In contrast to ATM-null mice, mice expressing a kinase-dead ATM variant exhibit embryonic lethality, associated with greater deficiency in homologous recombination.
Abstract
Ataxia telangiectasia (A-T) mutated (ATM) is a key deoxyribonucleic acid (DNA) damage signaling kinase that regulates DNA repair, cell cycle checkpoints, and apoptosis. The majority of patients with A-T, a cancer-prone neurodegenerative disease, present with null mutations in Atm. To determine whether the functions of ATM are mediated solely by its kinase activity, we generated two mouse models containing single, catalytically inactivating point mutations in Atm. In this paper, we show that, in contrast to Atm-null mice, both D2899A and Q2740P mutations cause early embryonic lethality in mice, without displaying dominant-negative interfering activity. Using conditional deletion, we find that the D2899A mutation in adult mice behaves largely similar to Atm-null cells but shows greater deficiency in homologous recombination (HR) as measured by hypersensitivity to poly (adenosine diphosphate-ribose) polymerase inhibition and increased genomic instability. These results may explain why missense mutations with no detectable kinase activity are rarely found in patients with classical A-T. We propose that ATM kinase-inactive missense mutations, unless otherwise compensated for, interfere with HR during embryogenesis.
Introduction
The ataxia telangiectasia (A-T) mutated (ATM) kinase plays a key role in the DNA damage response by phosphorylating numerous substrates that signal for DNA repair, cell cycle checkpoint activation, and apoptosis (Banin et al., 1998; Canman et al., 1998; Matsuoka et al., 2007; Bensimon et al., 2010; Olsen et al., 2010; Beli et al., 2012). The critical function of ATM is underscored by its high conservation in eukaryotes and its deficiency in the recessive, cancer-prone A-T disease (Savitsky et al., 1995). The exact mechanism by which ATM is activated in vivo still remains unclear (Pellegrini et al., 2006; Daniel et al., 2008), but one current model suggests that autophosphorylation triggers its activation (Bakkenist and Kastan, 2003; Kozlov et al., 2006, 2011; So et al., 2009). A prediction is that loss of ATM kinase activity would be equivalent to loss of the ATM protein. Indeed, patient cells reconstituted with kinase-inactive ATM (Bakkenist and Kastan, 2003) and treatment of cells with a pharmacological ATM kinase inhibitor (Hickson et al., 2004; Bredemeyer et al., 2006; Callén et al., 2007; Rainey et al., 2008; White et al., 2008) support the notion that inhibition of cellular ATM kinase activity mirrors complete ATM deficiency.
The classical form of A-T is most frequently caused by compound heterozygote-truncating mutations that result in a total loss of destabilized ATM protein (∼90% of cases; Gilad et al., 1996; Lakin et al., 1996; Li and Swift, 2000; Micol et al., 2011). Mouse models deficient in ATM have been invaluable for the study of A-T (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Difilippantonio and Nussenzweig, 2007; Lavin, 2008; Stracker and Petrini, 2011). In stark contrast to Atm-null mice (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996), we show here that single point mutations disrupting the kinase activity of ATM lead to early embryonic lethality in mice. These results reveal a previously unknown essential role for ATM kinase activity during embryonic development and provide a potential explanation for why kinase-inactive missense mutations do not constitute a significant portion of the molecular lesions in childhood A-T. Furthermore, our finding that inhibition of ATM kinase activity in vivo does not always equate to complete loss of ATM protein suggests that treatments using ATM inhibitors (ATMi’s) could be more toxic than previously anticipated.
Results and discussion
Mutations in the ATM kinase domain display severely impaired kinase activity in vitro and cause embryonic lethality in mice
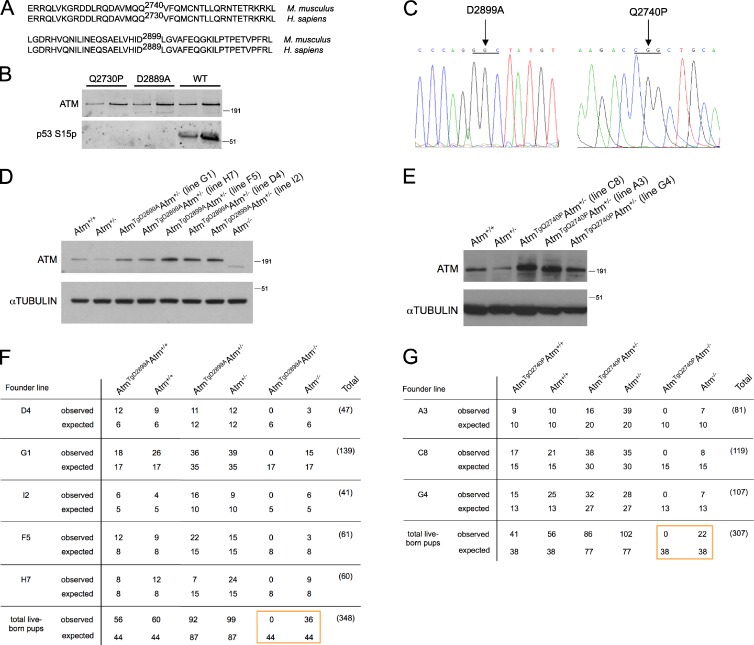
To determine whether the diverse in vivo functions of ATM are mediated solely by its kinase domain, we used bacterial artificial chromosome (BAC) recombineering to generate two kinase-inactive mouse models of ATM. To identify a single residue for mutagenesis without any high-resolution structural information for ATM, we took two approaches. In a structural modeling approach, we identified the p110 phosphoinositide 3-kinase domain as having a statistically significant protein sequence alignment with the ATM kinase domain and found that an essential catalytic aspartic acid residue in the active site of the p110 kinase structure aligns with D2889 of human ATM. Moreover, a p110 knockin mouse substituting this residue with alanine, p110δD910A/D910A, demonstrated that this mutation abolishes kinase activity in vitro and in vivo without affecting protein stability (Okkenhaug et al., 2002); therefore, we chose to generate the conserved D2899A mutation in the mouse (Fig. 1 A). For the second approach, we decided to model a rare missense variant from a patient with classical A-T, c.8189A>C (p.Gln2730Pro), whose cells displayed ATM protein expression without detectable kinase activity (Taylor and Byrd, 2005; Barone et al., 2009).
Figure 1.
ATM D2899A and Q2740P mutations display severely impaired kinase activity in vitro and cause embryonic lethality in mice. (A) Pairwise local alignments of human (Homo sapiens) and mouse (Mus musculus) ATM protein sequences surrounding sites of mutagenesis. (B) Kinase assay with 0.2 nM WT, D2889A, or Q2730P mutant dimeric human ATM, 3.6 nM MRN, 50 nM GST-p53 substrate, and linear DNA probed with an antibody to p53 S15 phosphorylation. A titration of ATM is shown for each sample. (C) Sequence chromatograms showing the amino acid mutations generated in mouse BAC RP24-122F10. (D and E) Thymocytes were harvested for lysate preparation and Western blot analysis. Molecular mass markers are in kilodaltons. (F and G) Tables showing the expected and observed genotypes of 3-wk-old pups from breeding AtmTgAtm+/− with Atm+/− mice. The boxes highlight the comparison between the total number of ATM kinase-inactive and Atm-null live-born pups.
Before generating BAC transgenic mouse models, we tested whether the D2889A and Q2730P mutations in human ATM are required for ATM kinase activity in a completely reconstituted system. Kinase assays with wild-type (WT), D2889A, and Q2730P mutant dimeric human ATM complexes were performed using p53 as a substrate (Lee and Paull, 2005). Although WT ATM dimers displayed p53 S15 phosphorylation in an MRN (MRE11-RAD50-NBS1)- and DNA-dependent manner, kinase activity of D2889A and Q2730P mutant dimers was completely abolished (Fig. 1 B). Thus, the D2889A and Q2730P mutations are required for human ATM kinase activity.
As both D2889 and Q2730 residues in human ATM are conserved in the mouse (Fig. 1 A), we recombineered the equivalent mutations in a WT BAC spanning the genomic mouse Atm locus to generate BAC transgenic mice as previously described (Pellegrini et al., 2006; Daniel et al., 2008). Both mutation sites were confirmed by sequencing (Fig. 1 C). Founder lines expressing mutant murine ATM from the BAC transgene were identified and bred to Atm+/−. Founder lines G1 and H7 of AtmTgD2899A mice expressed transgenic murine ATM near the level observed in Atm+/− mice, whereas founder lines F5, D4, and I2 overexpressed murine ATM (Fig. 1 D). For AtmTgQ2740P mice, founder line G4 expressed transgenic murine ATM near the level observed in Atm+/+ mice, whereas the C8 and A3 founder lines overexpressed ATM (Fig. 1 E). Similar ATM protein levels were observed from splenocytes and thymocytes of transgenic animals (Fig. 1, D and E; and not depicted).
We crossed AtmTgD2899AAtm+/− and AtmTgQ2740PAtm+/− mice with Atm+/− but, to our surprise, failed to generate transgenic animals on the Atm−/− background (Fig. 1, F and G). This failure to generate AtmTgAtm−/− live-born pups was observed across all founder lines of both mutations (Fig. 1, F and G) and was not the result of transgene integration on the chromosome containing endogenous Atm, as shown by FISH (Fig. S1). To begin to elucidate the stage of embryonic development that is impaired with deficient ATM kinase activity, we prepared embryos at embryonic day 9.5 (E9.5) from D2899A mutant mice. We found that AtmTgD2899AAtm−/− embryos were produced at a dramatically lower frequency than expected, indicating that D2899A mutant embryos die before E9.5 (Table 1). Transgenic animals with one or both WT copies of Atm were found at the expected frequency, indicating an absence of dominant-negative interfering activity. We conclude that the kinase-inactive ATM mutation leads to early embryonic lethality in mice.
Table 1.
Embryonic lethality of ATM D2899A mice occurs before E9.5
| Founder line | AtmTgD2899AAtm+/+ | Atm+/+ | AtmTgD2899AAtm+/− | Atm+/− | AtmTgD2899AAtm−/− | Atm−/− | Total genotyped | Absorb |
| G1 observed | 3 | 9 | 7 | 13 | 1 | 5 | 38 | 5 |
| G1 expected | 5 | 5 | 10 | 10 | 5 | 5 | ||
| H7 observed | 0 | 5 | 2 | 8 | 0 | 2 | 17 | 2 |
| H7 expected | 2 | 2 | 4 | 4 | 2 | 2 | ||
| D4 observed | 0 | 4 | 0 | 1 | 0 | 0 | 5 | 1 |
| D4 expected | 0.5 | 0.5 | 1 | 1 | 0.5 | 0.5 | ||
| F5 observed | 1 | 4 | 5 | 2 | 0 | 0 | 12 | 3 |
| F5 expected | 1.5 | 1.5 | 3 | 3 | 1.5 | 1.5 | ||
| I2 observed | 4 | 3 | 4 | 6 | 0 | 2 | 19 | 4 |
| I2 expected | 2 | 2 | 5 | 5 | 2 | 2 | ||
| Total E9.5 embryos observed | 11 | 25 | 18 | 30 | 1 | 9 | 91 | 15 |
| Total E9.5 embryos expected | 11 | 11 | 23 | 23 | 11 | 11 |
Shown are the expected and observed genotypes from breeding AtmTgD2899AAtm+/− with Atm+/− mice. Note the comparison between the total number of ATM kinase-inactive and Atm-null embryos.
We speculated that the severe defect during embryogenesis is the result of ATM kinase recruitment at DNA breaks, which may impair the function of other proteins by occluding their access to DNA damage. To support this, we found that a kinase-inactive human ATM D2870A mutant protein, expressed in cells that do not express endogenous ATM, is recruited to sites of laser-induced DNA damage (Fig. S2). Moreover, WT human ATM was similarly recruited to DNA damage sites in cells treated with the KU55933 ATMi (Fig. S2). These results are consistent with other studies in human cells showing that ATM kinase activity is dispensable for recruitment of epitope-tagged ATM to sites of DNA breaks after laser- or ionizing radiation (IR)–induced DNA damage (Barone et al., 2009; Davis et al., 2010). They are also consistent with results from Xenopus laevis egg extracts showing an increase in ATM association to damaged chromatin in the presence of caffeine or the KU55933 ATMi (You et al., 2007, 2009).
Double-stranded break (DSB)–induced activation and recruitment of ATM to chromatin is dependent on Nbs1 (Uziel et al., 2003; Difilippantonio et al., 2005). If recruitment of kinase-dead ATM to DNA breaks is toxic, we reasoned that we might be able to rescue viability by breeding with Nbs1ΔB/ΔB mice, a hypomorphic Nbs1 mutant mouse that exhibits a mild defect in ATM activation (Williams et al., 2002). However, no Nbs1ΔB/ΔBAtmTgD2899AAtm−/− mice were born (Table 2). Thus, a mutation that is predicted to reduce ATM activation and association with chromatin cannot rescue viability of ATM kinase-inactive mice.
Table 2.
Embryonic lethality of ATM D2899A (G1 founder line) mice is not rescued by disruption of Nbs1, Ku80, or 53bp1
| Genotype | AtmTgD2899AAtm+/+ | Atm+/+ | AtmTgD2899AAtm+/− | Atm+/− | AtmTgD2899AAtm−/− | Atm−/− |
| Nbs1+/+ observed | 9 | 10 | 14 | 10 | 0 | 11 |
| Nbs1+/+ expected | 7 | 7 | 15 | 15 | 7 | 7 |
| Nbs1ΔB/+ observed | 16 | 20 | 42 | 38 | 0 | 16 |
| Nbs1ΔB/+ expected | 15 | 15 | 30 | 30 | 15 | 15 |
| Nbs1ΔB/ΔB observed | 7 | 12 | 14 | 19 | 0 | 0 |
| Nbs1ΔB/ΔB expected | 7 | 7 | 15 | 15 | 7 | 7 |
| Ku80+/+ observed | 16 | 9 | 21 | 16 | 0 | 2 |
| Ku80+/+ expected | 10 | 3 | 19 | 6 | 10 | 3 |
| Ku80+/− observed | 21 | 16 | 41 | 26 | 0 | 9 |
| Ku80+/− expected | 19 | 6 | 39 | 13 | 19 | 6 |
| Ku80−/− observed | 9 | 1 | 17 | 3 | 0 | 0 |
| Ku80−/− expected | 10 | 3 | 19 | 6 | 10 | 3 |
| 53bp1−/− observed | 14 | 11 | 52 | 19 | 0 | 6 |
| 53bp1−/− expected | 19 | 6 | 38 | 13 | 19 | 6 |
Shown are the expected and observed genotypes of 3-wk-old pups from breeding Nbs1ΔB/+AtmTgD2899AAtm+/− with Nbs1ΔB/+Atm+/− mice (n = 238), intercrossing Ku80+/−AtmTgD2899AAtm+/− mice (n = 207), or intercrossing 53bp1−/−AtmTgD2899AAtm+/− mice (n = 102). Note the numbers of ATM kinase-inactive live-born pups in the different Ku80 backgrounds, the comparison between the number of ATM kinase-inactive and Atm-null live-born pups in the 53bp1-/- background, and the numbers of ATM kinase-inactive live-born pups in the different Nbs1 backgrounds.
Recently, a DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) mutant mouse model with three phosphorylation site substitutions revealed that the DNA-PKcs mutant protein causes a more severe phenotype than Dnapkcs-null mice (Zhang et al., 2011). Because DNA-PKcs has overlapping functions with ATM (Gurley and Kemp, 2001; Sekiguchi et al., 2001; Callén et al., 2009; Gapud and Sleckman, 2011), we reasoned that if ATM and DNA-PKcs are being recruited to DNA breaks and occluding access of other factors, disruption of the DNA-dependent protein kinase holoenzyme may partially alleviate the toxic effect of kinase-inactive ATM. To test this hypothesis, we crossed Ku80-null mice (Nussenzweig et al., 1996) with our kinase-inactive ATM mice. However, AtmTgD2899AAtm−/− live-born pups were not recovered in the Ku80−/− background (Table 2), suggesting that disruption of DNA-PKcs recruitment to DNA breaks does not rescue ATM kinase-inactive toxicity. Similarly, loss of 53BP1, which reduces nonhomologous end joining (NHEJ) and promotes resection of DSBs (Ward et al., 2004; Xie et al., 2007; Difilippantonio et al., 2008; Dimitrova et al., 2008; Bunting et al., 2010), also did not rescue the viability of AtmTgD2899AAtm−/− mice (Table 2).
Conditional ATM D2899A B lymphocytes display defects in development, genome stability, and sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitor
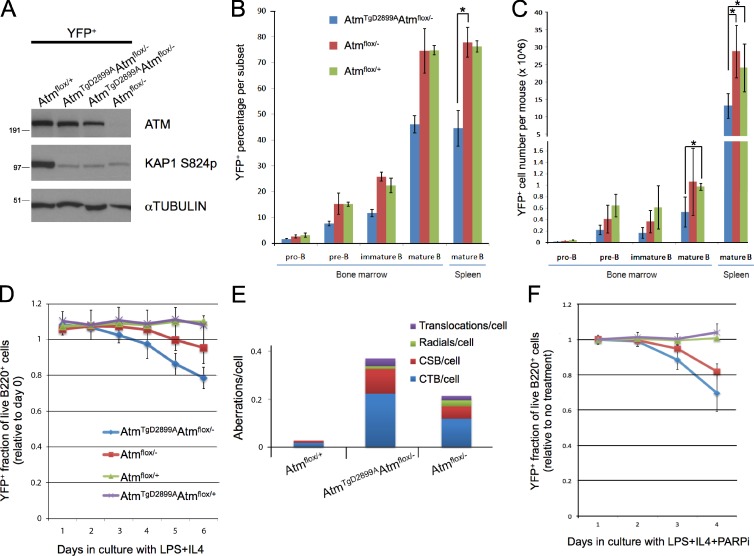
To overcome the embryonic lethality of AtmTgD2899AAtm−/− mice, we crossed a previously described conditional Atm knockout allele (Zha et al., 2008) and CD19-cre (Rickert et al., 1997) to our Atm transgenic mice for generating ATM kinase-inactive primary B cells. With this model, we investigated the effect of ATM kinase inhibition on DNA damage signaling, lymphocyte development, and genome stability. To monitor the frequency of Cre recombinase–expressing cells in these mice, we crossed in the Rosa26-stop-YFP allele (Srinivas et al., 2001) to generate Cd19cre/+Rosa26YFP/+AtmTgD2899AAtmflox/− mice (herein referred to as AtmTgD2899AAtmflox/−) and compared with littermate ATM-deficient control mice (Cd19cre/+Rosa26YFP/+Atmflox/−, herein referred to as Atmflox/−). To verify deletion efficiency, we sorted live YFP+ cells from AtmTgD2899AAtmflox/− and control splenic B cell cultures and demonstrated that, although Atmflox/− cells have no detectable ATM protein as measured by Western blotting, AtmTgD2899AAtmflox/− cells displayed levels similar to Atmflox/+ cells (Fig. 2 A). To test whether the D2899A mutation abrogates ATM kinase activity in vivo, splenic B cells were irradiated with 10 Gy, and KAP1 S824 phosphorylation was assessed by Western blotting. Although Atmflox/+ cells responded normally to γ irradiation with robust KAP1 S824 phosphorylation, AtmTgD2899AAtmflox/− and Atmflox/− B cells behaved similarly, displaying severely impaired phosphorylation (Fig. 2 A). We conclude that ATM D2899A mutant murine B cells are deficient in IR-induced ATM kinase activity in vivo similar to ATM-deficient cells.
Figure 2.
Conditional ATM D2899A mice display defects mildly more severe than ATM deficiency in B cell development, genome stability, and sensitivity to PARP inhibitor. (A) Western blot analysis of splenic B cells stimulated with LPS and RP105 for 72 h, FACS sorted for YFP expression, and harvested for lysate preparation after a 10-Gy irradiation. Shown for comparison are sorted cells from two individual AtmTgD2899AAtmflox/− mice. In addition to the labeled Atm genotype, note that mice are also Cd19cre/+Rosa26YFP/+, for which YFP expression (as a marker of Cre recombinase expression) is found in pro-B cells and becomes progressively higher through B cell development to the mature B cell stage. Molecular mass markers are in kilodaltons. (B) YFP+ frequency from freshly isolated B cells of the following subsets of bone marrow and spleen harvested and stained with antibodies to B220, CD43, and IgM for flow cytometric analysis. Pro-B (B220+CD43+IgM−), pre-B (B220+CD43−IgM−), immature B (B220+CD43−IgMint), mature B (B220+IgM+), and splenic mature B (B220+IgM+) are shown. *, P < 0.00001. (C) YFP+ absolute cell numbers from each subset in bone marrow and spleen. Legend same as in B. *, P < 0.05. (D) YFP+ fraction of live B220+ splenic B cells stimulated with LPS as a function of days in culture. Each data point represents the YFP+ frequency measured by flow cytometry, relative to day 0. For days 5 and 6, P < 0.0001 for AtmTgD2899AAtmflox/− versus Atmflox/+, and P < 0.05 for AtmTgD2899AAtmflox/− versus Atmflox/−. (E) Splenic YFP+ B cells stimulated with LPS, IL4, and RP105 for 72 h were harvested for metaphase chromosome preparation. FISH was performed on slides with a probe for telomeres and counterstained with DAPI. Frequency of aberrations per metaphase is shown for chromatid breaks (CTB), chromosome breaks (CSB), radials, and translocations (Atmflox/+: n = 105 from 1 mouse; AtmTgD2899AAtmflox/−: n = 313 from 3 mice; Atmflox/−: n = 200 from 2 mice). (F) YFP+ fraction of live B220+ splenic B cells stimulated with LPS and 1 µM PARP inhibitor. Each data point represents the YFP+ frequency measured by flow cytometry relative to no treatment of the same genotype. Legend same as in D. On days 3 and 4, P < 0.05 for AtmTgD2899AAtmflox/− versus Atmflox/−. Error bars represent standard deviation. PARPi, PARP inhibitor.
To assess the role of ATM kinase activity during B cell development, we performed flow cytometric analyses on freshly isolated bone marrow and splenocytes. AtmTgD2899AAtmflox/− mice displayed significant decreases in the frequency and number of YFP+ cells in developing and mature B cell subsets in bone marrow and spleen compared with the similar frequencies observed between Atmflox/− and Atmflox/+ mice (Fig. 2, B and C; and Fig. S3). Thus, different from ATM-deficient cells, Cre-expressing YFP+ cells with the D2899A mutant form of ATM as their sole ATM species are out competed in vivo by cells that express a WT copy of Atm. To test whether the D2899A mutation causes defects in the overall fitness of stimulated cells, we activated splenic B cells in culture with lipopolysaccharide (LPS) and IL4 and monitored YFP frequency of B220+ cells over time by flow cytometry. Although YFP frequency in live Atmflox/+ B cells remained constant over 6 d in culture, AtmTgD2899AAtmflox/− B cells displayed a reproducible decrease in YFP frequency, with Atmflox/− B cells showing an intermediate phenotype (Fig. 2 D). AtmTgD2899AAtmflox/+ B cells expressing WT ATM showed similar results as Atmflox/+ B cells (Fig. 2 D), supporting an absence of dominant-negative interfering activity in the D2899A mutant. Thus, although the D2899A point mutation in mice leads to severe defects in embryonic development, defects in B lymphocyte development and activation are only mildly more severe than ATM-deficient controls.
Chromosomal instability is a hallmark of both human and murine ATM-deficient cells (Xu et al., 1996; Shiloh, 2003; Callén et al., 2007) and is also a rapid consequence of treatment of WT cells with ATM kinase inhibitors (Hickson et al., 2004; Bredemeyer et al., 2006; Callén et al., 2007; Rainey et al., 2008; White et al., 2008). To test whether genetic ablation of the murine ATM kinase causes defects in genome stability, metaphase chromosome spreads of YFP+ cells sorted from AtmTgD2899AAtmflox/− and control B cell cultures were analyzed for chromosome abnormalities. As expected, ATM-deficient cells displayed a higher number of aberrations per cell compared with controls (Fig. 2 E). AtmTgD2899AAtmflox/− cells showed a 1.7-fold increase in aberrations per cell compared with ATM-deficient cells, marked by chromosome breaks and chromatid breaks (Fig. 2 E). We conclude that ATM D2899A mutant murine B cells exhibit more genomic instability than ATM-deficient control cells.
The increase in chromatid breaks observed in D2899A mutant cells suggested that these cells may have more severely impaired homologous recombination (HR). To test this, we used flow cytometry to assess the YFP frequency of live B220+ cells cultured with PARP inhibitor. Relative to the YFP frequency measured with no drug treatment, this assay recapitulated the reported sensitivity of ATM-deficient human cells to PARP inhibitor (Fig. 2 F; Bryant and Helleday, 2006; McCabe et al., 2006). Using YFP as a marker for Cre-expressing cells in competition with nondeleted cells, our analyses indicated that AtmTgD2899AAtmflox/− cells were mildly more sensitive to PARP inhibitor treatment compared with ATM-deficient cells (Fig. 2 F), suggesting a more significant defect in HR. We conclude that D2899A mutant murine B cells display more severe defects in genome stability and PARP inhibitor sensitivity compared with ATM-deficient cells.
Conditional ATM D2899A B lymphocytes display defects in variable, diversity, and joining (V(D)J) recombination and immunoglobulin class switching
Deficiency in murine ATM leads to impaired DNA end joining in lymphocytes undergoing V(D)J recombination (Bredemeyer et al., 2006). To test whether the D2899A mutation causes similar V(D)J recombination defects, we generated v-Abl (viral Abl) kinase-transformed pre-B cell lines (herein referred to as abl pre-B cells) from bone marrow of AtmTgD2899AAtmflox/− mice expressing an Eμ-Bcl2 transgene. These cells were transiently transfected with Cre recombinase to generate AtmTgD2899AAtmΔ/− abl pre-B cells and retrovirally infected with the pMX-INV recombination substrate to generate AtmTgD2899AAtmΔ/−:INV abl pre-B cells. To induce recombination activating gene (RAG)–mediated DSBs, Atm+/+:INV, AtmTgD2899AAtmΔ/−:INV, and Atm−/−:INV abl pre-B cells were treated with the Abl kinase inhibitor STI571. Southern blot analysis of pMX-INV rearrangement revealed that the defects in V(D)J recombination in AtmTgD2899AAtmΔ/−:INV abl pre-B cells were similar to those observed in Atm−/−:INV abl pre-B cells. Specifically, there was a decrease in normal coding joint formation with an accumulation of unrepaired coding ends and an increase in hybrid joint formation (Fig. S3). We conclude that ATM D2899A mutant murine lymphocytes display V(D)J recombination defects similar to Atm-null lymphocytes.
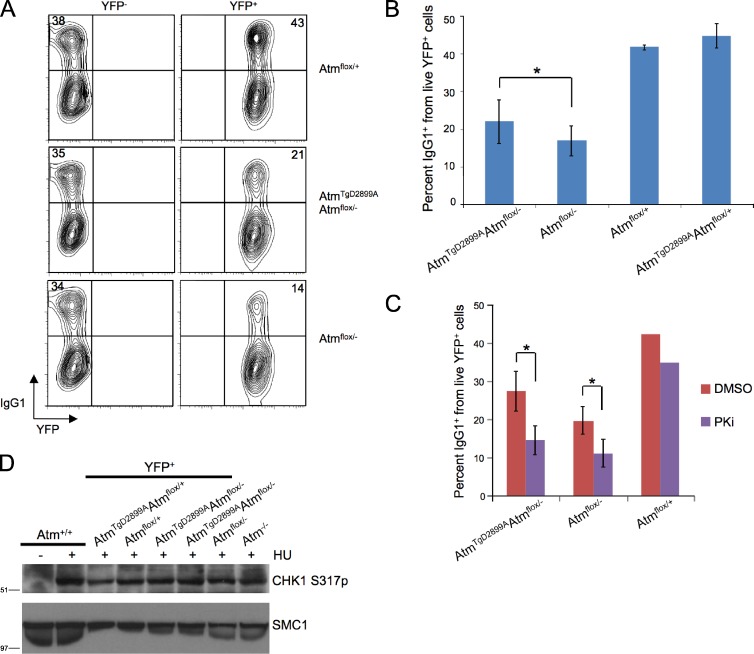
During an immune response, physiological DSBs at the IgH locus occur in B lymphocytes during class switch recombination (CSR; Stavnezer et al., 2008). ATM is required for efficient CSR and has been proposed to play a role as a DNA damage response factor in the synapsis of two broken switch regions (Pan et al., 2002; Lumsden et al., 2004; Reina-San-Martin et al., 2004). To test whether the D2899A mutation affects CSR, splenic B cells from AtmTgD2899AAtmflox/− and control mice were stimulated with LPS and IL4, and the frequency of YFP+ cells positive for surface IgG1 was assessed by flow cytometry. Consistent with previous studies using germline Atm−/− mice (Lumsden et al., 2004; Reina-San-Martin et al., 2004), B cells from Atmflox/− mice with CD19-cre led to a 2.5-fold decrease in IgG1 CSR, relative to the IgG1+ frequency in YFP+ cells from Atmflox/+ mice (Fig. 3, A and B). B cells from AtmTgD2899AAtmflox/− mice displayed a 1.9-fold defect in IgG1 CSR, albeit less severe than ATM-deficient controls (Fig. 3, A and B). We conclude that ATM D2899A mutant B cells display defects in CSR that are mildly less severe than ATM-deficient controls.
Figure 3.
Conditional ATM D2899A B cells display defects in immunoglobulin class switching without impairing DNA-PKcs or ATR. (A) Flow cytometric analysis of IgG1 expression on B cells stimulated with LPS and IL4 for 5 d. (B) Mean percentage of IgG1 class switching from multiple flow cytometric analyses as in B (*, P < 0.05). (C) Mean percentage of IgG1 class switching of YFP+ splenic B cells treated with DNA-PKcs inhibitor NU7026 (PKi) or DMSO (*, P < 0.05). (D) Splenic B cells stimulated with LPS and RP105 for 72 h were treated with 2 mM hydroxyurea (HU) for 2 h, FACS sorted for YFP expression, and harvested for lysate preparation and Western blot analysis. Shown for comparison are sorted cells from two individual AtmTgD2899AAtmflox/− mice along with Atm+/+ and Atm−/− unsorted B cells. Molecular mass markers are in kilodaltons. Error bars represent standard deviation.
DNA-PKcs and A-T and Rad3 related (ATR) are functional in conditional ATM D2899A B cells
Because DNA-PKcs and ATM have redundant roles in development and CSR (Gurley and Kemp, 2001; Sekiguchi et al., 2001; Callén et al., 2009), we directly tested whether DNA-PKcs kinase activity is functional during CSR in D2899A mutant B cells. To this end, we stimulated AtmTgD2899AAtmflox/− and control cells to undergo CSR in the presence or absence of the DNA-PKcs inhibitor NU7026 (Callén et al., 2009). In contrast to Atmflox/+ cells, inhibiting DNA-PKcs kinase activity in either AtmTgD2899AAtmflox/− or Atmflox/− B cells resulted in a nearly twofold reduction in IgG1 CSR (Fig. 3 C), a decrease consistent with PKcs inhibitor treatment of germline Atm−/− B cells (Callén et al., 2009). We conclude that DNA-PKcs is functional in conditional ATM D2899A mutant B cells.
A different phosphoinositide 3-kinase–like kinase, ATR, is required for cell cycle progression and genome stability during replicative stress (Brown and Baltimore, 2003; López-Contreras and Fernandez-Capetillo, 2010), and there is evidence that ATM can regulate ATR function. For example, ATM is required for ATR activation and CHK1 phosphorylation in response to IR; however, ATR-dependent CHK1 phosphorylation in response to replication-associated DNA damage occurs independently of ATM (Adams et al., 2006; Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006). To test whether ATR is functional in ATM D2899A mutant cells, we examined CHK1 S317 phosphorylation in YFP+ sorted AtmTgD2899AAtmflox/− and control cultured B cells treated with hydroxyurea. We found that hydroxyurea-induced CHK1 S317 phosphorylation was indistinguishable between mutant and control cells (Fig. 3 D). Double deficiency in the human ATM and ATR kinases leads to extensive chromosome fragmentation in the presence of the DNA replication inhibitor aphidicolin (Ozeri-Galai et al., 2008); however, AtmTgD2899AAtmflox/− cells were also indistinguishable from controls in response to aphidicolin in our previously described flow cytometric YFP frequency assay (unpublished data). Altogether, our results indicate that expression of kinase-inactive murine ATM does not abrogate DNA-PKcs or ATR function in B cells.
Although an explanation for the embryonic lethality in kinase-inactive ATM mice is yet to be determined, Atm-null mutations clearly can become embryonically lethal when combined with deficiencies in various DNA damage response pathways. For instance, Atm-null mice show embryonic lethality when disrupted in combination with Fancg−/− (Kennedy et al., 2007), Parp1−/− (E8.0; Ménisser-de Murcia et al., 2001), Parp2−/− (between E9.5 and newborn; Huber et al., 2004), Ku70−/− and Ku80−/− (earlier than E11.5), Dnapkcs−/− (E7.5–8.5; Gurley and Kemp, 2001; Sekiguchi et al., 2001; Gladdy et al., 2006), H2ax−/− (E11.5–12.5; Zha et al., 2008), and Nbs1ΔB/ΔB (E10; Williams et al., 2002). Some of these synthetic lethal interactions may arise from additive defects in DNA repair. For example, it is possible that Nbs1ΔB/ΔB mice are synthetically lethal with Atm−/− (Williams et al., 2002) because of the essential role of the MRN complex in DSB resection that is independent of its function in murine ATM activation (Buis et al., 2008). In dividing embryonic cells, an HR defect in Nbs1ΔB/ΔB mice may synergize with ATM deficiency to cause an accumulation of DNA damage above a tolerable threshold. Similarly, we found that D2899A mutant B cells were mildly more sensitive than ATM-deficient controls to PARP inhibition, and we hypothesize that this may be caused by a more severe decrease in resection. Although D2899A mutant B cells show a decrease in HR, they also show a mild increase in the frequency of class switching compared with ATM-deficient cells, suggestive of increased NHEJ activity. Based on these findings, we suggest that the mild imbalances in DNA repair pathways in AtmTgD2899AAtm−/− cells above those governed by ATM could result in very severe developmental defects, while having less dramatic outcomes in adult tissues.
Only rarely do A-T patients with the classical phenotype display relatively normal ATM protein levels without detectable kinase activity (Barone et al., 2009; Demuth et al., 2011; Jacquemin et al., 2012). We hypothesize that similar kinase-inactive mutations also cause embryonic lethality in humans and expect that patients with a similar genotype as the one modeled here must have additional mutations to bypass the embryogenesis defect. Indeed, there is clear precedence for secondary mutations that rebalance DSB repair pathways and alleviate early embryonically lethal phenotypes. For example, loss of the NHEJ factor 53BP1 rescues the early embryonic lethality observed in Brca1-null mice (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2012). Although loss of 53BP1 and Ku deficiency both increase HR (Pierce et al., 2001; Bunting et al., 2010, 2012), 53bp1−/−AtmTgD2899AAtm−/− mice and Ku80−/−AtmTgD2899AAtm−/− mice were not obtained (Table 2), suggesting that other mutations might compensate for kinase-inactive ATM protein. In sum, our results contribute a novel perspective to the role of ATM kinase activity during embryogenesis and raise caution regarding the consequences of ATMi treatment in cancer patients and in experimental systems.
Materials and methods
Generation of mice
The D2899A and Q2740P mutations were targeted using BAC recombineering as previously described (Yang and Sharan, 2003; Pellegrini et al., 2006; Daniel et al., 2008). In brief, mutations were introduced into the murine Atm BAC RP24-122F10, which consists of a 160-kb insert including 48.3 kb of sequence upstream and 17.9 kb of sequence downstream of the Atm initiation and stop codons, respectively, along with an engineered EcoRI site between exons 35 and 36 for a PCR-based method to distinguish between AtmTgAtm−/− and AtmTgAtm+/− genotypes as described previously (Pellegrini et al., 2006; Daniel et al., 2008). Both mutant BACs were used to generate transgenic mice. The presence of the transgene was determined by PCR as previously described (Pellegrini et al., 2006) using mouse ATM forward, 5′-AGCACAACCACACTGAATGC-3′, and SP6R, 5′-GTTTTTTGCGATCTGCCGTTTC-3′, primers. To distinguish between AtmTgAtm−/− and AtmTgAtm+/− genotypes, EcoRI digestion of the PCR-purified product from amplification using mouse ATM 5B, 5′-GCAGATCCTAAGTAGGTGAGCT-3′, and ATM WT reverse, 5′-CGAATTTGCAGGAGTTGCTGAG-3′, primers was performed. AtmTgAtm+/− mice yielded the predicted digest products running at 600, 400, and 200 bp, whereas AtmTgAtm−/− mice yielded the predicted products at 400 and 200 bp. Transgenic founders were crossed to Atm+/− mice. All experiments were performed in compliance with the Animal Welfare Act regulations and other federal statutes relating to animals and adhered to the principles set forth in the Guide for Care and Use of Laboratory Animals (National Research Council, 1996). J. Petrini (Memorial Sloan-Kettering Cancer Center, New York, NY) provided Nbs1ΔB mice. F. Alt (Harvard Medical School, Boston, MA) provided Atm conditional knockout mice.
Lymphocyte cultures, flow cytometry, and genome stability
Single-cell suspensions of ACK-treated bone marrow and splenocytes from 6–12-wk-old mice were stained with α-B220–FITC, α-IgM–R-phycoerythrin, and α-CD43–biotin followed by streptavidin-allophycocyanin. Cultured B cells were isolated from spleens of 6–12-wk-old mice by immunomagnetic depletion with anti-CD43 beads (Miltenyi Biotec) and stimulated with either 25 mg/ml LPS (Sigma-Aldrich), 5 ng/ml interleukin 4 (Sigma-Aldrich), and/or 2.5 µg/ml RP105 (BD) as indicated. For assaying class switching, cultured B cells were harvested and stained in single-cell suspensions with α-IgG1–biotin followed by streptavidin–R-phycoerythrin. Cells were acquired through a propidium iodide–negative live-lymphocyte gate with either a FACSCalibur (BD) or an LSR II (BD) flow cytometer. Live YFP+ cells were sorted on a cell sorter (FACSAria II; BD). Data were analyzed using FlowJo software (Tree Star). All statistical significance analyses were determined by a two-tailed t test assuming unequal variance. For genome stability analyses, cultured B cells were arrested at mitosis with 0.1 µg/ml colcemid (Roche) treatment for 1 h, live YFP+ cells were sorted by FACS, and metaphase chromosome spreads were prepared following standard procedures. FISH was performed on slides with a probe for telomere repeat–specific peptide nucleic acid conjugated to Cy3 fluorochrome (Panagene) and counterstained with DAPI. Metaphase images were acquired with an upright microscope (Axioplan 2; Carl Zeiss) equipped with a 63×, NA 1.4 objective lens (Plan-Apochromat; Nikon) and a monochrome charge-coupled device camera (ORCA-ER; Hamamatsu Photonics) using MetaMorph software (Molecular Devices). Abl cell lines were generated, and V(D)J recombination was assayed as described including the Atm+/+:INV (A70.3INV3) and Atm−/−:INV (Atm−/−INV14) lines (Bredemeyer et al., 2006). The pMX-INV retroviral recombination substrate contains GFP cDNA and human CD4 cDNA downstream of an internal ribosomal entry site. For Cre deletion in AtmTgAtmflox/− abl cells, cells were transfected with mouse stem cell virus–thy1.1-Cre plasmid DNA using the a transfection system (1,500 V/15 ms/4 pulses; Neon; Invitrogen). 24 h after transfection, cells were sorted using biotin anti-Thy1.1 antibodies (BD) and antibiotin microbeads according to the company’s protocol (Miltenyi Biotec). The sorted cells were subcloned by limiting dilution and identified using a 3′ ATM southern probe from a 635-bp fragment generated by PCR using 5′-GGCATCTGCTTGACTGCAGTAAATCAGGCGG-3′ and 5′-GGGGTACTGCAGCATAGGGCTGGAAGAGG-3′ primers with standard procedures as described previously (Zha et al., 2008).
Western blotting
Whole-cell lysates from single-cell suspensions of thymocytes or B lymphocytes were prepared as previously described (Difilippantonio et al., 2005), and Western blotting was performed as previously described (Daniel et al., 2008). In brief, 40 µg protein was loaded on 4–12% Bis-Tris gels (NuPAGE; Life Technologies) and probed with the following antibodies using HRP-conjugated secondary antibodies and ECL Western blotting detection reagents (GE Healthcare): ATM (clone 5C2; 1:400; Novus Biologicals), α-Tubulin (clone B512; 1:30,000; Sigma-Aldrich), KAP1 S824p (1:700; Bethyl Laboratories, Inc.), CHK1 S317p (1:500, Bethyl Laboratories, Inc.), and SMC1 (1:8,000; Novus Biologicals). Human ATM complexes were purified from 293T cells by sequential anti-Flag and anti-HA immunoprecipitation, and kinase assays were performed as previously described (Lee and Paull, 2005). In brief, kinase assays were performed in kinase buffer composed of 50 mM Hepes, pH 7.5, 50 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM ATP, and 1 mM DTT for 90 min at 30°C.
Online supplemental material
Fig. S1 shows that integration of the Atm D2899A mutant transgene in each founder occurs on a different chromosome than endogenous Atm using metaphase FISH. Fig. S2 shows that ATM kinase activity is dispensable for ATM recruitment to DNA breaks using laser microirradiation and immunofluorescence microscopy. Fig. S3 shows FACS histograms of YFP expression in bone marrow and spleen subsets and Southern blot analysis of digested genomic DNA assaying for rearrangement of the pMX-INV–integrated substrate within v-Abl–transformed pre-B cell lines, indicative of RAG-dependent DNA recombination. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201204035/DC1.
Supplementary Material
Acknowledgments
We thank Nancy Wong, Steve Jay, and Jeffrey Hammer for technical assistance. We thank John Petrini for Nbs1ΔB mice and Fred Alt for Atmflox mice.
This work was supported by funding from the Italian Association for Cancer Research to M. Pellegrini, from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research to A. Nussenzweig. The Center for Protein Research is partly supported by a generous donation from the Novo Nordisk Foundation. J.A. Daniel is the recipient of a National Institutes of Health Pathway to Independence K99/R00 Award.
Footnotes
Abbreviations used in this paper:
- A-T
- ataxia telangiectasia
- ATM
- A-T mutated
- ATMi
- ATM inhibitor
- ATR
- A-T and Rad3 related
- BAC
- bacterial artificial chromosome
- CSR
- class switch recombination
- DSB
- double-stranded break
- HR
- homologous recombination
- IR
- ionizing radiation
- LPS
- lipopolysaccharide
- NHEJ
- nonhomologous end joining
- PARP
- poly (ADP-ribose) polymerase
- PKcs
- protein kinase, catalytic subunit
- RAG
- recombination activating gene
- V(D)J
- variable, diversity, and joining
- WT
- wild type
References
- Adams K.E., Medhurst A.L., Dart D.A., Lakin N.D. 2006. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 25:3894–3904 10.1038/sj.onc.1209426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Banin S., Moyal L., Shieh S., Taya Y., Anderson C.W., Chessa L., Smorodinsky N.I., Prives C., Reiss Y., Shiloh Y., Ziv Y. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 281:1674–1677 10.1126/science.281.5383.1674 [DOI] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Barone G., Groom A., Reiman A., Srinivasan V., Byrd P.J., Taylor A.M. 2009. Modeling ATM mutant proteins from missense changes confirms retained kinase activity. Hum. Mutat. 30:1222–1230 10.1002/humu.21034 [DOI] [PubMed] [Google Scholar]
- Beli P., Lukashchuk N., Wagner S.A., Weinert B.T., Olsen J.V., Baskcomb L., Mann M., Jackson S.P., Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell. 46:212–225 10.1016/j.molcel.2012.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A., Schmidt A., Ziv Y., Elkon R., Wang S.Y., Chen D.J., Aebersold R., Shiloh Y. 2010. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal. 3:rs3 10.1126/scisignal.2001034 [DOI] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., Sleckman B.P. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615–628 10.1101/gad.1067403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Helleday T. 2006. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 34:1685–1691 10.1093/nar/gkl108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 135:85–96 10.1016/j.cell.2008.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 46:125–135 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Wong N., Zha S., Chen H.T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A., Nussenzweig M. 2009. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell. 34:285–297 10.1016/j.molcel.2009.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 281:1677–1679 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., Liu J., Wang R.H., Cao L.L., Wu J.J., Peng T.N., Chen J., Nussenzweig A., et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 35:534–541 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado M., Martinez-Pastor B., Murga M., Toledo L.I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. 2006. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203:297–303 10.1084/jem.20051923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Pellegrini M., Lee J.H., Paull T.T., Feigenbaum L., Nussenzweig A. 2008. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 183:777–783 10.1083/jcb.200805154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.J., So S., Chen D.J. 2010. Dynamics of the PI3K-like protein kinase members ATM and DNA-PKcs at DNA double strand breaks. Cell Cycle. 9:2529–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth I., Dutrannoy V., Marques W., Jr, Neitzel H., Schindler D., Dimova P.S., Chrzanowska K.H., Bojinova V., Gregorek H., Graul-Neumann L.M., et al. 2011. New mutations in the ATM gene and clinical data of 25 AT patients. Neurogenetics. 12:273–282 10.1007/s10048-011-0299-0 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Nussenzweig A. 2007. The NBS1-ATM connection revisited. Cell Cycle. 6:2366–2370 10.4161/cc.6.19.4758 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H.T., Reina San Martin B., Van Laethem F., Yang Y.P., Petukhova G.V., Eckhaus M., Feigenbaum L., et al. 2005. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 7:675–685 10.1038/ncb1270 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Gapud E., Wong N., Huang C.Y., Mahowald G., Chen H.T., Kruhlak M.J., Callen E., Livak F., Nussenzweig M.C., et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 456:529–533 10.1038/nature07476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N., Chen Y.C., Spector D.L., de Lange T. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 456:524–528 10.1038/nature07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089 10.1073/pnas.93.23.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapud E.J., Sleckman B.P. 2011. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle. 10:1928–1935 10.4161/cc.10.12.16011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S., Khosravi R., Shkedy D., Uziel T., Ziv Y., Savitsky K., Rotman G., Smith S., Chessa L., Jorgensen T.J., et al. 1996. Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet. 5:433–439 10.1093/hmg/5.4.433 [DOI] [PubMed] [Google Scholar]
- Gladdy R.A., Nutter L.M., Kunath T., Danska J.S., Guidos C.J. 2006. p53-independent apoptosis disrupts early organogenesis in embryos lacking both ataxia-telangiectasia mutated and Prkdc. Mol. Cancer Res. 4:311–318 10.1158/1541-7786.MCR-05-0258 [DOI] [PubMed] [Google Scholar]
- Gurley K.E., Kemp C.J. 2001. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr. Biol. 11:191–194 10.1016/S0960-9822(01)00048-3 [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 10.1158/0008-5472.CAN-04-2727 [DOI] [PubMed] [Google Scholar]
- Huber A., Bai P., de Murcia J.M., de Murcia G. 2004. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst.). 3:1103–1108 10.1016/j.dnarep.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Jacquemin V., Rieunier G., Jacob S., Bellanger D., d’Enghien C.D., Laugé A., Stoppa-Lyonnet D., Stern M.H. 2012. Underexpression and abnormal localization of ATM products in ataxia telangiectasia patients bearing ATM missense mutations. Eur. J. Hum. Genet. 20:305–312 10.1038/ejhg.2011.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37–45 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- Kennedy R.D., Chen C.C., Stuckert P., Archila E.M., De la Vega M.A., Moreau L.A., Shimamura A., D’Andrea A.D. 2007. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Invest. 117:1440–1449 10.1172/JCI31245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S.V., Graham M.E., Peng C., Chen P., Robinson P.J., Lavin M.F. 2006. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 25:3504–3514 10.1038/sj.emboj.7601231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S.V., Graham M.E., Jakob B., Tobias F., Kijas A.W., Tanuji M., Chen P., Robinson P.J., Taucher-Scholz G., Suzuki K., et al. 2011. Autophosphorylation and ATM activation: additional sites add to the complexity. J. Biol. Chem. 286:9107–9119 10.1074/jbc.M110.204065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin N.D., Weber P., Stankovic T., Rottinghaus S.T., Taylor A.M., Jackson S.P. 1996. Analysis of the ATM protein in wild-type and ataxia telangiectasia cells. Oncogene. 13:2707–2716 [PubMed] [Google Scholar]
- Lavin M.F. 2008. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9:759–769 10.1038/nrm2514 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 308:551–554 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Li A., Swift M. 2000. Mutations at the ataxia-telangiectasia locus and clinical phenotypes of A-T patients. Am. J. Med. Genet. 92:170–177 [DOI] [PubMed] [Google Scholar]
- López-Contreras A.J., Fernandez-Capetillo O. 2010. The ATR barrier to replication-born DNA damage. DNA Repair (Amst.). 9:1249–1255 10.1016/j.dnarep.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J.M., McCarty T., Petiniot L.K., Shen R., Barlow C., Wynn T.A., Morse H.C., III, Gearhart P.J., Wynshaw-Boris A., Max E.E., Hodes R.J. 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 200:1111–1121 10.1084/jem.20041074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- McCabe N., Turner N.C., Lord C.J., Kluzek K., Bialkowska A., Swift S., Giavara S., O’Connor M.J., Tutt A.N., Zdzienicka M.Z., et al. 2006. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66:8109–8115 10.1158/0008-5472.CAN-06-0140 [DOI] [PubMed] [Google Scholar]
- Ménisser-de Murcia J., Mark M., Wendling O., Wynshaw-Boris A., de Murcia G. 2001. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol. 21:1828–1832 10.1128/MCB.21.5.1828-1832.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol R., Ben Slama L., Suarez F., Le Mignot L., Beauté J., Mahlaoui N., Dubois d’Enghien C., Laugé A., Hall J., Couturier J., et al. ; CEREDIH Network Investigators 2011. Morbidity and mortality from ataxia-telangiectasia are associated with ATM genotype. J. Allergy Clin. Immunol. 128:382–389: e1 10.1016/j.jaci.2011.03.052 [DOI] [PubMed] [Google Scholar]
- Myers J.S., Cortez D. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281:9346–9350 10.1074/jbc.M513265200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 382:551–555 10.1038/382551a0 [DOI] [PubMed] [Google Scholar]
- Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., Peskett E., Pearce W., Meek S.E., Salpekar A., Waterfield M.D., et al. 2002. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 297:1031–1034 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Ozeri-Galai E., Schwartz M., Rahat A., Kerem B. 2008. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 27:2109–2117 10.1038/sj.onc.1210849 [DOI] [PubMed] [Google Scholar]
- Pan Q., Petit-Frére C., Lähdesmäki A., Gregorek H., Chrzanowska K.H., Hammarström L. 2002. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur. J. Immunol. 32:1300–1308 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Celeste A., Difilippantonio S., Guo R., Wang W., Feigenbaum L., Nussenzweig A. 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 443:222–225 10.1038/nature05112 [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Hu P., Han M., Ellis N., Jasin M. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237–3242 10.1101/gad.946401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey M.D., Charlton M.E., Stanton R.V., Kastan M.B. 2008. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res. 68:7466–7474 10.1158/0008-5472.CAN-08-0763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen H.T., Nussenzweig A., Nussenzweig M.C. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110 10.1084/jem.20041162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 268:1749–1753 10.1126/science.7792600 [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Ferguson D.O., Chen H.T., Yang E.M., Earle J., Frank K., Whitlow S., Gu Y., Xu Y., Nussenzweig A., Alt F.W. 2001. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. USA. 98:3243–3248 10.1073/pnas.051632098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 3:155–168 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- So S., Davis A.J., Chen D.J. 2009. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 187:977–990 10.1083/jcb.200906064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T.H., Petrini J.H. 2011. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12:90–103 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Byrd P.J. 2005. Molecular pathology of ataxia telangiectasia. J. Clin. Pathol. 58:1009–1015 10.1136/jcp.2005.026062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612–5621 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.S., Choi S., Bakkenist C.J. 2008. Irreversible chromosome damage accumulates rapidly in the absence of ATM kinase activity. Cell Cycle. 7:1277–1284 10.4161/cc.7.9.5961 [DOI] [PubMed] [Google Scholar]
- Williams B.R., Mirzoeva O.K., Morgan W.F., Lin J., Dunnick W., Petrini J.H. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12:648–653 10.1016/S0960-9822(02)00763-7 [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Yang Y., Sharan S.K. 2003. A simple two-step, ‘hit and fix’ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 31:e80 10.1093/nar/gng080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Bailis J.M., Johnson S.A., Dilworth S.M., Hunter T. 2007. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 9:1311–1318 10.1038/ncb1651 [DOI] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S., Sekiguchi J., Brush J.W., Bassing C.H., Alt F.W. 2008. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc. Natl. Acad. Sci. USA. 105:9302–9306 10.1073/pnas.0803520105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yajima H., Huynh H., Zheng J., Callen E., Chen H.T., Wong N., Bunting S., Lin Y.F., Li M., et al. 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 193:295–305 10.1083/jcb.201009074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.