ABSTRACT
Surface-associated bacterial structures known as biofilms are the target of intense antimicrobial research efforts. We recently identified several secreted proteins that are retained in the bacterial biofilm matrix by their association with the biofilm exopolysaccharide scaffold. Based on our findings, we hypothesized that these problematic bacterial structures might be reengineered to serve as reservoirs for surface-active secreted proteins of biomedical, bioengineering, or biotechnological importance. By piggybacking onto one of these scaffold-associated proteins, we were able to sequester a functional enzyme to the biofilm matrix. We hypothesize that this technology may have diverse applications in vaccine design, digestive disease, and bioremediation.
THE HYPOTHESIS
Bacterial biofilm formation is the process by which bacteria adhere to surfaces to form single or multilayer structures (1). These structures are found on biotic surfaces such as the epithelia of animals and on abiotic surfaces such as those of mineral deposits, soil, and air-water interfaces. Because of their affinity for surfaces, biofilms are ideal vehicles for presentation of vaccine antigens to epithelia and for delivery of enzymes of therapeutic or bioremediative importance to surfaces.
The bacterial biofilm matrix is comprised of exopolysaccharide, proteins, and DNA. While the biofilm exopolysaccharide has been described as the adhesive material that cements cell-surface and intercellular interactions, there is increasing evidence to support a different paradigm in which the biofilm exopolysaccharide serves as a scaffold for proteins that mediate these adhesive interactions (2, 3). Because of the abundance of these adhesive proteins in the biofilm matrix, we conceived of using this matrix as a reservoir for functional enzymes intended for delivery to surfaces. We and others have identified the three adhesive proteins Bap1, RbmA, and RbmC in the Vibrio cholerae biofilm matrix (2, 4-6). Here we demonstrate the feasibility of using RbmA in V. cholerae as a biofilm matrix-targeting moiety for proteins of biological significance. We hypothesize that a similar strategy could be used to harness the biofilm matrices of diverse bacteria for protein presentation and delivery.
DELIVERY OF A FUNCTIONAL ENZYME TO THE BIOFILM MATRIX
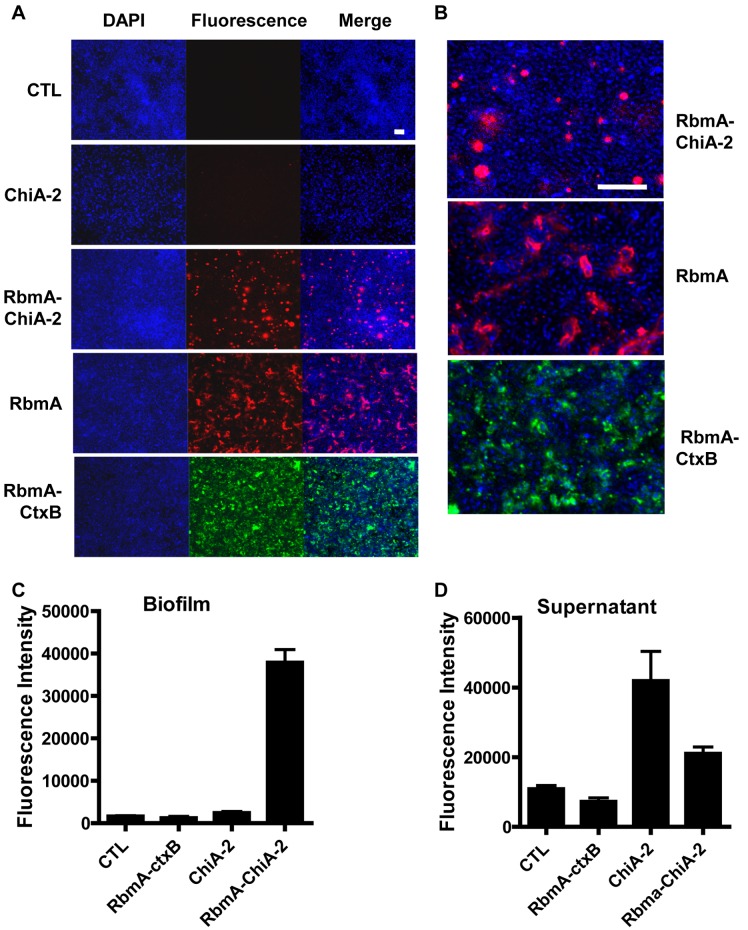
We reasoned that we might deliver a functional enzyme to a surface by fusing it to RbmA. To test this, we inserted a secreted V. cholerae chitinase between RbmA and a C-terminal FLAG tag (RbmA–ChiA-2–FLAG) and expressed this fusion protein from a plasmid in wild-type V. cholerae. As controls, we also generated wild-type V. cholerae carrying an empty vector or the same vector encoding RbmA alone, RbmA fused to the B subunit of cholera toxin (CtxB), or ChiA-2–FLAG. Biofilms were formed with these five strains, and immunofluorescence experiments were performed using an anti-FLAG antibody or an antibody to CtxB in the case of RbmA-CtxB. As shown in Fig. 1A and B, the RbmA–ChiA-2–FLAG fusion was concentrated in the biofilm, whereas the ChiA-2–FLAG protein alone was not visible. Like native RbmA, the RbmA-CtxB fusion protein surrounded cells in the biofilm (2). In contrast, the RbmA–ChiA-2 fusion protein had a more punctuate distribution.
FIG 1 .
An enzymatically active RbmA–ChiA-2–FLAG fusion protein is retained in the biofilm matrix. (A) Immunofluorescent imaging of the distribution of ChiA-2–FLAG, RbmA–ChiA-2–FLAG, RbmA-FLAG, or RbmA-CtxB in a biofilm formed by wild-type V. cholerae carrying a plasmid encoding each of these proteins. The proteins were visualized with an anti-FLAG antibody or anti-CtxB antibody in the case of RbmA-CtxB. Bacterial DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). As a control, a biofilm formed by wild-type V. cholerae carrying an empty vector was developed with an anti-FLAG antibody (CTL) (bar = 10 µM). (B) A magnified view of the distribution of RbmA–ChiA-2–FLAG in the biofilm (bar = 10 µM). (C and D) Chitinase activity measured in the biofilms (C) and supernatants (D) of wild-type V. cholerae carrying an empty vector (CTL) or a plasmid encoding RbmA-CtxB (RbmA-CtxB), ChiA-2–FLAG (ChiA-2), or RbmA–ChiA-2–FLAG (RbmA–ChiA-2). The chitinase activity in the biofilm of the strain expressing RbmA–ChiA-2–FLAG was significantly different from that in all other biofilms (P ≤ 0.0003). Similarly, chitinase activity in the supernatants of strains expressing either RbmA–ChiA-2–FLAG or ChiA-2–FLAG was significantly different from that of strains carrying the control vector (P = 0.007 and P = 0.0215, respectively) or the RbmA-CtxB fusion (P = 0.0025 or P = 0.0149, respectively). The difference in chitinase activity between the supernatants of the strains expressing RbmA–ChiA-2–FLAG and ChiA-2–FLAG was not statistically significant (P = 0.07).
To be useful in surface modification, enzymes directed to the biofilm matrix must retain their activity. Therefore, we assessed whether the biofilm-associated RbmA–ChiA-2 fusion protein retained enzymatic activity. We formed biofilms with wild-type V. cholerae expressing ChiA-2–FLAG or RbmA–ChiA-2–FLAG from a plasmid. As additional controls, wild-type strains carrying either an empty vector or a vector encoding RbmA-CtxB were included. Planktonic cells were removed, the biofilms were rinsed, and the chitinase activity of the biofilms and cell supernatants was tested using a luminescence-based assay. As shown in Fig. 1C, chitinase activity was approximately 15 times greater in the biofilm formed by the strain expressing the RbmA–ChiA-2–FLAG fusion than in that formed by the strain expressing ChiA-2–FLAG. Because chitinase is a secreted protein native to V. cholerae, chitinase activity was high in all the cell supernatants and particularly high in those expressing either ChiA-2–FLAG alone or the RbmA–ChiA-2–FLAG fusion protein (Fig. 1D). Lastly, fusion of RbmA to CtxB did not alter the basal level of chitinase activity observed in the biofilm or supernatant. These experiments demonstrate that our strategy can be used to retain active enzymes in the biofilm matrix. We hypothesize that this would allow targeting of enzymes to surfaces.
USE OF EXOPOLYSACCHARIDE-COATED PLANKTONIC CELLS AS PROTEIN PRESENTATION PLATFORMS
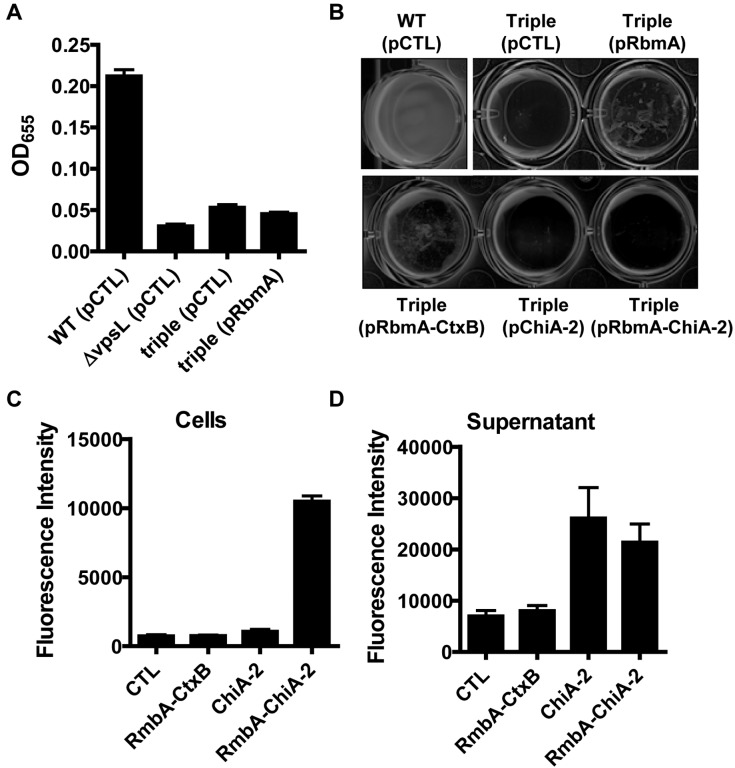
For applications in which the biofilm exopolysaccharide is to be used as a protein reservoir but not a surface targeting device, it may be more convenient to anchor the secreted protein to planktonic cells. We have previously shown that Bap1 and RbmC are found at the biofilm-surface interface and are important for anchoring the biofilm to surfaces, while RbmA is distributed throughout the biofilm and cements intercellular interactions (2). We hypothesized that a Δbap1ΔrbmAΔrbmC mutant would be defective in cell-surface and cell-cell interactions and, therefore, unable to form a biofilm. To test this, we created a triple mutant and assessed biofilm formation. As shown in Fig. 2A, in a standard biofilm assay, surface association by the triple mutant was comparable to that by an exopolysaccharide mutant. As expected, provision of a plasmid encoding RbmA did not rescue surface association by the triple mutant. Furthermore, the triple mutant did not form a pellicle, suggesting that intercellular interactions were also defective (Fig. 2B). Rescue of the triple mutant with native RbmA or an RbmA-CtxB fusion protein resulted in aggregation of cells, indicating some restoration of intercellular interactions (Fig. 2B). In contrast, neither ChiA-2 nor the RbmA–ChiA-2 fusion protein restored aggregation to the triple mutant. We conclude that, unlike the RbmA-CtxB fusion, the RbmA–ChiA-2 fusion is unable to fulfill the function of native RbmA. One possibility is that the irregular distribution of this fusion protein around cells prevents rescue of RbmA function.
FIG 2 .
A V. cholerae Δbap1ΔrbmAΔrbmC triple mutant does not make a biofilm but can recruit the chitinase activity of RbmA–ChiA-2–FLAG to the cell surface. (A and B) Quantification of biofilms formed by wild-type V. cholerae (WT), an exopolysaccharide mutant (ΔvpsL), and a Δbap1ΔrbmAΔrbmC mutant (triple) carrying an empty vector (pCTL) or a vector encoding RbmA (pRbmA) (A) and the pellicle formed by wild-type V. cholerae (WT) or the Δbap1ΔrbmAΔrbmC triple mutant carrying either an empty vector (pCTL) or plasmids encoding RbmA-FLAG (pRbmA), RbmA-CtxB (pRbmA-CtxB), ChiA-2–FLAG (pChiA-2), or RbmA–ChiA-2–FLAG (pRbmA–ChiA-2) (B). (C and D) Chitinase activity in the cellular fraction (C) and supernatants (D) of V. cholerae Δbap1ΔrbmAΔrbmC mutant carrying an empty vector or a plasmid encoding RbmA-CtxB, ChiA-2–FLAG, or RbmA–ChiA-2–FLAG. Chitinase activity in the cellular fraction of the mutant expressing RbmA–ChiA-2–FLAG was significantly different from those in strains expressing all other recombinant proteins (P < 0.0001). The chitinase activity in the supernatants of mutants expressing ChiA-2–FLAG and RbmA–ChiA-2–FLAG was significantly different from those of the mutant carrying an empty vector (P = 0.034 and P = 0.0172) and the mutant expressing RbmA-CtxB (P = 0.039 and P = 0.0215). The difference in chitinase activity between the supernatants of the strains expressing RbmA–ChiA-2–FLAG and ChiA-2–FLAG was not statistically significant (P = 0.535).
To determine whether the RbmA–ChiA-2–FLAG fusion could remain associated with planktonic cells, a triple mutant expressing RbmA–ChiA-2–FLAG was cultured with vigorous agitation, cells were pelleted, and the chitinase activity of both the cellular fraction and the supernatant was assayed. As shown in Fig. 2C and 2D, chitinase activity remained sequestered to cells expressing the RbmA–ChiA-2–FLAG fusion but not to cells expressing ChiA-2–FLAG or RbmA-CtxB. This suggests that RbmA–ChiA-2–FLAG remains cell associated even in the absence of a biofilm structure and in the face of considerable agitation. We hypothesize that the biofilm matrix exopolysaccharide is produced and exported by the triple mutant. Furthermore, we provide evidence that the interaction of RbmA fusion proteins with this matrix exopolysaccharide is robust. These experiments suggest that this protein presentation platform is also amenable to use with planktonic bacterial cells.
CONCLUSION
Here we demonstrate the feasibility of a novel application of the bacterial biofilm matrix exopolysaccharide as a scaffold for localization and presentation of proteins and for delivery of functional enzymes to biotic or abiotic surfaces. These “proof-of-principle” experiments have been performed with the diarrheal pathogen V. cholerae. While V. cholerae is not the appropriate vehicle for every application, results emerging from other laboratories suggest that the matrices of many bacterial biofilms incorporate structural proteins that may be used in a fashion similar to that presented here (7, 8-10). However, this remains to be tested.
We hypothesize that this approach will have broad applications. We envision that it can be used to deliver many types of enzymes to surfaces. For instance, a commensal bacterium such as Escherichia coli or a commonly used probiotic might be used to deliver lactase or pancreatic enzymes to the intestinal brush border of hosts deficient in these enzymes. A nonpathogenic bacterium colonizing the lung of a cystic fibrosis patient might be reengineered to deliver mucinase or alginase, thus helping to clear biofilm-associated Pseudomonas aeruginosa from the lung. Enzymes are often used in bioremediation. A third application could be the delivery of such enzymes to contaminated surfaces. Lastly, this suggests an economical and versatile new platform for delivery of protein antigens or immune adjuvants in whole-cell vaccines.
Bacterial biofilms, which are often described as slime, have been vilified in medicine and industry. Here we present evidence that the biofilm matrix can be exploited as a vehicle for concentration of enzymes and antigens on the surfaces of cells and as a delivery system targeting abiotic surfaces. This technology has broad implications that may include vaccine development, digestive dysfunction, bioremediation, and molecular biology. Furthermore, this genetic approach is self-renewing and, therefore, is more efficient and economical than chemical or biochemical approaches. With the discovery of additional structural proteins in the biofilm matrices of bacteria, this technology is poised for rapid implementation.
Bacterial strains, plasmids, and media.
V. cholerae O139 strain MO10 was used in all experiments (11). Vectors used for protein expression included an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter driving expression of the protein of interest with a C-terminal FLAG tag (pFLAG-CTC; Sigma-Aldrich). Bacteria were cultivated in Luria-Bertani (LB) broth supplemented with ampicillin (100 µg/ml). Because adequate protein expression was observed without induction, the growth medium was not supplemented with IPTG.
Construction of plasmids for protein expression.
The open reading frames (ORFs) of interest were amplified by PCR using primers that included the start and stop codons of each gene of interest. KpnI restriction sites were included in the primers used for amplification of chiA-2. These restriction sites were used to insert chiA-2 between rbmA and the FLAG tag in expression vector pFLAG-rbmA (2). KpnI and SalI restriction sites were used to fuse ctxB to the C-terminal end of rbmA pFLAG-rbmA. In this case, the FLAG tag was removed. All insertions were confirmed by sequence analysis.
Mutant construction.
The V. cholerae Δbap1ΔrbmCΔrbmA mutant was constructed as previously described (12) using the strain V. cholerae Δbap1ΔrbmC mutant (PW707) and the suicide plasmid pWM91ΔrbmA (2).
Biofilm assays.
A single colony of V. cholerae was inoculated into 1 ml of LB broth and allowed to grow to mid-exponential phase. The culture was then diluted in LB broth to yield an optical density at 655 nm (OD655) of 0.05 and divided into three disposable glass culture tubes (10 mm by 75 mm). These tubes were incubated statically at 27°C. After 24 h, planktonic cells were removed, and the OD655 of the cells was measured. Remaining biofilms were washed with 0.1 M phosphate-buffered saline (PBS) (pH 7.0) and then disrupted with 1-mm-diameter beads (Biospec). The OD655 of the resulting cell suspension was measured. For assays of biofilm integrity, biofilms were formed in 24-well plates as described above and then washed with PBS.
Immunofluorescence.
Immunofluorescence experiments were performed as previously described with the following modifications (2). To detect ChiA-2–FLAG and RbmA–ChiA-2–FLAG, an anti-FLAG M2 antibody (Sigma-Aldrich) (1:1,000 dilution) was used followed by incubation with DyLight 549 AffiniPure Rabbit anti-mouse IgG H + L (Jackson ImmunoResearch) (1:1,000 dilution). To detect the RbmA-CtxB fusion protein, an anti-CtxB antibody (Sigma) (1:1,000 dilution) followed with an Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen) was used. Confocal images were acquired at the Children’s Hospital, Boston, Imaging Core with an LSM700 microscope (Zeiss) equipped with a 63× objective and 405-, 488-, and 555-nm laser lines. A computer equipped with ZEN 2009 software was used to acquire and process images.
Chitinase assays.
For assays of protein activity, cells were cultured in LB broth supplemented with ampicillin at 27°C for approximately 5 h and then back diluted in the same medium to yield an OD655 of 0.05. For assays of activity within the biofilm, three 80-µl aliquots of each culture were transferred to the wells of a 96-well, black microtiter dish and three to wells of a polystyrene 96-well plate. Both plates were incubated statically at 27°C for 24 h. The planktonic fractions of the resulting cultures were removed, fractions from one well of the black plate and one well of the polystyrene plate were pooled, an OD655 was recorded, and the cell suspensions were centrifuged. A 5-µl volume of the supernatant was removed and assayed for chitinase activity. Biofilms remaining in the black 96-well plates were rinsed twice with PBS and assayed directly for chitinase activity. For chitinase assays using the Δbap1ΔrbmAΔrbmC mutant, cells were cultured in 1 ml of LB broth at 27°C with shaking overnight. An OD655 was recorded. The cell suspension was pelleted, and the supernatant was removed. Cells were rinsed once with PBS and then resuspended in an equal volume of PBS. A 5-μl volume of the cell suspension and 5 μl of the supernatant were assayed for chitinase activity. Chitinase activity was measured using a fluorometric chitinase assay kit (Sigma-Aldrich) according to the manufacturer’s protocol, including the following steps. Bacterial cells, biofilms, or supernatants were incubated in substrate buffer containing 4-methylumbelliferyl N,N′-diacetylchitobioside hydrate (0.2 mg/ml) for 20 min at 37°C in the dark prior to measurement of fluorescence with an Infinite 200 spectrophotometer (Tecan).
Statistical analysis.
Three experimental replicates were included in all quantitative experiments, and each experiment was repeated at least twice. Reported values represent the means of the results of three experimental replicate experiments, error bars represent standard deviations, and statistical significance was calculated using a Student’s t test.
ACKNOWLEDGMENTS
We thank Edward Ryan for helpful suggestions and Simon Dove and Daniel Smith for insightful discussions and careful reading of the manuscript.
This work was funded by NIH AI097612 to P.I.W.
Footnotes
Citation Absalon C, Ymele-Leki P, Watnick PI. 2012. The bacterial biofilm matrix as a platform for protein delivery. mBio 3(4):e00127-12. doi:10.1128/mBio.00127-12.
REFERENCES
- 1. Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7 e1002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nadell CD, Bassler BL. 2011. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl. Acad. Sci. U. S. A. 108:14181–14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fong JC, Karplus K, Schoolnik GK, Yildiz FH. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 188:1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fong JC, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189:2319–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diggle SP, et al. 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 8:1095–1104 [DOI] [PubMed] [Google Scholar]
- 8. Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 107:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tielker D, et al. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313–1323 [DOI] [PubMed] [Google Scholar]
- 10. Vidal O, et al. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waldor MK, Colwell R, Mekalanos JJ. 1994. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinant. Proc. Natl. Acad. Sci. U. S. A. 91:11388–11392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haugo AJ, Watnick PI. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]