Abstract
One of the major forces driving the birth of the field of cell biology was the application of electron microscopy to cells. Today, virtual nanoscopy has brought electron microscopy and the cell biology community to a new frontier in biological imaging and cell biological inquiry. The Journal of Cell Biology is pleased to announce that the JCB DataViewer is “going big” to host electron microscopy data at a whole new scale.
“See anything?”
© Robert Weber, The New Yorker Collection, www.cartoonbank.com
A new frontier: Gigapixel images at nanometer resolution
The field of cell biology emerged in the 1950s with the ability to observe—at nanometer resolution—cellular structures and their interconnections through the electron microscope. Electron microscopy continues to be a valuable tool for cell biological studies today. However, a limitation of this technology is the small field of view captured in a single image—only on the scale of a few microns square. Although these high-magnification images provide impressive resolution of the selected area of the cell, they are often difficult to put into the broader biological context of a cell or tissue. On the other hand, overview images taken at a lower magnification help to present the big picture but lack cellular detail.
Recent technological developments have enabled the integration of imaging and computational tools to allow “virtual nanoscopy,” the visualization of specimens at nanometer resolution but millimeter scale, a scale unheard of just a few years ago. These approaches allow the seamless zooming in and out—from subcellular- to tissue- or even organism-level views—without loss of resolution (Fig. 1). In the current issue of JCB, Faas et al. (2012) describe automated image acquisition and stitching technology using a standard laboratory transmission electron microscopy platform to generate large-scale electron microscopy datasets. By accurately stitching together more than 26,000 individual images at nanometer resolution, they generated virtual slides up to 1.5 mm (921,600 pixels) × 0.6 mm (380,928 pixels) for a total of 281 gigapixels (http://jcb-dataviewer.rupress.org/jcb/browse/5553). As a proof of principle, they applied this method to generate a map of the vast majority of a 5-day post-fertilization zebrafish embryo at 1.6-nm resolution per pixel (the equivalent of 16 million dpi). Importantly, the technology described by Faas et al. (2012) does not require a customized imaging platform, and it is applicable to a broad range of transmission electron microscopy samples, including samples that have been embedded in resin or cryo-vitrified and that are either unstained or have been immunolabeled using chemical or immunogold techniques. The resulting large-scale images allow the interrogation of morphological features over scales ranging from subcellular organelles and single cells to whole tissues to nearly an entire embryo. The ability to integrate information across cells and tissues in an unbiased manner provides an exceptional opportunity for discovery—from the cell biological to the organismal level.
Figure 1.
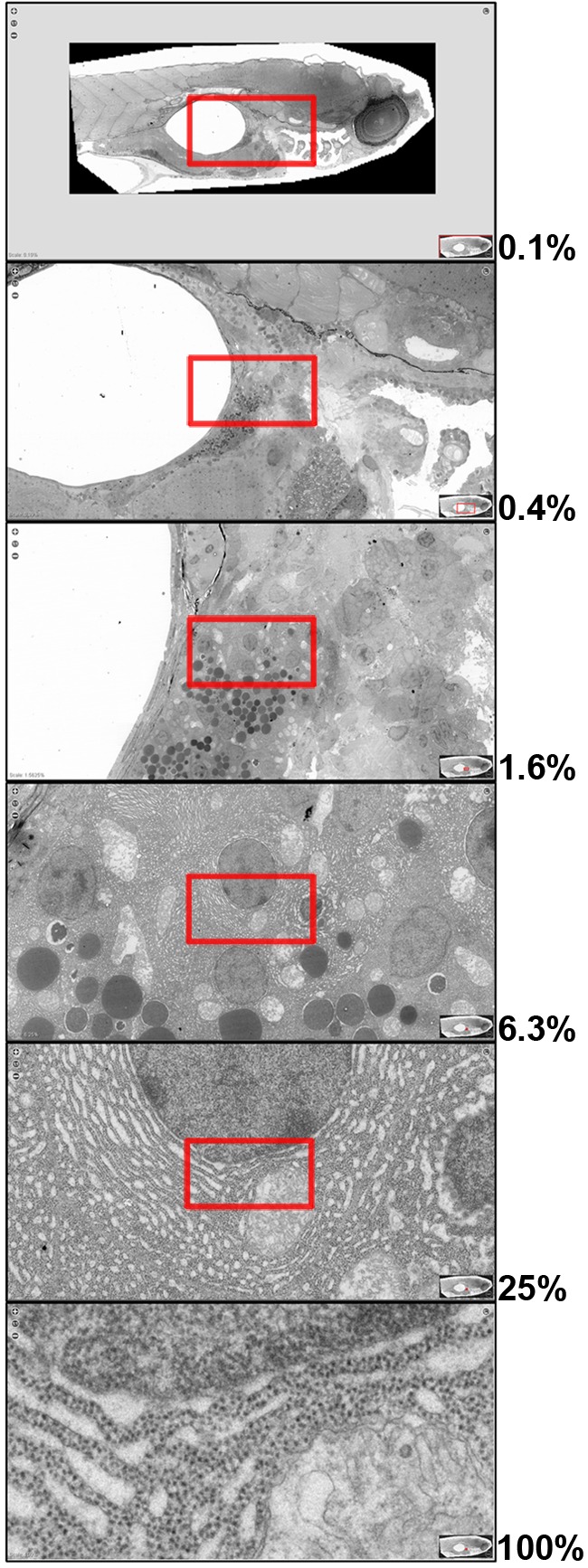
Electron microscopy from nanometer resolution to millimeter scale. Overlapping images at progressively higher magnification from a 281-gigapixel composite image of a 5-day post-fertilization zebrafish embryo reveal subcellular features within single-cell, tissue, and organismal contexts (image data © Faas et al., 2012). Red boxes indicate the region selected for each successive magnification step (indicated as percent magnification). The maximum original image resolution of 1.6 nm per pixel is shown as 100%.
Scaling up
If you can image it, you should be able to publish it. Conventional venues such as Supplemental Materials sections are not sufficient for publishing these large-scale image maps, however. To make publication of these data possible, JCB announces a major enhancement to the JCB DataViewer to host virtual nanoscopy data at the gigapixel scale. The JCB DataViewer was launched in 2008 to promote sharing of original data associated with JCB publications (Hill, 2008). Since then we have expanded the DataViewer to promote data analysis (deCathelineau et al., 2010) and hosting of entire high-content, image-based screens (Williams and Misteli, 2011). Now, if you are ready to go big with your imaging, the JCB DataViewer is ready to help you share your data with the cell biology community.
The next frontier
Despite ever-changing cell biological methods, imaging remains one of the key tools in our field. JCB remains committed to developing cutting-edge tools for the presentation of the data that drive progress in the field of cell biology. Looking forward, we know that further frontiers await us. Wherever the field of cell biology goes, JCB will continue to lead the way.
References
- deCathelineau A., Williams E.H., Misteli T., Rossner M. 2010. Friends, colleagues, authors, lend us your data. J. Cell Biol. 191:231–232 10.1083/jcb.201009056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas F.G.A., Avramut M.C., van den Berg B.M., Mommaas A.M., Koster A.J., Ravelli R.B.G. 2012. Virtual nanoscopy: Generation of ultra-large high resolution electron microscopy maps. J. Cell Biol. 198:457–469 10.1083/jcb.201201140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E. 2008. Announcing the JCB DataViewer, a browser-based application for viewing original image files. J. Cell Biol. 183:969–970 10.1083/jcb.200811132 [DOI] [Google Scholar]
- Williams E.H., Misteli T. 2011. New tools for JCB. J. Cell Biol. 194:663–664 10.1083/jcb.201108096 [DOI] [PMC free article] [PubMed] [Google Scholar]