Abstract
Summary: Microbial communities have an important role in natural ecosystems and have an impact on animal and human health. Intuitive graphic and analytical tools that can facilitate the study of these communities are in short supply. This article introduces Microbial Community Analysis GUI, a graphical user interface (GUI) for the R-programming language (R Development Core Team, 2010). With this application, researchers can input aligned and clustered sequence data to create custom abundance tables and perform analyses specific to their needs. This GUI provides a flexible modular platform, expandable to include other statistical tools for microbial community analysis in the future.
Availability: The mcaGUI package and source are freely available as part of Bionconductor at http://www.bioconductor.org/packages/release/bioc/html/mcaGUI.html
Contact: wade@kingcopeland.com; zabdo@uidaho.edu
Supplementary Information: Supplementary data and figures are available at Bioinformatics online.
1 INTRODUCTION
Several graphical user interface software (GUI) packages exist for the analysis of microbial community data. These programs include InVUE, which specializes in creating interactive graphical representations for large datasets (Ravel et al., 2011) and UniFrac, a web-based GUI that allows users to compare microbial communities using phylogenetic information (Lozupone et al., 2006). Many of these applications were built using programming languages such as C and C++. Currently, there are few GUIs specifically available for the analysis of microbial communities that also take advantage of the R programming language. This article introduces Microbial Community Analysis GUI (mcaGUI), an R-package built to help microbial biologists harness the power of statistical analysis tools provided in R without having to learn the R programming language. Using mcaGUI, researchers can perform statistical analyses and create various interactive and analytical data representations specific to microbial ecology.
What distinguishes mcaGUI from similar applications is its ability to directly input aligned sequence data that provide information about microbial community composition. This allows researchers to create custom abundance tables using both sample metadata and data on operational taxonomic units (OTU). mcaGUI is modular and expandable; it provides a platform that can be extended to include more functionality in the future.
2 METHODS
mcaGUI has a simple layout that is divided into three parts (Supplementary Fig. S1). The left side of the screen contains the variable browser, which shows all created variables or objects that contain imported data and outcomes of data manipulations such as richness estimates. On the top right is the tabbed browser where the data can be viewed in tabular form and analyses are performed. For most operations in mcaGUI, a new tab opens where the user can input additional arguments. On the bottom right is the command window where information and output of some operations is shown.
We implemented package OTUbase (Beck et al., 2011) to simplify loading sequence data into R. These data, along with all relevant information, are stored in an R object and can be used to construct custom abundance tables. OTU abundance tables can also be loaded directly but with limited customization options.
3 BIOLOGICAL APPLICATIONS
mcaGUI provides an interface to access a set of statistical tools to summarize and analyze microbial community data such as principal component analysis (PCA), cluster analysis and others (see Supplementary Table S1 for a detailed listing). A complete workflow with examples, along with detailed description of data input and output, can be found in the Supplementary material. Herein, we introduce three examples to illustrate the use of mcaGUI. The first illustrates how to input sequence data and create custom abundance tables. The second illustrates how to estimate microbial richness and evenness. The third illustrates how to perform PCA. These examples utilize the 454 pyrosequencing data presented in Sogin et al. (2006) that were used to study and compare the microbial community composition and diversity in seven oceanic sites. Elaborate details for each of these examples can be found in the Supplementary material.
3.1 Example 1: create a custom abundance table
To create a custom abundance table, the user must first input sequence data. This is done by navigating to the File Menu and clicking the Read OTUbase option (Supplementary Fig. S2). Supplementary Figure S3 shows an image with filled in values for reading in the Sogin et al. (2006) sequence data. Once the data are loaded, the user can navigate to the Data Menu and press the Abundance Table option (Supplementary Fig. S4). The user can customize the rows and columns of the abundance table by pressing the Update Feature and Sample Meta-Data button. Supplementary Figure S5 shows the dialogue box by which the user creates an abundance table on the genus level aggregated by site. The researcher can either save this custom abundance table or use it in further analyses within the mcaGUI interface.
3.2 Example 2: richness and diversity estimation
Richness and diversity can be estimated by navigating to the Analysis Menu and Clicking the Richness Estimation or Diversity Estimation buttons, respectively (Supplementary Fig. S6). An abundance table should be specified and the output table of estimates should be assigned to a new variable (Supplementary Fig. S7). Estimates can be accessed by double clicking this new variable in the variable browser (Supplementary Fig. S8). The researcher can then use built in tools to plot these estimates or export the results to a delimited data file that can be used with other programs.
3.3 Example 3: using PCA to view groups of correlated OTUs
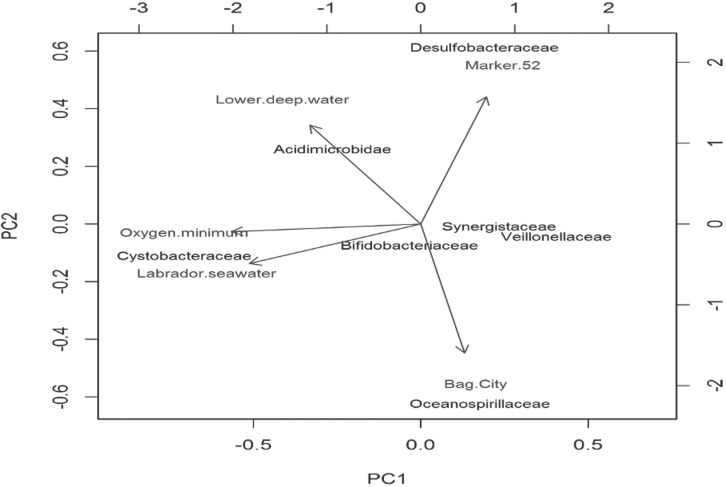
We perform a PCA analysis by navigating to the Analysis Menu, Multivariate and then PCA. Supplementary Figure S9 shows the dialogue window for PCA. Plot 2D displays a biplot with the first two PCA coordinates (Fig. 1). Figure 1 indicates that Acidimicrobidae is associated with lower deep water while Cystobacteraceae is associated with Labrador seawater, for example. Plot 3D shows an interactive biplot in the first three PCA coordinates. These plots allow the user to zoom in and out and rotate the axes to identify interesting patterns in the data. Selecting Scree Plot or Elbow plot, displays a graphic used to identify which PCA directions account for most of the variation in the data.
Fig. 1.
Example output of a Biplot created using mcaGUI. The data used is from Sogin et al. (2006) but the number of observations and explanatory variables were reduced to improve readability
4 THE GRAPHICAL VIEW
mcaGUI provides both basic and interactive graphics. When Bootstrap Interval Estimation is used (Supplementary material), mcaGUI returns histograms that show the bootstrap distribution of each statistic requested. When using PCA the user can view a biplot in the first three PCA directions and interactively explore the space. Other graphical tools include histograms, box plots, scatter plots, pairs-plots and cluster diagrams.
5 IMPLEMENTATION
Poor Man's GUI (Verzani, 2010) provided the base layout for mcaGUI. Additional functionality was added with the gWidgets (Verzani, 2007) and gWidgetsRGtk2 (Lawrence and Verzani 2011) R-packages. All GUI layouts were created using Widget and Handler combinations; a widget is an object that specifies a layout, and a handler is an action that is then used by the widget to run analyses. Utilizing these tools, new functionality can be added by linking pre-existing packages to this GUI.
Supplementary Material
ACKNOWLEDGEMENTS
We thank John Verzani for his development and assistance with Poor Man's GUI.
Funding: National Institutes of Health grant number NIH-NCRR COBRE # P20 RR16448, NIH-1UH2AI083264-01, NIH-U19 AI084044 and NIH-R24 RR023344-01A2.
Conflicts of Interest: none declared.
REFERENCES
- Beck D., et al. OTUBase: an R infrastructure package for taxonomic unit data. Bioinformatics. 2011;27:1700–1701. doi: 10.1093/bioinformatics/btr196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., et al. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M.L., et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. PNAS. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzani J. An Introduction to gWidgets. R News. 2007;7:26–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.