Abstract
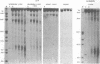
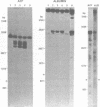
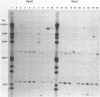
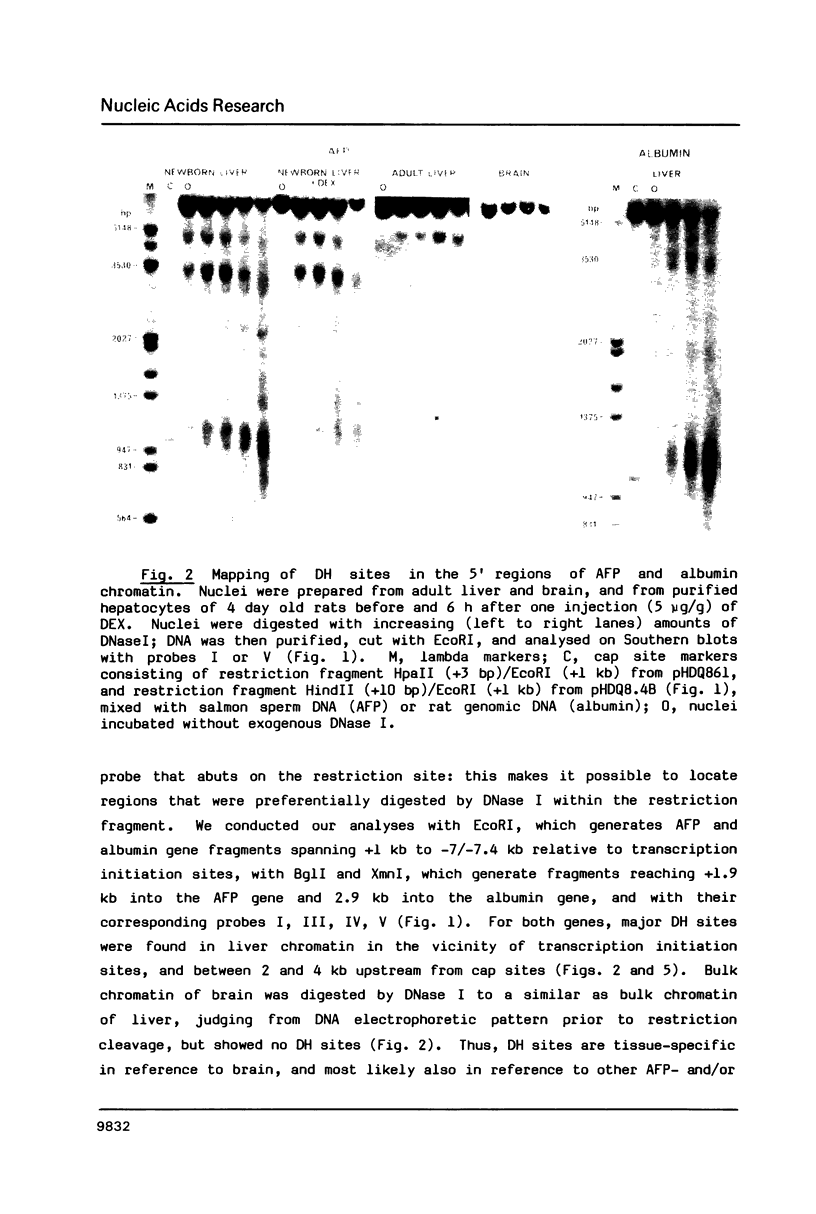
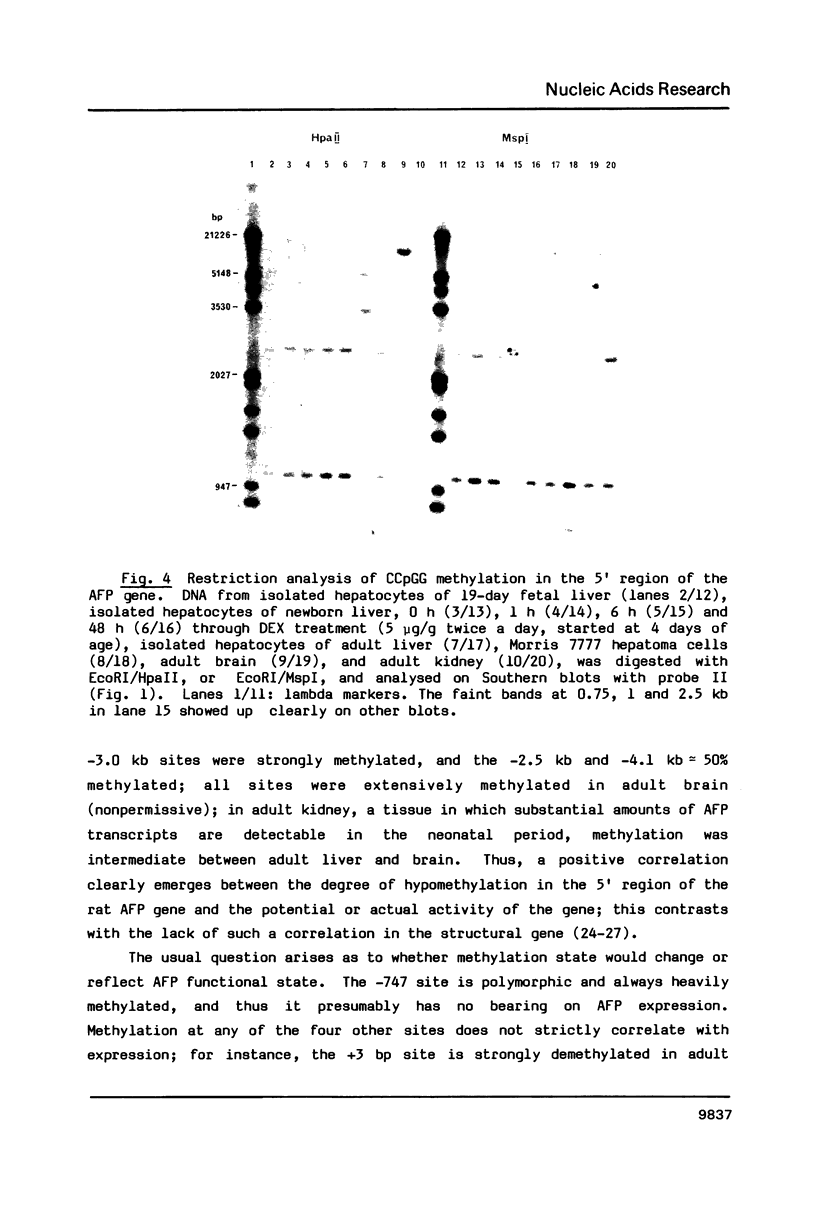
Three major regions of DNase I hypersensitivity (DH) were found in alpha 1-fetoprotein (AFP) chromatin of rat liver. DH site I is located at the transcription initiation site and associated with ongoing AFP transcription. DH site II is located 2.5 kb upstream from the cap site: it is developmental stage-dependent but dissociable from ongoing AFP transcription. DH site III, 3.7 kb upstream from the cap site, behaves as hepatocyte-constitutive. DH sites are present in similar regions of liver albumin chromatin. Dexamethasone-induced AFP gene repression is accompanied by the selective loss of AFP DH site I, a likely result of glucocorticoid receptors binding to a DNA recognition sequence located 5'-adjacent to DH site I. Sl nuclease-hypersensitive sites were found on naked superhelical AFP and albumin DNA, but do not appear to contribute DH sites in liver chromatin. The extent of hypomethylation of HpaII sites at the 5'-end of the AFP gene correlates positively with the level of potential and actual expression of the gene. We conclude that developmental and hormonal regulation of the AFP gene is confined within congruent to 4 kb of 5'-flanking DNA, and we discuss possible hierarchical interactions among DH sites, in relation to DNA methylation and replication.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews G. K., Dziadek M., Tamaoki T. Expression and methylation of the mouse alpha-fetoprotein gene in embryonic, adult, and neoplastic tissues. J Biol Chem. 1982 May 10;257(9):5148–5153. [PubMed] [Google Scholar]
- Balcarek J. M., McMorris F. A. DNase I hypersensitive sites of globin genes of uninduced Friend erythroleukemia cells and changes during induction with dimethyl sulfoxide. J Biol Chem. 1983 Sep 10;258(17):10622–10628. [PubMed] [Google Scholar]
- Baribault H., Leroux-Nicollet I., Marceau N. Differential responsiveness of cultured suckling and adult rat hepatocytes to growth-promoting factors: entry into S phase and mitosis. J Cell Physiol. 1985 Jan;122(1):105–112. doi: 10.1002/jcp.1041220116. [DOI] [PubMed] [Google Scholar]
- Belanger L., Hamel D., Lachance L., Dufour D., Tremblay M., Gagnon P. M. Hormonal regulation of alpha1 foetoprotein. Nature. 1975 Aug 21;256(5519):657–659. doi: 10.1038/256657a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R., Ingram R., Tilghman S. M. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986 Feb;6(2):477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Conkin K. F. Chromatin structure and de novo methylation of sperm DNA: implications for activation of the paternal genome. Science. 1985 May 31;228(4703):1061–1068. doi: 10.1126/science.2986289. [DOI] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982 Aug;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Ingram R. S., Scott R. W., Tilghman S. M. alpha-Fetoprotein and albumin genes are in tandem in the mouse genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4694–4698. doi: 10.1073/pnas.78.8.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kioussis D., Eiferman F., van de Rijn P., Gorin M. B., Ingram R. S., Tilghman S. M. The evolution of alpha-fetoprotein and albumin. II. The structures of the alpha-fetoprotein and albumin genes in the mouse. J Biol Chem. 1981 Feb 25;256(4):1960–1967. [PubMed] [Google Scholar]
- Koropatnick J., Andrews G., Duerksen J. D., Varshney U., Gedamu L. Mouse hepatic metallothionein-I gene cleavage by micrococcal nuclease is enhanced after induction by cadmium. Nucleic Acids Res. 1983 May 25;11(10):3255–3267. doi: 10.1093/nar/11.10.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R., Hammer R. E., Tilghman S. M., Brinster R. L. Developmental regulation of alpha-fetoprotein genes in transgenic mice. Mol Cell Biol. 1985 Jul;5(7):1639–1648. doi: 10.1128/mcb.5.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath L., Locker J. DNaseI sensitivity of the rat albumin and alpha-fetoprotein genes. Nucleic Acids Res. 1985 Jan 11;13(1):115–129. doi: 10.1093/nar/13.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath L., Locker J. Developmental changes in the methylation of the rat albumin and alpha-fetoprotein genes. EMBO J. 1983;2(3):317–324. doi: 10.1002/j.1460-2075.1983.tb01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T., Iyer B., Wu J. R., Lapeyre J. N., Becker F. F. Methylation of the alpha-fetoprotein gene in productive and nonproductive rat hepatocellular carcinomas. Cancer Res. 1984 Apr;44(4):1642–1647. [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Larue H., Bissonnette E., Bélanger L. Histone H1(0) expression during developmental growth of rat liver. Can J Biochem Cell Biol. 1983 Nov;61(11):1197–1200. doi: 10.1139/o83-154. [DOI] [PubMed] [Google Scholar]
- Latchman D. S., Brzeski H., Lovell-Badge R., Evans M. J. Expression of the alpha-fetoprotein gene in pluripotent and committed cells. Biochim Biophys Acta. 1984 Nov 22;783(2):130–136. doi: 10.1016/0167-4781(84)90004-6. [DOI] [PubMed] [Google Scholar]
- Marshall A. J., Burgoyne L. A. Interpretation of the properties of chromatin extracts from mammalian nuclei. Nucleic Acids Res. 1976 Apr;3(4):1101–1110. doi: 10.1093/nar/3.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahon J. L., Gal A., Erdos T., Sala-Trepat J. M. Differential DNase I sensitivity of the albumin and alpha-fetoprotein genes in chromatin from rat tissues and cell lines. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5031–5035. doi: 10.1073/pnas.81.16.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos C. J., Yaswen P., Panzica M., Fausto N. Methylation of the alphafetoprotein gene in cell populations isolated from rat livers during carcinogenesis. Nucleic Acids Res. 1985 Nov 25;13(22):8105–8118. doi: 10.1093/nar/13.22.8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J., Ruiz-Carrillo A. Fine analysis of the active H5 gene chromatin of chicken erythroid cells at different stages of differentiation. J Mol Biol. 1986 May 5;189(1):217–226. doi: 10.1016/0022-2836(86)90392-x. [DOI] [PubMed] [Google Scholar]
- Roche J., Gorka C., Goeltz P., Lawrence J. J. Association of histone H1(0) with a gene repressed during liver development. Nature. 1985 Mar 14;314(6007):197–198. doi: 10.1038/314197a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A. The histone H5 gene is flanked by S1 hypersensitive structures. Nucleic Acids Res. 1984 Aug 24;12(16):6473–6492. doi: 10.1093/nar/12.16.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M., Morinaga T., Urano Y., Watanabe K., Wegmann T. G., Tamaoki T. The human alpha-fetoprotein gene. Sequence organization and the 5' flanking region. J Biol Chem. 1985 Apr 25;260(8):5055–5060. [PubMed] [Google Scholar]
- Scott R. W., Tilghman S. M. Transient expression of a mouse alpha-fetoprotein minigene: deletion analyses of promoter function. Mol Cell Biol. 1983 Jul;3(7):1295–1309. doi: 10.1128/mcb.3.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell S., Nichols M., Becker F. F., Leffert H. L. Hepatocyte proliferation and alpha 1-fetoprotein in pregnant, neonatal, and partially hepatectomized rats. Cancer Res. 1974 Apr;34(4):865–871. [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Shimada T., Nienhuis A. W. Only the promoter region of the constitutively expressed normal and amplified human dihydrofolate reductase gene is DNase I hypersensitive and undermethylated. J Biol Chem. 1985 Feb 25;260(4):2468–2474. [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Turcotte B., Guertin M., Chevrette M., Bélanger L. Rat alpha 1-fetoprotein messenger RNA: 5'-end sequence and glucocorticoid-suppressed liver transcription in an improved nuclear run-off assay. Nucleic Acids Res. 1985 Apr 11;13(7):2387–2398. doi: 10.1093/nar/13.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Sakai M., Watanabe K., Tamaoki T. Tandem arrangement of the albumin and alpha-fetoprotein genes in the human genome. Gene. 1984 Dec;32(3):255–261. doi: 10.1016/0378-1119(84)90001-5. [DOI] [PubMed] [Google Scholar]
- Vedel M., Gomez-Garcia M., Sala M., Sala-Trepat J. M. Changes in methylation pattern of albumin and alpha-fetoprotein genes in developing rat liver and neoplasia. Nucleic Acids Res. 1983 Jul 11;11(13):4335–4354. doi: 10.1093/nar/11.13.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger C., Hollenberg S. M., Rosenfeld M. G., Evans R. M. Domain structure of human glucocorticoid receptor and its relationship to the v-erb-A oncogene product. Nature. 1985 Dec 19;318(6047):670–672. doi: 10.1038/318670a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- Young P. R., Tilghman S. M. Induction of alpha-fetoprotein synthesis in differentiating F9 teratocarcinoma cells is accompanied by a genome-wide loss of DNA methylation. Mol Cell Biol. 1984 May;4(5):898–907. doi: 10.1128/mcb.4.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]