintenance as well as in adult stem cell maintenance (Hermann et al., 2006).
Targeting CSC molecular pathways.
Based on the hypothesis that cancer cells might be more dependent on the activation of oncogenic pathways than their normal counterparts, a phenomenon termed “oncogene addiction” (Weinstein, 2002), pharmaceutical companies have applied major efforts to target signaling pathways activated in CSCs and cancer cells in general. For instance, the NFκB pathway has been successful targeted and allowed selective eradication of AML and final phase (blast crisis) CML LSCs. Guzman et al. (2005) treated in vitro leukemic cells with parthenolide, a drug known to directly bind IκB kinase (IKK) as well as to modify p50 and p65 subunits. This pretreatment induced higher levels of ROS, which led to a decrease of CD34+/CD38− leukemic populations and an impaired capacity of leukemic cells to engraft in NOD/SCID mice, which suggests that mainly LSCs were targeted (Guzman et al., 2005). Similarly, inhibition of NOTCH signaling using γ-secretase inhibitors in CD133-expressing medulloblastoma CSCs led to the diminution of CD133+ cells and correlated with impaired tumor engraftment in vivo (Fan et al., 2006). Last but not least, inhibition of the TGFβ pathway by bone morphogenetic proteins (BMPs) was reported to lead to the differentiation of brain CSCs and successive cure of the disease in a xenograft model, providing a first proof of principle for the possible efficacy of a “differentiation therapy” (Piccirillo et al., 2006).
Sensitization to therapy.
Several studies have aimed to sensitize CSCs to therapy. For instance, in colon cancer, IL-4 blockade successfully primed CSCs to chemotherapy (Francipane et al., 2008). In glioblastoma, a CHK kinase inhibitor increased the radio sensitivity of CSCs (Bao et al., 2006). Also, recent reports showing that dormant and chemotherapy-resistant normal stem cells can be activated and simultaneously sensitized to chemotherapy-mediated killing might provide a strategy for targeting dormant CSCs (Essers et al., 2009; Essers and Trumpp, 2010). Here a sequential treatment scheme would be used in which dormant CSCs would first be “primed” and activated, followed by chemotherapy, leading to complete elimination of the tumor including the initially dormant and resistant CSCs. Such a strategy was successfully applied in mouse models of AML and CML (Ito et al., 2008; Saito et al., 2010). Moreover, in a mouse model of acute promyelocytic leukemia (APL), LSCs expressing PML-RARα (pro-myelocytic leukemia-retinoic acid receptor α) could be forced out of self-renewal into differentiation by cooperative treatment of arsenic, cyclic AMP, and retinoic acid, leading to LSC clearance and impressive remission in patients (de Thé and Chen, 2010).
Novel strategies to target CSCs.
Some new hope might arise from the development of bi-specific or tri-specific antibodies that are able to specifically target cell populations. For instance, T cell recognition via an anti-CD3 antibody can be combined with cancer cell recognition via an additional antigen-binding site such as EPCAM (catumaxomab) or HER2 (ertumaxomab; Shen and Zhu, 2008; Hess et al., 2012).
In addition, high-throughput screening of drugs selectively inhibiting CSCs, rather than other tumor cells, have been successfully used to uncover compounds such as salinomycin. This antibiotic drug targets putative breast CSCs (grown as mammospheres in culture), as well as CLL LSCs, probably via inhibition of the WNT pathway (Gupta et al., 2009; Lu et al., 2011). However, the toxicity of this molecule on physiological stem cells remains unexplored.
Last but not least, recent discoveries concerning specific CSC biological properties might lead to the development of novel targeting strategies: for instance, glioma stem cells were found to be mechanistically distinct from their less tumorigenic counterparts regarding production of nitric oxide via nitric oxide synthase-2 (NOS2). NOS2 inhibition successfully slowed down tumor growth in a xenograft glioma model (Eyler et al., 2011). Similarly, glycine decarboxylase activity was found to be critical for non-small cell lung CSCs, which suggests that glycine metabolism might be a novel anti-CSC target (Zhang et al., 2012). However, in both cases, general toxicity of such treatments has not yet been thoroughly investigated.
In summary, therapeutic applications deriving from CSC studies are less straightforward than was initially anticipated, notably because of the large overlap in molecules expressed by CSCs and their respective normal stem cells that has so far hampered their use in the clinic due to collateral toxicity (Deonarain et al., 2009). Nevertheless, without the efficient eradication of CSCs, a long-term cure for many cancer patients appears unreachable. Innovative efforts will therefore be required from the field to achieve this goal in the not too distant future.
Evolution toward a more dynamic view of CSCs
Given the pace of research in the field and the diverse nature of the results, the classical view of CSCs needs to be updated. In this section, we propose a more dynamic model for CSCs and integrate recent results into this model.
Multiple genetic CSC clones within one tumor.
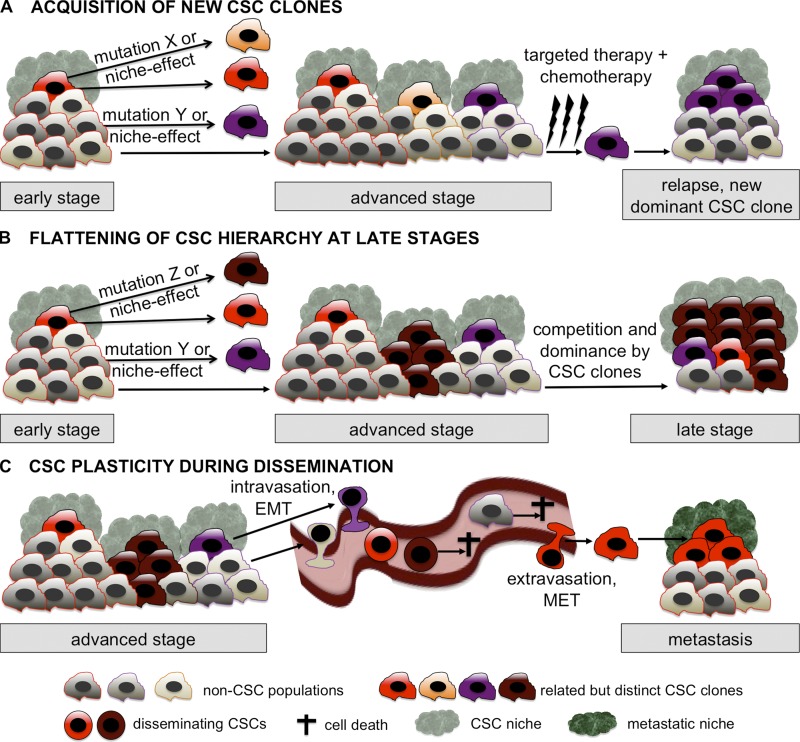
Recent studies in which several subregions of the primary tumor and metastases of pancreatic and kidney cancers were sequenced revealed an unexpected degree of intratumor heterogeneity (Campbell et al., 2010; Yachida et al., 2010; Gerlinger et al., 2012). This often highly underestimated heterogeneity may result from the fact that CSCs are genetically unstable. Indeed, in colon cancer, such genetic instability was reported, leading to the formation of new CSC clones deriving from an initial “parental” CSC clone (Odoux et al., 2008). In addition, two landmark studies in human acute lymphoblastic leukemia (ALL) studied the genetic architecture of LSCs during disease progression and after successive transplantations in xenografted mice. Both studies show that several LSC clones coexist within single patients. During disease progression, after therapy, and after serial xenotransplantation, different LSC clones can take over and initiate new tumors (Anderson et al., 2011; Notta et al., 2011). This might explain at least partially the “acquired resistance” to targeted therapy observed in some patients. That is, after successfully targeting sensitive CSC clones, already present resistant clones may take over, mimicking an acquisition of resistance to the applied treatment. As a consequence, tumors might not be faithfully represented by single-headed hierarchical structures bur rather might resemble more oligarchic structures, displaying multiple heads: if one head, i.e., a CSC clone, is cut off (by targeted therapy, for instance), another one might take over, repopulating the tumor. Thus, dynamic hierarchies might exist within a single CSC clone and its progeny (intra-CSC clone hierarchy), but also between different genetically diverse CSC clones that compete with each other (inter-CSC clone hierarchy; Fig. 2 A).
Figure 2.
The dynamic CSC concept. (A) Although early stage tumors might be governed by a single CSC clone, advanced stage tumors might contain several distinct but related clones, either arising from the initial CSC clone or from its differentiated progeny via mutations or via induction by the CSC niche. Targeted therapy and/or chemotherapy eliminate the tumor mass, possibly including some of the CSCs. At least one resistant CSC clone is then responsible (possibly after acquiring additional mutations) for tumor relapse. (B) Advanced stage tumors contain several distinct but related CSC clones. Some acquire enhanced self-renewal capabilities with simultaneously decreased differentiation. During tumor progression, the CSC clones compete with each other, leading to the dominance of at least one CSC clone with the subsequent loss of differentiated tumor progeny. Over time, this leads to a flattening of the hierarchical structure and to a selection of the most aggressive CSCs. Late stage tumors may thus be comprised almost exclusively of aggressive, multiresistant CSCs, a situation similar to the one proposed by the stochastic model. (C) Carcinoma CSCs display a dynamic phenotype during systemic dissemination: they are able to at least partially lose epithelial traits through EMT. It is most likely that only a subset of such disseminating CSCs are able to survive in the systemic circulation, extravasate, and reacquire epithelial features (MET) to seed in a new microenvironment and to initiate metastasis. All three scenarios illustrated in A, B, and C occur in parallel and/or during different phases of tumor progression.
CSCs need not be rare.
CSCs do not need to be rare, as shown by Kelly et al. (2007) using leukemia and lymphoma genetic mouse models. Indeed, CSC-enriched populations have been reported to represent extremely variable proportions of bulk tumor cells, ranging from 0.2% to 82.5% (Table 1). Moreover, the frequency of CSCs might increase during tumor progression, as recently shown by in vivo limiting dilution assays of grade 1 and grade 3 breast tumors (Pece et al., 2010).
During the last few years, the idea that CSCs must be rare, which is based on our knowledge on physiological stem cells, often led to the questioning of the CSC hypothesis. For instance, it was reported that melanoma stage III–IV tumorigenic cells are very abundant, representing up to 30% of the tumor bulk. Moreover, the screening of a very high number of cell surface markers failed to distinguish these tumorigenic cells from other tumor cells (Quintana et al., 2010). Based on these results, it was proposed that melanoma does not follow a CSC model but rather a stochastic model, where tumorigenicity is a random feature distributed among all tumor cells (Gupta et al., 2005, 2011; Joseph et al., 2008).
Although certainly possible, there are also alternative explanations for these observations. The difficulty in finding any relevant biomarkers for stage III–IV tumorigenic melanoma cells might be technically explained by the temporary loss of surface proteins caused by the use of trypsin. Without using this enzyme, CD271 could be identified as a melanoma CSC marker by two other groups (Boiko et al., 2010; Civenni et al., 2011). Alternatively, late-stage melanoma might consist of several distinct CSC clones displaying different cell surface biomarkers, which are all highly malignant. These CSC clones might even differ from patient to patient, as described for ALL (Anderson et al., 2011; Notta et al., 2011). Moreover, a likely scenario is a high selection and enrichment for CSC clones, which have retained little or no differentiation capacity. In this scenario, the initial hierarchy in the tumor is steadily decreased and flattened, leading to late-stage tumors, composed almost exclusively of heterogeneous CSC clones (Fig. 2 B).
Plasticity of CSC phenotype.
In recent years, the EMT process, in which epithelial polarized adherent cells are converted into mesenchymal cell–like depolarized migratory tumor cells, has been linked to the CSC phenotype and function in breast, pancreatic, and colorectal tumors. CSCs were found to express EMT-inducing transcription factors such as TWIST, SNAIL, and SLUG and, vice versa, EMT-undergoing cells were found enriched for CSC activities compared with cells not undergoing EMT (Mani et al., 2008; Thiery et al., 2009; Wellner et al., 2009; Cordenonsi et al., 2011; Rhim et al., 2012). The process can be controlled by a ZEB1/mir200 feedback loop mechanism (maintaining stemness), itself controlled by p53 (Shimono et al., 2009; Keck and Brabletz, 2011; Schubert and Brabletz, 2011). Overall, these findings suggest that epithelial markers might be down-regulated in carcinoma CSCs and therefore would be missed in typical screens, which generally exclude cells lacking epithelial markers such as EPCAM. Furthermore, it underlines the fact that epithelial CSCs might strongly modulate their phenotype during tumor progression: for example, a CSC detected in an early stage primary tumor might have a completely different phenotype from the one of a CSC circulating in the blood (Fig. 2 C). As a consequence of this plasticity, some of the reported CSC biomarkers might be relevant in some stages of tumor progression but obsolete in others.
Generating a new dynamic concept of CSCs.
These recent data and others suggest a more complex and dynamic CSC model integrating the three main additional features.
First, tumors are by definition genetically unstable entities. Therefore, the cellular composition of a tumor in an early stage disease, at relapse, or at late stages may display significant genetic differences including genetic heterogeneity. The available data suggest that in early neoplasms, only a single or very few CSC clones drive tumor growth. During disease progression, new CSC clones can arise either from existing CSC clones or from the differentiated progeny via mutation-mediated partial reprogramming. After eradication of the majority of tumor cells by chemotherapy and/or targeted approaches, one or few CSC clones may survive and expand, leading to a change in the clonal and cellular composition of the relapsing cancer (Fig. 2 A).
Second, the different genetically distinct CSC clones may also compete with each other, leading to the selection of CSC clones with high self-renewing activity and simultaneous loss of differentiation capacity. This would provide an explanation for the observed flattening of the cellular hierarchy within advanced stage tumors, which creates a situation similar to what is proposed by the stochastic model (Fig. 2 B). Third, CSC biomarkers can be unstable, as indicated by studies on EMT in solid tumors (Fig. 2 C). Thus, reported CSC biomarkers might only be relevant for a given tumor stage and therefore need to be validated for each case in conjunction with functional analyses.
Importantly, this dynamic CSC model suggests that only complex combinatorial treatments are likely to be efficacious in targeting late stage tumors or metastasis. First, mutations that are commonly present in all clones have to identified, for instance by whole genome sequencing of various regions within a single tumor mass (Yachida et al., 2010; Gerlinger et al., 2012). The associated deregulated pathways of these common mutations present in all subregions (likely including the various tumor subclones) need then to be targeted by pathway-specific strategies. Second, these strategies need to be complemented by therapies targeting additional CSC-specific features (see previous sections) to eliminate not only the non-CSC parts of each tumor clone, but also the various CSCs present in advanced stage tumors or metastasis.
CSCs and metastasis
Metastasis might be initiated by a subset of CSC clones.
Systemic dissemination and metastasis is responsible for most cancer-related deaths. To date, human metastasis-initiating cells (MICs) have not yet been prospectively identified. However, several lines of evidence indicate that MICs might be found within subpopulations of CSCs. First of all, carcinoma CSCs possess tumor-initiating capacity, which is a mandatory trait for the establishment of secondary tumors in distant organs. Second, they express EMT markers (Mani et al., 2008), which suggests that they are able to migrate and which makes them likely candidates for metastasis initiating activities.
More specifically, several reports suggest that MICs might be found within CSC populations: CD44+ breast cancer cells (displaying a breast CSC phenotype) were proposed to have enriched metastatic activities in xenograft studies (Al-Hajj et al., 2003; Liu et al., 2010). Similarly, CD44+/CXCR4+ cells, a subfraction of the putative pancreatic CSCs present at the invasive front of cell line–induced pancreatic tumors, were reported to be enriched for metastatic capabilities (Hermann et al., 2007). Along the same lines, CD26, in combination with the CSC marker CD133, has been proposed as a marker for colorectal MIC-enriched tumor populations in primary tumor xenograft experiments (Pang et al., 2010).
MICs might be late-stage disseminating CSC clones.
MICs might disseminate early during tumor progression or might derive from late-stage disseminating CSC clones (Klein, 2009). Through mapping of the genetic evolution of both pancreatic primary tumors and metastases by next generation sequencing, two recent studies identified the genealogy of metastatic clones (Campbell et al., 2010; Yachida et al., 2010). Both studies suggest that even if both primary tumors and metastases consist of heterogeneous clones, additional driver mutations are present in the metastasis-initiating clones compared with the clones present in the primary tumors. In addition, metastatic clones were found to arise rather late within primary pancreatic tumors (Yachida et al., 2010), even if the dissemination process itself might start rather early (Rhim et al., 2012); this suggests that functional MICs typically disseminate from advanced-stage tumors rather than from early stage primary tumors.
MICs must be found among disseminating tumor cells.
Because metastasis results from the successful dissemination of primary tumor cells into a distant organ, MICs have to be found among disseminating tumor cells. These include circulating tumor cells (CTCs) as well as disseminated tumor cells (DTCs) found, respectively, in the blood or in the bone marrow of carcinoma patients. In the case of breast cancer, variable proportions of CTCs and DTCs have been reported to display CSC phenotypes by immunocytochemistry (Balic et al., 2006; Theodoropoulos et al., 2010). However, the detection methods for carcinoma CTCs remain controversial. This is due to the use of EPCAM and cytokeratins (CK) as positive selection markers, as they might be down-regulated during EMT-mediated dissemination. Indeed, <40% of genetically YFP-labeled CTCs expressed CK19 or EPCAM in a mouse model for pancreatic ductal adenocarcinomas, which suggests that a large proportion of CTCs undergo EMT and lose expression of epithelial markers. Also, >40% of the detected YFP-labeled CTCs expressed the previously reported pancreatic CSC markers (CD44+/CD24+) and showed high clonogenicity in vitro, which suggests that at least some of the detected CTCs have self-renewing potential (Li et al., 2007; Rhim et al., 2012). Functional in vivo analyses of different subfractions of human CTCs or DTCs using xenograft assays are required to test whether phenotypic disseminating CSCs are indeed involved in metastasis initiation and, if so, which CSC subpopulation is responsible. However, these studies are technically challenging, as they require the development of optimized metastatic read-out assays starting from extremely low numbers of patient disseminating tumor cells.
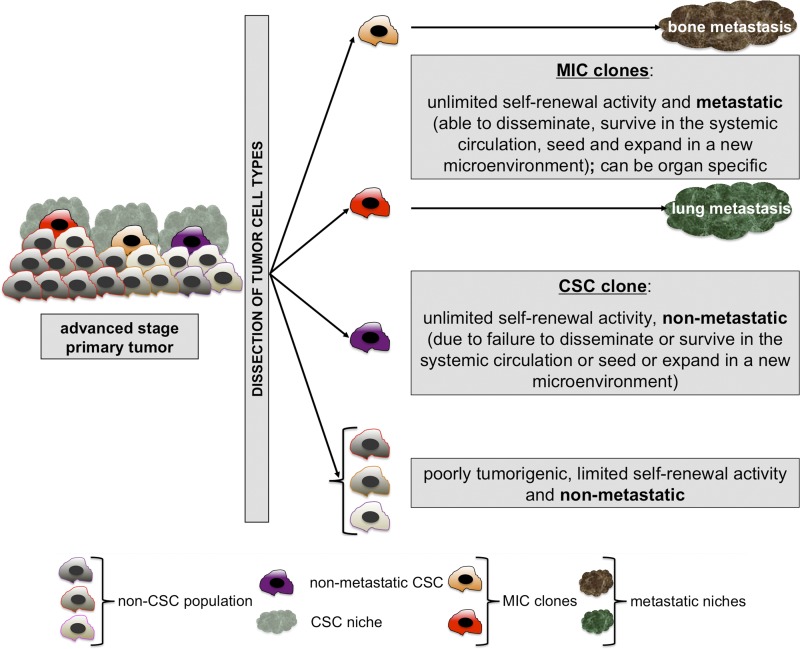
The metastasis initiating cell model.
MICs likely arise from subpopulations of late-stage CSC clones, hence the relevance for characterizing CSCs, even in late-stage tumors. In addition to the functional capacities of CSCs, MICs have to display metastatic capacities, meaning that they must disseminate, survive in the systemic circulation, extravasate at the metastatic site, and seed and grow in the new environment. Several metastatic clones might coexist during dissemination, exhibiting different site-specific migration and seeding capacities (Fig. 3). Indeed, Massagué and colleagues have proposed that, in the context of breast cancer, different metastatic populations can be distinguished according to their capacity to metastasize to the bone, the lung, or the brain in patients with systemic disease (Kang et al., 2003; Minn et al., 2005; Bos et al., 2009). This appears to be related to their capacity to directly or indirectly generate a metastatic niche (Psaila et al., 2006-2007; Erez and Coussens, 2011; Oskarsson et al., 2011; Malanchi et al., 2012).
Figure 3.
The “metastasis initiating cell” (MIC) concept. Advanced stage primary tumors are heterogeneous and multi-hierarchical structures. Upon dissociation and transplantation in immunocompromised hosts, MICs can be functionally distinguished from CSC clones by their metastatic ability in vivo (dissemination, survival in the systemic circulation, extravasation, seeding, and expansion in a new microenvironment, termed “metastatic niche”). MICs are adapted descendants and therefore subpopulations of CSCs with metastatic capacity and, possibly, organ specificity.
Conclusions
CSCs are a specific subset of transformed cells that are able to sustain primary tumor growth according to a hierarchical pattern. Strong evidence for the existence of such cells exists in many cancer types, although the model may not be appropriate for all cancer types and/or all stages. Like any model, the CSC concept needs to be constantly adapted to the currently available data and thus is steadily evolving. Recent findings uncovering high intratumor genetic heterogeneity have led to the observation that several CSC clones can coexist and compete with each other within a tumor. Furthermore, CSCs may have unstable phenotypes and genotypes, which makes it difficult to identify reliable and robust biomarkers for the development of targeted therapies. In addition, cell-extrinsic factors provided by the CSC niche might provide yet another dimension to the complexity of CSC regulation. Importantly, most cancer-related deaths are a consequence of metastasis development and not a direct consequence of primary tumor growth. Therefore, the characterization of MICs will become pivotal in the future in order to prevent and target so-far untreatable metastatic disease. Importantly, the identification of CSCs remains therapeutically relevant, even for late stages tumors, because MICs are likely a subset of these. Evolving the CSC concept will help to focus research toward developing improved therapies without risk of tumor recurrence and allow for the targeting of fatal late-stage cancers.
Acknowledgments
The authors would like to thank Drs. Thordur Oskarsson, Melania Tesio, Hind Medyouf, Michael Milsom, and Anja Schillert for helpful discussions.
This work was supported by the BioRN Spitzencluster “Molecular and Cell based Medicine” supported by the German Bundesministerium für Bildung und Forschung (BMBF), the EU-FP7 Program “EuroSyStem,” the SFB 873 funded by the Deutsche Forschungsgemeinschaft (DFG), and the Dietmar Hopp Foundation.
Footnotes
Abbreviations used in this paper:
- ALL
- acute lymphoblastic leukemia
- AML
- acute myeloid leukemia
- CML
- chronic myeloid leukemia
- CSC
- cancer stem cell
- CTC
- circulating tumor cell
- DTC
- disseminated tumor cell
- EMT
- epithelial-to-mesenchymal transition
- HSC
- hematopoietic stem cell
- LSC
- leukemic stem cell
- MIC
- metastasis-initiating cell
- NOD/SCID
- nonobese diabetic/severe combined immunodeficient
- ROS
- reactive oxygen species
- TKI
- tyrosine kinase inhibitor
References
- Aguirre-Ghiso J.A. 2007. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer. 7:834–846 10.1038/nrc2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akala O.O., Park I.K., Qian D., Pihalja M., Becker M.W., Clarke M.F. 2008. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature. 453:228–232 10.1038/nature06869 [DOI] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 100:3983–3988 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society 2011. Cancer Facts & Figures 2011. American Cancer Society, Atlanta: 58 pp [Google Scholar]
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469:356–361 10.1038/nature09650 [DOI] [PubMed] [Google Scholar]
- Balic M., Lin H., Young L., Hawes D., Giuliano A., McNamara G., Datar R.H., Cote R.J. 2006. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin. Cancer Res. 12:5615–5621 10.1158/1078-0432.CCR-06-0169 [DOI] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444:756–760 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- Barnes D.J., Melo J.V. 2006. Primitive, quiescent and difficult to kill: the role of non-proliferating stem cells in chronic myeloid leukemia. Cell Cycle. 5:2862–2866 10.4161/cc.5.24.3573 [DOI] [PubMed] [Google Scholar]
- Bissell M.J., Hines W.C. 2011. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17:320–329 10.1038/nm.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M.V. 2006. Target for cancer therapy: proliferating cells or stem cells. Leukemia. 20:385–391 10.1038/sj.leu.2404075 [DOI] [PubMed] [Google Scholar]
- Boiko A.D., Razorenova O.V., van de Rijn M., Swetter S.M., Johnson D.L., Ly D.P., Butler P.D., Yang G.P., Joshua B., Kaplan M.J., et al. 2010. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 466:133–137 10.1038/nature09161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Bos P.D., Zhang X.H., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A., Massagué J. 2009. Genes that mediate breast cancer metastasis to the brain. Nature. 459:1005–1009 10.1038/nature08021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabarcas S.M., Mathews L.A., Farrar W.L. 2011. The cancer stem cell niche—there goes the neighborhood? Int. J. Cancer. 129:2315–2327 10.1002/ijc.26312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B., Oh E.Y., Gaber M.W., Finklestein D., Allen M., et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11:69–82 10.1016/j.ccr.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Campbell P.J., Yachida S., Mudie L.J., Stephens P.J., Pleasance E.D., Stebbings L.A., Morsberger L.A., Latimer C., McLaren S., Lin M.L., et al. 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 467:1109–1113 10.1038/nature09460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.S., Espinosa I., Chao M., Wong D., Ailles L., Diehn M., Gill H., Presti J., Jr, Chang H.Y., van de Rijn M., et al. 2009. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. USA. 106:14016–14021 10.1073/pnas.0906549106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.W., Wang X., Diehn M., Shedden K., Chen G.Y., Sherlock G., Gurney A., Lewicki J., Clarke M.F. 2008. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 26:364–371 10.1634/stemcells.2007-0440 [DOI] [PubMed] [Google Scholar]
- Civenni G., Walter A., Kobert N., Mihic-Probst D., Zipser M., Belloni B., Seifert B., Moch H., Dummer R., van den Broek M., Sommer L. 2011. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 71:3098–3109 10.1158/0008-5472.CAN-10-3997 [DOI] [PubMed] [Google Scholar]
- Clevers H. 2011. The cancer stem cell: premises, promises and challenges. Nat. Med. 17:313–319 10.1038/nm.2304 [DOI] [PubMed] [Google Scholar]
- Colmone A., Amorim M., Pontier A.L., Wang S., Jablonski E., Sipkins D.A. 2008. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 322:1861–1865 10.1126/science.1164390 [DOI] [PubMed] [Google Scholar]
- Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A.R., Poletti A., et al. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 147:759–772 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486:346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., et al. 2007. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA. 104:10158–10163 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Fojo T., Bates S. 2005. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 5:275–284 10.1038/nrc1590 [DOI] [PubMed] [Google Scholar]
- de Thé H., Chen Z. 2010. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat. Rev. Cancer. 10:775–783 10.1038/nrc2943 [DOI] [PubMed] [Google Scholar]
- Deonarain M.P., Kousparou C.A., Epenetos A.A. 2009. Antibodies targeting cancer stem cells: a new paradigm in immunotherapy? MAbs. 1:12–25 10.4161/mabs.1.1.7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A.J., Cusan M., Rawat V.P., Reuter H., Krause A., Pott C., Quintanilla-Martinez L., Kakadia P., Kuchenbauer F., Ahmed F., et al. 2006. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 10:363–374 10.1016/j.ccr.2006.08.023 [DOI] [PubMed] [Google Scholar]
- Dick J.E. 2008. Stem cell concepts renew cancer research. Blood. 112:4793–4807 10.1182/blood-2008-08-077941 [DOI] [PubMed] [Google Scholar]
- Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. 2009. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 458:780–783 10.1038/nature07733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter S.M., Ball C.R., Hoffmann C.M., Nowrouzi A., Herbst F., Zavidij O., Abel U., Arens A., Weichert W., Brand K., et al. 2011. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 9:357–365 10.1016/j.stem.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Douville J., Beaulieu R., Balicki D. 2009. ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev. 18:17–25 10.1089/scd.2008.0055 [DOI] [PubMed] [Google Scholar]
- Druker B.J., Guilhot F., O’Brien S.G., Gathmann I., Kantarjian H., Gattermann N., Deininger M.W., Silver R.T., Goldman J.M., Stone R.M., et al. 2006. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355:2408–2417 10.1056/NEJMoa062867 [DOI] [PubMed] [Google Scholar]
- Ehninger A., Trumpp A. 2011. The bone marrow stem cell niche grows up: mesenchymal stem cells and macrophages move in. J. Exp. Med. 208:421–428 10.1084/jem.20110132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Eramo A., Lotti F., Sette G., Pilozzi E., Biffoni M., Di Virgilio A., Conticello C., Ruco L., Peschle C., De Maria R. 2008. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15:504–514 10.1038/sj.cdd.4402283 [DOI] [PubMed] [Google Scholar]
- Erez N., Coussens L.M. 2011. Leukocytes as paracrine regulators of metastasis and determinants of organ-specific colonization. Int. J. Cancer. 128:2536–2544 10.1002/ijc.26032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Trumpp A. 2010. Targeting leukemic stem cells by breaking their dormancy. Mol. Oncol. 4:443–450 10.1016/j.molonc.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Eyler C.E., Wu Q., Yan K., MacSwords J.M., Chandler-Militello D., Misuraca K.L., Lathia J.D., Forrester M.T., Lee J., Stamler J.S., et al. 2011. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 146:53–66 10.1016/j.cell.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Matsui W., Khaki L., Stearns D., Chun J., Li Y.M., Eberhart C.G. 2006. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 66:7445–7452 10.1158/0008-5472.CAN-06-0858 [DOI] [PubMed] [Google Scholar]
- Francipane M.G., Alea M.P., Lombardo Y., Todaro M., Medema J.P., Stassi G. 2008. Crucial role of interleukin-4 in the survival of colon cancer stem cells. Cancer Res. 68:4022–4025 10.1158/0008-5472.CAN-07-6874 [DOI] [PubMed] [Google Scholar]
- Fuchs E. 2009. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 137:811–819 10.1016/j.cell.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Nowak J.A. 2008. Building epithelial tissues from skin stem cells. Cold Spring Harb. Symp. Quant. Biol. 73:333–350 10.1101/sqb.2008.73.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366:883–892 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs K.D., Jr, Jager A., Crespo O., Goltsev Y., Trejo A., Richard C.E., Nolan G.P. 2012. Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell. 10:210–217 10.1016/j.stem.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C.G., Liu S., et al. 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 1:555–567 10.1016/j.stem.2007.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh A.M., Coffill C.R., Lane D.P. 2011. The role of mutant p53 in human cancer. J. Pathol. 223:116–126 10.1002/path.2784 [DOI] [PubMed] [Google Scholar]
- Goldman J.M. 2010. Chronic myeloid leukemia: a historical perspective. Semin. Hematol. 47:302–311 10.1053/j.seminhematol.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Goldman J.M., Green A.R., Holyoake T., Jamieson C., Mesa R., Mughal T., Pellicano F., Perrotti D., Skoda R., Vannucchi A.M. 2009. Chronic myeloproliferative diseases with and without the Ph chromosome: some unresolved issues. Leukemia. 23:1708–1715 10.1038/leu.2009.142 [DOI] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.P., Minn A.J., Kang Y., Siegel P.M., Serganova I., Cordón-Cardo C., Olshen A.B., Gerald W.L., Massagué J. 2005. Identifying site-specific metastasis genes and functions. Cold Spring Harb. Symp. Quant. Biol. 70:149–158 10.1101/sqb.2005.70.018 [DOI] [PubMed] [Google Scholar]
- Gupta P.B., Onder T.T., Jiang G., Tao K., Kuperwasser C., Weinberg R.A., Lander E.S. 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 138:645–659 10.1016/j.cell.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S. 2011. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 146:633–644 10.1016/j.cell.2011.07.026 [DOI] [PubMed] [Google Scholar]
- Guzman M.L., Rossi R.M., Karnischky L., Li X., Peterson D.R., Howard D.S., Jordan C.T. 2005. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 105:4163–4169 10.1182/blood-2004-10-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Coussens L.M. 2012. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 21:309–322 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- Heider K.H., Kuthan H., Stehle G., Munzert G. 2004. CD44v6: a target for antibody-based cancer therapy. Cancer Immunol. Immunother. 53:567–579 10.1007/s00262-003-0494-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann D.M., Kilic E., Spudich A., Krämer S.D., Wunderli-Allenspach H., Bassetti C.L. 2006. Role of drug efflux carriers in the healthy and diseased brain. Ann. Neurol. 60:489–498 10.1002/ana.21012 [DOI] [PubMed] [Google Scholar]
- Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., Heeschen C. 2007. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 1:313–323 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Herschkowitz J.I., Simin K., Weigman V.J., Mikaelian I., Usary J., Hu Z., Rasmussen K.E., Jones L.P., Assefnia S., Chandrasekharan S., et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8:R76 10.1186/gb-2007-8-5-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J., Ruf P., Lindhofer H. 2012. Cancer therapy with trifunctional antibodies: linking innate and adaptive immunity. Future Oncol. 8:73–85 10.2217/fon.11.138 [DOI] [PubMed] [Google Scholar]
- Ho M.M., Ng A.V., Lam S., Hung J.Y. 2007. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 67:4827–4833 10.1158/0008-5472.CAN-06-3557 [DOI] [PubMed] [Google Scholar]
- Hope K.J., Jin L., Dick J.E. 2004. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5:738–743 10.1038/ni1080 [DOI] [PubMed] [Google Scholar]
- Huang E.H., Hynes M.J., Zhang T., Ginestier C., Dontu G., Appelman H., Fields J.Z., Wicha M.S., Boman B.M. 2009. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69:3382–3389 10.1158/0008-5472.CAN-08-4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K., Mintz B. 1976. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc. Natl. Acad. Sci. USA. 73:549–553 10.1073/pnas.73.2.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F., Yoshida S., Saito Y., Hijikata A., Kitamura H., Tanaka S., Nakamura R., Tanaka T., Tomiyama H., Saito N., et al. 2007. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 25:1315–1321 10.1038/nbt1350 [DOI] [PubMed] [Google Scholar]
- Ishizawa K., Rasheed Z.A., Karisch R., Wang Q., Kowalski J., Susky E., Pereira K., Karamboulas C., Moghal N., Rajeshkumar N.V., et al. 2010. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 7:279–282 10.1016/j.stem.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Bernardi R., Morotti A., Matsuoka S., Saglio G., Ikeda Y., Rosenblatt J., Avigan D.E., Teruya-Feldstein J., Pandolfi P.P. 2008. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 453:1072–1078 10.1038/nature07016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. 2011. Global cancer statistics. CA Cancer J. Clin. 61:69–90 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., Dick J.E. 2006. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12:1167–1174 10.1038/nm1483 [DOI] [PubMed] [Google Scholar]
- Jin L., Lee E.M., Ramshaw H.S., Busfield S.J., Peoppl A.G., Wilkinson L., Guthridge M.A., Thomas D., Barry E.F., Boyd A., et al. 2009. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 5:31–42 10.1016/j.stem.2009.04.018 [DOI] [PubMed] [Google Scholar]
- Joseph N.M., Mosher J.T., Buchstaller J., Snider P., McKeever P.E., Lim M., Conway S.J., Parada L.F., Zhu Y., Morrison S.J. 2008. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell. 13:129–140 10.1016/j.ccr.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Siegel P.M., Shu W., Drobnjak M., Kakonen S.M., Cordón-Cardo C., Guise T.A., Massagué J. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 3:537–549 10.1016/S1535-6108(03)00132-6 [DOI] [PubMed] [Google Scholar]
- Keck T., Brabletz T. 2011. Under stress: p53 controls EMT and stemness in pancreatic epithelial cells. Cell Cycle. 10:1715 10.4161/cc.10.11.15645 [DOI] [PubMed] [Google Scholar]
- Kelly P.N., Dakic A., Adams J.M., Nutt S.L., Strasser A. 2007. Tumor growth need not be driven by rare cancer stem cells. Science. 317:337 10.1126/science.1142596 [DOI] [PubMed] [Google Scholar]
- Klein C.A. 2009. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer. 9:302–312 10.1038/nrc2627 [DOI] [PubMed] [Google Scholar]
- Kong Y., Yoshida S., Saito Y., Doi T., Nagatoshi Y., Fukata M., Saito N., Yang S.M., Iwamoto C., Okamura J., et al. 2008. CD34+CD38+CD19+ as well as CD34+CD38-CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia. 22:1207–1213 10.1038/leu.2008.83 [DOI] [PubMed] [Google Scholar]
- Krause D.S., Lazarides K., von Andrian U.H., Van Etten R.A. 2006. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat. Med. 12:1175–1180 10.1038/nm1489 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., Wicha M., Clarke M.F., Simeone D.M. 2007. Identification of pancreatic cancer stem cells. Cancer Res. 67:1030–1037 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- Li X., Lewis M.T., Huang J., Gutierrez C., Osborne C.K., Wu M.F., Hilsenbeck S.G., Pavlick A., Zhang X., Chamness G.C., et al. 2008. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 100:672–679 10.1093/jnci/djn123 [DOI] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M.A., Serrano M. 2009a. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 460:1136–1139 10.1038/nature08290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Bao S., Wu Q., Wang H., Eyler C., Sathornsumetee S., Shi Q., Cao Y., Lathia J., McLendon R.E., et al. 2009b. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 15:501–513 10.1016/j.ccr.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Patel M.R., Prescher J.A., Patsialou A., Qian D., Lin J., Wen S., Chang Y.F., Bachmann M.H., Shimono Y., et al. 2010. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. USA. 107:18115–18120 10.1073/pnas.1006732107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., Dean M. 2007. Targeted therapy for cancer stem cells: the patched pathway and ABC transporters. Oncogene. 26:1357–1360 10.1038/sj.onc.1210200 [DOI] [PubMed] [Google Scholar]
- Lu D., Choi M.Y., Yu J., Castro J.E., Kipps T.J., Carson D.A. 2011. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA. 108:13253–13257 10.1073/pnas.1110431108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee J.A., Piskounova E., Morrison S.J. 2012. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 21:283–296 10.1016/j.ccr.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I., Peinado H., Kassen D., Hussenet T., Metzger D., Chambon P., Huber M., Hohl D., Cano A., Birchmeier W., Huelsken J. 2008. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 452:650–653 10.1038/nature06835 [DOI] [PubMed] [Google Scholar]
- Malanchi I., Santamaria-Martínez A., Susanto E., Peng H., Lehr H.A., Delaloye J.F., Huelsken J. 2012. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 481:85–89 10.1038/nature10694 [DOI] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massagué J. 2005. Genes that mediate breast cancer metastasis to lung. Nature. 436:518–524 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Weissman I.L., Akashi K. 2000. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc. Natl. Acad. Sci. USA. 97:7521–7526 10.1073/pnas.97.13.7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshaver B., van Rhenen A., Kelder A., van der Pol M., Terwijn M., Bachas C., Westra A.H., Ossenkoppele G.J., Zweegman S., Schuurhuis G.J. 2008. Identification of a small subpopulation of candidate leukemia-initiating cells in the side population of patients with acute myeloid leukemia. Stem Cells. 26:3059–3067 10.1634/stemcells.2007-0861 [DOI] [PubMed] [Google Scholar]
- Murphy M.J., Wilson A., Trumpp A. 2005. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 15:128–137 10.1016/j.tcb.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Nguyen L.V., Vanner R., Dirks P., Eaves C.J. 2012. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 12:133–143 [DOI] [PubMed] [Google Scholar]
- Notta F., Mullighan C.G., Wang J.C., Poeppl A., Doulatov S., Phillips L.A., Ma J., Minden M.D., Downing J.R., Dick J.E. 2011. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 469:362–367 10.1038/nature09733 [DOI] [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., Dick J.E. 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445:106–110 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- Odoux C., Fohrer H., Hoppo T., Guzik L., Stolz D.B., Lewis D.W., Gollin S.M., Gamblin T.C., Geller D.A., Lagasse E. 2008. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res. 68:6932–6941 10.1158/0008-5472.CAN-07-5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz-Wilson K.I., Philips S.T., Yilmaz O.H., Ames H.M., Li L., Crawford B.D., Gauvin A.M., Lucas P.C., Sitwala K., Downing J.R., et al. 2009. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 16:137–148 10.1016/j.ccr.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian-Rousseau V. 2010. CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer. 46:1271–1277 10.1016/j.ejca.2010.02.024 [DOI] [PubMed] [Google Scholar]
- Oskarsson T., Acharyya S., Zhang X.H., Vanharanta S., Tavazoie S.F., Morris P.G., Downey R.J., Manova-Todorova K., Brogi E., Massagué J. 2011. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 17:867–874 10.1038/nm.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang R., Law W.L., Chu A.C., Poon J.T., Lam C.S., Chow A.K., Ng L., Cheung L.W., Lan X.R., Lan H.Y., et al. 2010. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 6:603–615 10.1016/j.stem.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Pantel K., Alix-Panabières C., Riethdorf S. 2009. Cancer micrometastases. Nat Rev Clin Oncol. 6:339–351 10.1038/nrclinonc.2009.44 [DOI] [PubMed] [Google Scholar]
- Park D., Sykes D.B., Scadden D.T. 2012. The hematopoietic stem cell niche. Front. Biosci. 17:30–39 10.2741/3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D.J., Taussig D., Simpson C., Allen K., Rohatiner A.Z., Lister T.A., Bonnet D. 2005. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 23:752–760 10.1634/stemcells.2004-0292 [DOI] [PubMed] [Google Scholar]
- Pece S., Tosoni D., Confalonieri S., Mazzarol G., Vecchi M., Ronzoni S., Bernard L., Viale G., Pelicci P.G., Di Fiore P.P. 2010. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 140:62–73 10.1016/j.cell.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. 2000. Molecular portraits of human breast tumours. Nature. 406:747–752 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Perrotti D., Jamieson C., Goldman J., Skorski T. 2010. Chronic myeloid leukemia: mechanisms of blastic transformation. J. Clin. Invest. 120:2254–2264 10.1172/JCI41246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S.G., Reynolds B.A., Zanetti N., Lamorte G., Binda E., Broggi G., Brem H., Olivi A., Dimeco F., Vescovi A.L. 2006. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 444:761–765 10.1038/nature05349 [DOI] [PubMed] [Google Scholar]
- Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P., Weissman I.L., Clarke M.F., Ailles L.E. 2007. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 104:973–978 10.1073/pnas.0610117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaila B., Kaplan R.N., Port E.R., Lyden D. 2006-2007. Priming the ‘soil’ for breast cancer metastasis: the pre-metastatic niche. Breast Dis. 26:65–74 [DOI] [PubMed] [Google Scholar]
- Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., Morrison S.J. 2008. Efficient tumour formation by single human melanoma cells. Nature. 456:593–598 10.1038/nature07567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E., Shackleton M., Foster H.R., Fullen D.R., Sabel M.S., Johnson T.M., Morrison S.J. 2010. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 18:510–523 10.1016/j.ccr.2010.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran D., Schubert M., Taubert I., Eckstein V., Bellos F., Jauch A., Chen H., Bruckner T., Saffrich R., Wuchter P., Ho A.D. 2012. Heterogeneity of leukemia stem cell candidates at diagnosis of acute myeloid leukemia and their clinical significance. Exp. Hematol. 40:155–165.e1 10.1016/j.exphem.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., Weissman I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature. 414:105–111 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H., et al. 2012. EMT and dissemination precede pancreatic tumor formation. Cell. 148:349–361 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechelmann H., Sauter A., Golze W., Hanft G., Schroen C., Hoermann K., Erhardt T., Gronau S. 2008. Phase I trial with the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head and neck squamous cell carcinoma. Oral Oncol. 44:823–829 10.1016/j.oraloncology.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Romagosa C., Simonetti S., López-Vicente L., Mazo A., Lleonart M.E., Castellvi J., Ramon y Cajal S. 2011. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 30:2087–2097 10.1038/onc.2010.614 [DOI] [PubMed] [Google Scholar]
- Ross D.M., Hughes T.P., Melo J.V. 2011. Do we have to kill the last CML cell? Leukemia. 25:193–200 10.1038/leu.2010.197 [DOI] [PubMed] [Google Scholar]
- Saito Y., Uchida N., Tanaka S., Suzuki N., Tomizawa-Murasawa M., Sone A., Najima Y., Takagi S., Aoki Y., Wake A., et al. 2010. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat. Biotechnol. 28:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. 2011. In vitro generation of human cells with cancer stem cell properties. Nat. Cell Biol. 13:1051–1061 10.1038/ncb2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T., Murphy G.F., Frank N.Y., Yamaura K., Waaga-Gasser A.M., Gasser M., Zhan Q., Jordan S., Duncan L.M., Weishaupt C., et al. 2008. Identification of cells initiating human melanomas. Nature. 451:345–349 10.1038/nature06489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert J., Brabletz T. 2011. p53 Spreads out further: suppression of EMT and stemness by activating miR-200c expression. Cell Res. 21:705–707 10.1038/cr.2011.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J., Weissman I.L. 2010. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2:640–653 10.1002/wsbm.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell S., Pierce G.B. 1994. Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers. Lab. Invest. 70:6–22 [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141:69–80 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Zhu Z. 2008. Catumaxomab, a rat/murine hybrid trifunctional bispecific monoclonal antibody for the treatment of cancer. Curr. Opin. Mol. Ther. 10:273–284 [PubMed] [Google Scholar]
- Shiao S.L., Ganesan A.P., Rugo H.S., Coussens L.M. 2011. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25:2559–2572 10.1101/gad.169029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S.P., Chiao E., et al. 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 138:592–603 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa Y., Pedersen E.A., Havens A.M., Jung Y., Mishra A., Joseph J., Kim J.K., Patel L.R., Ying C., Ziegler A.M., et al. 2011. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J. Clin. Invest. 121:1298–1312 10.1172/JCI43414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Sládek N.E., Kollander R., Sreerama L., Kiang D.T. 2002. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother. Pharmacol. 49:309–321 10.1007/s00280-001-0412-4 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Taussig D.C., Pearce D.J., Simpson C., Rohatiner A.Z., Lister T.A., Kelly G., Luongo J.L., Danet-Desnoyers G.A., Bonnet D. 2005. Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood. 106:4086–4092 10.1182/blood-2005-03-1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig D.C., Miraki-Moud F., Anjos-Afonso F., Pearce D.J., Allen K., Ridler C., Lillington D., Oakervee H., Cavenagh J., Agrawal S.G., et al. 2008. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 112:568–575 10.1182/blood-2007-10-118331 [DOI] [PubMed] [Google Scholar]
- Tehranchi R., Woll P.S., Anderson K., Buza-Vidas N., Mizukami T., Mead A.J., Astrand-Grundström I., Strömbeck B., Horvat A., Ferry H., et al. 2010. Persistent malignant stem cells in del(5q) myelodysplasia in remission. N. Engl. J. Med. 363:1025–1037 10.1056/NEJMoa0912228 [DOI] [PubMed] [Google Scholar]
- Theodoropoulos P.A., Polioudaki H., Agelaki S., Kallergi G., Saridaki Z., Mavroudis D., Georgoulias V. 2010. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 288:99–106 10.1016/j.canlet.2009.06.027 [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 478:255–259 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Wiestler O.D. 2008. Mechanisms of Disease: cancer stem cells—targeting the evil twin. Nat. Clin. Pract. Oncol. 5:337–347 [DOI] [PubMed] [Google Scholar]
- Turhan A.G., Lemoine F.M., Debert C., Bonnet M.L., Baillou C., Picard F., Macintyre E.A., Varet B. 1995. Highly purified primitive hematopoietic stem cells are PML-RARA negative and generate nonclonal progenitors in acute promyelocytic leukemia. Blood. 85:2154–2161 [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 460:1145–1148 10.1038/nature08285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F., Asselin-Labat M.L., Shackleton M., Forrest N.C., Lindeman G.J., Visvader J.E. 2008. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 68:7711–7717 10.1158/0008-5472.CAN-08-1949 [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., Clevers H. 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71:241–260 10.1146/annurev.physiol.010908.163145 [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12:468–476 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Visvader J.E. 2011. Cells of origin in cancer. Nature. 469:314–322 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- Weinstein I.B. 2002. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 297:63–64 10.1126/science.1073096 [DOI] [PubMed] [Google Scholar]
- Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., et al. 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11:1487–1495 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Wulf G.G., Wang R.Y., Kuehnle I., Weidner D., Marini F., Brenner M.K., Andreeff M., Goodell M.A. 2001. A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood. 98:1166–1173 10.1182/blood.V98.4.1166 [DOI] [PubMed] [Google Scholar]
- Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467:1114–1117 10.1038/nature09515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.F., Ho D.W., Ng M.N., Lau C.K., Yu W.C., Ngai P., Chu P.W., Lam C.T., Poon R.T., Fan S.T. 2008. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 13:153–166 10.1016/j.ccr.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318:1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zhang M., Behbod F., Atkinson R.L., Landis M.D., Kittrell F., Edwards D., Medina D., Tsimelzon A., Hilsenbeck S., Green J.E., et al. 2008. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 68:4674–4682 10.1158/0008-5472.CAN-07-6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S., Soh B.S., Sun L.L., Tai B.C., Nga M.E., et al. 2012. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 148:259–272 (published erratum appears in Cell. 2012. 148:1066) 10.1016/j.cell.2011.11.050 [DOI] [PubMed] [Google Scholar]
- Zöller M. 2011. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer. 11:254–267 10.1038/nrc3023 [DOI] [PubMed] [Google Scholar]