Seg1 establishes a platform for the assembly of eisosomes and is important for determining their length.
Abstract
Eisosomes are stable domains at the plasma membrane of the budding yeast Saccharomyces cerevisiae and have been proposed to function in endocytosis. Eisosomes are composed of two main cytoplasmic proteins, Pil1 and Lsp1, that form a scaffold around furrow-like plasma membrane invaginations. We show here that the poorly characterized eisosome protein Seg1/Ymr086w is important for eisosome biogenesis and architecture. Seg1 was required for efficient incorporation of Pil1 into eisosomes and the generation of normal plasma membrane furrows. Seg1 preceded Pil1 during eisosome formation and established a platform for the assembly of other eisosome components. This platform was further shaped and stabilized upon the arrival of Pil1 and Lsp1. Moreover, Seg1 abundance controlled the shape of eisosomes by determining their length. Similarly, the Schizosaccharomyces pombe Seg1-like protein Sle1 was necessary to generate the filamentous eisosomes present in fission yeast. The function of Seg1 in the stepwise biogenesis of eisosomes reveals striking architectural similarities between eisosomes in yeast and caveolae in mammals.
Introduction
Cells subdivide their plasma membrane into regions with specialized functions. One way to achieve this compartmentalization is to construct diffusion barriers within the plasma membrane and furnish the resulting surface domains with unique compositions by means of dedicated membrane trafficking pathways (Nakada et al., 2003; Schuck and Simons, 2004; Caudron and Barral, 2009; Steed et al., 2010). Another way to segregate plasma membrane components is based on the propensity of certain lipids, namely sterols and sphingolipids, to form microdomains by preferential association (Lingwood and Simons, 2010). These microdomains, called lipid rafts, can be clustered into larger assemblies by specialized protein scaffolds. Mammalian scaffolding proteins of this type are the caveolins. These integral membrane proteins bind cholesterol, polymerize into stable protein lattices, and shape the plasma membrane into 60–80-nm-deep cup-like caveolae that serve as sites of clathrin-independent endocytosis (Parton and Simons, 2007; Hansen and Nichols, 2009; Bastiani and Parton, 2010).
The yeast Saccharomyces cerevisiae possesses plasma membrane domains that share many fundamental features with caveolae (Ziółkowska et al., 2012). Their principal protein components are Pil1 and Lsp1, two highly similar cytoplasmic proteins that are each present at an abundance of ∼100,000 copies per cell (Ghaemmaghami et al., 2003). The two proteins bind to one another and generate 20–50 immobile plasma membrane-associated assemblies in every cell, with each assembly containing on the order of 2,000–5,000 subunits of both Pil1 and Lsp1 (Walther et al., 2006). Pil1/Lsp1 assemblies are evenly distributed over the plasma membrane and maintain a minimal distance from each other (Moreira et al., 2009). The sites at which Pil1/Lsp1 associate with the plasma membrane correspond to furrow-like membrane invaginations that are ∼50 nm deep and 200–300 nm long (Strádalová et al., 2009). These invaginated membrane patches appear to be enriched in ergosterol, the major yeast sterol (Grossmann et al., 2007), and they require sphingolipids for proper organization (Grossmann et al., 2006; Fröhlich et al., 2009). Pil1/Lsp1 have been suggested to participate in endocytosis, but this connection remains to be clarified (Walther et al., 2006; Grossmann et al., 2008; Brach et al., 2011).
The Pil1/Lsp1 assemblies have been named “eisosomes” (Walther et al., 2006), whereas the ergosterol-enriched membrane patches that colocalize with Pil1/Lsp1 have been called “membrane compartment of arginine permease Can1” (MCC; Malínská et al., 2003, 2004; Grossmann et al., 2008; Malínsky et al., 2010). The terms eisosome and MCC likely describe connected parts of the same cellular structure. First, in cells lacking Pil1, MCC-associated transmembrane proteins disperse in the plasma membrane and furrow-like invaginations disappear (Walther et al., 2006; Grossmann et al., 2007, 2008; Fröhlich et al., 2009; Strádalová et al., 2009). The integrity of the MCC therefore depends on the eisosome protein Pil1. Second, disruption of the MCC, for example by sphingolipid depletion, is relayed to Pil1 by phosphorylation, and causes a large fraction of Pil1 to dissociate from the plasma membrane (Walther et al., 2007; Luo et al., 2008; Fröhlich et al., 2009). The integrity of eisosomes therefore depends on an intact MCC. Third, Pil1 and Lsp1 both contain membrane-shaping BAR domains, bind to liposomes in vitro, and self-assemble into filaments whose dimensions match those of plasma membrane furrows in vivo (Karotki et al., 2011; Olivera-Couto et al., 2011; Ziółkowska et al., 2011). Eisosome components in cells are therefore likely to directly interact with and scaffold the plasma membrane. In view of these links, we suggest treating the whole subcellular structure as a single entity, consisting of a furrow-like plasma membrane domain, the transmembrane proteins that partition into this domain, and the proteins that form a scaffolding lattice on its cytoplasmic face. In this paper, we shall use the term eisosome in this sense.
The proper assembly of eisosomes critically depends on Pil1. In its absence, Lsp1 is mostly cytoplasmic, whereas Pil1 retains its normal distribution in cells lacking Lsp1 (Walther et al., 2006). Furthermore, without Pil1, eisosome components partially collapse into a small number of clusters, referred to as eisosome remnants (Walther et al., 2006; Grossmann et al., 2007). Eisosome remnants correspond to large aberrant plasma membrane invaginations (Walther et al., 2006; Strádalová et al., 2009). Reducing or raising the levels of Pil1 yields a lower number of normal eisosomes or a normal number of larger eisosomes, respectively (Moreira et al., 2009). These observations indicate that there is a lower limit for eisosome size and an upper limit for eisosome number. The molecular mechanisms imposing these limits are unknown.
To better understand the architecture and ultimately the function of eisosomes, we have previously conducted a screen to identify genes involved in eisosome formation (Fröhlich et al., 2009). Several of the identified genes had no known function. Here, we study one of these poorly characterized genes, YMR086W, which encodes a large coiled-coil protein without recognizable functional domains. Based on the observation that its homologue in the yeast Ashbya gossypii is important for eisosome stability, YMR086W has been named SEG1 for “stability of eisosomes guaranteed” (Seger et al., 2011). We find that the Seg1 protein facilitates eisosome assembly and controls eisosome shape.
Results
Seg1 is required for proper eisosome architecture
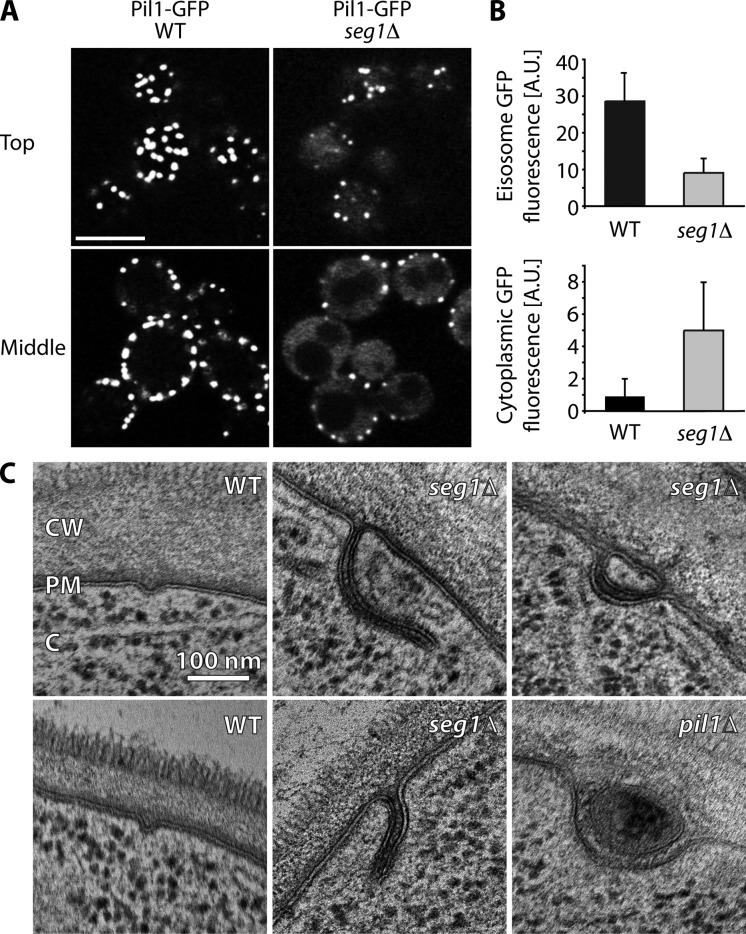
Our previous screen had shown that cells lacking Seg1 fail to properly localize Pil1-GFP, which indicates a defect in eisosome formation (Fröhlich et al., 2009). To analyze this phenotype in detail, we first imaged Pil1-GFP in wild-type and seg1Δ cells. In the absence of Seg1, cells displayed a reduced number of eisosomes, as defined by Pil1-GFP patches at the plasma membrane (Fig. 1 A). In addition, the Pil1-GFP signal of remaining eisosomes was decreased and the cytoplasmic Pil1-GFP signal was increased (Fig. 1 B). These findings show that Seg1 is required for efficient incorporation of Pil1-GFP into eisosomes.
Figure 1.
Seg1 is required for proper eisosome architecture. (A) Confocal images of Pil1-GFP in wild-type (WT) and seg1Δ cells. Representative top views and mid sections are shown. Bar, 5 µm. (B) Quantification of Pil1-GFP signal per eisosome (eisosome GFP fluorescence) and Pil1-GFP signal in the cytoplasm (cytoplasmic GFP fluorescence) in WT and seg1Δ cells. A.U., arbitrary units. Error bars indicate standard deviations. (C) Electron micrographs of WT, seg1Δ, and pil1Δ cells. CW, cell wall; PM, plasma membrane; C, cytoplasm.
Next, we analyzed the plasma membrane morphology of wild-type, seg1Δ, and pil1Δ cells by electron microscopy. Consistent with earlier studies (Moor and Mühlethaler, 1963; Strádalová et al., 2009), wild-type cells showed plasma membrane furrows ∼30 nm deep, 30 nm wide and 200 nm long (Fig. 1 C, left; see Fig. S1 A for serial sections). In contrast, seg1Δ cells had deep, irregularly shaped plasma membrane invaginations (Fig. 1 C, middle and top right; see Fig. S1 B for serial sections). These invaginations were sometimes reminiscent of eisosome remnants seen in pil1Δ cells (Fig. 1 C, bottom right), but were generally smaller. These findings show that Seg1 is required for proper plasma membrane morphology. It appears likely that the aberrant invaginations observed in seg1Δ cells by electron microscopy correspond to the remaining Pil1-GFP patches seen in these cells by light microscopy. Collectively, Seg1 is needed for two aspects of eisosome architecture: the assembly of Pil1-GFP into membrane-associated complexes of characteristic size and the local molding of the plasma membrane into well-defined furrows.
Seg1 is an eisosome component
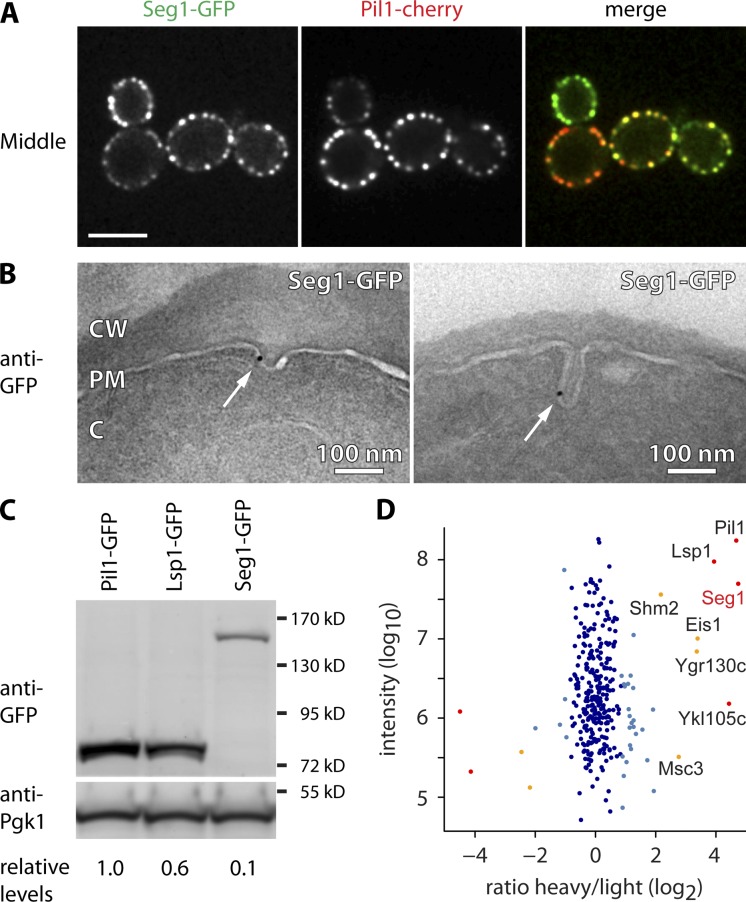
Seg1 has been shown to colocalize with Lsp1 and interact with Pil1/Lsp1 (Deng et al., 2009). Accordingly, Seg1-GFP colocalized with Pil1-cherry (Fig. 2 A). We next used immunogold labeling with an anti-GFP antibody to localize Seg1-GFP by immunoelectron microscopy. As expected, the immunogold marked plasma membrane invaginations characteristic of eisosomes (Fig. 2 B). Although the labeling was specific, its density was quite low, possibly because the GFP epitope is rendered largely inaccessible by the eisosomal protein lattice.
Figure 2.
Seg1 is an eisosome component. (A) Confocal mid sections of cells expressing Seg1-GFP and Pil1-cherry. Bar, 5 µm. (B) Electron micrographs of Seg1-GFP cells labeled with anti-GFP antibody and gold-conjugated protein A. Arrows indicate gold particles. CW, cell wall; PM, plasma membrane; C, cytoplasm. (C) Western blot of GFP and Pgk1 from cells expressing Pil1-GFP, Lsp1-GFP, or Seg1-GFP from their endogenous loci. Numbers indicate GFP levels relative to Pgk1 and normalized to Pil1-GFP. (D) Mass spectrometric analysis of Seg1 affinity-purified from heavy-labeled cells expressing Seg1-TEV-GFP and light-labeled, untagged control cells. The averaged peptide intensity is plotted against the ratio of heavy/light. Significant outliers are colored in red (P < 10−11), orange (P < 10−4), or light blue (P < 0.05). Other identified proteins are colored in dark blue.
Using quantitative Western blotting, we compared the levels of Seg1, Pil1, and Lsp1 expressed as GFP fusions from their endogenous chromosomal loci and found that Seg1 is about 10-fold less abundant than Pil1 or Lsp1 (Fig. 2 C). Because a single eisosome contains 2,000–5,000 molecules of each Pil1 and Lsp1 (Walther et al., 2006), there are likely 200–500 Seg1 molecules per eisosome.
To identify Seg1 interaction partners, we quantitatively analyzed Seg1 immunoprecipitates using SILAC (stable isotope labeling with amino acids in cell culture). We immunopurified Seg1 from cells that expressed Seg1-TEV-GFP and had been metabolically labeled with heavy isotope lysine. The resulting eluate was mixed with that from a mock purification using untagged control cells grown in the presence of normal, light isotope lysine. Finally, the ratio of heavy/light lysine was determined for each protein identified by mass spectrometry. A high heavy/light ratio for a given protein indicates enrichment in the metabolically labeled sample and hence interaction with Seg1. By this measure, Seg1 interacts with the known eisosome proteins Pil1, Lsp1, Eis1/Ymr031c, and Ygr130c, as well as the Seg1 paralogue Seg2/Ykl105c (Fig. 2 D). These results confirm that Seg1 is an eisosome protein.
Seg1 precedes Pil1 during eisosome assembly
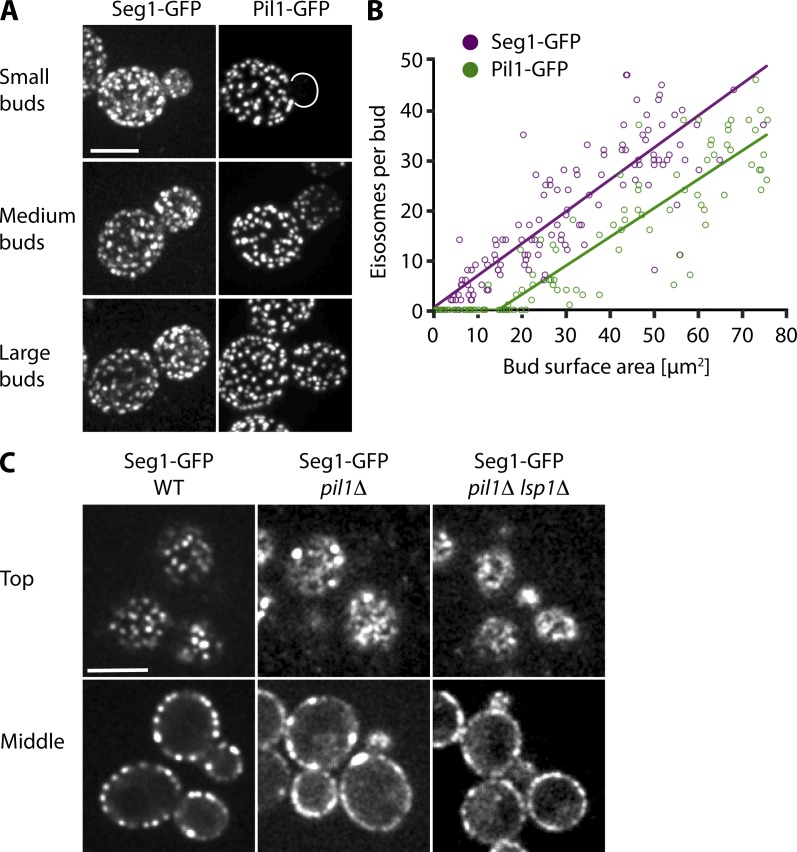
To begin to investigate the role of Seg1 in eisosome formation, we analyzed the incorporation of Seg1 into nascent eisosomes during yeast budding. Growing buds are initially devoid of eisosomes as marked by Pil1 and Lsp1. Once a bud exceeds a critical size, it is colonized by newly assembled eisosomes. Colonization occurs in a polarized fashion, starting from the bud neck (Moreira et al., 2009). However, when we imaged the deposition of Seg1, we observed Seg1-GFP already in small buds, where it was diffusely distributed and formed heterogeneous patches at the plasma membrane (Fig. 3 A, top). Medium-sized buds were evenly colonized by Seg1-GFP patches, whereas Pil1-GFP patches exhibited the characteristic polarized distribution observed previously (Fig. 3 A, middle). Large buds showed a uniform pattern for both Seg1-GFP and Pil1-GFP patches (Fig. 3 A, bottom). These observations indicate that Seg1 deposition precedes that of Pil1. We also attempted to image Seg1 and Pil1 in the same cells by fusing them to different fluorescent proteins. However, these experiments were rendered uninterpretable by the different maturation times of the fluorophores so that the protein fused to the faster maturing fluorescent protein always seemed to enter growing buds first. To refine our results, we quantified Seg1-GFP patches in buds of different sizes and plotted their number against bud surface area. Consistent with earlier measurements (Moreira et al., 2009), Pil1-GFP patches were absent in buds with a surface area <15 µm2, showing a lag phase for Pil1 deposition (Fig. 3 B). In contrast, there was no lag phase for the formation of Seg1-GFP patches, which indicates that deposition of Seg1 does not require a minimum bud size. These results confirm that Seg1 becomes part of eisosome precursors before the arrival of Pil1.
Figure 3.
Seg1 precedes Pil1 during eisosome assembly. (A) Projections from confocal stacks of cells expressing Seg1-GFP (left) or Pil1-GFP (right). Representative images of small, medium, and large buds are shown. (B) Number of Seg1-GFP and Pil1-GFP patches per bud (determined from projections as in A), plotted against bud surface area and fitted using a biphasic model (see Materials and methods). (C) Confocal images of WT, pil1Δ, and pil1Δ lsp1Δ cells expressing Seg1-GFP. Representative top views and mid sections are shown. Bars, 5 µm.
Seg1 facilitates eisosome assembly
The diffuse distribution of Seg1 in small buds lacking Pil1 suggested that uniform and stable assembly of Seg1 requires Pil1. To test this idea, we analyzed Seg1-GFP in pil1Δ cells. Consistent with Pil1 being critical for eisosome biogenesis, Seg1-GFP displayed an uneven distribution at the plasma membrane with a few remaining patches (Fig. 3 C, middle). Additional deletion of Lsp1 had no effect, nor did deletion of Lsp1 alone (Fig. 3 C, right; and not depicted).
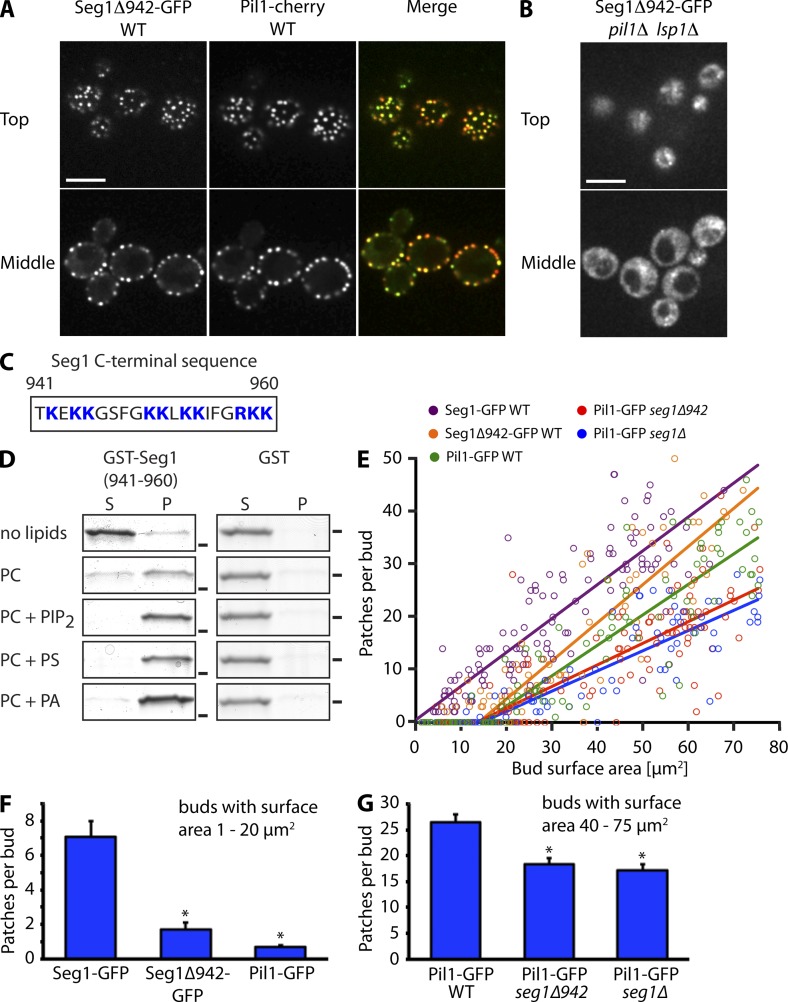
Given that Seg1 has no predicted transmembrane domains, its plasma membrane association in pil1Δ cells and in small buds lacking Pil1 was unexpected. So far, eisosome proteins without transmembrane domains, such as Lsp1 and Pkh2, have been found mainly in the cytoplasm in the absence of Pil1 (Walther et al., 2006, 2007). We noticed that the C terminus of Seg1 contains clusters of basic residues (Fig. 4 C). To test if this region mediates plasma membrane association, we analyzed the localization of Seg1Δ942-GFP, which lacks the last 18 amino acids of Seg1. The truncated Seg1 localized mostly to eisosomes in wild-type cells, as judged by colocalization with Pil1-cherry (Fig. 4 A). In addition, cells expressing untagged Seg1Δ942 as the only copy of Seg1 had a normal steady-state distribution of Pil1-GFP (unpublished data). However, Seg1Δ942-GFP was completely cytoplasmic in pil1Δ lsp1Δ cells, demonstrating that the basic C terminus targets Seg1 to the plasma membrane in the absence of Pil1/Lsp1 (Fig. 4 B). To test directly if the C terminus of Seg1 is able to bind lipids, we fused it to GST and assayed binding of recombinant GST-Seg1(941–960) to liposomes of varying composition. GST-Seg1(941–960) showed binding to liposomes consisting exclusively of phosphatidylcholine, but binding was enhanced by addition of the negatively charged lipids phosphatidylinositol-(4,5)-bisphosphate (PIP2), phosphatidylserine, or phosphatidic acid (Fig. 4 D). Therefore, the C terminus of Seg1 is sufficient to bind lipids, at least in vitro. We conclude that Seg1 is kept at the plasma membrane by two interactions. The first requires Pil1/Lsp1 and may involve direct binding to Pil1 or Lsp1, whereas the second is independent of Pil1/Lsp1 and requires the polybasic C terminus of Seg1.
Figure 4.
Targeting of Seg1 to small buds via its lipid-binding C terminus is important for efficient eisosome assembly. (A) Confocal images of wild-type (WT) cells expressing Seg1Δ942-GFP and Pil1-cherry. Bar, 5 µm. (B) Confocal images of pil1Δ lsp1Δ cells expressing Seg1Δ942-GFP. Bar, 5 µm. (C) Schematic of the C terminus of Seg1. Positively charged residues are in blue. (D) Coomassie-stained SDS-PAGE gels from spin-down assays of GST-Seg1(941–960) and GST with liposomes containing phosphatidylcholine (PC), or PC with either 1.5% PIP2, 30% phosphatidylserine (PS), or 30% phosphatidic acid (PA). S, supernatant; P, pellet. Bars indicate the position of the 26 kD marker band. (E) Number of Seg1-GFP, Seg1Δ942-GFP, and Pil1-GFP patches per bud, plotted against bud surface area and fitted as in Fig. 3 B. The data for Seg1-GFP and Pil1-GFP from Fig. 3 B are included for reference. (F) Mean number of Seg1-GFP, Seg1Δ942-GFP, and Pil1-GFP patches in buds with a surface area of 1–20 µm2. Error bars indicate SEM, with n = 44, 39, and 37. Asterisks indicate significant difference to Seg1-GFP (P < 10−5). (G) Mean number of Pil1-GFP patches in buds with a surface area of 40–75 µm2 in WT, seg1Δ942, and seg1Δ cells. Error bars indicate SEM, with n = 45, 39, and 59. Asterisks indicate significant difference to Pil1-GFP in WT cells (P < 10−5).
Next, we tested whether deposition of Seg1 in small buds lacking Pil1 is mediated by its C terminus. We measured the formation of Seg1Δ942-GFP patches in growing buds and found that the truncated protein was excluded from small buds almost as stringently as Pil1-GFP. Fitting of the data revealed a critical bud size for patch formation of 14 µm2 compared with 0 µm2 for Seg1-GFP and 15 µm2 for Pil1-GFP (Fig. 4 E). Accordingly, the mean number of patches formed by Seg1Δ942-GFP in buds with a surface area of 1−20 µm2 was significantly lower than that of Seg1-GFP and similar to that of Pil1-GFP (Fig. 4 F). This result shows that the C terminus is important for targeting of Seg1 to small buds. To examine the role of Seg1 targeting in eisosome assembly, we compared deposition of Pil1-GFP in buds of wild-type, seg1Δ, and seg1Δ942 cells. Formation of Pil1-GFP patches in the buds of seg1Δ cells was diminished (Fig. 4 E). The same was true for cells expressing Seg1Δ942 as the only copy of Seg1. This result was confirmed by determining the mean number of Pil1-GFP patches in buds with a surface area of 40–75 µm2, which revealed a reduced number of patches in seg1Δ and seg1Δ942 cells (Fig. 4 G). Thus, the arrival of Seg1 in small buds by means of its lipid-binding C terminus is important for the subsequent incorporation of Pil1-GFP into nascent eisosomes.
Our results suggest the following order of events during eisosome assembly: first, the C-terminus of Seg1 mediates Pil1/Lsp1-independent targeting to the plasma membrane in small buds, where Seg1 assembles into loose patches. Pil1/Lsp1 then arrives at these patches and stabilizes them into well-defined eisosomes. Whether all Seg1 patches become eisosomes or some represent unproductive intermediates remains to be established. Because Seg1Δ942 supports a normal steady-state distribution of Pil1, Seg1 is ultimately dispensable for the targeting of Pil1/Lsp1 to the plasma membrane. Nevertheless, the early arrival of Seg1 is important for efficient eisosome assembly, perhaps by ensuring that no assembly is initiated at sites devoid of Seg1.
Seg1 controls eisosome shape
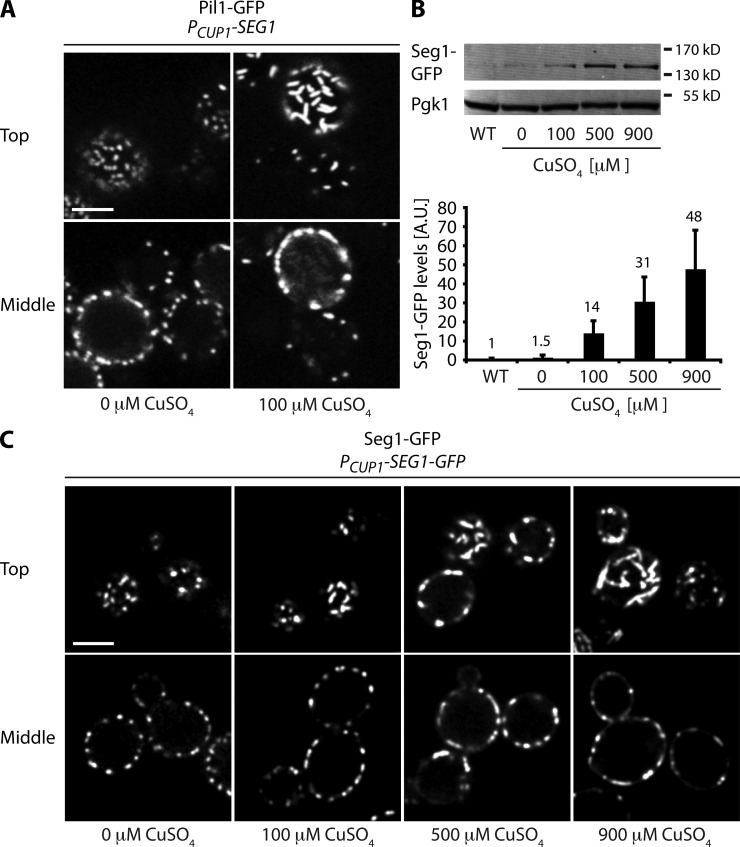
If Seg1 indeed helps organize eisosome assembly, raising Seg1 levels might change eisosome morphology. We therefore placed Seg1 under the control of the copper-inducible CUP1 promoter and followed eisosome formation using Pil1-GFP. The CUP1 promoter is leaky (Janke et al., 2004), and the amount of Seg1 produced even in the absence of copper was sufficient for normal eisosome formation (Fig. 5 A, left). However, after overnight growth in the presence of 100 µM CuSO4 to overexpress Seg1, mother cells had striking, rod-shaped eisosomes that were aligned parallel to the plane of the membrane (Fig. 5 A, right, top cell). Young daughter cells that still shared the cytoplasm with their mothers showed reduced eisosome density yet had normal, round eisosomes (Fig. 5 A, right, bottom cell). Because the amount of Pil1-GFP is unchanged by Seg1 overexpression (Fig. S2 A), eisosome overassembly in mother cells may hamper formation of new eisosomes in daughter cells.
Figure 5.
Seg1 can direct the formation of rod-shaped eisosomes. (A) Confocal images of Pil1-GFP cells expressing Seg1 from the CUP1 promoter. Cells were grown overnight in the absence or presence of 100 µM CuSO4. (B) Western blotting and quantification of Seg1-GFP levels relative to Pgk1 in cells constitutively expressing Seg1-GFP from the SEG1 promoter (WT) or in cells inducibly expressing Seg1-GFP from the CUP1 promoter. The latter cells were grown overnight in the presence of 0, 100, 500, or 900 µM CuSO4. Seg1-GFP levels are in arbitrary units (A.U.). Values above the bars indicate fold change compared with WT. Error bars indicate standard deviations from three independent experiments. (C) Confocal images of cells expressing Seg1-GFP from the CUP1 promoter grown overnight in the presence of 0, 100, 500, or 900 µM CuSO4. Representative top views and mid sections are shown. Bars, 3 µm.
To test if Seg1 itself assembles into elongated structures when overproduced, we tagged Seg1 with GFP and replaced the SEG1 promoter with the CUP1 promoter. Because of the leakiness of the CUP1 promoter, growth in the absence of copper yielded Seg1-GFP levels somewhat higher than those in cells expressing Seg1-GFP from the endogenous SEG1 promoter (Fig. 5 B). Growth in medium with up to 900 µM CuSO4 yielded up to 50-fold higher expression levels. Seg1-GFP in the uninduced condition showed a normal distribution (Fig. 5 C, left, compare with Fig. 2 A). However, as we raised the copper concentration, Seg1-GFP structures elongated and eventually became filamentous (Fig. 5 C, right). These results suggest that Seg1 can control the shape of eisosomes.
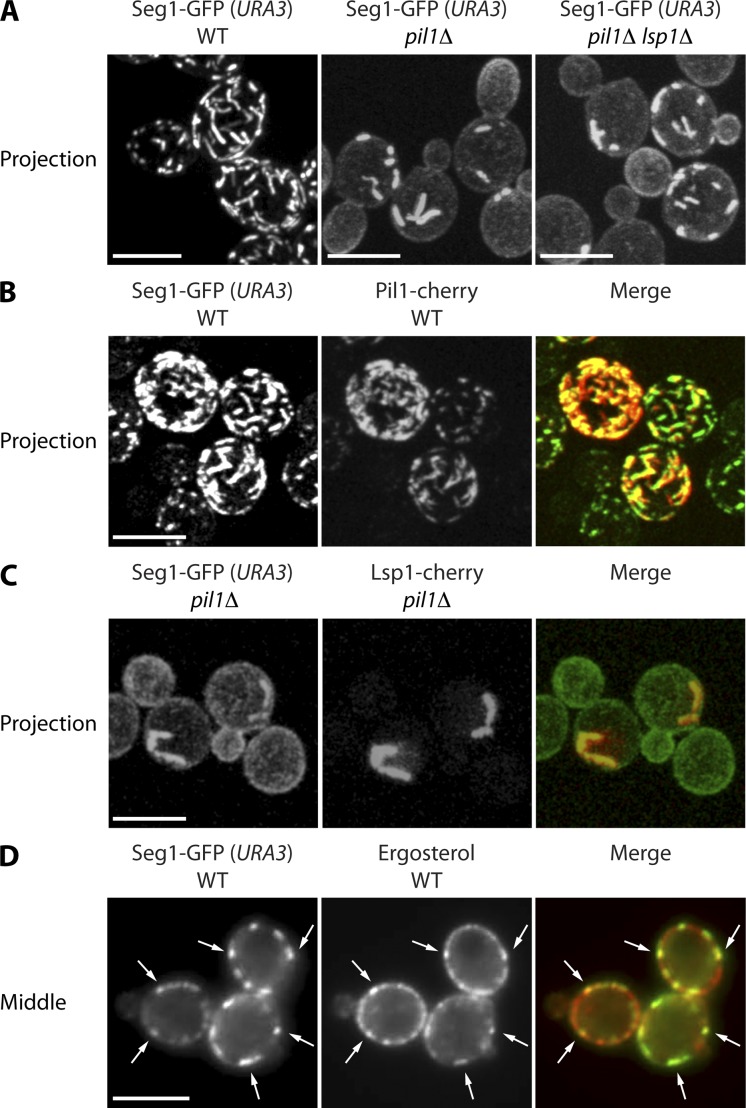
To further explore the properties of elongated eisosomes in cells overproducing Seg1, we generated strains that constitutively express Seg1-GFP at high levels, thus obviating the need for growth with CuSO4. We deleted the endogenous SEG1 gene and integrated a Seg1-GFP construct including the SEG1 promoter into the URA3 locus. The SEG1 promoter is more active in this location, resulting in approximately ninefold higher Seg1-GFP levels compared with strains expressing Seg1-GFP from the native SEG1 locus (Fig. S2 B). These elevated levels were sufficient to generate rod-shaped eisosomes, as is best appreciated in 2D projections from confocal stacks (Fig. 6 A, left). Next, we tested whether formation of Seg1-GFP rods requires Pil1 or Lsp1. Deleting Pil1 yielded rods that were thicker but also shorter and less abundant (Fig. 6 A, middle). The increased thickness may account for the decrease in rod length and number because sequestration of Seg1-GFP into thick rods may reduce free Seg1-GFP below the concentrations necessary to drive elongation of existing rods or assembly of new ones. Additional deletion of Lsp1 had no effect (Fig. 6 A, right). These results show that overexpressed Seg1 can assemble into plasma membrane-associated rods independently of Pil1/Lsp1. In addition, they suggest that Pil1 can shape Seg1 rods by restricting their width. This finding reinforces the notion that Pil1 and Seg1 collaborate during eisosome assembly in that Seg1 provides an early platform that is reshaped upon incorporation of Pil1.
Figure 6.
Seg1 rods form without Pil1 and contain eisosome components. (A) Projections from confocal stacks of wild-type (WT), pil1Δ, and pil1Δ lsp1Δ cells lacking endogenous Seg1 and expressing Seg1-GFP from the URA3 locus. (B) Projections of WT cells expressing Pil1-cherry, lacking endogenous Seg1, and expressing Seg1-GFP from the URA3 locus. (C) Projections of pil1Δ cells expressing Lsp1-cherry, lacking endogenous Seg1, and expressing Seg1-GFP from the URA3 locus. (D) Epifluorescence images of WT cells lacking endogenous Seg1, expressing Seg1-GFP from the URA3 locus, and stained with filipin to visualize ergosterol. Arrows indicate colocalization of Seg1-GFP and filipin. Bars, 5 µm.
We next asked whether Seg1-GFP rods are entirely artificial structures or likely to bear informative resemblance to native eisosomes. To this end, we first tested whether Seg1-GFP rods colocalize with other eisosome components. Consistent with the results obtained with copper-induced overexpression of untagged Seg1, Seg1-GFP rods completely reorganized the intracellular distribution of Pil1-cherry, which was now found in the same rods (Fig. 6 B). Lsp1-cherry also localized to Seg1 rods, in both otherwise wild-type and pil1Δ cells (Figs. 6 C and S3 A). The relocalization to Seg1 rods in wild-type cells was expected because Lsp1 binds to and therefore follows Pil1. The localization of Lsp1 to Seg1 rods in the absence of Pil1, however, was surprising. Lsp1 has so far only been found in the cytoplasm and in eisosome remnants in cells lacking Pil1 (Walther et al., 2006). The fact that overproduction of Seg1 prevents Lsp1 from becoming cytoplasmic and redirects it into Seg1 rods points to a Pil1-independent interaction of Lsp1 and Seg1. Notably, Lsp1 is unable to shape Seg1-GFP rods into long, thin filaments as Pil1 does, despite closely resembling Pil1 in structure and abundance. Finally, we analyzed the distribution of ergosterol by filipin staining and found that ergosterol patches colocalize with elongated eisosomes in Seg1-GFP–overproducing cells (Fig. 6 D). Interestingly, the localization of ergosterol to Seg1-GFP rods was abolished in pil1Δ cells (Fig. S3 B).
Our results show that overexpressed Seg1 forms membrane-associated rod-like structures, even in the absence of Pil1. These structures contain other eisosome components, including Pil1, Lsp1, and ergosterol. Thus, formation of Seg1 rods recapitulates aspects of normal eisosome assembly and reveals a role for Seg1 in controlling eisosome shape. Localization of Lsp1 to Seg1 rods is independent of Pil1, whereas enrichment of ergosterol at these sites requires Pil1, highlighting that Pil1 and Seg1 coordinate different steps of eisosome assembly.
Seg1 controls eisosome length
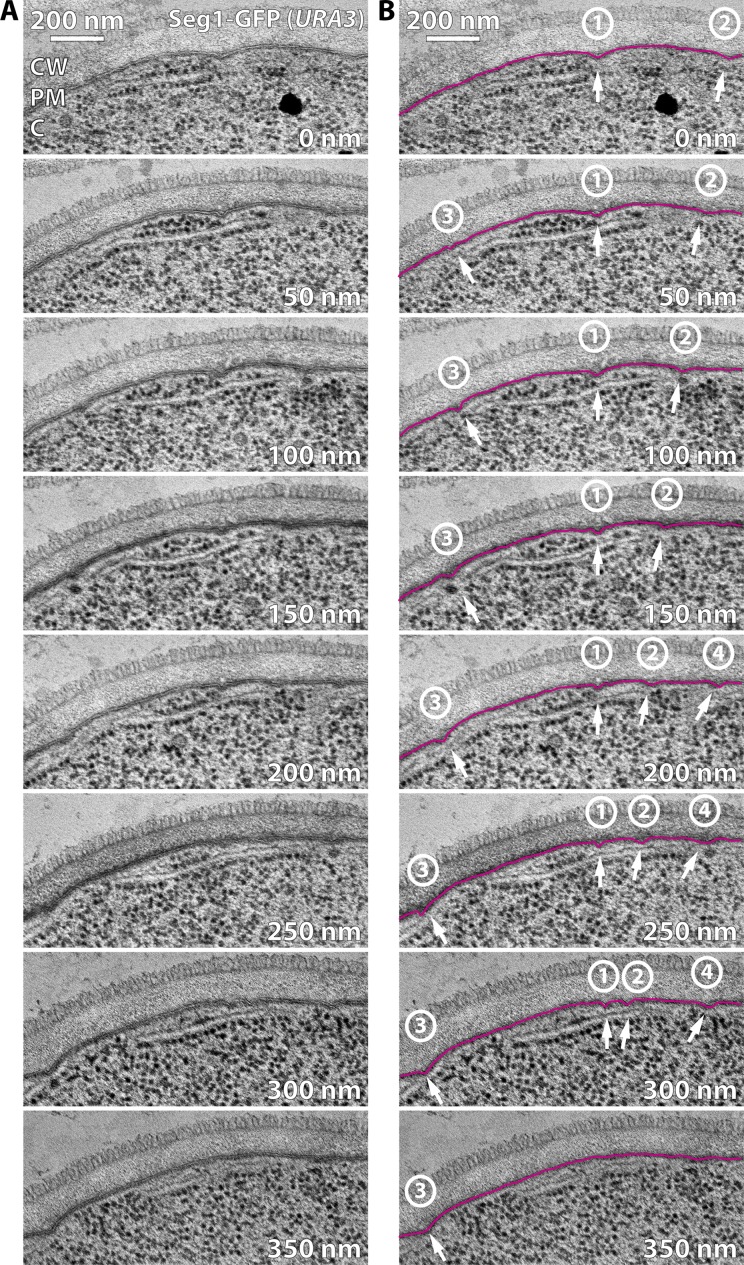
Next, we analyzed Seg1-GFP–overproducing cells by electron microscopy to determine if Seg1 rods affect plasma membrane morphology. We observed plasma membrane furrows of normal width and depth. However, these furrows were encountered much more frequently than in cells with normal Seg1 levels (Fig. 7, A and B). Importantly, serial sections revealed that plasma membrane furrows in cells overproducing Seg1-GFP were unusually long (Fig. 8). The elongation of plasma membrane furrows likely accounts for their more frequent appearance in single thin sections because it increases the probability that furrows are captured in any given section. Quantification from serial sections showed that the furrows are 510 ± 130 nm long (n = 10), which is substantially longer than the 200 nm observed in cells with normal Seg1 levels.
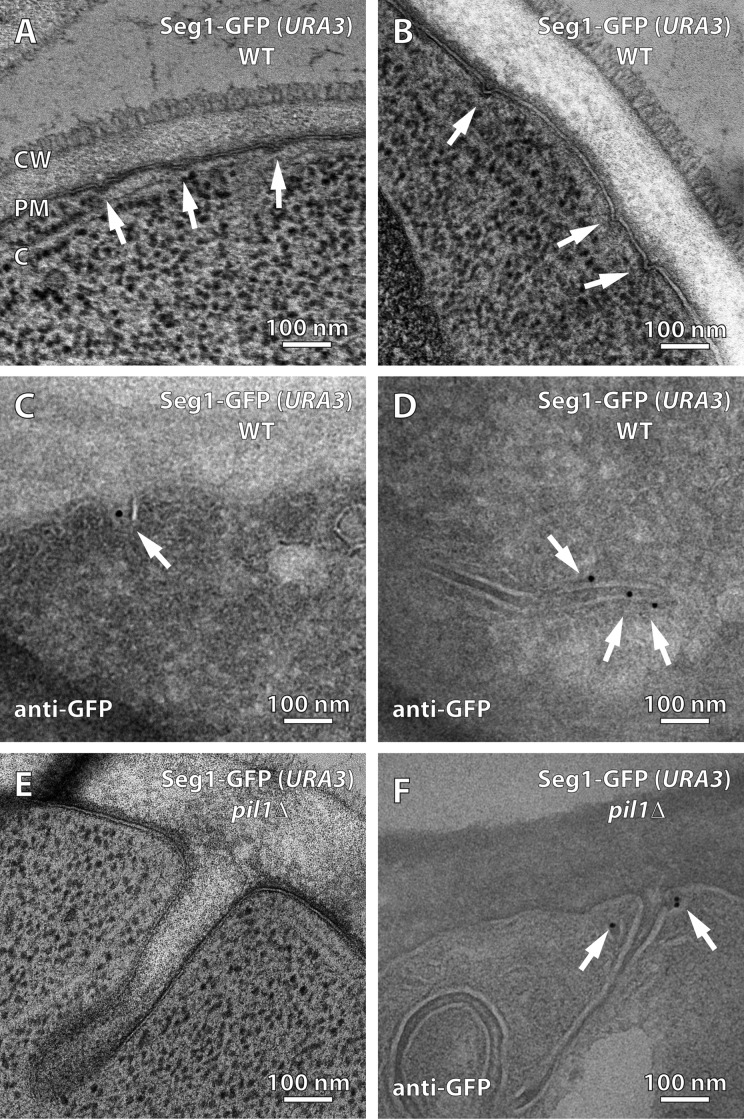
Figure 7.
Seg1 can direct the formation of plasma membrane invaginations. (A and B) Electron micrographs of wild-type (WT) cells lacking endogenous Seg1 and expressing Seg1-GFP from the URA3 locus. Arrows indicate plasma membrane invaginations. (C and D) Electron micrographs of the same cells labeled with anti-GFP antibody and gold-conjugated protein A. Arrows indicate gold particles. (E) Electron micrograph of pil1Δ cells lacking endogenous Seg1 and expressing Seg1-GFP from the URA3 locus. (F) Electron micrograph of the same cells labeled with anti-GFP antibody and gold-conjugated protein A. Arrows indicate gold particles. CW, cell wall; PM, plasma membrane; C, cytoplasm.
Figure 8.
Seg1 can direct the formation of long plasma membrane furrows. (A) Electron micrographs of sequential 50-nm sections from a seg1Δ cell expressing Seg1-GFP from the URA3 locus. The 200-nm image corresponds to the one shown in Fig. 6 A. CW, cell wall; PM, plasma membrane; C, cytoplasm. (B) Same micrographs as in A but the plasma membrane is traced in magenta and invaginations are indicated by arrows. Numbers denote the four furrows that can be followed in this series.
To confirm that the Seg1-GFP rods seen in Seg1-overproducing cells by light microscopy and the elongated furrows observed in these cells by electron microscopy represent the same cellular structures, we used immunoelectron microscopy. We found that Seg1-GFP indeed still localized to plasma membrane invaginations (Fig. 7 C; also see Fig. S4). Grazing sections, which afford a top view of the cell surface, provided particularly clear evidence for both the elongation of plasma membrane furrows by Seg1-GFP overexpression and their specific labeling with an anti-GFP antibody (Fig. 7 D).
We also analyzed the plasma membrane morphology of pil1Δ cells overproducing Seg1-GFP, which display thick Seg1-GFP rods (Fig. 6). Accordingly, electron microscopy revealed large plasma membrane invaginations that were wider and much deeper than those in Seg1-GFP–overproducing wild-type cells (Fig. 7 E). Immunoelectron microscopy confirmed that Seg1-GFP localized to these invaginations (Fig. 7 F). Intriguingly, Seg1-GFP was typically seen adjacent to the neck of these large invaginations, which may reflect a role for Seg1 in the inward bending of the plasma membrane.
In conclusion, Seg1-GFP–overproducing cells generate Seg1 rods that contain other eisosome components and shape the plasma membrane into elongated but otherwise normal furrows. These findings suggest that Seg1 rods are neither random aggregates nor eisosome remnants but true eisosomes, albeit with an altered shape. Thus, Seg1 specifically controls the geometry of eisosomes by determining their length.
Seg1-like Sle1 is required for filamentous eisosomes in Schizosaccharomyces pombe
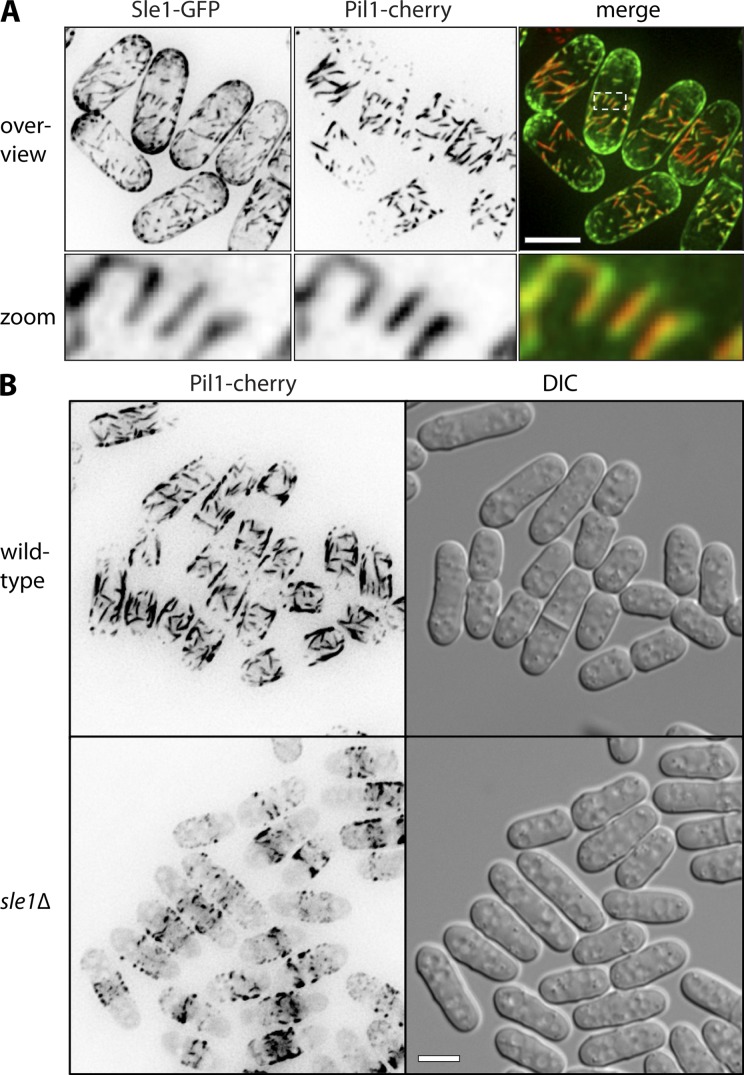
The elongated eisosomes resulting from Seg1 overexpression are reminiscent of fission yeast eisosomes, which appear as elongated filaments (Kabeche et al., 2011). We therefore wondered if a Seg1-like protein in fission yeast might facilitate the assembly of elongated eisosomes in these cells. We could not identify any fission yeast gene with clear sequence homology to S. cerevisiae SEG1, but we examined the uncharacterized gene SPAC1A6.07 for two reasons. First, SPAC1A6.07 is a large coiled-coil protein with a polybasic C terminus (Fig. S5 A). Second, a fragment of this protein localized to eisosome-like structures in a large-scale localization study (Ding et al., 2000). We confirmed that SPAC1A6.07 is an eisosome protein as judged by colocalization with Pil1-cherry in the middle of the cells, where mature filamentous eisosomes are found (Fig. 9 A). SPAC1A6.07 was also present at the cell tips. We mapped the eisosome-targeting domain of SPAC1A6.07 to an N-terminal region that is necessary and sufficient for colocalization with Pil1. In the absence of this region, the polybasic C terminus is required for general plasma membrane localization (Fig. S5 B). Thus, SPAC1A6.07 contains separate eisosome and plasma membrane targeting domains. Based on these similarities to S. cerevisiae Seg1, we have renamed this protein Sle1, for Seg1-like eisosome protein 1.
Figure 9.
Sle1/SPAC1A6.07 is an S. pombe eisosome protein required for filamentous eisosomes. (A) Colocalization of Sle1 and Pil1. Images are inverted maximum projections from deconvolved z planes in the top half of cells. (B) Localization of Pil1-cherry in wild-type and sle1Δ cells. Images are inverted projections as in A. Bars, 5 µm.
If Sle1 functions in eisosome length control in S. pombe, its ablation would be expected to shorten eisosomes. Indeed, Pil1-cherry filaments were disrupted in sle1Δ cells, showing that proper assembly of elongated eisosomes requires Sle1 (Fig. 9 B). Thus, Sle1 appears to function in S. pombe in a similar manner as Seg1 in S. cerevisiae, which suggests that basic features of eisosome biogenesis and architecture have been conserved between the two yeasts, despite their evolutionary divergence more than 1 billion years ago (Heckman et al., 2001).
Discussion
We have shown that Seg1 is required for proper eisosome assembly, that it precedes Pil1/Lsp1 during the formation of eisosomes, and that Seg1 levels determine eisosome length. We propose that the membrane domains generated by Seg1 serve as assembly platforms for Pil1/Lsp1, which are then converted into mature eisosomes. Hence, eisosomes arise through the coordinated assembly of mutually dependent components. Without Pil1, aberrant eisosome remnants form. Without Seg1, eisosomes assemble less efficiently and contain less Pil1. Thus, Seg1 also helps to determine the previously postulated minimum size of normal eisosomes (Moreira et al., 2009).
How could Seg1 facilitate eisosome assembly? One possibility is that Seg1 regulates Pil1 phosphorylation. Nce102 controls Pkh1/2 kinases, which can phosphorylate Pil1 on multiple sites, causing eisosome disassembly (Walther et al., 2007; Fröhlich et al., 2009). We tested the role of Seg1 in the Nce102–Pkh1/2–Pil1 phosphorylation pathway by disrupting SEG1 in cells expressing Pil1(4A)-GFP as the only copy of Pil1. If eisosome disassembly in seg1Δ cells were caused by increased Pil1 phosphorylation, nonphosphorylatable Pil1(4A) eisosomes should be resistant to SEG1 disruption. However, Pil1(4A), like wild-type Pil1, was partially cytoplasmic in the absence of Seg1 (unpublished data). Therefore, Seg1 is not a regulator of Pil1 phosphorylation at previously identified sites. A second possibility is that Seg1 links Pil1/Lsp1 to the plasma membrane. However, cells expressing only truncated Seg1Δ942, which cannot associate with the plasma membrane without Pil1/Lsp1, show a normal steady-state distribution of Pil1 (Fig. 4 A). Therefore, Seg1 is not a tether for Pil1/Lsp1, and its C terminus is not strictly necessary for eisosome assembly. The lipid-binding C terminus does, however, ensure the early presence of Seg1 at sites of eisosome formation and makes the generation of eisosomes more efficient, possibly by restraining aberrant assembly without the participation of Seg1. A third possibility is that Seg1 remodels the plasma membrane to assist eisosome assembly. The elongated furrows produced by overexpressed Seg1 suggest that Seg1 can induce membrane bending. Pil1/Lsp1 alone are able to bind and tubulate liposomes in vitro (Karotki et al., 2011), but the generation of membrane furrows in vivo may involve additional proteins. An attractive speculation is that Seg1 initiates plasma membrane invagination and in this way prepares the deposition of Pil1/Lsp1. The subsequent assembly of the Pil1/Lsp1 lattice, which forms a half cylinder (Karotki et al., 2011), would exert a constricting force and give the membrane its final shape. Without prior membrane remodeling by Seg1, Pil1/Lsp1 may produce less stable eisosomes, resulting in the observed partial localization of Pil1 to the cytoplasm. This scenario is consistent with work on the A. gossypii Seg1, which is dispensable for the initial targeting of Pil1 to regions of eisosome formation but required for its sustained membrane association (Seger et al., 2011). How Seg1 specifically controls eisosome length remains to be discovered but may involve Seg1 polymers that serve as a ruler. Furthermore, there must be additional morphogenic factors because irregularly shaped plasma membrane invaginations persist in a quadruple mutant lacking Pil1, Lsp1, Seg1, and Seg2 (unpublished data).
The yeast gene most closely related to SEG1 is SEG2/YKL105C. Like SEG1, SEG2 encodes a large coiled-coil protein with a polybasic C terminus. The Seg2 protein directly or indirectly interacts with Seg1 (Fig. 2 D). Similar to Seg1-GFP, Seg2-GFP localizes to eisosomes and requires the basic C terminus of Seg2 for plasma membrane association in the absence of Pil1/Lsp1 (unpublished data). Nevertheless, we found that disruption of SEG2 does not impair eisosome assembly and only slightly exacerbates the seg1 mutant phenotype. Furthermore, Seg2 protein levels are 10-fold lower than those of Seg1, or 100-fold lower than those of Pil1. Thus, Seg2 is an eisosome component but likely plays only a minor role in eisosome assembly.
Our study extends the intriguing similarities between eisosomes and caveolae. Until recently, the caveolins (caveolin-1/2/3) were thought to be the sole structural proteins of caveolae. Caveolins assume hairpin structures in the membrane, assemble into large protein lattices, and shape cholesterol/sphingolipid-rich membranes into cuplike caveolae by wedging and scaffolding (Shibata et al., 2009). This picture has become more complex with the discovery of the cavins (cavin-1/2/3/4; Hansen and Nichols, 2010). Cavins are cytosolic coiled-coil proteins that form large complexes with one another, contain polybasic regions, and bind phosphatidylserine (Burgener et al., 1990; Hill et al., 2008; Bastiani et al., 2009). Interestingly, caveolins cluster phosphatidylserine (Wanaski et al., 2003) and may thereby create multivalent binding platforms for the cavins. Depleting or removing cavin-1 or cavin-2 causes loss of caveolae (Hill et al., 2008; Liu et al., 2008; Hansen et al., 2009; McMahon et al., 2009). In the absence of cavin-1, caveolin-1 diffuses in the plasma membrane, indicating that cavins immobilize caveolins at invaginated caveolar membranes (Hill et al., 2008). Cavin-2 overexpression induces long plasma membrane tubules (Hansen et al., 2009). Caveolin-1 overexpression also causes tubule formation, which can be suppressed by raising cavin-1 levels (Verma et al., 2010). Thus, proper caveola morphology depends on the balance between caveolins and cavins. During caveola biogenesis, caveolin complexes arrive at the plasma membrane first, where they organize domains rich in cholesterol, sphingolipids, and possibly phosphatidylserine. Incipient caveolae are then stabilized by cavin complexes (Hayer et al., 2010). Finally, Pacsin 2, a BAR domain protein, has recently been found to participate in caveola biogenesis (Hansen et al., 2011; Senju et al., 2011).
These new findings reveal principles of construction that are shared by caveolae and eisosomes. Both domains consist of characteristic plasma membrane invaginations coated with heteromultimeric protein scaffolds. Both caveolae and eisosomes self-assemble in a stepwise fashion, with caveolins and Seg1 arriving first, followed by cavins and Pil1/Lsp1. Generation of the proper plasma membrane shape requires balanced levels of mutually dependent components in both cases, as is evident from the contorted morphologies produced by overexpression of cavin-2, caveolin-1, or Seg1. In addition, caveolar and eisosome shape generation involves BAR domain proteins, namely Pacsin 2 and Pil1/Lsp1. Finally, both caveolae and eisosomes are domains rich in sterols and sphingolipids and may use negatively charged lipids, such as phosphatidylserine and phosphatidylinositol-4,5-bisphosphate (Fujita et al., 2009), for the recruitment of some of their protein components, including cavins and Seg1. Caveolae and eisosomes therefore represent a remarkable example of convergent evolution, in which unrelated proteins assemble into corresponding structures by means of strikingly similar architectural principles.
How the form of eisosomes relates to their functions remains to be resolved. Paradoxically, eisosomes have been proposed to act as endocytic portals similar to caveolae (Walther et al., 2006), to constitute membrane domains protected from endocytosis (Grossmann et al., 2008), and to have no role in endocytosis at all (Brach et al., 2011). The elongated and easily visible eisosomes generated by Seg1 overexpression may prove useful in investigating the controversial spatial organization of yeast endocytosis. We anticipate that our still limited understanding of eisosome function will improve rapidly as we elaborate new ways of manipulating eisosome architecture.
Materials and methods
S. cerevisiae strains
Strains used in this study are listed in Table S1. Most chromosomal integrations and replacements were introduced by homologous recombination using PCR products (Longtine et al., 1998; Janke et al., 2004). To generate strains expressing Seg1-GFP from the URA3 locus, the SEG1-GFP coding sequence including 536 upstream base pairs was PCR-amplified from strain KEM130 and cloned between the SacI and HindIII sites of pRS306 (Sikorski and Hieter, 1989). The resulting vector pRS306-Seg1-GFP was integrated into the URA3 gene.
S. cerevisiae culture
Strains were cultured at 30°C in complete synthetic (SC) medium with 2% dextrose. For labeling with light and heavy lysine, cells were grown overnight for at least 10 doubling times in 100 ml of SC medium containing 30 mg/liter normal l-lysine or l-lysine-U-13C6, 15N2, respectively, until cultures had reached OD600 = 0.7. For induction of copper-controlled expression, strains were grown to early log phase (OD600 = 0.2–0.3) and diluted into medium containing up to 900 µM CuSO4 such that they reached early log phase again after overnight culture.
Western blotting
Strains were grown to mid log phase (OD600 = 0.5); cell lysates were prepared in 8 M urea, 2% SDS, and 50 mM Hepes, pH 7.4; and protein concentrations were determined by bicinchoninic acid protein assay (Thermo Fisher Scientific). Equal amounts of protein were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. GFP fusion proteins were detected with mouse anti-GFP antibody 7.1/13.1 (Roche). Pgk1 was detected with mouse anti-Pgk1 antibody 22C5 (Invitrogen). After incubation with primary antibodies, membranes were probed with alkaline phosphatase–conjugated secondary antibodies (EMD Millipore) and incubated with enhanced chemifluorescence substrate (GE Healthcare). Fluorescence was detected and bands were quantified with a Typhoon 9400 variable mode imager equipped with Image Quant software (GE Healthcare).
Proteomics
Protein extraction, affinity purification, sample processing, and mass spectrometry were performed as described previously (Aguilar et al., 2010). In brief, equivalent amounts of protein from wild-type cells (strain TWY70) labeled with normal light l-lysine and Seg1-TEV-GFP cells (strain TWY1118) labeled with heavy l-lysine-U-13C6, 15N2 were incubated with anti-GFP antibody conjugated to magnetic nanobeads (Miltenyi Biotech). Bound proteins were eluted by tobacco etch virus (TEV) protease cleavage. Eluates from the two strains were mixed, reduced, alkylated, and digested with endoproteinase LysC. The resulting peptide mixtures were separated by HPLC and analyzed using an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific).
Light microscopy
Strains were grown to mid-log phase and cells were mounted onto coverslips coated with Concanavalin A. Images were taken at room temperature on a laser-scanning confocal microscope (LSM510; Carl Zeiss) and an inverted microscope (TE2000U; Nikon) with a Yokogawa CSU22 spinning disk confocal from Solamere Technology (provided by the Nikon Imaging Center, University of California, San Francisco, CA), controlled by Micro-manager (Edelstein et al., 2010), or a Deltavision Imaging System (Applied Precision; Kabeche et al., 2011). Images were processed using ImageJ software. Cytoplasmic and eisosomal Pil1-GFP fluorescence were quantified according to Fröhlich et al. (2009). Bud surface areas were quantified from confocal stacks according to Moreira et al. (2009). Buds were treated as spheroids, and bright field images capturing the middle of a bud were used to measure bud length (the distance from bud neck to bud tip) and width. Surface area was calculated using S = 2πa2 + 2π(ab/e)sin−1 e, where a is bud length, b is bud width, and e = [√]1 − (b2/a2). The number of GFP patches per bud was determined from 3D reconstructions generated from fluorescent images from the same confocal stacks. The number of patches was plotted against bud surface area and data were fitted using a biphasic model that assumes a lag phase followed by a linear increase of patch number with bud size. The two fitted parameters were the critical bud size for patch formation, which marks the end of the lag phase, and the slope of the subsequent increase. To visualize ergosterol, cells were washed with 50 mM potassium phosphate, pH 5.5, stained with 2 µg/ml filipin (Sigma-Aldrich) for 5 min, washed again, and imaged at room temperature with a wide-field microscope (Axiovert 200M; Carl Zeiss).
Electron microscopy
For regular electron microscopy, strains were grown to early log phase in yeast extract peptone dextrose (YPD) medium containing 1% dextrose. Cells were processed as described previously (Schuck et al., 2009). In brief, cells were harvested by filtration, rapidly frozen using an EM PACT high-pressure freezer (Leica), freeze substituted in fixative (1% osmium tetroxide, 0.1% uranyl acetate, and 3% water in acetone) using an EM AFS2 freeze substitution system (Leica), and embedded in epon resin. 50–90-nm-thin sections were cut, stained with uranyl acetate and Reynold’s lead citrate, and viewed with a transmission electron microscope (Tecnai 12; FEI). For immunoelectron microscopy, strains were grown to mid-log phase in YPD medium containing 2% dextrose, concentrated by filtration, chemically fixed, treated with periodic acid, embedded in gelatin, and infused with sucrose according to Griffith et al. (2008). Blocks were frozen in liquid nitrogen, and 75-nm-thin cryo-sections were cut with a cryo-ultramicrotome (Ultracut UCT with EM FCS; Leica) at −110°C and placed on Formvar-coated nickel grids. For immunolabeling, sections were incubated with polyclonal rabbit anti-GFP antibodies (Abcam), followed by incubation with protein A-10 nm gold (CMC, Universitair Medisch Centrum Utrecht). After contrasting with 0.4% (wt/vol) uranyl acetate in 2 M methyl-cellulose and embedding in the same solution, sections were examined with a transmission electron microscope (CM120; Philips).
Liposome binding assay
The 20 C-terminal amino acids of Seg1 were cloned into pGEX-pP-2 (GE Healthcare). The resulting GST-Seg1(941–960) fusion protein was expressed in E. coli strain BL21DE3RIPL by IPTG induction, purified over a glutathione-Sepharose column in buffer A (150 mM sodium chloride, 50 mM Tris, pH 7.6, 2.5% glycerol, 3 mM β-mercaptoethanol, and 1 mM PMSF) and concentrated on a S200 Superdex column (GE Healthcare). Lipids (Avanti Polar Lipids, Inc.) were mixed (pure phosphatidylcholine, or phosphatidylcholine with 1.5% PIP2, 30% phosphatidylserine, or 30% phosphatidic acid), dried under an argon stream, dissolved in buffer A at 9 mM, subjected to five freeze–thaw cycles, and extruded at 65°C through a 200-nm pore-size polycarbonate filter using a mini extruder (Avanti Polar Lipids, Inc.). GST-Seg1(941–960) or GST (Sigma-Aldrich) at 3 µM were incubated in the presence or absence of 4 mM liposomes in 40 µl buffer A at room temperature for 20 min. Samples were centrifuged with an OptimaTXL ultracentrifuge (Beckman) using a TLA.100 rotor at 47,000 rpm at 4°C for 30 min. Supernatants and pellets were collected, adjusted to equal volumes, and analyzed by SDS-PAGE and Coomassie blue staining.
S. pombe strains and techniques
Standard S. pombe media and methods were used (Moreno et al., 1991). Gene tagging and deletion were performed using PCR and homologous recombination (Bähler et al., 1998). Strains JM1262 (pil1-cherry::NATR h−) and JM1467 (sle1Δ::KANR pil1-cherry::NATR leu1-32) were used in this study. For localization of Sle1 constructs, the coding sequence was subcloned into pREP41 containing a C-terminal GFP tag, and the resulting plasmids were transformed into strain JM1467. Expression was induced by growth in minimal medium lacking thiamine for 20 h before imaging.
Online supplemental material
Fig. S1 shows electron micrographs of serial thin sections of wild-type and seg1Δ cells. Fig. S2 shows Pil1-GFP and Seg1-GFP levels in Seg1-overexpression strains. Fig. S3 shows localization of Lsp1-cherry and ergosterol to Seg1-GFP rods. Fig. S4 shows immunogold labeling of GFP in cells overexpressing Seg1-GFP. Fig. S5 shows domain analysis of S. pombe Sle1. Table S1 list the S. cerevisiae strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201202097/DC1.
Supplementary Material
Acknowledgments
We thank Mei-Lie Wong and Jon Mulholland for help with electron microscopy, Kurt Thorn at the Nikon Imaging Center at University of California, San Francisco, for help with light microscopy, Matthias Mann for providing the instruments for mass spectrometric analysis, Katja Gotthard for help with protein purification, Lena Karotki for help with spin-down assays, and Martin Kampmann and Hana El-Samad for help with curve fitting. We are grateful to Blanche Schwappach for support and to Dietmar Riedel and Dirk Wenzel at the Max Planck for Biophysical Chemistry Göttingen for providing the equipment for immuno-EM.
S. Schuck was supported by a postdoctoral fellowship from the Human Frontier Science Program. T.C. Walther and F. Fröhlich acknowledge support from the German Research Council (DFG) and the Minna-James-Heineman Foundation. J.B. Moseley is supported by grants from the National Institutes of Health (P30GM092357) and the American Cancer Society (#IRG-82-003-26), and is a Pew Scholar in the Biomedical Sciences. This work was supported by grants to P. Walter from the National Institutes of Health (R01GM32384). P. Walter is an Investigator of the Howard Hughes Medical Institute.
References
- Aguilar P.S., Fröhlich F., Rehman M., Shales M., Ulitsky I., Olivera-Couto A., Braberg H., Shamir R., Walter P., Mann M., et al. 2010. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat. Struct. Mol. Biol. 17:901–908 10.1038/nsmb.1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Bastiani M., Parton R.G. 2010. Caveolae at a glance. J. Cell Sci. 123:3831–3836 10.1242/jcs.070102 [DOI] [PubMed] [Google Scholar]
- Bastiani M., Liu L., Hill M.M., Jedrychowski M.P., Nixon S.J., Lo H.P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M.R., et al. 2009. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185:1259–1273 10.1083/jcb.200903053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach T., Specht T., Kaksonen M. 2011. Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J. Cell Sci. 124:328–337 10.1242/jcs.078519 [DOI] [PubMed] [Google Scholar]
- Burgener R., Wolf M., Ganz T., Baggiolini M. 1990. Purification and characterization of a major phosphatidylserine-binding phosphoprotein from human platelets. Biochem. J. 269:729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F., Barral Y. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell. 16:493–506 10.1016/j.devcel.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Deng C., Xiong X., Krutchinsky A.N. 2009. Unifying fluorescence microscopy and mass spectrometry for studying protein complexes in cells. Mol. Cell. Proteomics. 8:1413–1423 10.1074/mcp.M800397-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.Q., Tomita Y., Yamamoto A., Chikashige Y., Haraguchi T., Hiraoka Y. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 5:169–190 10.1046/j.1365-2443.2000.00317.x [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:Unit 14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich F., Moreira K., Aguilar P.S., Hubner N.C., Mann M., Walter P., Walther T.C. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185:1227–1242 10.1083/jcb.200811081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Cheng J., Tauchi-Sato K., Takenawa T., Fujimoto T. 2009. A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA. 106:9256–9261 10.1073/pnas.0900216106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Griffith J., Mari M., De Mazière A., Reggiori F. 2008. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 9:1060–1072 10.1111/j.1600-0854.2008.00753.x [DOI] [PubMed] [Google Scholar]
- Grossmann G., Opekarová M., Novakova L., Stolz J., Tanner W. 2006. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot. Cell. 5:945–953 10.1128/EC.00206-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G., Opekarová M., Malínsky J., Weig-Meckl I., Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 10.1038/sj.emboj.7601466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G., Malínsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W.B., Opekarová M., Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183:1075–1088 10.1083/jcb.200806035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Nichols B.J. 2009. Molecular mechanisms of clathrin-independent endocytosis. J. Cell Sci. 122:1713–1721 10.1242/jcs.033951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Nichols B.J. 2010. Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol. 20:177–186 10.1016/j.tcb.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Hansen C.G., Bright N.A., Howard G., Nichols B.J. 2009. SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 11:807–814 10.1038/ncb1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Howard G., Nichols B.J. 2011. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 124:2777–2785 10.1242/jcs.084319 [DOI] [PubMed] [Google Scholar]
- Hayer A., Stoeber M., Bissig C., Helenius A. 2010. Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic. 11:361–382 10.1111/j.1600-0854.2009.01023.x [DOI] [PubMed] [Google Scholar]
- Heckman D.S., Geiser D.M., Eidell B.R., Stauffer R.L., Kardos N.L., Hedges S.B. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science. 293:1129–1133 10.1126/science.1061457 [DOI] [PubMed] [Google Scholar]
- Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 132:113–124 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kabeche R., Baldissard S., Hammond J., Howard L., Moseley J.B. 2011. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol. Biol. Cell. 22:4059–4067 10.1091/mbc.E11-07-0605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karotki L., Huiskonen J.T., Stefan C.J., Ziółkowska N.E., Roth R., Surma M.A., Krogan N.J., Emr S.D., Heuser J., Grünewald K., Walther T.C. 2011. Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 195:889–902 10.1083/jcb.201104040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Liu L., Brown D., McKee M., Lebrasseur N.K., Yang D., Albrecht K.H., Ravid K., Pilch P.F. 2008. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 8:310–317 10.1016/j.cmet.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Luo G., Gruhler A., Liu Y., Jensen O.N., Dickson R.C. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444 10.1074/jbc.M709972200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská K., Malínský J., Opekarová M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 14:4427–4436 10.1091/mbc.E03-04-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská K., Malínsky J., Opekarová M., Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117:6031–6041 10.1242/jcs.01493 [DOI] [PubMed] [Google Scholar]
- Malínsky J., Opekarová M., Tanner W. 2010. The lateral compartmentation of the yeast plasma membrane. Yeast. 27:473–478 10.1002/yea.1772 [DOI] [PubMed] [Google Scholar]
- McMahon K.A., Zajicek H., Li W.P., Peyton M.J., Minna J.D., Hernandez V.J., Luby-Phelps K., Anderson R.G. 2009. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 28:1001–1015 10.1038/emboj.2009.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor H., Mühlethaler K. 1963. Fine structure of frozen-etched yeast cells. J. Cell Biol. 17:609–628 10.1083/jcb.17.3.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira K.E., Walther T.C., Aguilar P.S., Walter P. 2009. Pil1 controls eisosome biogenesis. Mol. Biol. Cell. 20:809–818 10.1091/mbc.E08-03-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Nakada C., Ritchie K., Oba Y., Nakamura M., Hotta Y., Iino R., Kasai R.S., Yamaguchi K., Fujiwara T., Kusumi A. 2003. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5:626–632 10.1038/ncb1009 [DOI] [PubMed] [Google Scholar]
- Olivera-Couto A., Graña M., Harispe L., Aguilar P.S. 2011. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell. 22:2360–2372 10.1091/mbc.E10-12-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Simons K. 2007. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8:185–194 10.1038/nrm2122 [DOI] [PubMed] [Google Scholar]
- Schuck S., Simons K. 2004. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117:5955–5964 10.1242/jcs.01596 [DOI] [PubMed] [Google Scholar]
- Schuck S., Prinz W.A., Thorn K.S., Voss C., Walter P. 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187:525–536 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger S., Rischatsch R., Philippsen P. 2011. Formation and stability of eisosomes in the filamentous fungus Ashbya gossypii. J. Cell Sci. 124:1629–1634 10.1242/jcs.082487 [DOI] [PubMed] [Google Scholar]
- Senju Y., Itoh Y., Takano K., Hamada S., Suetsugu S. 2011. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 124:2032–2040 10.1242/jcs.086264 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Hu J., Kozlov M.M., Rapoport T.A. 2009. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25:329–354 10.1146/annurev.cellbio.042308.113324 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E., Balda M.S., Matter K. 2010. Dynamics and functions of tight junctions. Trends Cell Biol. 20:142–149 10.1016/j.tcb.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Strádalová V., Stahlschmidt W., Grossmann G., Blazíková M., Rachel R., Tanner W., Malínsky J. 2009. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 122:2887–2894 10.1242/jcs.051227 [DOI] [PubMed] [Google Scholar]
- Verma P., Ostermeyer-Fay A.G., Brown D.A. 2010. Caveolin-1 induces formation of membrane tubules that sense actomyosin tension and are inhibited by polymerase I and transcript release factor/cavin-1. Mol. Biol. Cell. 21:2226–2240 10.1091/mbc.E09-05-0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., Walter P. 2006. Eisosomes mark static sites of endocytosis. Nature. 439:998–1003 10.1038/nature04472 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Aguilar P.S., Fröhlich F., Chu F., Moreira K., Burlingame A.L., Walter P. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955 10.1038/sj.emboj.7601933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanaski S.P., Ng B.K., Glaser M. 2003. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry. 42:42–56 10.1021/bi012097n [DOI] [PubMed] [Google Scholar]
- Ziółkowska N.E., Karotki L., Rehman M., Huiskonen J.T., Walther T.C. 2011. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 18:854–856 10.1038/nsmb.2080 [DOI] [PubMed] [Google Scholar]
- Ziółkowska N.E., Christiano R., Walther T.C. 2012. Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 22:151–158 10.1016/j.tcb.2011.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.