Abstract
Summary: Site-directed mutagenesis is frequently used by scientists to investigate the functional impact of amino acid mutations in the laboratory. Over 10 000 such laboratory-induced mutations have been reported in the UniProt database along with the outcomes of functional assays. Here, we explore the performance of state-of-the-art computational tools (Condel, PolyPhen-2 and SIFT) in correctly annotating the function-altering potential of 10 913 laboratory-induced mutations from 2372 proteins. We find that computational tools are very successful in diagnosing laboratory-induced mutations that elicit significant functional change in the laboratory (up to 92% accuracy). But, these tools consistently fail in correctly annotating laboratory-induced mutations that show no functional impact in the laboratory assays. Therefore, the overall accuracy of computational tools for laboratory-induced mutations is much lower than that observed for the naturally occurring human variants. We tested and rejected the possibilities that the preponderance of changes to alanine and the presence of multiple base-pair mutations in the laboratory were the reasons for the observed discordance between the performance of computational tools for natural and laboratory mutations. Instead, we discover that the laboratory-induced mutations occur predominately at the highly conserved positions in proteins, where the computational tools have the lowest accuracy of correct prediction for variants that do not impact function (neutral). Therefore, the comparisons of experimental-profiling results with those from computational predictions need to be sensitive to the evolutionary conservation of the positions harboring the amino acid change.
Contact: s.kumar@asu.edu
1 INTRODUCTION
Site-directed mutagenesis followed by functional assays has enabled scientists to directly profile the functional differences among proteins that only differ by a single amino acid (Hutchison et al., 1978; Ling and Robinson, 1997; Yan et al., 2009). Through such experiments, one can identify sequence domains critical for specific functions and evaluate the degree of functional change induced by a mutation (Tao and Cornish, 2002). Frequently the functional outcomes of laboratory experiments have also been compared with predictions yielded by computational tools that annotate whether or not a single amino acid mutation will impact protein function. If they are successful, computational tools would offer a cost-efficient way to prioritize variants to explore in the laboratory. However, investigators have reported a wide range of success of computational tools from perfect accuracy (e.g. Zou et al., 2011) to rather poor results [e.g. <60% accuracy (Di et al., 2009)]. These differences might occur due to limited sample sizes (number of mutations) investigated in individual studies that are focused on one protein or members of a protein family (e.g. Hao et al., 2010). The availability of information on over 10 000 laboratory-induced mutations provides an opportunity to (1) measure the performance of computational tools for accuracy in diagnosing laboratory mutations and (2) compare them with the performance observed for a large collection of naturally occurring human variants with and without disease implications.
Here, we report a computational analysis of 10 913 protein point mutations from 2372 proteins investigated in a variety of functional assays in the laboratory, which are available online in the UniProt resource (Magrane and Consortium, 2011). Although a large number of computational tools are available, we focused our investigation on three tools: Condel, which is reported to have the highest accuracy (González-Pérez and López-Bigas, 2011); PolyPhen-2, which has widespread and long-term usage in the field (Adzhubei et al., 2010); and SIFT (Kumar et al., 2009a), which is intended to guide laboratory experiments and has also been used for diagnosing protein variation for many years.
2 METHODS
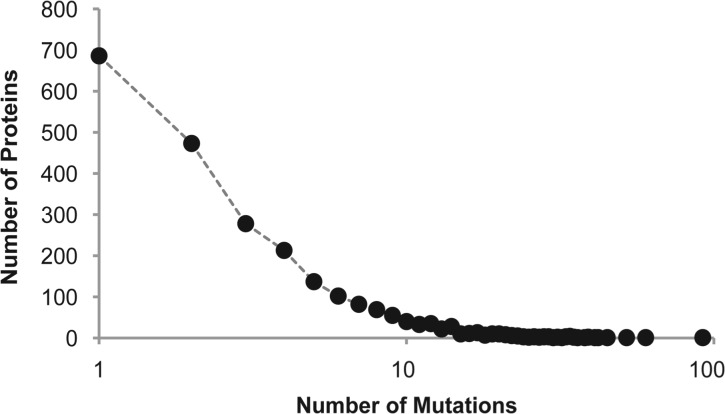
UniProt database (www.uniprot.org) is currently the largest repository of mutations of human proteins that have been experimentally induced by mutagenesis (Magrane and Consortium, 2011). We used this resource to retrieve all available laboratory-induced mutations with experimental descriptions and effects in which exactly one mutation was tested for each polypeptide in order to avoid confounding evolutionary interpretations. The final dataset contained 10 913 laboratory-induced mutations from 2372 proteins (Fig. 1).
Fig. 1.
A frequency distribution showing the number of different mutants explored in the laboratory for 2372 human proteins, as reported in the UniProt database. The median (mean) number of mutations analyzed is 3 (4.6)
Based on the experimental outcomes, we designated all laboratory-induced mutations with any measurable effect on function in the laboratory to be lab-damaging mutations. All others were given a lab-neutral designation. In this dataset, the ratio of damaging to neutral mutations was 6:1. In a survey of the severity of functional impact of lab-damaging mutations, we found that a subset of lab-damaging mutations (42%) was reported to completely abolish the function assayed. We refer to them as lab-abolishing mutations.
Each laboratory-induced mutation was subjected to Condel, PolyPhen-2 and SIFT analyses using their respective web servers (bg.upf.edu/condel/analysis; genetics.bwh.harvard.edu/pph2 and sift.jcvi.org). These tools did not produce diagnoses (neutral or non-neutral) for some mutations, therefore we reported accuracies for a subset (in parentheses) of all laboratory-induced mutations for Condel (10 765), PolyPhen-2 (10 814) and SIFT (7172). In particular, SIFT servers failed to produce diagnosis for ∼ 35% of the laboratory-induced mutations because SIFT's pre-computed database only provides results for variants that have been reported in dbSNP (Sherry et al., 2001) in order to increase the speed. We found that the accuracy of Condel and PolyPhen-2 for the subset of mutations with SIFT diagnosis was <1% different from those obtained by using all mutations, so we presented and compared all accuracies directly.
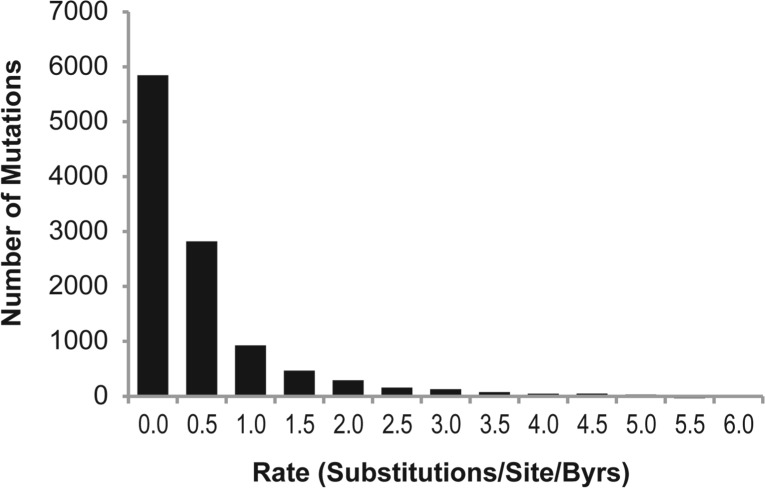
In order to assess the degree of (evolutionary) selective pressure, we estimated the absolute evolutionary rate (r) for each position containing laboratory-induced mutations by using the approach of Kumar et al. (2009b) where evolutionary rate is calculated using the appropriate protein sequence alignment from the UCSC Genome Browser (Rhead et al., 2010) and a timetree of 46 species (Kumar and Hedges, 2011). The evolutionary rate has the unit of the number of amino acid substitutions per site per billion years. A histogram showing the distribution of evolutionary rates for positions harboring laboratory-induced mutations is shown in Figure 2. For simplicity, we divided these amino acid positions into three categories based on the estimated evolutionary rates: ultra-conserved (r = 0), well-conserved (0 < r ≤ 1) and less-conserved (r > 1).
Fig. 2.
The distribution of long-term evolutionary rates (r) of positions containing 10 913 laboratory mutations analyzed in this study
3 RESULTS AND DISCUSSION
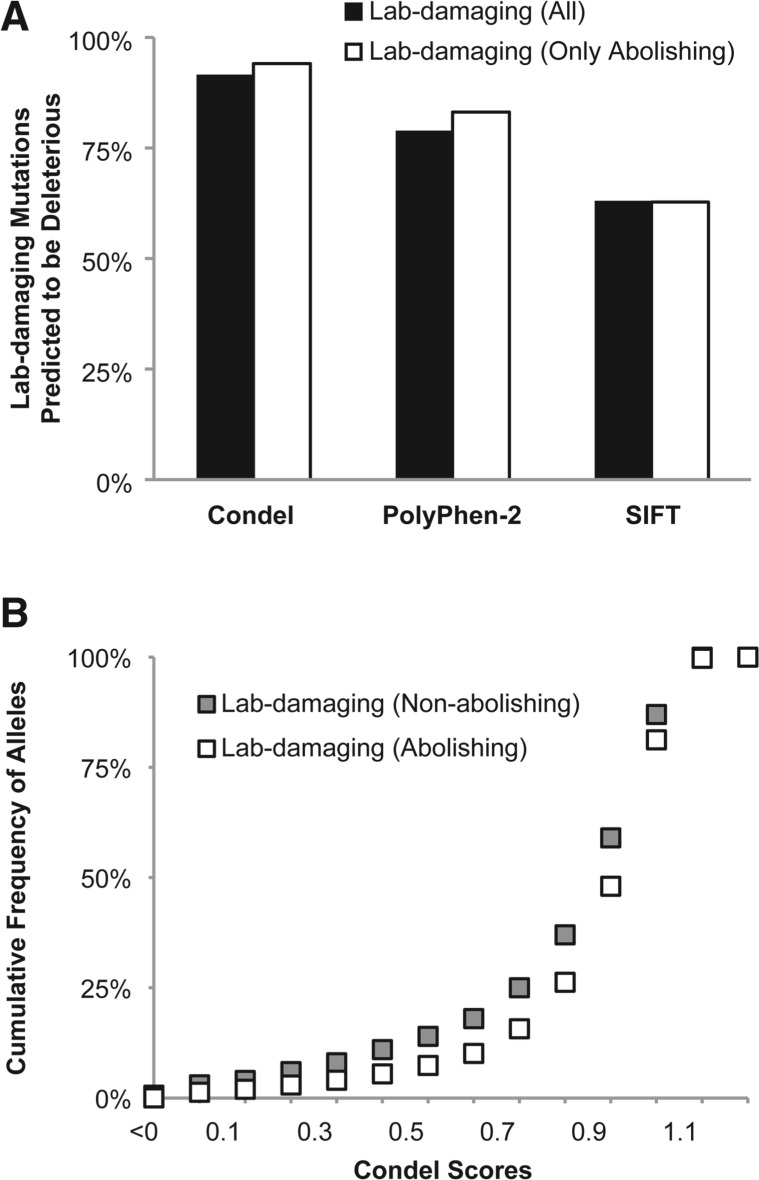
The true positive rate (non-neutral diagnosis) for lab-damaging mutations was generally high for all three methods (Fig. 3A). Condel provided the highest accuracy with a 92% correct diagnosis rate, whereas PolyPhen-2 and SIFT showed lower accuracies (79 and 63%, respectively). More than 95% of lab-damaging mutations received non-neutral diagnosis by at least one of the tools, and all three tools produced correct concordant diagnoses for slightly <50% mutations. A similar pattern of prediction accuracy was obtained for 3969 lab-abolishing mutations, which are a subset of lab-damaging mutations (Fig. 3A).
Fig. 3.
Accuracy of computational tools in predicting the functional impact of laboratory-induced mutations that alter the protein function. (A) Proportion of mutations correctly diagnosed to be non-neutral by Condel, PolyPhen-2 and SIFT. The results are shown for all lab-damaging mutations (filled bars) and only those damaging mutations that abolish the protein function (open bars). (B) The cumulative frequency distribution of lab-damaging mutations that abolish (open squares) and do not abolish function (gray squares) at various levels of deleteriousness as measured by their Condel scores
The functional impact scores that measure the deleteriousness of laboratory-induced mutations produced by Condel were quite similar for lab-damaging mutations that completely abolished protein function and the remainder of lab-damaging mutations that exhibited partial protein function (Fig. 3B). On average they differed by 6% (0.85 and 0.79, respectively), which is small but statistically significant (two-tailed Z-test; P < 0.01). Overall, computational tools performed well in diagnosing laboratory-induced mutations with function effects. In addition, these accuracies are comparable to those reported for human disease-associated variants (Adzhubei et al., 2010; González-Pérez and López-Bigas, 2011; Kumar et al., 2011).
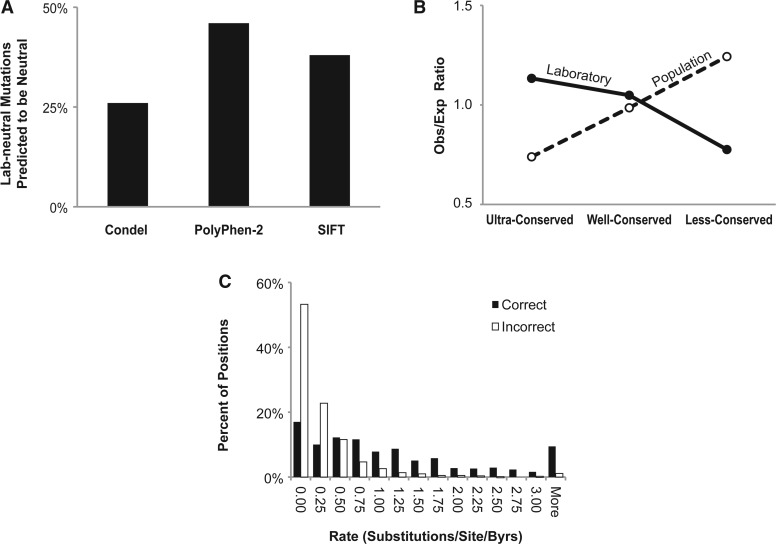
In contrast, computational tools performed poorly on the lab-neutral mutation set. Only 26, 46 and 38% of mutations were correctly diagnosed to be neutral by Condel, PolyPhen-2 and SIFT, respectively (Fig. 4A). All three methods produced the correct diagnosis for <10% of the mutations, with only 35% laboratory-induced mutations correctly diagnosed by two or more methods (i.e. consensus approach). Therefore, computational tools are unable to aid in correctly diagnosing mutations found to have no functional impact in the laboratory, even when we used a strict rule for laboratory neutrality where only mutations with no measureable functional effects are given the lab-neutral designation. These results are rather surprising, because computational tools are known to be highly accurate in diagnosing neutral polymorphisms found in the human population (Adzhubei et al., 2010; González-Pérez and López-Bigas, 2011; Kumar et al., 2009a; Ng and Henikoff, 2006).
Fig. 4.
The accuracies of computational tools and evolutionary properties of mutations. (A) Fraction of lab-neutral mutations predicted correctly by Condel, PolyPhen-2 and SIFT. (B) Observed-to-expected ratios of the lab-neutral mutations (closed circles, solid line) and human population variants (open circles, dashed line) in three evolutionary conservation categories. (C) Histogram of evolutionary rates of lab-neutral mutations that were diagnosed correctly (black bars) and incorrectly (white bars) by PolyPhen-2; similar results are observed for Condel and SIFT. For panel B, 456,426 protein variants were obtained from the 1000 Genomes Project (Consortium, 2010). Relative proportions of positions in each category was estimated by considering evolutionary rates of all amino acid positions found in proteins containing at least lab-induced mutation or population variant, as appropriate. These relative proportions were then used to generate expected numbers of lab-neutral mutations in each category
We explored different possibilities to potentially explain the observed patterns. First, the laboratory mutation sets are expected to contain an overabundance of changes to alanine because of alanine scanning, which is a cost-efficient alternative to testing every possible non-native amino acid (Bromberg and Rost, 2008). Indeed, nearly half of all lab-neutral mutations are changes to alanine. Therefore, we estimated the accuracy of computational tools after excluding all such mutations. This resulted in <3% change in accuracy of computational tools and, thus, does not explain the observed pattern.
Second, we evaluated the accuracies of computational tools for amino acid changes caused by single base pair mutations. This is important because we found that one-third of the laboratory-induced protein mutations in our dataset were a result of multiple base pair changes in the same codon, which is not common in naturally observed variants in human populations. Amino acid differences with multiple base pair mutations have much lower (>2-fold) amino acid substitution probabilities (e.g. BLOSUM62 scores) when compared with those with single base pair mutations. This fact might explain the reduced performance of computational tools that have been optimized using data from human disease-associated and neutral variants, e.g. HumVar2 (Adzhubei et al., 2010; González-Pérez and López-Bigas, 2011). The accuracies of individual computational tools differed by <10% in comparisons of single- and multi-nucleotide variants. Therefore, the inclusion of amino acid mutations caused by multi-nucleotide changes does not explain the observed pattern of low prediction accuracy of computational tools.
Third, we examined the preponderance of lab-neutral mutations in ultra-, well- and less-conserved positions because we have previously found that the accuracy of PolyPhen in correctly diagnosing neutral mutations is the lowest at the most highly conserved amino acid positions (Kumar et al., 2009b). If lab-neutral mutations were overrepresented at ultra-conserved positions, this might explain the reduced accuracy of computational tools for lab-neutral mutations as well. Indeed, there is a significant overabundance of lab-neutral mutations at highly conserved positions, which is exactly opposite of the pattern observed for human polymorphisms (Subramanian and Kumar, 2006; Fig. 4B). Figure 4C clearly shows that the lab-neutral mutations found at highly conserved positions are very difficult to diagnose correctly, whereas the accuracy of diagnosis is higher for lab-neutral mutations found at fast evolving positions. The inaccuracy of computational tools in diagnosing mutations with no discernible functional change in the laboratory points to the need for high-throughput laboratory profiling to identify truly neutral mutations at evolutionarily conserved positions.
Of course, one could argue that the identification of truly neutral mutations in the laboratory is much harder than identifying damaging mutations, because the numbers of laboratory functional assays are limited by current technology and by our knowledge of all functions of a protein. This would mean that a protein mutation that is not disruptive for a known set of assayed functions would be designated a neutral status in the laboratory, although it may be disruptive to an untested function or in vivo. These possibilities can only be tested when information on in vivo assays and additional functional knowledge becomes available.
However, our analyses clearly show that the differences in successful prediction of neutral mutations in the laboratory and those observed in nature can be resolved to a large extent by taking an evolutionary-aware approach. For example, all 92 damaging mutations of the beta-globin gene (HBB) in the UniProt resource occur at ultra- and well-conserved positions, for which both Condel and PolyPhen-2 produce 100% correct diagnosis. This consistency occurs because computational tools have high rates of correct diagnosis for damaging mutations at ultra- and well-conserved positions. On the other hand, 48 lab-neutral mutations of cystathionine beta synthase proteins also occur at ultra- and well-conserved positions. They are expected to be misdiagnosed by computational tools, which is indeed the case (100% incorrect by PolyPhen-2 and Condel). Therefore, by using the evolutionary conservation of positions mutated in the laboratory, it will now be possible for scientists to better understand the reasons for the discordance between the outcomes of laboratory experiments and computational predictions. This would ultimately improve the combined use of experimental and computational techniques to survey the functional impacts of millions of protein mutations that we are expected to encounter as we acquire increasing numbers of sequences from human exomes.
ACKNOWLEDGEMENTS
We thank Li Liu, Mia Champion, Alan Filipski and Crystal Hepp for helpful discussions and comments on an initial version of the manuscript. We are grateful to Kristyn Gerold and Maxwell Sanderford for extensive assistance with data analysis.
Funding: The US National Library of Medicine (NIH LM10834 to S.K.) and National Institute of General Medicine (NIH training grant GM071798, Brenda Hogue).
Conflict of interest: none declared.
REFERENCES
- Adzhubei I., et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., Rost B. Comprehensive in silico mutagenesis highlights functionally important residues in proteins. Bioinformatics. 2008;24:i207–i212. doi: 10.1093/bioinformatics/btn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G.P. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y.M., et al. Prediction of deleterious non-synonymous single-nucleotide polymorphisms of human uridine diphosphate glucuronosyltransferase genes. AAPS J. 2009;11:469–480. doi: 10.1208/s12248-009-9126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score. Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D., et al. Phenotype prediction of nonsynonymous single nucleotide polymorphisms in human phase II drug/xenobiotic metabolizing enzymes: perspectives on molecular evolution. Sci. China Life. Sci. 2010;53:1252–1262. doi: 10.1007/s11427-010-4062-9. [DOI] [PubMed] [Google Scholar]
- Hutchison C.A., et al. Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 1978;253:6551–6560. [PubMed] [Google Scholar]
- Kumar P., et al. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009a;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kumar S., et al. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009b;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., et al. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27:377–386. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B. TimeTree2: species divergence times on the iPhone. Bioinformatics. 2011;27:2023–2024. doi: 10.1093/bioinformatics/btr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M.M., Robinson B.H. Approaches to DNA mutagenesis: an overview. Anal Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- Magrane M. UniProt Consortium. UniProt Knowledgebase: a hub of integrated protein data. 2011:bar009. doi: 10.1093/database/bar009. Database (Oxford), 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics. Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Rhead B., et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Cornish V.W. Milestones in directed enzyme evolution. Curr. Opin. Chem. Biol. 2002;6:858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- Yan Z., et al. Progress and prospects: techniques for site-directed mutagenesis in animal models. Gene Ther. 2009;16:581–588. doi: 10.1038/gt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., et al. Mutation prediction by PolyPhen or functional assay, a detailed comparison of CYP27B1 missense mutations. Endocrine. 2011;40:14–20. doi: 10.1007/s12020-011-9489-7. [DOI] [PubMed] [Google Scholar]