Abstract
The ataxia telangiectasia mutated (ATM) protein kinase regulates the cellular response to deoxyribonucleic acid (DNA) double-strand breaks by phosphorylating numerous players in the extensive DNA damage response network. Two papers in this issue (Daniel et al. 2012. J. Cell Biol. http://dx.doi.org/10.1083/jcb201204035; Yamamoto et al. 2012. J. Cell Biol. http://dx.doi.org/10.1083/jcb201204098) strikingly show that, in mice, the presence of a catalytically inactive version of ATM is embryonically lethal. This is surprising because mice completely lacking ATM have a much more moderate phenotype. The findings impact on basic cancer research and cancer therapeutics.
Maintenance of genomic stability is essential for prevention of undue cell death or neoplasia (Cassidy and Venkitaraman, 2012). Critical DNA lesions, such as double-strand breaks (DSBs), activate the DNA damage response (DDR)—a widespread signaling network that involves DNA repair, activation of cell cycle checkpoints, and extensive modulation of gene expression and many metabolic pathways (Ciccia and Elledge, 2010; Hiom, 2010). DSBs are induced by ionizing radiation, radiomimetic chemicals, and endogenous oxygen radicals. They accompany replication fork stalling and are formed and resealed in meiotic recombination and the rearrangement of the antigen receptor genes during the development of the immune system. Major repair pathways for DSBs are error-prone nonhomologous end joining (NHEJ) or high-fidelity homologous recombination repair (HRR; Holthausen et al., 2010; Lieber, 2010). The broad, powerful signaling network evoked by DSBs begins with rapid accumulation at DSB sites of a large group of proteins dubbed “sensors” or “moderators” and continues with the activation of several protein kinases (“transducers”) with partially redundant functions that relay the signal to numerous downstream effectors, which are typically key players in the various DDR branches (Lovejoy and Cortez, 2009; Ciccia and Elledge, 2010; Lukas et al., 2011).
The primary transducer of the DSB alarm is the serine-threonine kinase ataxia telangiectasia (A-T) mutated (ATM; Banin et al., 1998; Canman et al., 1998), which is activated in response to DSB induction (Bakkenist and Kastan, 2003) and goes on to phosphorylate a plethora of substrates (Matsuoka et al., 2007; Bensimon et al., 2010). ATM belongs to a conserved family of phosphoinositide 3-kinase–like protein kinases (PIKKs) that includes, among others, two other major DDR transducers: the catalytic subunit of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and ATR (ataxia telangiectasia and Rad3 related). These three kinases maintain close functional relationships (Lovejoy and Cortez, 2009). Recent evidence suggests that ATM’s broad capacity as a protein kinase enables it to regulate other processes, such as oxidative stress levels (Guo et al., 2010), and play a role in cytoplasmic, non-DDR arenas, among them mitochondrial homeostasis (Yang et al., 2011; Valentin-Vega and Kastan, 2012; Valentin-Vega et al., 2012).
Human germline mutations that abrogate cellular responses to DNA damage cause severe genomic instability syndromes (Jeppesen et al., 2011). The ATM gene is mutated in the genomic instability syndrome, A-T (Savitsky et al., 1995). A-T is characterized by progressive neurodegeneration, immunodeficiency, cancer predisposition, genomic instability, and sensitivity to DSB-inducing agents (McKinnon, 2012). The disease is caused by null ATM mutations, and the patients usually exhibit complete loss of the ATM protein (Gilad et al., 1996).
Studies of ATM-dependent processes typically rely on human wild-type versus A-T cells, ATM knockdown using RNAi, reconstitution of ATM-deficient cells by ectopic expression of wild-type or kinase-dead ATM protein, or treating cultured cells with ATM inhibitors. Laboratories using these experimental systems have long felt that the physiological consequences of ATM loss as opposed to harboring inactive ATM may not be similar (Choi et al., 2010). The papers by Daniel et al. and Yamamoto et al. (both in this issue) provide solid evidence of this notion and mark a turning point in our view of ATM’s mode of function. Both works are based on manipulating the Atm gene in the mouse.
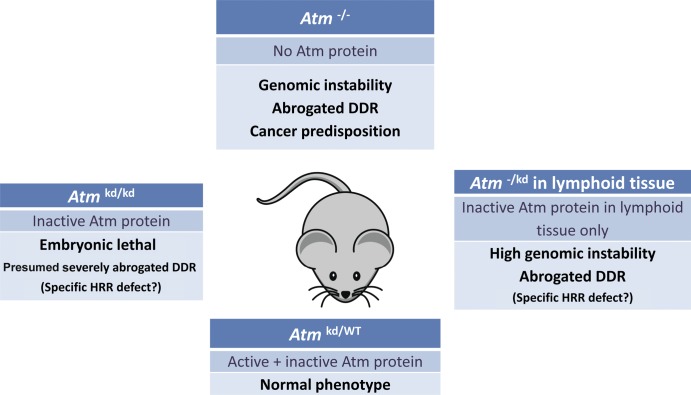
Atm knockout mice have long been around. These mice exhibit most of the symptoms of A-T, including low body weight, sterility, radiosensitivity, and cancer predisposition, but neurodegeneration is considerably less marked in these animals compared with that observed in human A-T patients (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Borghesani et al., 2000). Thus, before cancer emergence and without exposure to radiation, the murine Atm−/− phenotype is relatively moderate. Using mutant Atm transgene expression in an Atm−/− background (Daniel et al., 2012) and via direct knockin (Yamamoto et al., 2012), the two groups generated new mouse strains that lack Atm activity; rather than being devoid of Atm, these animals express physiological levels of catalytically inactive (kinase dead) protein. Strikingly, in both laboratories, this genotype led to early embryonic lethality, with inherent genomic instability that was higher than that observed in Atm−/− animals (Fig. 1). Conditional expression of the mutant protein in the immune system reduced the efficiency of V(D)J (variable, diversity, and joining) recombination and immunoglobulin class switching—two processes that involve the NHEJ pathway of DSB repair and require active ATM for optimal function. However, this reduction was comparable to that caused by absence of Atm. Collectively, the data from both laboratories suggest that the HRR pathway of DSB repair, rather than NHEJ, may be affected to a greater extent by the presence of inactive Atm compared with the effect obtained after Atm loss.
Figure 1.
Phenotypic comparison of mouse Atm genotypes. Mice expressing an inactive protein as their sole source of Atm die in utero (Daniel et al., 2012; Yamamoto et al. 2012). Heterozygotes resemble wild-type (WT) animals, indicating lack of a dominant-negative effect. HRR, homologous recombination repair; kd, kinase dead.
This dramatic phenotype is presumably caused by severe malfunction of the DDR, attesting once again to its importance in early development. The critical role of the DDR in development has been documented in the past (Phillips and McKinnon, 2007), but the novelty of the current studies lies in the profound difference between Atm loss and the presence of catalytically inactive Atm. The same likely applies in humans as well: A-T patients typically exhibit ATM loss, and in rare cases of catalytically inactive ATM in patients, its level is low enough to allow for viability. A similar observation was made recently by Zhang et al. (2011) with another member of the PIKK family—DNA-PKcs. This group found that mice expressing a mutant version of DNA-PKcs, lacking three phosphorylation sites associated with its activation, die shortly after birth as a result of bone marrow failure. It is interesting to note that in contrast to this, abolishing three phosphorylation sites in mouse Atm, whose equivalents in human ATM are phosphorylated during its activation (Bakkenist and Kastan, 2003; Kozlov et al., 2006), did not result in any discernible phenotype (Pellegrini et al., 2006; Daniel et al., 2008).
It appears, therefore, that the presence of physiological levels of inactive Atm severely interferes with the DDR, certainly more than its absence. Why could this be? Although the exact mechanism of this phenomenon is unknown, some assumptions can be made. ATM is recruited to DSB sites (Andegeko et al., 2001) and is therefore present in the huge nuclear foci spanning these sites. Many ATM-mediated phosphorylations occur within these protein conglomerates. Importantly, the recruitment of kinase-dead Atm to sites of DNA damage was found by Daniel et al. (2012) and Yamamoto et al. (2012) to occur normally. It is possible that the presence of catalytically inactive Atm within these DDR hubs severely disturbs the ability of the cell to respond to the damage. Presumably, it interferes with the ordered temporal dynamics of events within these protein factories (Lukas et al., 2011). Deeper understanding of the spatial organization of these protein assemblies (Chapman et al., 2012) and the temporal hierarchy of events within them may elucidate ATM’s role not only as an enzyme but also as a protein moiety in these structures. Of note, ATM is a large protein of 3,056 residues, of which ∼10% constitute its active site. The regulatory functions of the remaining 90% of this polypeptide are largely elusive. In a broader sense, these studies convincingly show, at the organismal level, that loss of an enzyme versus having it residing inactive in the cell can be worlds apart. In this context, it would be interesting to monitor the development of malignancies in those animals expressing the mutant Atm in their lymphoid system. This is particularly important because the malignancies observed in Atm−/− mice, similar to A-T patients, are primarily lymphoid.
The implications for ATM-related translational research are notable. ATM has naturally been considered a potential target to be inactivated in tumor cells to selectively sensitize them to radiotherapy (Begg et al., 2011; Basu et al., 2012; Golding et al., 2012). The advent of efficient ATM inhibitors (Hickson et al., 2004; Golding et al., 2009) has further spurred these hopes. The good news is that the effect of these inhibitors on cellular radiosensitivity (and, probably, general well being) might be more profound than previously estimated, provided that these small molecules could be targeted specifically into the malignant cells. On the other hand, exposure of normal, proliferating body tissues to ATM inhibitors may be undesirable, depending on the type of tissue. Such exposure of normal tissue to ATM inhibition, even if brief, could lead to substantial genomic instability—a potential driving force toward new malignancy.
Acknowledgments
We are grateful to Ayelet Klartag and Adva Levy-Barda for valuable comments.
Y. Shiloh is a Research Professor of the Israel Cancer Research Fund.
References
- Andegeko Y., Moyal L., Mittelman L., Tsarfaty I., Shiloh Y., Rotman G. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224–38230 [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Banin S., Moyal L., Shieh S., Taya Y., Anderson C.W., Chessa L., Smorodinsky N.I., Prives C., Reiss Y., Shiloh Y., Ziv Y. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 281:1674–1677 10.1126/science.281.5383.1674 [DOI] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Basu B., Yap T.A., Molife L.R., de Bono J.S. 2012. Targeting the DNA damage response in oncology: past, present and future perspectives. Curr. Opin. Oncol. 24:316–324 10.1097/CCO.0b013e32835280c6 [DOI] [PubMed] [Google Scholar]
- Begg A.C., Stewart F.A., Vens C. 2011. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer. 11:239–253 10.1038/nrc3007 [DOI] [PubMed] [Google Scholar]
- Bensimon A., Schmidt A., Ziv Y., Elkon R., Wang S.Y., Chen D.J., Aebersold R., Shiloh Y. 2010. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal. 3:rs3 10.1126/scisignal.2001034 [DOI] [PubMed] [Google Scholar]
- Borghesani P.R., Alt F.W., Bottaro A., Davidson L., Aksoy S., Rathbun G.A., Roberts T.M., Swat W., Segal R.A., Gu Y. 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. USA. 97:3336–3341 10.1073/pnas.050584897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 281:1677–1679 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- Cassidy L.D., Venkitaraman A.R. 2012. Genome instability mechanisms and the structure of cancer genomes. Curr. Opin. Genet. Dev. 22:10–13 10.1016/j.gde.2012.02.003 [DOI] [PubMed] [Google Scholar]
- Chapman J.R., Sossick A.J., Boulton S.J., Jackson S.P. 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Gamper A.M., White J.S., Bakkenist C.J. 2010. Inhibition of ATM kinase activity does not phenocopy ATM protein disruption: implications for the clinical utility of ATM kinase inhibitors. Cell Cycle. 9:4052–4057 10.4161/cc.9.20.13471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Pellegrini M., Lee J.H., Paull T.T., Feigenbaum L., Nussenzweig A. 2008. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 183:777–783 10.1083/jcb.200805154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Pellegrini M., Lee B.-S., Guo Z., Filsuf D., Belkina N.V., You Z., Paull T.T., Sleckman B.P., Feigenbaum L., Nussenzweig A. 2012. Loss of ATM kinase activity leads to embryonic lethality in mice. J. Cell Biol. 198:295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089 10.1073/pnas.93.23.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S., Khosravi R., Shkedy D., Uziel T., Ziv Y., Savitsky K., Rotman G., Smith S., Chessa L., Jorgensen T.J., et al. 1996. Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet. 5:433–439 10.1093/hmg/5.4.433 [DOI] [PubMed] [Google Scholar]
- Golding S.E., Rosenberg E., Valerie N., Hussaini I., Frigerio M., Cockcroft X.F., Chong W.Y., Hummersone M., Rigoreau L., Menear K.A., et al. 2009. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 8:2894–2902 10.1158/1535-7163.MCT-09-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding S.E., Rosenberg E., Adams B.R., Wignarajah S., Beckta J.M., O’Connor M.J., Valerie K. 2012. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle. 11:1167–1173 10.4161/cc.11.6.19576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. 2010. ATM activation by oxidative stress. Science. 330:517–521 10.1126/science.1192912 [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 10.1158/0008-5472.CAN-04-2727 [DOI] [PubMed] [Google Scholar]
- Hiom K. 2010. Coping with DNA double strand breaks. DNA Repair (Amst.). 9:1256–1263 10.1016/j.dnarep.2010.09.018 [DOI] [PubMed] [Google Scholar]
- Holthausen J.T., Wyman C., Kanaar R. 2010. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst.). 9:1264–1272 10.1016/j.dnarep.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Jeppesen D.K., Bohr V.A., Stevnsner T. 2011. DNA repair deficiency in neurodegeneration. Prog. Neurobiol. 94:166–200 10.1016/j.pneurobio.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S.V., Graham M.E., Peng C., Chen P., Robinson P.J., Lavin M.F. 2006. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 25:3504–3514 10.1038/sj.emboj.7601231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79:181–211 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C.A., Cortez D. 2009. Common mechanisms of PIKK regulation. DNA Repair (Amst.). 8:1004–1008 10.1016/j.dnarep.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- McKinnon P.J. 2012. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 7:303–321 10.1146/annurev-pathol-011811-132509 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Celeste A., Difilippantonio S., Guo R., Wang W., Feigenbaum L., Nussenzweig A. 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 443:222–225 10.1038/nature05112 [DOI] [PubMed] [Google Scholar]
- Phillips E.R., McKinnon P.J. 2007. DNA double-strand break repair and development. Oncogene. 26:7799–7808 10.1038/sj.onc.1210877 [DOI] [PubMed] [Google Scholar]
- Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 268:1749–1753 10.1126/science.7792600 [DOI] [PubMed] [Google Scholar]
- Valentin-Vega Y., Kastan M.B. 2012. A new role for ATM: Regulating mitochondrial function and mitophagy. Autophagy. 8:840–841 10.4161/auto.19693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B. 2012. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 119:1490–1500 10.1182/blood-2011-08-373639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Wang Y., Jiang W., Liu X., Dubois R.L., Lin C.-S., Ludwig T., Bakkenist C.J., Zha S. 2012. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J. Cell Biol. 198:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.Q., Halaby M.J., Li Y., Hibma J.C., Burn P. 2011. Cytoplasmic ATM protein kinase: an emerging therapeutic target for diabetes, cancer and neuronal degeneration. Drug Discov. Today. 16:332–338 10.1016/j.drudis.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Zhang S., Yajima H., Huynh H., Zheng J., Callen E., Chen H.T., Wong N., Bunting S., Lin Y.F., Li M., et al. 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 193:295–305 10.1083/jcb.201009074 [DOI] [PMC free article] [PubMed] [Google Scholar]