Abstract
The 6q25.1 locus was first identified via a genome-wide association study (GWAS) in Chinese women and marked by single nucleotide polymorphism (SNP) rs2046210, approximately 180 Kb upstream of ESR1. There have been conflicting reports about the association of this locus with breast cancer in Europeans, and a GWAS in Europeans identified a different SNP, tagged here by rs12662670. We examined the associations of both SNPs in up to 61,689 cases and 58,822 controls from forty-four studies collaborating in the Breast Cancer Association Consortium, of which four studies were of Asian and 39 of European descent. Logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI). Case-only analyses were used to compare SNP effects in Estrogen Receptor positive (ER+) versus negative (ER−) tumours. Models including both SNPs were fitted to investigate whether the SNP effects were independent. Both SNPs are significantly associated with breast cancer risk in both ethnic groups. Per-allele ORs are higher in Asian than in European studies [rs2046210: OR (A/G) = 1.36 (95% CI 1.26–1.48), p = 7.6×10−14 in Asians and 1.09 (95% CI 1.07–1.11), p = 6.8×10−18 in Europeans. rs12662670: OR (G/T) = 1.29 (95% CI 1.19–1.41), p = 1.2×10−9 in Asians and 1.12 (95% CI 1.08–1.17), p = 3.8×10−9 in Europeans]. SNP rs2046210 is associated with a significantly greater risk of ER− than ER+ tumours in Europeans [OR (ER−) = 1.20 (95% CI 1.15–1.25), p = 1.8×10−17 versus OR (ER+) = 1.07 (95% CI 1.04–1.1), p = 1.3×10−7, pheterogeneity = 5.1×10−6]. In these Asian studies, by contrast, there is no clear evidence of a differential association by tumour receptor status. Each SNP is associated with risk after adjustment for the other SNP. These results suggest the presence of two variants at 6q25.1 each independently associated with breast cancer risk in Asians and in Europeans. Of these two, the one tagged by rs2046210 is associated with a greater risk of ER− tumours.
Introduction
A genome-wide association study (GWAS) in Chinese women by Zheng et al. [1] identified a novel breast cancer susceptibility locus at 6q25.1. The most strongly associated single nucleotide polymorphism (SNP) was rs2046210, with an estimated Odds ratio (OR) [per-allele A/G] = 1.29 (95% confidence interval (CI) 1.21–1.37, p = 10−15). SNP rs2046210 did not show a clear association in GWAS carried out in women of European ancestry, and replication studies indicated its effect, if any, was weaker in Europeans [OR (per allele A/G) = 1.04 (95% CI 0.99–1.08), p = 0.09 in a combined analysis of European studies [2]]. More recent studies in European women suggested stronger associations with other SNPs in the region: Turnbull et al. [3] found the most significantly associated SNP to be rs3757318, which is only weakly correlated with rs2046210 in Europeans (r2 = 0.09 from in HapMap2 CEU), while Stacey et al. [2] suggested that SNPs closer to ESR1 may be more strongly associated. It is as yet unclear whether this difference in breast cancer associated SNPs between Asians and Europeans indicates the presence of a single or multiple causative variant(s) at this locus. If there is only one, it is unlikely to be highly correlated with the best tags identified from either the Asian or European GWAS and could potentially be a common variant with a small effect or a rarer one with a larger effect on breast cancer risk.
In this, by far the largest study to date, we investigate associations with SNP rs2046210, as well as with SNP rs12662670 in forty-four case-control studies within the Breast Cancer Association Consortium (BCAC). These two SNPs have been genotyped in a total of 120,511 female subjects, of which 110,265 subjects are of European ancestry and 8,559 are Asian. SNP rs2046210 is the best tag from the original Asian GWAS [1] and SNP rs12662670 is an easier to genotype surrogate for SNP rs3757318 - the best tag SNP at the 6q25.1 locus from a European GWAS [3]. Our aims were to compare the effects of these tags in well-powered studies of both Asian and European ancestry and to test if these known SNP associations are shared by the different ethnic groups. We have been successful in achieving these aims and our analyses provide additional insights into the nature of this locus.
Materials and Methods
Ethics Statement
Approval of the studies was obtained from the ethics committees listed in Table S1. All studies conform to the Declaration of Helsinki and all study participants gave written informed consent.
Study Populations
Data from forty-five BCAC case-control studies from Australia, Europe, North America, and South-East Asia were available for inclusion in this analysis (see Table S1 for a description of the individual studies). To be eligible for BCAC, studies needed to include at least 500 cases of invasive breast cancer and 500 controls, with DNA samples available for genotyping. The controls needed to be broadly from the same population as the cases (http://www.srl.cam.ac.uk/consortia/bcac/about/about.html). Some studies selected cases preferentially on the basis of age and/or family history.
All studies provided information on disease status (58,822 controls/62,061 invasive cases/2,769 in-situ cases/1,435 cases of unknown invasiveness), age at diagnosis or interview and ethnicity (Asian/European/other). Forty studies also provided information on estrogen receptor (ER) status for a total of 40,508 cases (9,878 Estrogen receptor negative (ER−)/30,630 Estrogen Receptor positive (ER+)).
Laboratory Methods
In most studies SNPs were assayed by Taqman™ (Applied Biosystems, Foster City, USA). Primers, probes and master mix were ordered in a single batch and alliquots shipped to each study. Reactions were performed according to manufacturer’s instructions, using the following thermal cycling profile 95°C for 10 mins followed by: [92°C for 15 secs, 60°C for 1 min] for 40–60 cycles.
SNP rs12662670 was chosen as the most easily assayable surrogate for the best European GWAS hit, rs3757318, for which no working Taqman™ assay could be designed. These two SNPs are correlated at r2 = 0.89 in the European samples used in Turnbull et al. [3] although the correlations in populations of Asian ancestry are somewhat weaker (r2 = 0.72 and r2 = 0.66 in HapMap2 JPT and CHB samples, respectively).
The primer and probe sequences used were:
For SNP rs2046210
Forward primerTGCCTCAACTGTCTTGTGAATCTTT
Reverse primerCTACTGTAGAATCATTTTCCTCACACATACA
G allele probeVIC ACAGTCACATACGCATCTA
A allele probe FAM CAGTCACATACACATCTA
For SNP rs12662670
Forward primerCTAACGAAGGCAGAGCAAAAAGAAA
Reverse primerCACACATGCATGACACGTAAATCTT
T allele probeVIC ATTAAATTCTTGTAAGTTTCC
G allele probe FAM AATTCTTGTCAGTTTCC
Four studies (ACP, GESBC, kConFab/AOCS and MARIE) used the Sequenom iPLEX MassARRAY™ system (Sequenom, San Diego, CA, USA) with oligonucleotide design performed using MassARRAY Assay Design software (version 3.1).
SNPs were genotyped in three different BCAC genotyping phases along with other SNPs of interest to the consortium (see Tables S2a and S2b for information on the respective phases for SNPs rs2046210 and rs12662670). All studies followed standard quality control guidelines (for details see http://www.srl.cam.ac.uk/consortia/bcac/about/about.html). Data were excluded for any sample that failed genotyping for >20% of the SNPs typed in a given phase of genotyping. All study data were excluded for any SNP with overall call rate <95% or duplicate concordance <94% (based on at least 2% of samples in each study being genotyped in duplicate) or departure of genotype distribution from Hardy-Weinberg equilibrium in controls (p<0.005). In addition, all genotyping centres assayed an identical plate of 80 control DNA samples (referred to as the Coriell plate; which also included 14 internal duplicates) and had to achieve call rates and duplicate concordance >98% in order for their data to be included. Data for both SNPs from one study (NBCS) were excluded from further analyses after quality control rules were applied. Quality control data for the individual studies are shown in Tables S2a and S2b. Thus, for SNP rs2046210 forty out of forty-one assayed studies (56,607 cases/49,559 controls), and for SNP rs12662670 thirty-three out of thirty-four assayed studies (47,251 cases/40,161 controls) were included in the statistical analysis.
Statistical Analyses
ORs were estimated using logistic regression. In order to provide reliable estimates of effect sizes, study-specific effect estimates of ORs were derived only for those studies that provided at least 100 cases and controls for the respective (sub-) group of interest.
The primary analysis estimated ORs for the main effect of the SNP, adjusted for the studies that provided data for the respective analysis (i.e. S-1 indicator variables were entered the logistic regression model, where S was number of studies that provided data for the respective analysis). ORs adjusted for both study and age were essentially identical and we did not therefore present the age-adjusted analyses. Per allele ORs were estimated under the assumption of a log-additive mode of inheritance, i.e. the SNP was coded according to the number of minor alleles 0, 1 or 2. Additionally, ORs by genotype were calculated, i.e. two indicator variables indicating the presence of the heterozygous genotype and the genotype homozygous for the minor allele, respectively, were entered the model. The primary p-values were derived by means of a Wald-Test assuming a log-additive mode of inheritance (one degree of freedom). Following Laird and Mosteller, heterogeneity of per allele ORs between studies was assessed by the p-value derived from the Q statistic [4] and using I2. Tests were two-sided.
Genetic main effects by ER status were estimated using case-control logistic regression and restricting the case sample to ER+ or ER− cases, respectively. To test for significant differences between main effects of rs2046210 or rs12662670 in ER+ versus ER− cases, logistic regression analyses were conducted in cases only. In these case-only analyses, the binary ER status was the outcome/dependent variable and the respective SNP and the indicator variables representing the studies were the independent variables.
Variation in OR by age was evaluated by testing for an interaction between age-group (<40, 40 to 49, 50 to 59, ≥60) and SNP, separately for each subgroup defined by ethnicity and ER status. Thus, the multiplicative SNP by age-group interaction term entered the model in addition to the main effect terms for SNP, age-group and study.
To investigate whether the association with breast cancer risk could be explained by one SNP or whether both SNPs had independent effects on disease risk, we fitted logistic regression models which included both SNPs, in addition to indicator variables for the studies, as independent variables in the model. Analyses were carried out separately for Europeans and Asians and for ER− versus ER+ cases and controls. Additionally, haplotype analyses were performed using logistic regression models that included the estimated two-marker haplotypes (coded according to a log-additive model) except for the reference haplotype (i.e., the most frequent haplotype) and the indicator variables for study. Haplotypes were estimated using the expectation-maximization algorithm.
All analyses, were performed using R version 2.11.0 [5] and the R packages meta, rmeta and haplo.stats.
Results
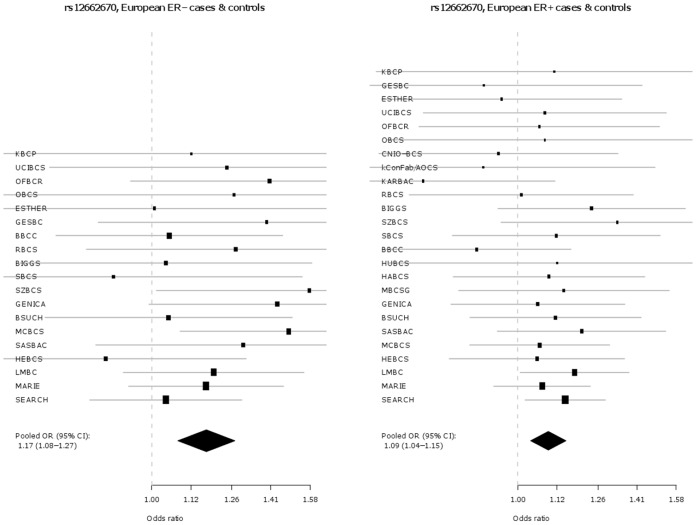
Key characteristics for each participating study are shown in Table S1. In addition to the originally discovered SNP rs2046210, SNP s12662670 was genotyped as a surrogate for the best tag from Turnbull et al. (rs3757318) [3], for which no working Taqman™ assay could be designed. The genotype distributions by ethnicity and study for SNPs rs2046210 and rs12662670 in cases and controls are given in Tables S3a and S3b. The associations of each SNP are presented in Table 1 and as Forest plots in Figure 1 and 2 , separately for Europeans and Asians. Both SNPs are significantly associated with breast cancer risk in both ethnic groups. However, the per-allele OR associated with the minor A allele of SNP rs2046210 is higher in Asian populations [OR (A/G) = 1.36 (95% CI 1.26–1.48), p = 7.6×10−14] than in Europeans [OR (A/G) = 1.09 (95% CI 1.07–1.11), p = 6.8×10−18] and this difference is statistically significant [pheterogeneity = 1.4×10−7]. SNP rs12662670 shows a similar pattern, with a higher OR associated with the minor G allele in Asian studies [OR (G/T) = 1.29 (95% CI 1.19–1.41), p = 1.2×10−9] than in Europeans [OR (G/T) = 1.12 (95% CI 1.08–1.17), p = 3.8×10−9] and again this difference is statistically significant [pheterogeneity = 0.002]. In each case there is no evidence for departure from a log-additive model (a co-dominant mode of inheritance).
Table 1. Association of rs2046210 and rs12662670 with breast cancer.
| Ethnicity | Number of cases/controls | Odds ratio (95% confidence interval) | P-valuea | ||
| per alleleb | Heterozygotec | Homozygotec | |||
| rs2046210 | |||||
| Analyses adjusted for study | |||||
| Overall | 54,647/49,559 | 1.10 (1.08–1.13) | 1.11 (1.08–1.14) | 1.22 (1.17–1.27) | 3.30×10−25 |
| Europeans | 49,634/46,679 | 1.09 (1.07–1.11) | 1.09 (1.06–1.12) | 1.18 (1.13–1.23) | 6.76×10−18 |
| Asians | 2983/2332 | 1.36 (1.26–1.48) | 1.39 (1.23–1.56) | 1.83 (1.54–2.18) | 7.60×10−14 |
| Analyses adjusted for study and rs12662570 | |||||
| Overall | 40,384/33,750 | 1.08 (1.05–1.11) | 1.08 (1.05–1.12) | 1.16 (1.10–1.23) | 3.06×10−9 |
| Europeans | 36,396/31,105 | 1.08 (1.05–1.11) | 1.08 (1.04–1.12) | 1.16 (1.09–1.22) | 1.78×10−8 |
| Asians | 3416/2420 | 1.17 (1.02–1.36) | 1.18 (0.99–1.41) | 1.37 (1.02–1.85) | 0.028 |
| rs12662670 | |||||
| Analyses adjusted for study | |||||
| Overall | 42,654/40,166 | 1.15 (1.12–1.19) | 1.15 (1.10–1.19) | 1.40 (1.23–1.58) | 2.52×10−16 |
| Europeans | 38,723/37,400 | 1.12 (1.08–1.17) | 1.13 (1.09–1.18) | 1.12 (0.94–1.34) | 3.83×10−9 |
| Asians | 3273/2451 | 1.29 (1.19–1.41) | 1.24 (1.10–1.39) | 1.77 (1.46–2.14) | 1.18×10−9 |
| Analyses adjusted for study and rs2046210 | |||||
| Overall | 40,384/33,750 | 1.10 (1.06–1.15) | 1.10 (1.05–1.15) | 1.25 (1.08–1.45) | 5.86×10−6 |
| Europeans | 36,396/31,105 | 1.07 (1.02–1.12) | 1.09 (1.03–1.14) | 1.01 (0.83–1.23) | 2.91×10−3 |
| Asians | 3416/2420 | 1.21 (1.04–1.40) | 1.16 (0.97–1.38) | 1.55 (1.12–2.13) | 0.012 |
Results are presented overall and separately for Europeans and Asians. Pooled analyses adjusted for study only as well as adjusted for rs12662670 or rs2046210, respectively, in addition to study were performed.
P-value derived from the log-additive model.
Odds ratio per minor allele (A allele for rs2046210, G allele for rs12662670).
Odds ratio relative to the major allele homozygous (GT) genotype.
Figure 1. Association of rs2046210 with breast cancer in Europeans versus Asians.
Figure 2. Association of rs12662670 with breast cancer in Europeans versus Asians.
Logistic regression models, which include both SNPs, indicate that the two SNPs are independently associated in both Europeans (p = 1.8×10−8 for rs2046210, p = 2.9×10−3 for rs12662670; Table 1 ) and Asians (p = 0.028 for rs2046210, p = 0.012 for rs12662670). In each ethnicity the estimated ORs for each SNP, after adjustment for the other SNP, are of similar magnitudes: For rs2046210 in Europeans OR (A/G) = 1.08 (95% CI 1.05–1.11) and for rs12662670 in Europeans OR (G/T) = 1.07 (95% CI 1.02–1.12). For rs2046210 in Asians OR (A/G) = 1.17 (95% CI 1.02–1.36) and for rs12662670 in Asians OR (G/T) = 1.21 (95% CI 1.04–1.40). Similar effect estimates are also obtained for haplotypes carrying one minor allele though estimates do not reach statistical significance for the very rare haplotype carrying the major (G) allele of rs2046210 along with the minor (G) allele of rs12662670 ( Table 2 ). Of note, from the four observed haplotypes, effects are strongest and highly statistically significant for the haplotype carrying both minor alleles: In Europeans OR (AG) = 1.16 (95% CI 1.11–1.21). In Asians OR (AG) = 1.42 (95% CI 1.30–1.56).
Table 2. Association of haplotypes composed of rs2046210 and rs12662670 with breast cancer.
| Ethnicity | Number of cases/controls | rs2046210 | rs12662670 | Haplotype frequency | Odds ratio (95% confidence interval)a | P-valueb |
| Overall | 40,384/33,750 | Ac | T | 0.26 | 1.08 (1.05–1.11) | 1.55×10−9 |
| G | Gc | 0.01 | 1.10 (0.95–1.27) | 0.189 | ||
| Ac | Gc | 0.09 | 1.23 (1.18–1.28) | 2.20×10−4 | ||
| Gd | Td | 0.64d | – | – | ||
| Europeanse | 36,396/31,105 | Ac | T | 0.27 | 1.08 (1.05–1.11) | 1.59×10−8 |
| G | Gc | 0.01 | 1.02 (0.86–1.21) | 0.832 | ||
| Ac | Gc | 0.07 | 1.16 (1.11–1.21) | 4.35×10−11 | ||
| Gd | Td | 0.65d | – | – | ||
| Asianse | 3416/2420 | Ac | T | 0.09 | 1.23 (1.05–1.45) | 1.15×10−2 |
| G | Gc | 0.04 | 1.23 (0.94–1.61) | 0.134 | ||
| Ac | Gc | 0.27 | 1.42 (1.30–1.56) | 4.44×10−14 | ||
| Gd | Td | 0.60d | – | – |
Results are presented overall and separately for Europeans and Asians. Pooled analyses adjusted for study were performed.
Odds ratio per haplotype compared to the reference haplotype (i.e., the most frequent haplotype).
P-value derived from the log-additive model.
Minor allele.
Reference haplotype.
The OR estimates for in-situ cancer are similar to those for invasive cancer for both SNPs in Europeans, although, due to small numbers, the effect of rs12662670 on in-situ tumours does not reach statistical significance (Table S4). For each Asian study, the number of in-situ cases is less than 100 and so effect estimates are inaccurate but do not differ from those for invasive cancer (data not shown).
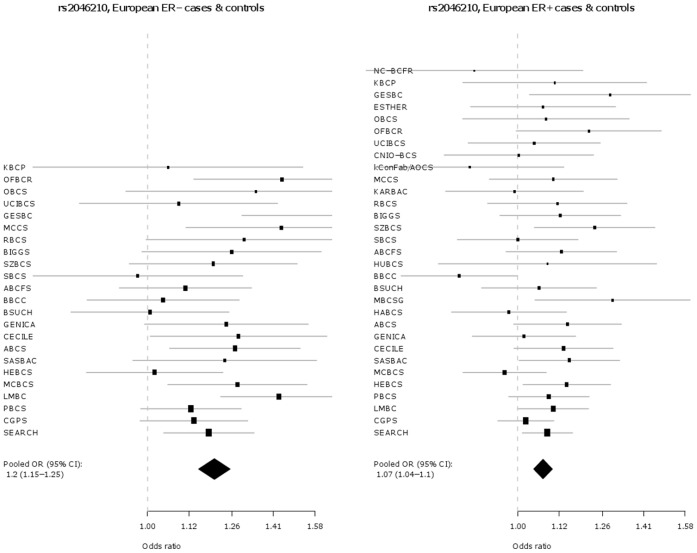
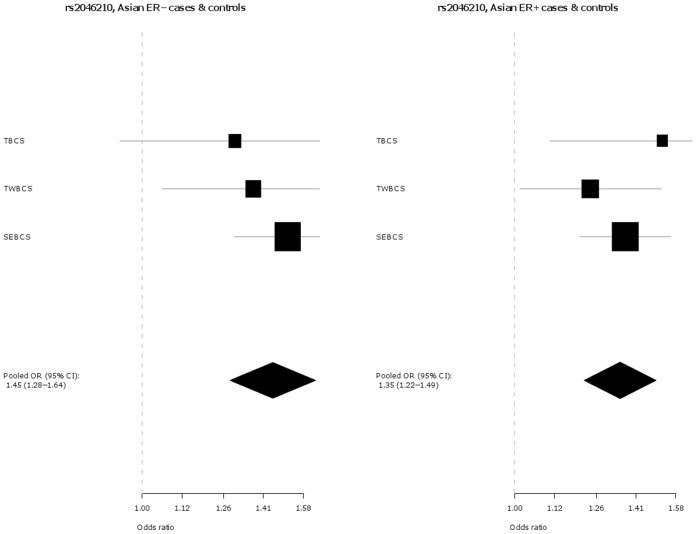
The associations of these two SNPs with tumour sub-types defined by ER status (ER+ and ER−) were also investigated and are presented in Table 3 and Figures 3 , 4, 5 , and 6 . In Europeans, SNP rs2046210 is associated with a greater OR for ER− than ER+ tumours: OR (ER−) = 1.20 (95% CI 1.15–1.25), p = 1.8×10−17 vs. OR (ER+) = 1.07 (95% CI 1.04–1.1), p = 1.3×10−7, pheterogeneity = 5.1×10−6. This difference remains significant after adjustment for rs12662670. A similar, although non-significant, difference is observed in European women for SNP rs12662670 ( Table 3 ). In the Asian studies, however, there is no clear evidence of a differential association by tumour receptor status for either SNP ( Table 3 ).
Table 3. Association of rs2046210 and rs12662670 with risk of ER−*/ER+** breast cancer.
| Ethnicity | Estrogen receptor status | Number of cases/controls | Odds ratio (95% confidence interval)a | P-valueb | P-heterogeneityc |
| rs2046210 | |||||
| Analyses adjusted for study | |||||
| Overall | ER−* | 7126/35,833 | 1.23 (1.18–1.28) | 6.90×10−25 | |
| ER+ ** | 5914/32,977 | 1.09 (1.06–1.12) | 1.66×10−11 | 5.93×10−7 | |
| Europeans | ER−* | 807/2334 | 1.20 (1.15–1.25) | 1.77×10−17 | |
| ER+ ** | 24,596/42,664 | 1.07 (1.04–1.10) | 1.26×10−7 | 5.10×10−6 | |
| Asians | ER−* | 22,336/39,856 | 1.45 (1.29–1.64) | 1.70×10−9 | |
| ER+ ** | 1307/2334 | 1.35 (1.22–1.49) | 1.09×10−8 | 0.513 | |
| Analyses adjusted for study and rs12662670 | |||||
| Overall | ER−* | 5526/25,627 | 1.19 (1.13–1.26) | 1.26×10−10 | |
| ER+ ** | 19,502/32,010 | 1.07 (1.03–1.10) | 7.26×10−5 | 1.01×10−3 | |
| Europeans | ER−* | 4497/23,090 | 1.20 (1.13–1.26) | 4.54×10−10 | |
| ER+ ** | 17,653/29,365 | 1.07 (1.03–1.10) | 2.16×10−4 | 4.63×10−4 | |
| Asians | ER−* | 975/2420 | 1.16 (0.94–1.44) | 0.169 | |
| ER+ ** | 1649/2420 | 1.13 (0.94–1.35) | 0.194 | 0.777 | |
| rs12662670 | |||||
| Analyses adjusted for study | |||||
| Overall | ER−* | 5422/28,201 | 1.23 (1.15–1.32) | 6.16×10−9 | |
| ER+ ** | 4563/26,198 | 1.14 (1.09–1.19) | 3.05×10−8 | 0.074 | |
| Europeans | ER−* | 808/1890 | 1.17 (1.08–1.27) | 1.90×10−4 | |
| ER+ ** | 20,095/34,460 | 1.09 (1.04–1.15) | 6.08×10−4 | 0.070 | |
| Asians | ER−* | 18,519/32,349 | 1.37 (1.20–1.56) | 3.30×10−6 | |
| ER+ ** | 1394/1890 | 1.35 (1.21–1.51) | 1.58×10−7 | 0.691 | |
| Analyses adjusted for study and rs2046210 | |||||
| Overall | ER−* | 5526/25,627 | 1.10 (1.01–1.20) | 0.028 | |
| ER+ ** | 19,502/32,010 | 1.09 (1.03–1.15) | 2.14×10−3 | 0.87 | |
| Europeans | ER−* | 4497/23,090 | 1.03 (0.94–1.14) | 0.516 | |
| ER+ ** | 17,653/29,365 | 1.05 (0.99–1.11) | 0.110 | 0.92 | |
| Asians | ER−* | 975/2420 | 1.33 (1.07–1.65) | 0.011 | |
| ER+ ** | 1649/2420 | 1.26 (1.04–1.52) | 0.017 | 0.30 | |
Results are presented overall as well as separately for Europeans and Asians. Pooled analyses adjusted for the studies were performed. A log-additive genetic model was assumed.
Estrogen receptor negative.
Estrogen receptor positive.
Odds ratio per minor allele (A allele for rs2046210, G allele for rs12662670) derived from case-control logistic regression restricted to ER+ or ER− cases, respectively, and the whole control sample.
P-value derived from the log-additive model derived from case-control logistic regression restricted to ER+ or ER− cases, respectively, and the whole control sample.
P-value for heterogeneity between estimates of genetic main effects derived from case-only analysis.
Figure 3. Association of rs2046210 with breast cancer in European ER.
−*versus ER+*cases and controls. *Estrogen receptor negative; **Estrogen receptor positive.
Figure 4. Association of rs2046210 with breast cancer in Asian ER.
−*versus ER+**cases and controls. *Estrogen receptor negative; **Estrogen receptor positive.
Figure 5. Association of rs12662670 with breast cancer in European ER.
−*versus ER+**cases and controls. *Estrogen receptor negative; **Estrogen receptor positive.
Figure 6. Association of rs12662670 with breast cancer in Asian ER.
−*versus ER+*cases and controls. *Estrogen receptor negative; **Estrogen receptor positive.
We further investigated whether the magnitudes of these SNP associations on tumour sub-types differed by age at diagnosis/interview (see Table S5). In Asian studies the data are too sparse to give meaningful results. In the combined ethnicities and the European studies alone, the magnitudes of the observed associations are greater in younger women.
Fourteen of the European studies had been designed to over-sample cases with a family history of breast cancer (see Table S1), which could have led to an overestimation of the ORs relative to those expected in a population-based case-control study. However, exclusion of these studies does not materially affect the estimated ORs for either SNP (see Table S6).
Discussion
In this large collaborative study of up to 61,689 cases and 58,822 controls, we demonstrate a highly statistically significant association between the A allele of rs2046210 and increased breast cancer risk in women of both Asian and European ancestry, thus extending the association previously observed in Asian populations. Consistent with previous reports [1]–[3], the effect sizes are significantly greater in Asians than in Europeans. Our study also reveals that the G allele of SNP rs12662670 is significantly associated with increased breast cancer risk in both ethnicities. SNP rs12662670 is used here as surrogate for SNP rs3757318 - the most strongly associated SNP at this locus in the European GWAS described by Turnbull et al. [3]. In addition, and also in contrast to Stacey et al. [2], we find that the OR for rs12662670 is greater in Asians than in Europeans ( Table 1 , Figure 1 and 2 ). In contrast to previous reports, our study indicates that both SNPs (rs2046210 and rs12662670) may be independently associated with breast cancer risk – in models including both SNPs, both maintain significant ORs after adjustment for the other. Haplotype analyses result in effect estimates for the AT and GG haplotypes, which carry only one minor allele, very similar to those of the single SNP analyses for the respective minor alleles. Furthermore, haplotype analyses show a clearly stronger effect of the AG haplotype, carrying both minor alleles, compared to the effects of the AT and GG haplotypes, further supporting the hypothesis that there may be two different causative variants, one on each haplotype carrying only one minor allele and both on the haplotype carrying both minor alleles (i.e., the stronger effect of the AG haplotype compared to the AT and GG haplotypes may be explained by the joint effect of the two minor alleles on the AG haplotype). However, the alternative conclusion that a single causative variant may exist that is intermediate between the two SNPs phylogenetically, i.e. on the AG haplotype and on some of the AT haplotypes, cannot yet be completely excluded, since this could also be an explanation for the stronger effect of the AG haplotype compared to the AT and GG haplotypes.
We also find evidence that SNP rs2046210 is more strongly associated with ER− than ER+ disease in both European and Asian women. In the present study this differential association with receptor status is statistically significant in European studies (and remains after adjustment for rs12662670) but is not quite significant in Asians which may be due to a lack of power attributable to the comparatively small number of Asian individuals involved in our study ( Table 3 ). However this same SNP had previously been reported to be more strongly associated with ER− tumours in the original Chinese cases [1] as well as in a recent replication study in Chinese women [6]. In line with these reports, a meta-analysis (14,231 cases, 10,244 controls) on this SNP-disease association by ER status in Asians, incorporating published results as well as those presented here, reveals a significant difference in OR associated with ER− versus ER+ tumour risk [OR (A/G - ER− ) = 1.37 (95% CI 1.30–1.44), p = 3.7×10−33 vs. OR (A/G- ER+) = 1.27 (95% CI 1.22–1.34), p = 2.2×10−24; pheterogeneity = 0.04]. A stronger association of SNP rs2046210 with ER− tumours is also consistent with the report from the Consortium of Modifiers of BRCA1/2 (CIMBA) [7] that the same allele is associated with an increased Hazard Ratio of breast cancer in BRCA1 mutation carriers (who predominantly develop ER− tumours). The CIMBA study also observed that this allele conferred increased Hazard Ratios among younger mutation carriers while we observed similar trends for greater SNP ORs at younger age groups (Table S5). By contrast, the CIMBA consortium reported that SNP rs9397435 (the tag they used for rs12662670; r2 = 0.61, r2 = 0.50 and r2 = 0.85 in HapMap2 CEU, JPT and CHB samples, respectively) shows evidence of modification of risk in both BRCA1 and BRCA2 mutation carriers (who mainly develop ER− and ER+ tumours respectively) [7] whilst similarly, we find that SNP rs12662670 is associated with increased risks of both ER− and ER+ tumours.
Previous fine-scale mapping publications on this locus [2], [8] have sought a single variant to explain the associations seen with all SNPs in the region: Stacey et al. [2] proposed SNP rs9397435 as a possible single causative variant since it was more strongly associated than rs2046210 in women of European, African and Asian ancestry. We are not able to comment on this variant, as it has not been genotyped in BCAC. However, our findings suggest there could be two independent associations at this locus: one, better tagged by SNP rs2046210, predisposing to ER− tumours and the second, better tagged by rs12662670, conferring similar risks of both tumour types. Although physically close, SNPs rs2046210 and rs12662670 are not highly correlated with each other, particularly in Europeans (in BCAC r2 = 0.12 in Europeans and r2 = 0.56 in Asians) and all four possible combinations (haplotypes) of these two SNPs clearly exist.
Examination of linkage disequilibrium plots of the regions surrounding these two SNPs in Europeans ( Figure 7 ) reveals little, if any, physical overlap between SNPs highly correlated (r2>0.9) with rs2046210 and those with rs12662670. If there were a single causal variant, directly responsible for the associations seen with both SNPs, it would need to be correlated with both SNPs. Such a variant has not been yet identified (e.g. by the 1000 Genomes Project). It would presumably be relatively rare. An alternative, and we think, more plausible, explanation for the pattern of associations may be the existence of two independent causative variants, one correlated with rs2046210 and another correlated with rs12662670. If this is the case, the former variant may be more strongly associated with ER− breast cancer than the latter. The reason why both SNPs confer higher relative risks in Asians than in Europeans is unclear. Within the BCAC studies, ER− tumours are relatively more prevalent among Asian (36%) compared to European cases (23%), but this is not sufficient to explain the higher ORs in Asians, since the effects persist after stratification by ER status. It remains possible that the higher relative risks are due to differential patterns of linkage disequilibrium if the, as yet, unidentified causal variants are not strongly correlated with the SNPs identified to date. These questions may be resolved by comprehensive re-sequencing of this locus and fine scale mapping to identify the causal variant (or variants) responsible for the observed breast cancer risks. One aim of the iCOGS Project [9], which is currently underway, is to address these questions. However it is possible that these observed differences between Asians and Europeans may reflect interactions with lifestyle risk factors or other unlinked genetic loci. Another possible explanation is that the estimated SNP effects in Asians are inflated given the phenomenon known as the “winner’s curse”, i.e. the suboptimal power of the pool of Asian studies (due to the small number of Asian individuals) together with the commonly used requirement for a published association to pass a certain pre-defined p-value threshold may have resulted in biased SNP effect estimates [10], [11].
Figure 7. Linkage disequilibrium blocks in the ESR1 region.
Five SNPs tagged (at r2>0.9) by rs12662670 and three by rs2046210 are marked by arrows (dark and light grey respectively); rs12662670 and rs2046210 are marked by stars; rs3757318 and rs9397435 are marked by points; blocks were generated using data from the 1000 Genomes Project and HapMap; blocks include all single nucleotide polymorphisms with a minor allele frequency >0.05. The directions of translation of ESR1 and C6orf97 are marked and other genes in the locus are listed.
Although there are eleven genes within 1 Mb of this locus, attention has focused on the ESR1 gene, whose transcription start site is located approximately 180 Kb downstream of SNP rs2046210. ESR1 encodes ERα and has long been implicated in breast carcinogenesis. However, it is possible that the proximity of this SNP to ESR1 may be providing a false lead – both SNPs (rs2046210 and rs12662670) lie in the flanking region of C6orf97 and there are numerous other genes in close physical proximity (see Figure 7 ). It is notable however, that SNPs mapping to this region have also been identified in GWAS for bone mineral density – another phenotype in which estradiol metabolism is clearly implicated [12], [13]. Furthermore, a recent paper [14] demonstrates that a number of genes, including ESR1 and C6orf97 are co-regulated at this locus although the functions of most of these co-regulated genes have not yet been elucidated. The SNP associations, presented here, may provide a basis to explore the biological role of this locus in estrogen signalling and cancer development in more detail.
Taken together our findings suggest the possibility of the presence of two different causative variants at the 6q25.1 locus and indicate that fine-scale mapping efforts aimed at finding a single variant accounting for associations with both marker SNPs, may not be successful.
Supporting Information
Characteristics of 45 case-control studies within the Breast Cancer Association Consortium (BCAC).
(DOC)
Characteristics of the study populations genotyped for rs2046210 (a) and rs12662670 (b).
(DOC)
Genotype frequencies of SNP rs2046210 (a) and rs12662670 (b) in the different studies.
(DOC)
Association of rs2046210 and rs12662670 with in-situ/invasive breast cancer.
(DOC)
Association of rs2046210 and rs12662670 with risk of ER−*/ER+**breast cancer.
(DOC)
Association of rs2046210 and rs12662670 with breast cancer.
(DOC)
Acknowledgments
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. In particular we thank Jonathan Morrison (BCAC); Maggie Angelakos, Judi Maskiell and Gillian Dite (ABCFS); Richard van Hien, Sten Cornelissen and the NKI-AVL Family Cancer Clinic (ABCS); all the participants in the Thai Breast Cancer Study within the Asia Cancer Programme (ACP) initiative; special thanks also go to the Thai Ministry of Public Health, doctors and nurses who helped with the data collection process; finally, we would like to thank Ms Sunee Swangsri, our Thai study co-ordinator (ACP); furthermore, we thank Eileen Williams, Elaine Ryder-Mills and Kara Sargus (BBCS); Niall McInerney, Gabrielle Colleran, Andrew Rowan and Angela Jones (BIGGS); Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez and Anna González-Neira (CNIO-BCS); Hartwig Ziegler, Sonja Wolf and Volker Hermann (ESTHER); all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out; in particular, we thank The Wellcome Trust Case Control Consortium (see the WTCCC website for a full list of contributing investigators) (FBCS); furthermore, we thank Bernd Frank, Rita K. Schmutzler and Claus R. Bartram (GC-HBOC); Christina Justenhoven, Beate Pesch, Thomas Brüning, Volker Harth, Sylvia Rabstein, Christina Baisch and Hans-Peter Fischer (GENICA); Ursula Eilber and Tanya Koehler (GESBC); Johann H. Karstens (HABCS, HMBCS, HUBCS); Kirsimari Aaltonen, Irja Erkkilä (HEBCS); Eija Myöhänen and Helena Kemiläinen (KBCP); Heather Thorne, Eveline Niedermayr, the AOCS Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb) and the ACS Management Group (A Green, P Parsons, N Hayward, P Webb, D Whiteman) (KConFab/AOCS); Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts (LMBC); Tracy Slanger, Elke Mutschelknauss, Ramona Salazar, S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese and K. Smit (MARIE); Marco Pierotti, Bernard Peissel and Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale Tumori, Bernardo Bonanni of the Istituto Europeo di Oncologia, Loris Bernard and the personnel of the Cancer Genetics Testing laboratory at the IFOM-IEO campus (MCBCS); all the people contributing to the MCCS, including the original investigators and the diligent team who recruited the participants and who continue working on follow up; finally, we would like to express our gratitude to the many thousands of Melbourne residents who continue to participate in the study (MCCS); furthermore, we thank the study participants of the MSKCC study (MSKCC); Meeri Otsukka and Kari Mononen for their contribution to OBCS (OBCS); the participants in the Ontario Familial Breast Cancer Registry; Teresa Selander and Nayana Weerasooriya from Cancer Care for their contributions to OFBCR (OFBCR); Michael Stagner and Pei Chao from Information Management Services (Sliver Spring, MD, USA), for data management support (PBCS); Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk and the Erasmus MC Family Cancer Clinic (RBCS); Helen Cramp, Dan Connley, Sabapathy Balasubramanian and Sue Higham for their contribution to patient recruitment and data collection (SBCS); the SEARCH and EPIC teams (SEARCH); Irene Masunaka (UCIBCS); John Hampton from University of Wisconsion and Michael Stagner and Pei Chao from Information Management Services (Sliver Spring, MD, USA), for data management support (US3SS).
Funding Statement
This work was partly supported by the European Community’s Seventh Framework Programme (COGS; http://cogseu.org/) [grant agreement number 223175, grant number HEALTH-F2-2009-223175]. The Breast Cancer Association Consortium (BCAC) is funded by Cancer Research-UK (http://science.cancerresearchuk.org/) [C1287/A10118 and C1287/A12014]. Meetings of the BCAC have been funded by the European Union COST programme (http://www.srl.cam.ac.uk/consortia/cost/index.html) [BM0606]. D.F.E. is a Principal Research Fellow of Cancer Research-UK. A.C.A. is a Cancer Research-UK Senior Cancer Research Fellow. A.M.D has been funded by Cancer Research- UK [C8197/A10865] and The Joseph Mitchell Trust. The Australian Breast Cancer Family Study (ABCFS) and Northern California Breast Cancer Family Registry (NC-BCFR) work was supported by the United States National Cancer Institute, National Institutes of Health (NIH) [RFA-CA-06-503]; and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR); and Principal Investigators, including Cancer Care Ontario [U01 CA69467]; Northern California Cancer Center [U01 CA69417]; University of Melbourne [U01 CA69638]. Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia; the New South Wales Cancer Council; the Victorian Health Promotion Foundation (Australia); and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The Amsterdam Breast Cancer Study (ABCS) study was supported by the Dutch Cancer Society [NKI 2001–2423; 2007–3839]; and the Dutch National Genomics Initiative. The Asian Cancer Program (ACP) study was supported by the Breast Cancer Research Trust. The Bavarian Breast Cancer Cases and Controls (BBCC) study was partly supported by ELAN-Fond of the University Hospital of Erlangen. The British Breast Cancer Study (BBCS) study is supported by Cancer Research UK; and Breakthrough Breast Cancer; and acknowledges National Health Service (NHS) funding to the National Institute for Health Research (NIHR) Biomedical Research Centre; and the National Cancer Research Network (NCRN). Breast Cancer in Galway Genetic Study (BIGGS) is supported by National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre [to E.S.]; Guy’s & St. Thomas’ National Health Service Foundation Trust in partnership with King’s College London [to E.S.]; and the Oxford Biomedical Research Centre [to I.T.]. The Breast Cancer Study of the University Clinic Heidelberg (BSUCH) study was supported by the Dietmar-Hopp Foundation; and the Helmholtz Society. The CECILE Breast Cancer Study (CECILE) study was supported by Fondation de France (2007), French National Institute of Cancer [INCa grant 2007, 2008, 2009]; French Agency for Environmental and Occupational Health [ANSES (ex–AFSSET) grant 2008 and 2009]; the French League Against Cancer; and National Research Agency (ANR). The Copenhagen General Population Study (CGPS) was supported by the Chief Physician Johan Boserup and Lise Boserup Fund; the Danish Medical Research Council; and Herlev Hospital. The Spanish National Cancer Centre Breast Cancer Study (CNIO-BCS) was supported by the Genome Spain Foundation; the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario [PI081120 to J.B., PI081583 to R.L.M.]. The California Teachers Study (CTS) was supported by the California Breast Cancer Act of 1993; National Institutes of Health [R01 CA77398]; the Lon V Smith Foundation [LVS39420]; and the California Breast Cancer Research Fund [contract 97–10500]. Collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. The ESTHER Breast Cancer Study (ESTHER) study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The ICR Familial Breast Cancer Study (FBCS) is supported by funds from Cancer Research UK [C8620/A8372; C8620/A8857]; a US Military Acquisition (ACQ) Activity, Era of Hope Award [W81XWH-05-1-0204]; the Institute of Cancer Research (UK) a Medical Research Council (UK) Clinical Research Fellowship [to C.T.]. The FBCS acknowledges National Health Service (NHS) funding to the Royal Marsden/Institute of Cancer Research, National Institute for Health Research (NIHR) Specialist Cancer. The German Consortium for Hereditary Breast & Ovarian Cancer (GC-HBOC) was supported by Deutsche Krebshilfe [107054]; the Center of Molecular Medicine, Cologne; the Helmholtz society; and the Dietmar Hopp Foundation [to B.B.]. The Gene Environment Interaction and Breast Cancer in Germany (GENICA) network was supported by the Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and the University of Tübingen, Germany [to C.J. and H.B.]; the Department of Internal Medicine, Evangelische Kliniken Bonn GmbH, Johanniter Krankenhaus, Bonn, Germany [to Y.D.K. and C.B.]; the Institute of Pathology, University of Bonn, Bonn, Germany [to HP.F.]; Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [to U.H.]; the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany [to T.B., B.P., S.R., A.S. and V.H.]. The Genetic Epidemiology Study of Breast Cancer by Age 50 (GESBC) was supported by the Deutsche Krebshilfe e. V. [70492]; and genotyping in part by the state of Baden-Württemberg through the Medical Faculty of the University of Ulm [P.685]. The Hannover Breast Cancer Study (HABCS) study was supported by an intramural grant from Hannover Medical School; and by a grant from the German Research Foundation [DFG, Do761/2-1]. The Helsinki Breast Cancer Study (HEBCS) study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland [132473]; the Finnish Cancer Society; and the Sigrid Juselius Foundation. The Hannover-Minsk Breast Cancer Study (HMBCS) was supported by short-term fellowships from the German Academic Exchange Program [to N.B.]; and the Friends of Hannover Medical School [to N.B.]. The Hannover-Ufa Breast Cancer Study (HUBCS) was supported by a grant from the German Federal Ministry of Research and Education [RUS08/017]. The Karolinska Breast Cancer Study (KARBAC) was supported by the Swedish Cancer Society and the Stockholm Cancer Society. The Kuopio Breast Cancer Project (KBCP) was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo; the Finnish Cancer Organizations; the Academy of Finland; and by the strategic funding of the University of Eastern Finland. Kathleen Cuningham Foundation Consortium for research into Familial Breast Cancer/Australian Ovarian Cancer Study (KConFab/AOCS) is supported by grants from the National Breast Cancer Foundation; the National Health and Medical Research Council (NHMRC); the Queensland Cancer Fund; the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia; and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684; 288704; 454508]. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729]; the Cancer Council of Tasmania and Cancer Foundation of Western Australia; and the NHMRC [199600]. G.C.T. and P.W. are supported by the NHMRC. Leuven Multidisciplinary Breast Centre (LMBC) is supported by the ‘Stichting tegen Kanker’ [232–2008; 196–2010]. The Mammary Carcinoma Risk Factor Investigation (MARIE) study was supported by the Deutsche Krebshilfe e.V. [70–2892-BR I]; the Hamburg Cancer Society; the German Cancer Research Center; and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. Milan Breast Cancer Study Group (MBCSG) was supported by grants from Ministero della Salute (Extraordinary National Cancer Program 2006 “Alleanza contro il Cancro”, and “Progetto Tumori Femminili”) [to P.R.]; Ministero dell’Universita’ e Ricerca (RBLAO3-BETH) [to P.R.]; Fondazione Italiana per la Ricerca sul Cancro (Special Project “Hereditary tumors”); Associazione Italiana per la Ricerca sul Cancro (4017) [to P.P.]; and by funds from Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5×1000”). The Mayo Clinic Breast Cancer Study (MCBCS) was supported by National Institutes of Health grants [R01CA122340; R01 CA128978]; and a Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201]. Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth; and Cancer Council Victoria. The MCCS was further supported by Australian National Health and Medical Research Council (NHMRC) grants [209057; 251553; 504711]; and by infrastructure provided by Cancer Council Victoria. The Memorial Sloan-Kettering Cancer Center (MSKCC) was supported by the Breast Cancer Research Foundation; the Sharon Levine Corzine Fund; the Niehaus Cancer Research Initiative; the Andrew Sabin Family Fund; the Charles Krasne Research Fund; and the Norman and Carol Stone Research Initiative. The Norwegian Breast Cancer Study (NBCS) was supported by grants from the Norwegian Research council [155218/V40, 175240/S10 to A.L.B.D.; FUGE-NFR 181600/V11 to V.N.K.]; and a Swizz Bridge Award [to A.L.B.D.]. The Oulu Breast Cancer Study (OBCS) was supported by research grants from the Finnish Cancer Foundation; the Sigrid Juselius Foundation; the Academy of Finland; the University of Oulu; and the Oulu University Hospital. The Ontario Familial Breast Cancer Registry (OFBCR) work was supported by the National Cancer Institute, National Institutes of Health [RFA # CA- 06–503]; and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR); and Principal Investigators, including Cancer Care Ontario [U01 CA69467]; Northern California Cancer Center [U01 CA69417]; and University of Melbourne [U01 CA69638]; and by Cancer Care Ontario. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the BCFR. The National Cancer Institute Polish Breast Cancer Study (PBCS) was supported by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The Rotterdam Breast Cancer Study (RBCS) was funded by the Dutch Cancer Society [DDHK 2004–3124; DDHK 2009–4318]. The Singapore and Sweden Breast Cancer Study (SASBAC) was funded by the Märit and Hans Rausing Initiative Against Breast Cancer; the Agency for Science, Technology and Research of Singapore (ASTAR); the United States National Institutes of Health (NIH); and the Susan G. Komen Breast Cancer Foundation. The Sheffield Breast Cancer Study (SBCS) was funded by Yorkshire Cancer Research; and the Breast Cancer Campaign. The Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH) study is funded by a programme grant from Cancer Research UK [C490/A11021] and laboratory infrastructure by [C8197/A10123]. The Seoul Breast Cancer Study (SEBCS) was supported by the Korea Health 21 R&D Project [AO30001], Ministry of Health and Welfare, Republic of Korea. The IHCC-Szczecin Breast Cancer Study (SZBCS) was supported by Grant PBZ_KBN_122/P05/2004; and the Polish Foundation of Science [to K.J.]. Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University. The International Agency for Research on Cancer-Thai Breast Cancer Study (TBCS) was funded by The National Cancer Institute Thailand. The Taiwanese Breast Cancer Study (TWBCS) is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. The University of California, Irvine, Breast Cancer Study (UCIBCS) component of this research was supported by the National Institute of Health [CA58860; CA92044]; and the Lon V Smith Foundation [LVS39420]. The UK Breakthrough Generations Study (UKBGS) is funded by Breakthrough Breast Cancer. The US Three State Study (US3SS) study was supported by Massachusetts [R01CA47305 to K.M.E.]; Wisconsin [R01 CA47147 to P.A.N.]; and New Hampshire [R01CA69664 to L. T.-E.] centers; and Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zheng W, Long J, Gao YT, Li C, Zheng Y, et al. (2009) Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet 41: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stacey SN, Sulem P, Zanon C, Gudjonsson SA, Thorleifsson G, et al. (2010) Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet 6: e1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, et al. (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laird NM, Mosteller F (1990) Some statistical methods for combining experimental results. Int J Technol Assess Health Care 6: 5–30. [DOI] [PubMed] [Google Scholar]
- 5.R Development Core Team (2005) R: A language and environment for statistical computing. Vienna, Austria, R Foundation for Statistical Computing. [Google Scholar]
- 6. Long J, Shu XO, Cai Q, Gao YT, Zheng Y, et al. (2010) Evaluation of breast cancer susceptibility Loci in chinese women. Cancer Epidemiol Biomarkers Prev 19: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antoniou AC, Kartsonaki C, Sinilnikova OM, Soucy P, McGuffog L, et al. (2011) Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet 20(16): 3304–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai Q, Wen W, Qu S, Li G, Egan KM, et al. (2011) Replication and Functional Genomic Analyses of the Breast Cancer Susceptibility Locus at 6q25.1 Generalize Its Importance in Women of Chinese, Japanese, and European Ancestry. Cancer Res 71: 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Collaborative Oncological Gene-Environment Study (COGS) Project. Available: http://www.cogseu.org. Accessed 2012 Jul 10.
- 10. Ioannidis JPA (2008) Why most discovered true associations are inflated. Epidemiology 19(5): 640–648. [DOI] [PubMed] [Google Scholar]
- 11. Kraft P (2008) Curses – winner’s and otherwise – in genetic epidemiology. Epidemiology 19(5): 649–651. [DOI] [PubMed] [Google Scholar]
- 12. Li WF, Hou SX, Yu B, Li MM, Ferec C, et al. (2010) Genetics of osteoporosis: accelerating pace in gene identification and validation. Hum Genet 127: 249–285. [DOI] [PubMed] [Google Scholar]
- 13. Xie H, Sun M, Liao XB, Yuan LQ, Sheng ZF, et al. (2010) Estrogen receptor-alpha36 mediates a bone-sparing effect of 17beta-estrodiol in postmenopausal women. J Bone Miner Res 26: 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunbier AK, Anderson H, Ghazoui Z, Lopez-Knowles E, Pancholi S, et al. (2011) ESR1 Is Co-Expressed with Closely Adjacent Uncharacterised Genes Spanning a Breast Cancer Susceptibility Locus at 6q25.1. PLoS Genet 7: e1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of 45 case-control studies within the Breast Cancer Association Consortium (BCAC).
(DOC)
Characteristics of the study populations genotyped for rs2046210 (a) and rs12662670 (b).
(DOC)
Genotype frequencies of SNP rs2046210 (a) and rs12662670 (b) in the different studies.
(DOC)
Association of rs2046210 and rs12662670 with in-situ/invasive breast cancer.
(DOC)
Association of rs2046210 and rs12662670 with risk of ER−*/ER+**breast cancer.
(DOC)
Association of rs2046210 and rs12662670 with breast cancer.
(DOC)