Abstract
In the 7 years since dynamin was first isolated from bovine brain in search of novel microtubule-based motors, our understanding of this enzyme has expanded significantly. We now know that brain dynamin belongs to a family of large GTPases, which mediate vesicle trafficking. Furthermore, this enzymatic activity is markedly increased through association with microtubules, acidic phospholipids, and certain regulatory proteins that contain Src homology 3 (SH3) domains. From functional, genetic, and cellular manipulations, it is now generally accepted that dynamin participates in the endocytic uptake of receptors, associated ligands, and plasma membrane following an exocytic event. These observations have confirmed at least one function of dynamin that was predicted from seminal studies on a pleiotropic mutant, shibirets (shits) in Drosophila melanogaster. Of equal interest is the finding that there are multiple dynamin gene products, including two that are expressed in a tissue-specific manner, and they share marked homology with a larger family of distinct but related proteins. Therefore, it is attractive to speculate that the different dynamins may participate in related cellular functions, such as distinct endocytic processes and even secretion. In turn, dynamin could play an important role in cell growth, cell spreading, and neurite outgrowth. The purpose of this review is to enumerate on the expansive dynamin literature and to discuss the nomenclature, expression, and putative functions of this growing and interesting family of proteins.
Early Insights from the Drosophila melanogaster Mutant, shibirets (shits)
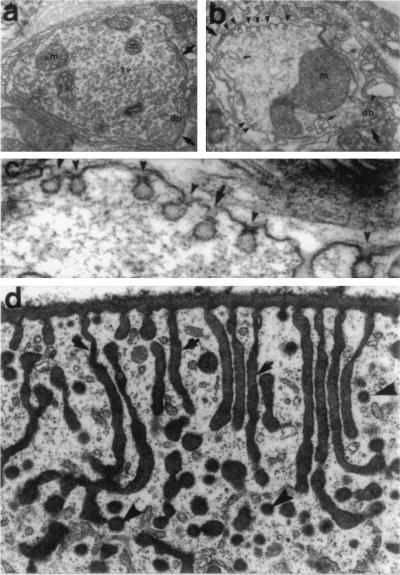
shits flies were initially isolated as temperature-sensitive paralytic mutants (1). The temperature-dependent paralysis that occurs in these flies results from a depletion of neurotransmitter-containing vesicles at nerve terminals (2). Ultrastructural studies have revealed that this reduction in the number of synaptic vesicles (SVs) is caused by an impairment in vesicle membrane recycling following exocytosis, resulting in an accumulation of “collared pits” at the presynaptic plasma membrane (refs. 3–5; Fig. 1 a–c). Interestingly, these invaginations accumulate in a number of other cell types in these flies, indicating a general defect in endocytosis (refs. 5 and 6; Fig. 1d).
Figure 1.
Endocytosis in neurons and epithelial cells is structurally altered in the Drosophila mutant shits. (a and b) Electron micrographs of a typical coxal synapse of a shits fly at 19°C (permissive temperature), which is characterized by numerous SVs, mitochondria (M), and a presynaptic dense body (DB). (b) A similar synapse at the restrictive temperature of 29°C. Note the loss of SVs and the appearance of several small invaginations of the plasma membrane, which lack clathrin but possess an electron-dense ring or collar around their necks (arrowheads). (c) A higher magnification of a synapse containing collared pits at 29°C. (d) In contrast to neuronal cells, long labyrinthine channels are formed in the cortical region of a shits garland cell at the restrictive temperature that lack collars. Cells have been impregnated with tannic acid to show that these structures (arrows) are continuous with the plasma membrane and end with a clathrin-coated pit (arrowheads). Few discrete coated vesicles are seen. [a–c were reproduced with permission from Koenig and Ikeda (4) (Copyright 1989, Soc. Neurosci.); and d was reproduced with permission from Kosaka and Ikeda (5) (Copyright 1983, Rockefeller Univ. Press).]
Shortly after the initial purification of dynamin from bovine brain, a cDNA encoding the full rat brain dynamin was isolated by Obar et al. (7) using an anti-dynamin polyclonal antibody. Subsequently, sequence analysis of the D. melanogaster shibire gene revealed an 81% similarity to the brain dynamin cDNA (8, 9). The finding that dynamin is the homologue of the shibire gene product was a significant step toward understanding the functional role of these proteins. Important information on the role of dynamin as an endocytic GTPase can be inferred from the sequence analyses of the two shibire alleles, shits1 and shits2, which occur within or near the GTP-binding motif and render the protein nonfunctional at the restrictive temperature (8). Similar mutations in dynamin overexpressed in mammalian cells inhibit receptor-mediated endocytosis and produce the shits phenotype (10–13). Thus, the extensive degree of structural similarity between dynamin and the shibire gene product suggests that they share functional homology and, as discussed below, substantiates recent experiments testing the involvement of dynamin in receptor-mediated endocytosis.
Identification of Novel Dynamin-Encoding Genes
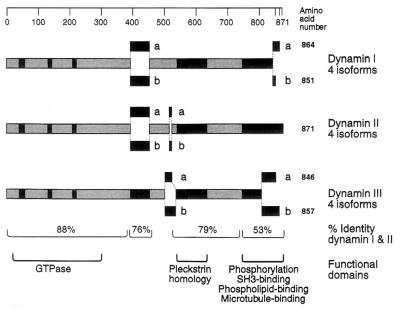
Because dynamin and its encoding cDNA were both originally isolated from mammalian brain, it was thought to be a neuron-specific protein. Early studies using Western and Northern blot analysis appeared to confirm this prediction (14). Subsequent studies using antibodies and cDNA probes to the enzymatic head domain of dynamin suggested that a highly related protein may be ubiquitous in distribution (15). Therefore, several laboratories initiated a search for additional dynamin-encoding genes that could be expressed in other tissues. Our laboratory (15) and Sontag et al. (16) independently isolated and characterized a novel dynamin-encoding cDNA that is ubiquitously expressed in all rat tissues examined. Sequence analyses revealed that this gene encodes for a protein of 868 aa with a calculated molecular mass of 98.2 kDa and is 79% identical at the protein level to the previously described rat brain dynamin. At the same time, Nakata et al. (17) isolated an additional dynamin cDNA from rat testis. The complete open reading frame encoded for a novel protein of 848 aa, which is approximately 80% identical to the two other dynamin isoforms. Northern blot analysis and in situ hybridization experiments had showed that this gene is expressed exclusively in Sertoli cells of the testis, although a recent study by Cook et al. (18) has demonstrated that this gene is also expressed in the brain and the lung. The degree of identity between the rat brain-specific dynamin and the other dynamin family members is exceptionally high at the amino terminus, which contains the tripartite GTP-binding motif (Fig. 2) and is highly conserved with the shibire gene product. In addition, dynamins also contain a pleckstrin homology motif and a proline-rich carboxyl-terminal tail. To distinguish among the different dynamin isoforms, we will follow the nomenclature proposed by Sontag et al. (ref. 16; Fig. 2) in which the neuron-specific dynamin is called dynamin I (DynI) and the ubiquitously expressed isoform is called dynamin II (DynII). Therefore, the dynamin expressed exclusively in the testis, lung, and brain is referred to as dynamin III (DynIII). Subsequently, additional dynamin isoforms that are yet to be identified would be termed DynIV and so on. Sontag et al. (16) have isolated four different alternatively spliced cDNAs encoding for both DynI and DynII. Cook et al. (18) have indicated that DynIII is also expressed as four different spliced forms. Thus, it is likely that there are at least 12 different dynamin isoforms present in the adult rat brain. The above nomenclature proposes that different splice sites be identified by letters in an amino- to carboxyl-terminal order (e.g., DynIab corresponds to a protein that contains two insertions located at the amino- and the carboxyl-terminal regions; ref. 16).
Figure 2.
Domain structure of the dynamin family. The products of three distinct mammalian genes have been characterized and give rise to multiple proteins that are generated by alternative splicing of the mRNA. DynI is expressed in neuronal tissue, DynII is expressed in all tissues, and DynIII is expressed in the brain, lung, and testis. Dynamin isoform nomenclature makes use of the alternative splice sites; thus the 96-kDa form of DynI is either DynIaa or DynIba, while the 94-kDa form is DynIab or DynIbb. [Reproduced with permission from Robinson et al. (19) (Copyright 1994, Elsevier Science).]
Multiple Dynamins for Distinct Endocytic Pathways
As described above, there are at least three different dynamin genes with several alternatively spliced forms (Fig. 2). This poses the question as to why are there so many dynamins and whether they perform unique or redundant cellular functions. Because DynII is expressed in all cells and tissues that have been examined (15, 16), it is attractive to predict that it participates in a ubiquitous process such as clathrin-mediated endocytosis (CME). At present, the evidence implicating a dynamin in CME is extensive. Morphological studies of epithelial tissues from shits flies show an accumulation of clathrin-coated pits (refs. 5 and 6; Fig. 1d). This observation is complemented by recent studies in cultured mammalian cells transfected with various dynamin mutations (10–13). Although these later experiments transfected neuronal DynI into epithelial cells, the effects on CME and cellular morphology were dramatic and convincing. Endocytic uptake of labeled transferrin in these cells was markedly reduced, and long tubular endocytic structures (Fig. 3a), reminiscent of those seen in shits flies (Fig. 1), were present. In many instances, clathrin cages appeared to associate with the cytoplasmic ends of these membranous invaginations, which were labeled with dynamin antibodies, as viewed by light and electron microscopy (12). In contrast to these observations, immunoblot analysis comparing the distribution of clathrin and dynamin immunoreactivity in subcellular fractions from rat brain does not demonstrate a convincing colocalization (20). Instead, there is a prevalence of dynamin in a heterogeneous membrane pellet and a reduction in the plasma membrane-coated vesicle fraction. Furthermore, of the numerous antibodies made to the conserved and isoform-specific domains of the dynamins, only one to date (12) has been demonstrated to localize with clathrin in mammalian cells. Thus, while the majority of data supports the involvement of the ubiquitously expressed DynII in CME, it is also possible that dynamins are involved with other non-clathrin-mediated cellular events.
Figure 3.
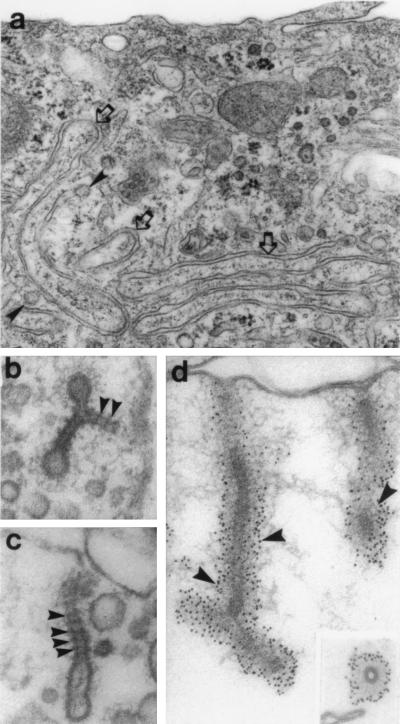
Manipulation of mammalian cells to induce endocytic alterations reminiscent of the shits phenotype. (a) Transmission electron microscopy of transfected HeLa cells overexpressing a mutant DynI protein. In these cells, long membranous invaginations which are continuous with the plasma membrane can be seen (arrows). (b and c) Transmission electron microscopy of isolated rat brain synaptosomes incubated with GTP[γS]. Under these conditions, numerous membrane invaginations can be observed extending from the synaptosomal membrane. Numerous dense staining striations similar to the collars observed in shits can be seen encircling the neck of each invagination (arrowheads). These striations are labeled extensively with a monoclonal antibody to DynI (d). Numerous immunogold particles can be seen around the shaft of each tubule (arrowheads). [a was reproduced with permission from Damke et al. (12) (Copyright 1994, Rockefeller Univ. Press); and b–d were reproduced with permission from Takei et al. (20) (Copyright 1995, Macmillan Magazines).]
The role of DynII in other endocytic processes, such as fluid phase endocytosis (FPE) or macropinocytosis, is less clear due partly to our incomplete understanding of these cellular events. For example, numerous studies have indicated that both FPE and macropinocytosis are distinct pathways that do not require clathrin, while others suggest that these processes are either linked or fully dependent upon CME (21, 22). Insights into the contribution of dynamin to FPE have been provided by studies on shits flies, either in the intact organism (3, 5, 23) or in cells isolated in culture (24). In both models, the restrictive temperature prevents internalization of fluid phase markers, such as horseradish peroxidase (3, 5), fluorescent plasma membrane lipids, and dextran (24). These observations are interesting because they contrast with the studies described above in transfected cells, which were defective in CME but not FPE (10–13). This suggests that these two endocytic processes are not intimately linked, as some have predicted, and highlights a significant contrast between the different experimental systems. Such a discrepancy could be accounted for by differences in the origin of the cells examined (fly neurons versus mammalian epithelium) or an incomplete inhibition of endogenous dynamins by the overexpressed mutant forms. It is possible that a DynII-independent mechanism of FPE may be increased in cultured mammalian cells to compensate for the inhibition of CME. Indeed, analysis of cultured cells overexpressing a temperature-sensitive mutant of dynamin (13) show a significant decrease in FPE (>50%) at early time points after shifting to the restrictive temperature. At later time points, FPE recovers and is increased to levels beyond that observed in control cells. This suggests that CME is responsible for a significant portion of the total FPE in a cell. However, once CME is inhibited, an additional clathrin- and dynamin-independent pathway is amplified to compensate.
While DynII is likely to have an important endocytic role in all cells, it is reasonable to predict that DynI and DynIII, which are generally limited to either brain or testis and brain, respectively, support more specialized membrane trafficking events in these tissues. In the testis, DynIII has been localized to Sertoli cells, which surround and “nurse” the developing spermatogonia through an active and elaborate endocytic processes (17). These Sertoli cells also absorb residual cytoplasm discarded by the nearly mature sperm. Thus, it is not surprising that this tissue may require the action of a second isoform. Similarly, neurons possess unique endocytic mechanisms that may use DynI to support the rapid and efficient retrieval of synaptic plasma membrane following synaptic transmission.
Because the mechanisms by which SVs recycle are undefined, the participation of DynI in this process is unclear. There are at least three current models for SV recycling. Two models implicate clathrin-mediated vesicle budding, either with or without an endosomal intermediate, while a third model describes a clathrin-independent mechanism by which SVs establish a “fusion pore” to incompletely fuse with and then release from the presynaptic membrane. This “kiss and run” model (25) is attractive, since it provides a mechanistic explanation for how SVs might recycle rapidly without having to take on the more laborious and time-consuming task of assembling clathrin baskets. In addition, a fourth recycling model has been provided by recent electrophysiological studies of endocytosis in cultured pituitary cells demonstrating that limited surface membrane can be retrieved rapidly (<4 sec) after secretion into structures that are 10 times larger than coated vesicles (26). Thus, it is likely that multiple vesicle recycling mechanisms are used in neurons, and they depend on the cell type and the form or intensity of stimulation.
It is unclear which of the endocytic mechanisms listed above are dependent upon the action of both dynamin and clathrin. An electrophysiological study measuring changes in membrane capacitance during stimulated secretion in isolated adrenal chromaffin cells has shown that rapid endocytosis, but not exocytosis, is ablated when either one of two different purified dynamin antibodies are included in the patch pipette (27). Interestingly, manipulations that normally disrupt clathrin function, such as potassium depletion, changes in pH, and clathrin antibodies, had no effect on rapid endocytosis in these cells. In contrast, a recent study by Takei et al. (20) has provided extraordinary electron microscopy images showing both clathrin and dynamin on membranous invaginations in isolated rat brain synaptosomes treated with GTP[γS]. In these preparations, one can clearly see densely stained, regularly spaced striations around the neck of the long invaginations (Fig. 3 b–d). These striations label with a monoclonal antibody to dynamin, while clathrin antibodies stain some of the bulbous vesicular ends, suggesting a proximal, if not intimate, relationship between the two proteins. The formation of these striated tubules is remarkably similar to the collared pits observed in the nerve terminals of shits flies at the restrictive temperature (Fig. 1 a and c). In a recent extension of this in vitro study (28), striated dynamin-coated membrane tubules are also seen on internalized endosomal compartments that are either continuous or discontinuous with the presynaptic plasma membrane. These findings support the first two models for SV recycling described above, which implicate an endosomal intermediate from which SVs recycle via a clathrin-based budding mechanism. There is an inconsistency, however, between these observations in regard to the relationship between clathrin and dynamin. In shits flies, the vast majority (>95%) of the presynaptic plasmalemmal invaginations seen at the restrictive temperature have putative dynamin collars yet are not clathrin-coated. In dramatic contrast, all of the pit structures observed in the fly oocytes and nephrocytes are coated yet lack collars (refs. 5 and 6; J. H. Koenig, personal communication). These are reminiscent of the coated structures that lack collars observed in the transfected cultured mammalian cells expressing mutant dynamins (12). Whether the regulated mechanisms for rapid membrane recycling in neurons require DynI, but not clathrin, while the slower constitutive uptake of cell surface receptors, such as those for low density lipoprotein, polymeric IgA, and transferrin, use both clathrin and DynII needs to be tested. However, the shits mutant phenotype is presumed to be the result of a single aberrant dynamin gene product (2, 8, 9) and thereby makes the distinct neuronal and epithelial phenotypes more difficult to explain in D. melanogaster.
Dynamin Interaction with Multiple Subcellular Components
Numerous biochemical studies have indicated that dynamins bind to several different cellular proteins (29). Pleckstrin homology and proline-rich domains in the primary sequence of the dynamins (19) implicate them in tyrosine kinase receptor signal transduction pathways (30). These two distinct domains, which can putatively mediate protein–protein interactions, suggest that the dynamins may be involved in cell growth and differentiation. In addition, the carboxyl-terminal region has been shown to bind to a number of other subcellular components.
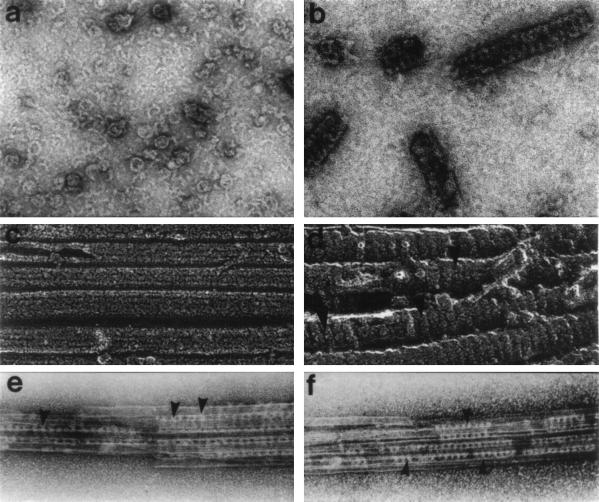
The first dynamin-associated protein to be identified was tubulin. Indeed, brain dynamin was originally isolated by its nucleotide-sensitive interaction with microtubules (Mts; ref. 31), a property usually exhibited only by true Mt-based motor enzymes. While the evidence supporting a biological interaction between dynamin and Mts is both substantial and provocative, it is far from absolute. Electron microscopy studies show that dynamin binds and crosslinks Mts (refs. 14, 31, and 32; Fig. 4 e and f). This crosslinking appears to represent a specific interaction based on a conserved periodicity that is exhibited by the well-characterized conventional Mt-associated proteins. Furthermore, like a true Mt-associated protein, dynamin promotes the assembly of purified sea urchin egg tubulin in vitro (34). Finally, tubulin polymer stimulates a substantial 75-fold increase in the GTPase activity of brain dynamin (refs. 31, 36, and 37; Table 1). This stimulation appears to be specific, since it is not mimicked by other acidic cytoskeletal proteins, such as actin, clathrin, and vimentin (31). Despite these observations, the physiological relevance of the interaction between tubulin and the dynamin proteins remains controversial for several reasons: first, it is not clear if Mts do (27, 38) or do not (39, 40) support the early steps of endocytosis; second, dynamin has not been found to colocalize with Mts in vivo (14, 41); and third, cultured cells transfected to overexpress mutant forms of dynamin do not show any alteration in Mt organization (10, 12). Certainly future in vivo studies on the interaction of dynamin with Mts will prove informative.
Figure 4.
DynI assembles into stacked ring structures in the absence or presence of Mts. (a) Negative-stain electron microscopy of individual disassembled dynamin oligomers in a low-salt buffer. With an increase in dynamin protein concentration (b), the oligomers self-associate to form elongated stacks of rings. Similar stacked-ring arrangements are seen when purified taxol-stabilizing Mts (c) are incubated with purified brain dynamin and viewed by quick-freeze deep-etch electron microscopy (d). Dynamin can also cross-link Mts as shown by negatively stained preparations of taxol-stabilized Mts with purified brain dynamin. Dynamin molecules can be seen as periodic striations connecting adjacent Mts into bundles (e and f). [a and b were reproduced with permission from Hinshaw and Schmid (33) (Copyright 1995, Macmillan Magazines); c and d were reproduced with permission from Maeda et al. (31) (Copyright 1992, Am. Soc. Cell Biol.); and e and f were reproduced with permission from Shpetner and Vallee (34) (Copyright 1989, Cell Press).]
Table 1.
Effect of SH3 domain-containing fusion proteins on dynamin GTPase activity
| Addition | GTPase activity, mol/min per mg |
|---|---|
| None | 9.9 ± 2.0 |
| GST | 17.0 ± 0.1 |
| c-Src SH3 | 85.0 ± 12.4 |
| GRB2 | 131.9 ± 11.9 |
| GRB2 SH3 (N) | 16.4 ± 0.9 |
| GRB2 SH3 (C) | 12.0 ± 1.4 |
| Mts | 658.2 ± 8.3 |
Dynamin GTPase activity was assayed in the presence of either the indicated glutathione S-transferase (GST) fusion proteins (0.08 mg/ml) or Mts (0.12 mg/ml). Errors indicate the range of duplicate data points. N, N-terminal SH3 domain; C, C-terminal SH3 domain. [Reproduced with permission from Herskovits et al. (36) (Copyright 1993, Proc. Natl. Acad. Sci. USA).]
In addition to Mts, both acidic phospholipids and endogenous rat brain vesicles stimulate the GTPase activity of brain dynamin through an interaction with the basic carboxyl terminus (37). Detailed analysis of this phospholipid-stimulated activity revealed positive cooperativity with regard to enzyme concentration, indicating that the interaction of more than one dynamin molecule is necessary for the full activation promoted by phospholipids (42, 43). As discussed later, it is attractive to speculate that dynamin binding to the plasma membrane, concomitant with self-assembly, activates this enzyme to sever nascent endocytic invaginations.
In addition to a structural mechanochemical function, it has been predicted that dynamin participates in cell signaling cascades. As stated earlier, the carboxyl terminus of dynamin contains a proline-rich domain, which is important for its interaction with Src homology 3 (SH3) domain-containing proteins. Gout et al. (44) have demonstrated by affinity chromatography that the SH3 domains of multiple signaling proteins preferentially bind dynamin from brain extracts. This work also shows that dynamin binds selectively and with high affinity to the SH3 domains of phospholipase Cγ, growth factor receptor-bound protein 2 (Grb2), and the regulatory subunit of phosphatidylinositol-3-kinase (p85α), but not to several other SH3 domain-containing proteins. Certain SH3 domain-containing fusion proteins stimulate the GTPase activity of brain dynamin (Table 1). This is likely to be mediated by consensus SH3 domain binding regions located at the carboxyl terminus, which consist of several proline-rich sequences (44). Using synthetic peptides spanning these regions for binding and competition studies, multiple groups have identified several different regions responsible for binding to all of the substrates with the same specificity displayed by the native protein (44–47).
The interaction of dynamin with these various SH3 domain-containing proteins suggests that it is involved in tyrosine kinase receptor-mediated signal transduction. In support of this hypothesis, dynamin has recently been shown to immunoprecipitate with a number of tyrosine kinase growth factor receptors, including the epidermal growth factor receptor (47) and the insulin receptor/insulin receptor substrate I complex (48) via joint interactions with Grb2. Dynamin is complexed with Grb2 in the absence or presence of growth factors, but becomes associated with the respective tyrosine-phosphorylated receptor only upon growth factor stimulation (46, 47). In addition, Scaife et al. (45) report that the association of dynamin with phospholipase Cγ and the platelet-derived growth factor receptor is further increased with growth factor stimulation. These results indicate that dynamin may interact with distinct signaling cascades in response to different agonists. It should be noted however, that like its putative interactions with Mts, dynamin has yet to be localized in vivo with any of the signaling proteins described above. Furthermore, despite the numerous studies done on the interactions of the dynamins with signaling proteins, there are presently no mechanistic or functional studies demonstrating the participation of a dynamin in any cell signaling cascade. We do not know if such a “signaling” function would represent the regulation of a focused, mechanical process (i.e., initiation of endocytic membrane scission) or participation in a more global, classical signaling transduction pathway (i.e., Ca2+ regulation or transcriptional activation).
Regulation and Assembly of Dynamin
Despite the numerous studies implicating the dynamin family in endocytosis and signaling, many questions remain unresolved. Mainly, what precise mechanochemical processes do the dynamins support, and how are these functions initiated and regulated? The recent observation that purified DynI, like tubulin and actin, can self-assemble into helical filaments, although in a GTP-independent manner, has provided an important clue into how it might interact with membranes during an endocytic event (ref. 33; Fig. 4 a and b). When combined with the study by Takei et al. (20), which was discussed earlier, it is likely that the electron-dense striations situated on endocytic membranes in the shits flies represent assembled dynamin. Thus, it is attractive to speculate that assembly of dynamin rings around the “neck” of endocytic invaginations is followed by a GTP-dependent contraction which induces membrane severing (33, 49). Consistent with this model, multiple observations demonstrate that an inhibition of dynamin GTPase activity reduces endocytic membrane budding. For example, mutations in the GTP-binding domains of dynamin lead to an accumulation of membrane invaginations in both shits (3–6) and transfected cells (10–13), as does addition of the nonhydrolyzable analogue GTP[γS] (20). How and when this GTPase activity is stimulated during endocytosis is not clear, but as discussed previously, it can be increased in vitro by multiple components including dynamin itself (42, 43), Mts (14, 31, 34), membranes (37), and SH3 domain-containing proteins (36, 44). Because these cytoplasmic constituents are situated at or near the invaginating endosome, they are likely to be true biological activators.
It is also likely that the functions of the dynamin proteins can be modulated further through posttranslational modifications such as phosphorylation. In vitro studies have demonstrated that protein kinase C phosphorylates DynI and leads to a stimulation of GTPase activity, while the calcium-dependent phosphatase calcineurin can reverse this stimulation (50–52). These studies show that only DynI appears to be a substrate for these enzymes and provides the first evidence supporting differences in function between the dynamin isoforms. These biochemical in vitro observations are further supported by a study using isolated synaptosome preparations, which suggests that phosphorylation may alter interactions between the dynamins and membranes. These observations show: (i) brain dynamin predominantly associates with membranes in a dephosphorylated form, while a small percentage remains cytosolic and phosphorylated (51); (ii) activation of calcineurin during calcium-induced depolarization of nerve terminals corresponds with the dephosphorylation of DynI (52); and (iii) the action of calcineurin reverses the effects of protein kinase C phosphorylation and inhibits the DynI GTPase activity, allowing it to reassociate with synaptic membranes (52). Taken together, these data support a model in which DynI cycles from cytosol to membrane during synaptic transmission, yet it is unclear how to interpret these findings in relationship to those described above. For example, the observation that phospholipids and membranes stimulate the GTPase activity of brain dynamin (42) appears to conflict with observations that an activated phosphorylated dynamin remains cytosolic. Certainly, future studies which define the synergism between dynamin phosphorylation and its interaction with other cellular components will provide insight into how this enzyme participates in membrane budding events.
Multiple Functions for the Dynamin Family
It is well documented that the dynamins are members of an extended family of large GTP-binding proteins (53), which include the yeast vacuolar protein sorting protein, Vps1p (54), the Mx subfamily, which confers cells with selective resistance to viral infection (55), and the mitochondrial protein MGM1 (56). While our understanding of the cellular functions performed by MGM1 and Mx are limited, recent studies on Vps1p mutants have proven informative. Vps1p possesses an amino terminus that shares 66% homology to the dynamins and contains the tripartite GTP-binding motif, while the carboxyl terminus is believed to convey targeting to specific membrane compartments (57). These compartments are likely to include the trans-Golgi and the vacuole, since Vps1p mutants missort both resident Golgi endoproteases and vacuolar membrane proteins to the plasma membrane where they are reendocytosed and indirectly sent to the vacuole (58, 59). It is unclear whether a homologue is present in mammalian cells or if Vps1p is the yeast dynamin equivalent. Although searches for a mammalian Vps1p gene have proven inconclusive to date, another dynamin-like gene (DNM1) has been recently identified in yeast (60). Although this gene product appears to be involved in an endocytic process, it is more similar to Vps1p, since these proteins do not contain a pleckstrin homology domain or the proline-rich carboxyl terminus characteristic of the mammalian dynamins.
While substantial experimentation indicates that dynamins function at the plasma membrane, it is enticing to predict that a dynamin isoform or family member could participate in membrane dynamics during other membrane trafficking processes (Fig. 5). It has become increasingly clear that endocytosis and secretion share common mechanisms that support membrane budding and fusion. Indeed, multiple proteins, such as the adaptins, clathrin, caveolin, and the ARF family, have been implicated in processes at both the Golgi and plasma membrane (62–67). Therefore, it is not surprising to learn that multiple antibodies made to the conserved amino-terminal domain of the dynamins stain the Golgi apparatus in cultured cells, as do antibodies to yeast Vps1p (61). It is certainly attractive to speculate that either a dynamin isoform or related protein acts as a “pinchase” at the trans-Golgi surface to liberate nascent, clathrin-coated secretory vesicles. This prediction is inconsistent, however, with observations made in isolated cells of shits flies or cultured mammalian cells transfected with mutant DynI. In these systems, endocytosis is perturbed, yet several specific secretory processes are unaltered, including normal trafficking of VSV-G protein or fluorescent Golgi lipids to the plasma membrane (24), transport of nascent lysosomal proteins (11), and localization of the Golgi-associated γ-adaptin (10). It is possible that some processes in these manipulated cells are more sensitive to altered dynamins than others, as demonstrated by the differential effects on CME versus FPE (10, 12, 13). Furthermore, these transfection studies may not affect all of the many dynamin-like proteins in a cell. Certainly, future functional studies testing the role of dynamins in the secretory pathway will be useful.
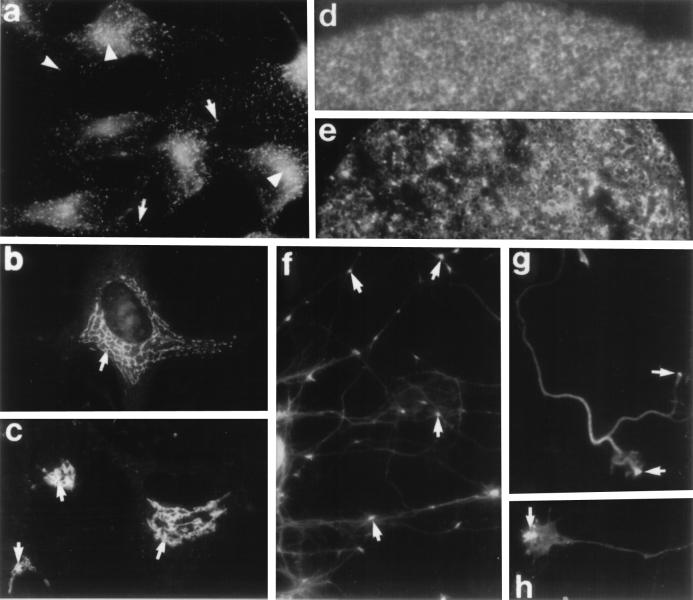
Figure 5.
Cellular localizations of the dynamin superfamily by immunofluorescence microscopy. (a) Cultured HeLa cells stained with a monoclonal antibody to dynamin. Numerous punctate structures (arrows) are seen throughout the cytoplasm as well as a prominent juxtanuclear labeling (arrowheads). (b) Immunostaining of cultured human fibroblasts stained with an affinity-purified polyclonal antibody made to a region conserved among the dynamins. An extensive reticular Golgi network can be seen surrounding the nucleus. (c) Cultured human fibroblasts stained with a polyclonal antibody (gift of T. Stevens, University of Oregon) to the yeast GTP-binding protein Vps1p. Similar to the staining pattern seen with the dynamin antibodies, elaborate reticular Golgi structures (arrows) are labeled in each cell. (d and e) Immunolocalization of dynamin in sea urchin eggs using an antibody made to a dynamin-related protein purified from these eggs. (d) Dynamin localization in the cortex of these eggs appears as diffuse and cytoplasmic in unfertilized eggs. (e) Immediately after fertilization, the honeycomb cortical granule staining pattern is disrupted, and two populations of brightly staining spots begin to appear throughout the periphery, suggesting an association with cortical membrane. (f) Immunostaining of cultured rat hippocampal neurons with affinity-purified antibodies to conserved domains of dynamin show bright punctate staining at specific locations along extended neurites (arrows). (g and h) Higher magnification images of growth cones from these cells show a restricted, yet prominent, dynamin localization at the very tip of these processes (arrows). [a was reproduced with permission from Damke et al. (12) (Copyright 1994, Rockefeller Univ. Press); b and c were reproduced with permission from Henley and McNiven (61) (Copyright 1996, Rockefeller Univ. Press); d and e were reproduced with permission from Faire and Bonder (36) (Copyright 1993, Academic Press); and f–h were reproduced with permission from E. Torre and M.A.M. (unpublished observations).
In addition to an endocytic function and a possible role in secretion, observations in several different cell models have indirectly implicated dynamin in the regulation and maintenance of cell shape. For example, neurons dissociated from shits larvae are unable to grow neurites or growth cones when maintained at the restrictive temperature (68). In support of these findings, we have demonstrated that a reduction in the intracellular level of DynI in cultured rat hippocampal neurons through antisense oligonucleotide treatment results in a significant impairment of neurite formation (69). In addition, we have found that DynI gene expression is increased during neurite growth in culture, while that of DynII remains unchanged (15). Finally, cultured mammalian cells that have been transfected to overexpress mutant DynI show marked alterations in shape and cytoskeletal organization (12). Whether these phenomena are induced by direct perturbations in cell signaling cascades or cytoskeletal dynamics or, alternatively, simply represent the indirect effects of an impaired endocytic pathway remains to be determined.
Conclusion
Here we have provided an overview of an expansive number of studies that lend insight into the diversity, localization, function, and enzymology of the dynamin family of large GTPases. It is now clear that dynamins are expressed in all cells and perform an important and universal function during endocytosis. There are three closely related dynamin genes and corresponding products that are alternatively spliced. These comprise one component of a broad superfamily of tissue-specific proteins that support membrane budding and trafficking events. Despite the many advances in this field, important questions remain to be addressed. There are likely to be other dynamin genes yet to be identified, and the distributions and functions of these isoforms in various cell types need to be defined. Finally, whether the function of the dynamins extends beyond endocytosis to participate in cell signaling cascades or other membrane budding events remains to be established.
Acknowledgments
The authors are grateful to the laboratories of Drs. T. Sudhof, S. Schmid, P. DeCamilli, K. Ikeda, J. Koenig, R. Vallee, N. Hirokawa, and E. Bonder for providing figures and information used in this review. Special thanks to Drs. Sandy Schmid, Pietro De Camilli, Robert Margolis, and Robin Scaife for their review of the manuscript and helpful comments. This study was funded by National Institutes of Health Grants AA09227 and DK44650 to M.A.M.
Footnotes
Abbreviations: SV, synaptic vesicle; CME, clathrin-mediated endocytosis; FPE, fluid phase endocytosis; Mt, microtubule.
References
- 1.Grigliatti T A, Hall L, Rosenbluth R, Suzuki D T. Mol Gen Genet. 1973;120:107–114. doi: 10.1007/BF00267238. [DOI] [PubMed] [Google Scholar]
- 2.Poodry C A. Dev Biol. 1990;138:464–472. doi: 10.1016/0012-1606(90)90212-2. [DOI] [PubMed] [Google Scholar]
- 3.Kosaka T, Ikeda K. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- 4.Koenig J H, Ikeda K. J Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosaka T, Ikeda K. J Cell Biol. 1983;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig J H, Ikeda K. Cell Tissue Res. 1990;262:233–244. doi: 10.1007/BF00309878. [DOI] [PubMed] [Google Scholar]
- 7.Obar R A, Collins C A, Hammarback J A. Nature (London) 1990;347:256–261. doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- 8.van der Bliek A M, Meyerowitz E M. Nature (London) 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen M S, Obar R A, Schroeder C C, Austin T W, Poodry C A, Wadsworth S C, Vallee R B. Nature (London) 1993;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 10.Herskovits J S, Burgess C C, Obar R A, Vallee R B. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damke H, Baba T, Warnock D E, Schmid S L. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damke H, Baba T, van der Bliek A M, Schmid S L. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaife R, Margolis R L. J Cell Biol. 1990;111:3023–3033. doi: 10.1083/jcb.111.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook T A, Urrutia R, McNiven M A. Proc Natl Acad Sci USA. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sontag J M, Fykse E M, Ushkaryov Y, Liu J P, Robinson P J, Südhof T C. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- 17.Nakata T, Takemura R, Hirokawa N. J Cell Sci. 1993;105:1–5. doi: 10.1242/jcs.105.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Cook, T., Mesa, K. & Urrutia, R. (1996) J. Neurochem. 67, in press. [DOI] [PubMed]
- 19.Robinson P J, Liu J P, Powell K A, Fykse E M, Südhof T C. Trends Neurosci. 1994;17:348–353. doi: 10.1016/0166-2236(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 20.Takei K, McPherson P S, Schmid S L, De Camilli P. Nature (London) 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 21.Sandvig K, van Deurs B. Trends Cell Biol. 1994;4:275–277. doi: 10.1016/0962-8924(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 22.Watts C, Marsh M. J Cell Sci. 1992;103:1–8. doi: 10.1242/jcs.103.1.1a. [DOI] [PubMed] [Google Scholar]
- 23.Kessell I, Holst B D, Roth T F. Proc Natl Acad Sci USA. 1989;86:4968–4972. doi: 10.1073/pnas.86.13.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radhakrishna H, Pagano R E, Machamer C E, Roth T F. J Cell Biol. 1993;4:211a. (abstr.). [Google Scholar]
- 25.Fesce R, Grohovaz F, Valtorta F, Meldolesi J. Trends Cell Biol. 1994;4:1–4. doi: 10.1016/0962-8924(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 26.Thomas P, Lee A K, Wong J G, Almers W. J Cell Biol. 1994;124:667–675. doi: 10.1083/jcb.124.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artalejo C R, Henley J R, McNiven M A, Palfrey H C. Proc Natl Acad Sci USA. 1995;92:8328–8332. doi: 10.1073/pnas.92.18.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takei K, Mundigl O, Daniell L, DeCamilli P. J Cell Biol. 1996;133:1237–1250. doi: 10.1083/jcb.133.6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallee R B, Okamoto P M. Trends Cell Biol. 1995;5:43–47. doi: 10.1016/s0962-8924(00)88937-0. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 31.Shpetner H S, Vallee R B. Nature (London) 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- 32.Maeda K, Nakata T, Noda Y, Sato-Yoshitake R, Hirokawa N. Mol Biol Cell. 1992;3:1181–1194. doi: 10.1091/mbc.3.10.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinshaw J E, Schmid S L. Nature (London) 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 34.Shpetner H S, Vallee R B. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 35.Faire K, Bonder E M. Dev Biol. 1993;159:581–594. doi: 10.1006/dbio.1993.1266. [DOI] [PubMed] [Google Scholar]
- 36.Herskovits J S, Shpetner H S, Burgess C C, Vallee R B. Proc Natl Acad Sci USA. 1993;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuma P L, Stachniak M C, Collins C A. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- 38.Jin M, Snider M D. J Biol Chem. 1993;268:18390–18397. [PubMed] [Google Scholar]
- 39.Caron J M, Jones A L, Kirschner M W. J Cell Biol. 1985;101:1763–1772. doi: 10.1083/jcb.101.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolkoff A W, Klausner R D, Ashwell G, Harford J. J Cell Biol. 1984;98:375–381. doi: 10.1083/jcb.98.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noda Y, Nakata T, Hirokawa N. Neuroscience. 1993;55:113–127. doi: 10.1016/0306-4522(93)90459-s. [DOI] [PubMed] [Google Scholar]
- 42.Tuma P L, Collins C A. J Biol Chem. 1994;269:30842–30847. [PubMed] [Google Scholar]
- 43.Warnock D E, Terlecky L J, Schmid S L. EMBO J. 1995;14:1322–1328. doi: 10.1002/j.1460-2075.1995.tb07118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, Campbell I D, Waterfield M D. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 45.Scaife R, Gout I, Waterfield M D, Margolis R L. EMBO J. 1994;13:2574–2582. doi: 10.1002/j.1460-2075.1994.tb06547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miki H, Miura K, Matuoka K, Nakata T, Hirokawa N, Orita S, Kaibuchi K, Takai Y, Takenawa T. J Biol Chem. 1994;269:5489–5492. [PubMed] [Google Scholar]
- 47.Seedorf K, Kostka G, Lammers R, Bashkin P, Daly R, Burgess W H, van der Bliek A M, Schlessinger J, Ullrich A. J Biol Chem. 1994;269:16009–16014. [PubMed] [Google Scholar]
- 48.Ando A, Yonezawa K, Gout I, Nakata T, Ueda H, Hara K, Kitamura Y, Noda Y, Takenawa T, Hirokawa N. EMBO J. 1994;13:3033–3038. doi: 10.1002/j.1460-2075.1994.tb06602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly R B. Nature (London) 1995;374:116–117. doi: 10.1038/374116a0. [DOI] [PubMed] [Google Scholar]
- 50.Robinson P J, Sontag J M, Liu J P, Fykse E M, Slaughter C, McMahon H, Südhof T C. Nature (London) 1993;365:163–166. doi: 10.1038/365163a0. [DOI] [PubMed] [Google Scholar]
- 51.Liu J P, Powell K A, Südhof T C, Robinson P J. J Biol Chem. 1994;269:21043–21050. [PubMed] [Google Scholar]
- 52.Liu J P, Sim A R, Robinson P J. Science. 1994;265:970–973. doi: 10.1126/science.8052858. [DOI] [PubMed] [Google Scholar]
- 53.Collins C A. Trends Cell Biol. 1991;1:57–60. doi: 10.1016/0962-8924(91)90090-v. [DOI] [PubMed] [Google Scholar]
- 54.Rothman J H, Raymond C K, Gilbert T, O’Hara P J, Stevens T H. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- 55.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 56.Jones B A, Fangman W L. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- 57.Vater C A, Raymond C K, Ekena K, Howald-Stevenson I, Stevens T H. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilsbach K, Payne G S. EMBO J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nothwehr S F, Conibear E, Stevens T H. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gammie A E, Kurihara L J, Vallee R B, Rose M D. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henley J R, McNiven M A. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M S. Curr Opin Cell Biol. 1994;6:538–544. doi: 10.1016/0955-0674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 63.Kurzchalia T V, Dupree P, Monier S. FEBS Lett. 1994;346:88–91. doi: 10.1016/0014-5793(94)00466-8. [DOI] [PubMed] [Google Scholar]
- 64.Dupree P, Parton R G, Raposo G, Kurzchalia T V, Simons K. EMBO J. 1993;12:1597–1605. doi: 10.1002/j.1460-2075.1993.tb05804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmid S L. BioEssays. 1992;14:589–596. doi: 10.1002/bies.950140903. [DOI] [PubMed] [Google Scholar]
- 66.Welsh C F, Moss J, Vaughan M. Mol Cell Biochem. 1994;138:157–166. doi: 10.1007/BF00928458. [DOI] [PubMed] [Google Scholar]
- 67.Donaldson J G, Klausner R P. Curr Opin Cell Biol. 1994;6:527–532. doi: 10.1016/0955-0674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 68.Kim Y, Wu C. J Neurosci. 1987;7:3245–3255. doi: 10.1523/JNEUROSCI.07-10-03245.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torre E, McNiven M A, Urrutia R. J Biol Chem. 1994;269:32411–32417. [PubMed] [Google Scholar]