Abstract
During routinely screening (50.000 milk samples on an annual basis) 14 MRSA ST398 strains were identified in the period of January 2008 to September 2008 in 14 different dairy herds located in the provinces Overijssel and Gelderland, The Netherlands. Molecular analysis was performed by Cfr9I PFGE, ST398-specific diagnostic PCR, spa typing, SCCmec typing and Panton-Valentine Leukocidin (PVL) gene PCR. The molecular analyses of 14 MRSA (one MRSA strain per herd) strains revealed that all strains belong to ST398 with 3 closely related spa types (t011, t108 and t889, all commonly found in pigs) and carry 2 different SCCmec types, IVa and V. All MRSA strains were resistant to two or more classes of antibiotics and also PVL negative. The majority of farms (n = 9, 64%) harboured combined livestock with both cows and pigs present. Our study contributes to the growing evidence that MRSA ST398 is transmitted among various animal species and can be considered as an etiological agent of mastitis in dairy cows.
Keywords: Bovine mastitis, Intramammary infection, Staphylococcus aureus, Dairy cows, MRSA, ST398
Findings
Recently, the emergence of methicilin-resistant Staphylococcus aureus sequence type 398 (MRSA ST398) has been reported worldwide. Most strains were isolated from farm animals as well as meat products meant for human consumption [1-5]. It was supposed that dissemination of ST398 strains was restricted to animals in general and pigs in particular [6]. For instance, a high prevalence of MRSA ST398 was documented for the pig population in The Netherlands. Thirty-nine percent of pigs appeared to be colonized with MRSA in their nares in 2006 [6]. MRSA ST398 strains are non-typeable by SmaI Pulsed-Field Gel Electrophoresis (PFGE) and have related spa types. In this study which include routine survey in milk samples during a 9 months period, we describe mastitis-associated MRSA ST398 in Dutch dairy cows.
Materials and methods
Survey in 14 dairy herds
From January 2008 to September 2008 approximately 38,000 milk samples (50.000 milk samples/year) were obtained from different herds in The Netherlands. Herds were either participating in a national MRSA prevalence study (3 herds) or submitting routine diagnostic milk samples to the veterinary diagnostic laboratory of GD Animal Health Service Deventer (The Netherlands). All milk samples were obtained from cows with (sub)clinical mastitis of at least one quarter (somatic cell count ≥ 200,000 cells/mL). Milk samples were inoculated on blood-agar plates (Becton and Dickinson Company, the Netherlands). Fourteen MRSA strains from 14 dairy herds located in the provinces Overijssel and Gelderland were identified by their morphology (yellow haemolytic colonies) on blood-agar plates (Becton and Dickinson Company, The Netherlands), a positive Staphaurex Plus test (bioMérieux, Lyon, France), cefoxitin 30 μg disk diffusion method and the presence of the mecA-gene by PCR.
Antimicrobiol susceptibilty testing
Methicillin resistance of all strains was confirmed using the disk diffusion method on Mueller-Hinton agar plates (Becton Dickinson and Company, Sparks, MD) and cefoxitin (30 μg) disks according to CLSI criteria [7]. For all MRSA strains antibiotic susceptibility testing of amoxicillin with clavulanic acid, cloxacillin, penicillin, ampicillin, amoxicillin, cefaperazone, cefquinome, neomycin, pirlimycin was performed according to standard disk diffusion methods [7,8]. Antibiotic susceptibility testing of benzylpenicillin, oxacillin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, moxifloxacin, erythromycin, clindamycin, linezolid, teicoplanin, vancomycin, tetracycline, fosfomycin, nitfurantoin, fusidic acid, mupirocin, rifampicin and trimethoprim-sulfamethoxazole was performed using Vitek2® (bioMérieux, France).
Genetic typing of strains
All 14 isolated MRSA were nontypeable by SmaI PFGE (NT-MRSA). The NT-MRSA ST398 strains were characterized by Cfr9I PFGE (a neoschizomer of SmaI) and identified with a ST398 specific diagnostic PCR [9]. PFGE fingerprints were interpreted using Bionumerics software (version 3.0; Applied Maths, Gent, Belgium). The presence of the mecA-gene and the assignment of the SCCmec types was determined using PCR [10,11]. The Short Sequence Repeat (SSR) region of the spa gene was sequenced using primers spa-1, 5'- TAA.AGA.CGA.TCC.TTC.GGT.GAG.C -3'; spa-2, 5'- CAG.CAG.TAG.TGC.CGT.TTG.CTT -3'. The spa sequences were determined and the spa types were assigned through the spa type database (http://www.spaserver.ridom.de) [12]. PVL gene PCR on MRSA strains was performed as described previously [13].
Results
Antimicrobiol susceptibilty testing
All 14 MRSA strains were resistant to amoxicillin with clavulanic acid, oxacillin, cloxacillin, penicillin, benzylpenicillin, ampicillin, amoxicillin, cefoperazone, cefquinome and tetracylin. Four strains (28.5%) were resistant to amoxicillin with clavulanic acid, oxacillin, cloxacillin, penicillin, benzylpenicillin, ampicillin, cefoperazone, cefquinome and tetracylin, trimethoprim-sulfamethoxazole(co-trimoxazole), tobramycin and gentamicin.
One strain (7%) was resistant to neomycin. One strain (7%) was resistant to pirlimycin. One strain (7%) was resistant to erytromycin and clindamycin (Table 1).
Table 1.
Resistance profiles of the 14 bovine MRSA ST398 strains
| Strain nr | Resistance profiles |
|---|---|
| Rww 221 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 222 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 223 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, COT, GEN, TOB |
| Rww 224 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, NEO |
| Rww 225 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 226 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 227 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, COT, GEN,TOB |
| Rww 228 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, COT, GEN, TOB |
| Rww 229 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 230 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 231 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
| Rww 232 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, CL, ERY |
| Rww 233 |
CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET, COT, GEN, TOB, PIR |
| Rww 234 | CF, CP,PEN,PENG, OXA, AMP, AUG, CLOX, TET |
Note: CF: cefquinom; CP: cefoperazone; PEN: penicillin; PENG: benzylpenicillin; OXA: oxacillin; AMP: ampicillin; AUG: amoxicillin/clavulanate; CLOX: cloxacillin; TET: tetracylin; COT: trimethoprim-sulfamethoxazole (co-trimoxazole); GEN: gentamicin; TOB: tobramycin; NEO: neomycin; PIR: pirlimycin.
Genetic typing of strains
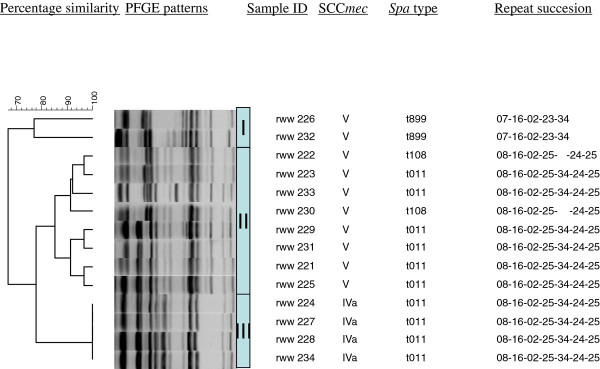
All 14 MRSA strains belong to ST398 and were PVL negative. Based on Cfr9I PFGE, the MRSA strains were subdivided in 3 clusters (I, II, III) with 3 closely related spa types t011, t108 and t889 (Figure 1). Spa types t108 and t889 differ from spa type t011 in 1 and 2 repeat units, respectively. Two SCC mec types, IVa (4/14) and V (10/14), were found among the mastitis strains. SCCmec type IVa was restricted to PFGE cluster III, SCCmec Type V was found in both PFGE cluster I and II.
Figure 1.
Cluster analysis of the PFGE data from 14 bovine Staphylococcus aureus ST 398 strains. On top of the dendrogram, the percentage similarity is indicated. Strains clusters are identified by Roman numerous (I, II, III). In column to the right, the Cfr9I gel picture, the sample code, the SCCmec, spa type and precise repeat content of spa are given.
Conclusion
During a 9 months period of routine survey of milk samples, 14 dairy farms were found to be positive for MRSA, being the first description of MRSA intramammary infections in The Netherlands. All MRSA strains were determined as MRSA ST398, and were resistant to two or more classes of antibiotics and shared the same genetic background as pig-associated MRSA strains [6]. Recently the highly prevalent MRSA clone ST398 causing clinical and subclinical mastitis has been described in Belgian cows [14]. Nearly 10% of the Belgian dairy farms included in the surveillance were affected by MRSA ST398 SCCmec types, IVa (5/10) and V (5/10), spa type t011 (10/11) and t567 (1/11), which could be the same in The Netherlands given the high prevalence of MRSA ST398 in the Dutch pig population with overlapping spa and SCCmec types [6]. We have identified infectious MRSA ST398 with 3 closely related spa types t011 (10/14), t108 (2/14) and t889 (2/14) and two SCCmec types, IVa (4/14) and V (10/14) from intramammary infections in Dutch dairy cows. The majority of the farms (n = 9, 64%) included in this study harboured combined livestock with both cows and pigs present. Our study contributes to the growing evidence that MRSA ST398 could be transmitted among various animal species and can be considered as an etiological agent of bovine intramammary infections.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All the authors have read and approved the final manuscript.
Contributor Information
Mehri Tavakol, Email: m.tavakol@erasmusmc.nl.
Richard GM Olde Riekerink, Email: richard@thecowdoctor.com.
Otlis C Sampimon, Email: Otlis.Sampimon@pfizer.com.
Willem JB van Wamel, Email: w.vanwamel@erasmusmc.nl.
Alex van Belkum, Email: alex.vanbelkum@biomerieux.com.
Theo JGM Lam, Email: T.Lam@gddeventer.com.
References
- Nemati M, Hermans K, Lipinska U, Denis O, Deplano A, Struelens M, Devriese LA, Pasmans F. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob Agents Chemother. 2008;52(10):3817–3819. doi: 10.1128/AAC.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati M, Hermans K, Devriese LA, Maes D, Haesebrouck F. Screening of genes encoding adhesion factors and biofilm formation in Staphylococcus aureus isolates from poultry. Avian Pathol. 2009;38(6):513–517. doi: 10.1080/03079450903349212. [DOI] [PubMed] [Google Scholar]
- Van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo IHM, Diederen BMW, Savelkoul PHM, Woudenberg JHC, Roosendaal R, van Belkum A, Lemmens-den Toom N, Verhulst C, van Keulen PHJ, Kluytmans AJW. Methicillin-resistant Staphylococcus aureus in meat products, the Netherlands. Emerg Infect Dis. 2007;13:1753–1755. doi: 10.3201/eid1311.070358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C, Strommenger B, Witte W, Stanek C. Clusters of infections in Horses with MRSA ST1, ST25, and ST398 in a veterinary hospital. Microb Drug Resist. 2008;14:307–310. doi: 10.1089/mdr.2008.0845. [DOI] [PubMed] [Google Scholar]
- De Neeling AJ, Van Den Broek MJM, Spalburg MC, Van Santen-Verheuvel MG, Dam-Deisz WDC, Boshuizen HC, Van De Giessen AW, Van Duikeren E, Huisdens XW. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2007;122:66–72. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- CLSI, 2009. Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard – Tenth Edition. M2-A10.
- CLSI, 2011. Performance standards for antimicrobial susceptibility testing; 21th informational supplement. M100–S21.
- Van Wamel WJB, Hansenová Maňásková S, Fluit AC, Verbrugh HA, de Neeling J, van Duijkeren E, van Belkum A. Short term micro-evolution and PCR-detection of methicillin resistant and -susceptible Staphylococcus aureus sequence type 398. Eur J Clin Microbiol Infect Dis. 2010;29:119–122. doi: 10.1007/s10096-009-0816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neeling AJ, van Leeuwen WJ, Schouls LM, Schot CS, Veen-Rutgers A, Beunders AJ, Buiting AG, Hol C, Ligtvoet EE, Petit PL, Sabbe LJ, van Griethuysen AJ, van Embden JD. Resistance of staphylococci in The Netherlands: surveillance by an electronic network during 1989-1995. J Antimicrob Chemother. 1998;41:93–101. doi: 10.1093/jac/41.1.93. [DOI] [PubMed] [Google Scholar]
- Boye K, Bartels MD, Andersen IS, Moller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect. 2007;13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- Rothganger J, Weniger M, Weniger T, Mellmann A, Harmsen D. Ridom TraceEdit: a DNA trace editor and viewer. Bioinformatics. 2006;22:493–494. doi: 10.1093/bioinformatics/btk002. [DOI] [PubMed] [Google Scholar]
- Severin JA, Lestari KES, Melles DC, Martijn P, Peeters JK, Snijders S, Usman H, Duerink DO, van Belkum A, Verbrugh HA. Unusually High Prevalence of Panton-Valentine Leukocidin Genes among Methicillin-Sensitive Staphylococcus aureus Strains Carried in the Indonesian Population. J Clin Microbiol. 2008;46:1989–1995. doi: 10.1128/JCM.01173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol. 2010;144(1-2):166–171. doi: 10.1016/j.vetmic.2009.12.044. [DOI] [PubMed] [Google Scholar]