Abstract
In Rhizobia the Irr protein is an important regulator for iron-dependent gene expression. We studied the role of the Irr homolog RSP_3179 in the photosynthetic alpha-proteobacterium Rhodobacter sphaeroides. While Irr had little effect on growth under iron-limiting or non-limiting conditions its deletion resulted in increased resistance to hydrogen peroxide and singlet oxygen. This correlates with an elevated expression of katE for catalase in the Irr mutant compared to the wild type under non-stress conditions. Transcriptome studies revealed that Irr affects the expression of genes for iron metabolism, but also has some influence on genes involved in stress response, citric acid cycle, oxidative phosphorylation, transport, and photosynthesis. Most genes showed higher expression levels in the wild type than in the mutant under normal growth conditions indicating an activator function of Irr. Irr was however not required to activate genes of the iron metabolism in response to iron limitation, which showed even stronger induction in the absence of Irr. This was also true for genes mbfA and ccpA, which were verified as direct targets for Irr. Our results suggest that in R. sphaeroides Irr diminishes the strong induction of genes for iron metabolism under iron starvation.
Introduction
As cofactor of several enzymes and regulatory proteins iron is an essential element for living organisms. However, since reduced iron potentiates oxygen toxicity by the production of hydroxyl radicals in the Fenton reaction, life in the presence of oxygen requires a strict regulation of iron metabolism. Several regulators of iron metabolism have been investigated in bacteria. In Escherichia coli and in many other bacteria the Fur protein is a major regulator for iron-dependent gene expression (reviewed in e.g. [1]). Fur binds to DNA at a specific sequence (Fur box) [2] and acts as transcriptional repressor in the presence of iron. Iron regulation in alpha-proteobacteria mainly occurs by regulators different from this type of Fur protein, namely by Irr, RirA and IscR (reviewed in [3]). RirA and IscR both belong to the Rrf2 superfamily of transcriptional regulators. In E. coli IscR mainly regulates the Fe-S cluster biogenesis genes (suf genes) [4], [5]. To date its function in alpha-proteobacteria has not been investigated. RirA represses more than 80 transcriptional units in Rhizobium and Sinorhizobium in high iron conditions [6], [7]. Another Fur-like protein detected in alpha-proteobacteria is rather involved in the regulation of Mn2+ transport and was therefore designated “Mur” (manganese uptake regulator) [8], [9], [10], [11]. In Rhodobacter sphaeroides Mur has not only a role in manganese homeostasis but also affects regulation of iron metabolism [12].
An important function in regulating iron metabolism in Rhizobia could be attributed to the Irr protein (reviewed in [3]). Irr proteins are found in members of the Rhizobiales, Rhodobacterales and few other genera, and form a distinct sub-branch of the Fur superfamily [13]. In Rhizobiales most iron-dependent genes are regulated by Irr [14], [15], [16], [17]. In all species investigated to date Irr represses genes under iron depletion, which is opposite to the function of RirA. Irr binds to conserved sequences, the so-called Irr boxes or ICE motifs (iron control elements) close to the promoters of its target genes. At high iron concentrations Irr is degraded in Bradyrhizobium japonicum but not in Rhizobium leguminosarum [18]. In B. japonicum this degradation is mediated by its interaction with heme, whose intracellular concentration increases with external iron availability [19]. Heme can directly interact with the Irr protein but interaction is more efficient if heme is delivered by the heme synthesis enzyme ferrochelatase [20]. Reactive oxygen species (ROS) seem to promote the heme dependent degradation of Irr [21].
R. sphaeroides is a facultative photosynthetic bacterium, which can generate ATP by anoxygenic photosynthesis but can also generate ATP from aerobic or anaerobic respiration. At high oxygen concentration the formation of photosynthetic complexes is repressed. When the oxygen tension in the environment drops, photosynthesis genes are induced. Several proteins involved in redox-dependent gene regulation have been identified in R. sphaeroides [22], [23] and also the response to different ROS has been studied intensively [24], [25], [26], [27], [28], [29], [30], [31]. A transcriptome study revealed that many genes of iron metabolism are induced in response to hydrogen peroxide [30]. Only a few of these genes are under control of the intensively studied OxyR regulator [31]. Another transcriptome study revealed only very limited overlap of the response of R. sphaeroides to iron limitation and to oxidative stress [12]. The regulatory link between oxidative stress response and iron metabolism remains to be elucidated for alpha-proteobacteria.
A bioinformatic analysis based on experimental data from Rhizobia predicted iron responsive regulators and DNA target sequences for such regulators in alpha-proteobacteria including Rhodobacter [13] and suggested putative Irr (RSP_3179), Fur/Mur (RSP_2494), Fur/Zur (RSP_3569) and IscR (RSP_0443) regulators in R. sphaeroides. No RirA homolog is found in Rhodobacterales. A strain lacking the Fur/Mur protein was clearly impeded in growth by iron limitation indicating a role of Fur/Mur in regulation of iron metabolism. This was supported by transcriptome data [12].
Here we present an analysis of the role of the Irr protein (RSP_3179) on iron metabolism and resistance to oxidative stress in the photosynthetic alpha-proteobacterium R. sphaeroides.
Results
The Irr Protein of R. sphaeroides does not Influence Growth Under Iron Limitation but Resistance to Oxidative Stress
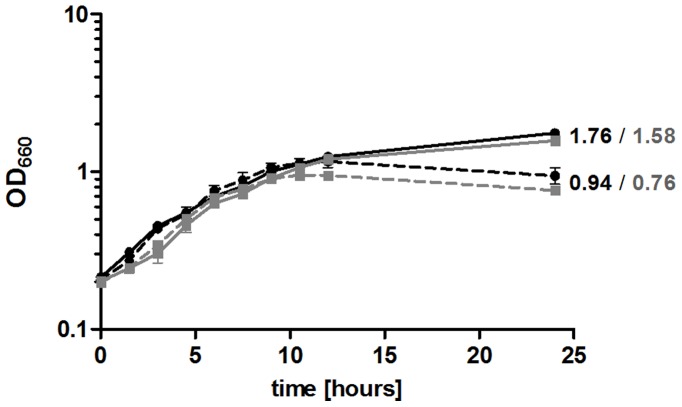
Exponential phase cultures of the R. sphaeroides wild type 2.4.1 and the 2.4.1Δirr deletion strain were subjected to iron limitation as described previously [12]. R. sphaeroides was cultivated without adding external Fe(III)citrate to the malate minimal medium but with the iron chelator 2,2′-dipyridyl. Cultivation was performed under microaerobic conditions to exclude the possibility that change in expression of genes for iron metabolism is caused by oxidative stress as observed previously for R. sphaeroides [30]. Growth curves confirmed our previous observation [12] that the wild type stops growing at an earlier time point during transition to stationary phase when iron is limiting (Fig. 1). The growth of the 2.4.1Δirr mutant (Table S4) was similar to that of the wild type. The mutant stopped growing a little bit earlier under iron limitation but this difference was not significant (Fig. 1). The irr deletion strain was clearly less impeded in growth by iron limitation than the 2.4.1Δfur/mur mutant that was characterized previously [12]. It can be excluded that the growth phenotypes are due to dipyridyl toxicity because cultures grown under decreased iron availability without added iron chelator are also impaired in growth (data not shown). Excess amounts of iron (20 µM Fe(III)citrate) did not influence the growth behavior of R. sphaeroides (data not shown). Furthermore, the wild type and the irr deletion strain were tested for porphyrin accumulation during growth in iron-deficient media. Spectral analyses did not reveal detectable levels of protoporphyrin in supernatants of R. sphaeroides cultures grown under iron limitation (data not shown). These results suggest that, in contrast to studies in Brad. japonicum and Brucella abortus, R. sphaeroides Irr is not involved in the down-regulation of heme biosynthesis under iron limitation. This view is supported by real-time RT-PCR data for the hemB and hemH genes (Fig. S1). R. sphaeroides Irr lacks the heme regulatory motif (HRM) that is associated with binding to heme and the turnover of the protein in Brad. japonicum. However, as heme can interact with an Irr protein lacking this motif in Bru. abortus [32] and R. leguminosarum [18], binding of heme to R. sphaeroides Irr was analyzed. The absorption spectrum of heme in the presence and absence of purified recombinant Irr was recorded. The 388 nm absorption peak of heme shifted to 430 nm in the presence of Irr (Fig. S2), supporting that heme binds to R. sphaeroides Irr under the conditions tested. We also used the R. sphaeroides IscR protein, which was purified with the identical protocol in this assay and did not result in a shift of the heme absorbance.
Figure 1. Growth curves of the R. sphaeroides wild type (black) and the 2.4.1Δirr mutant (gray) under normal iron (continuous line) and under iron limitation (dashed line) conditions are shown.
The optical density at 660 nm (OD660) of microaerobically grown R. sphaeroides cultures was determined over time. The data represent the mean of at least three independent experiments and error bars indicate standard error of the mean.
The sensitivity of the 2.4.1Δirr mutant to hydrogen peroxide and singlet oxygen was tested by zone inhibition assays. Singlet oxygen was generated by applying methylene blue to the filter disks and illumination. For both types of ROS the inhibition zones were significantly smaller for the 2.4.1Δirr mutant, indicating increased resistance (Fig. 2 A and B). When the irr gene was expressed in trans in strain 2.4.1Δirr (pRKirr), a wild type-like phenotype could be restored (Fig. 2 A and B, gray bars).
Figure 2. Sensitivity of R. sphaeroides wild type, 2.4.1Δirr mutant and complemented mutant to (photo-) oxidative stress.
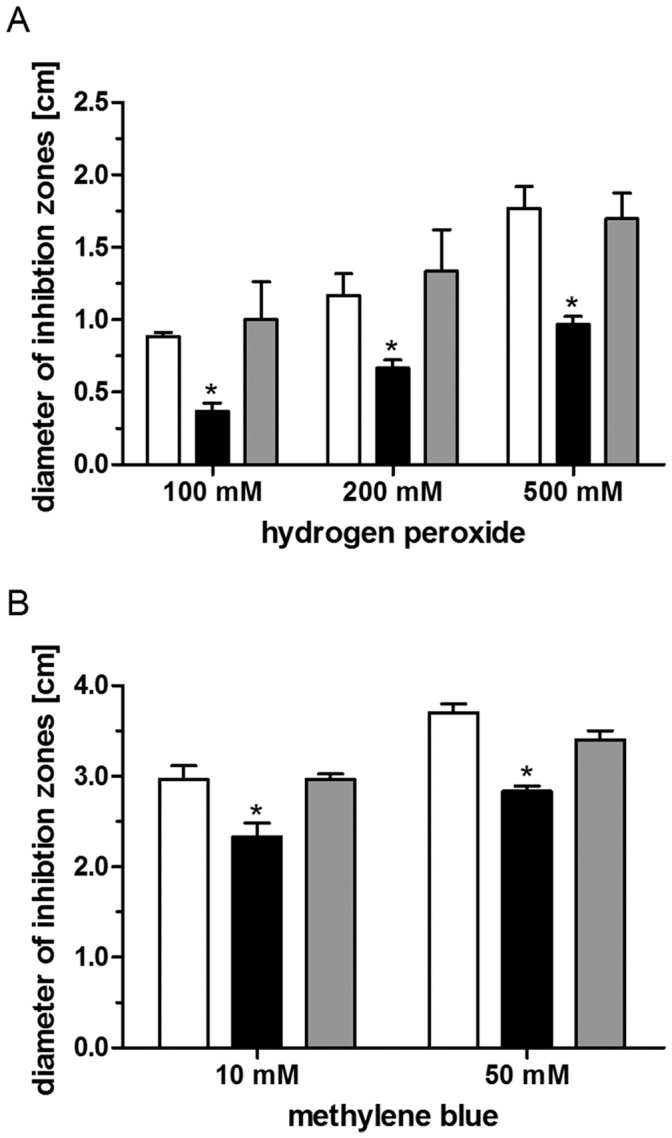
Inhibition of growth of the wild type (white bars), the irr deletion mutant (black bars) and the complemented mutant (gray bars) to hydrogen peroxide (A) and methylene blue (B) as determined by inhibition zone assays. Each bar represents the mean of at least three independent experiments and error bars indicate standard deviation. Levels of significance are indicated as follows: *P≤0.01.
Paraquat was used to generate superoxide stress. However, the inhibition zones were too diffuse for reliable quantification. Therefore growth was followed in liquid cultures containing 250 µM paraquat. No significant difference in growth between the wild type and the 2.4.1Δirr mutant was observed (data not shown).
Higher Resistance to ROS in the 2.4.1Δirr Mutant Correlates with Elevated katE Expression Under Non-stress Conditions
Catalases make a major contribution to resistance against ROS since they rapidly detoxify hydrogen peroxide. In R. sphaeroides wild type katE expression is strongly induced within one minute after hydrogen peroxide addition, while katC does not respond to this stress [29]. The katE gene (RSP_2779) is under control of the OxyR regulator and a lack of OxyR leads to increased sensitivity to hydrogen peroxide [29]. Our microarray analyses did not reveal reliable data for katE expression (low A values). In order to see, whether the increased resistance of the 2.4.1Δirr mutant correlates with katE expression, real-time RT-PCR was applied to compare katE expression levels to those of the wild type. When cultures were grown under microaerobic conditions (approx. 30 µM oxygen) without the addition of hydrogen peroxide, katE mRNA levels were about 6 fold higher in strain 2.4.1Δirr compared to the wild type. 20 min after addition of hydrogen peroxide both strains showed almost identical katE expression levels (Fig. 3A). Fig. 3B shows the change in katE expression in both strains after addition of hydrogen peroxide. While katE mRNA levels in the wild type where about 80 fold increased 20 min after hydrogen peroxide addition compared to untreated cultures, katE levels were only slightly increased in the 2.4.1Δirr mutant. We conclude that the mutant lacking the Irr protein has already very high katE levels under non-stress conditions, which are similar to the levels the wild type reaches only after hydrogen peroxide addition. Additionally, the expression of bfd (RSP_1547), RSP_1090 and tonB (RSP_0922) with and without added hydrogen peroxide was assessed using real-time RT-PCR. The behavior of these OxyR-dependent genes was similar to the expression pattern as described for katE (data not shown).
Figure 3. Relative expression of katE (RSP_2779) in R. sphaeroides wild type and 2.4.1Δirr mutant.
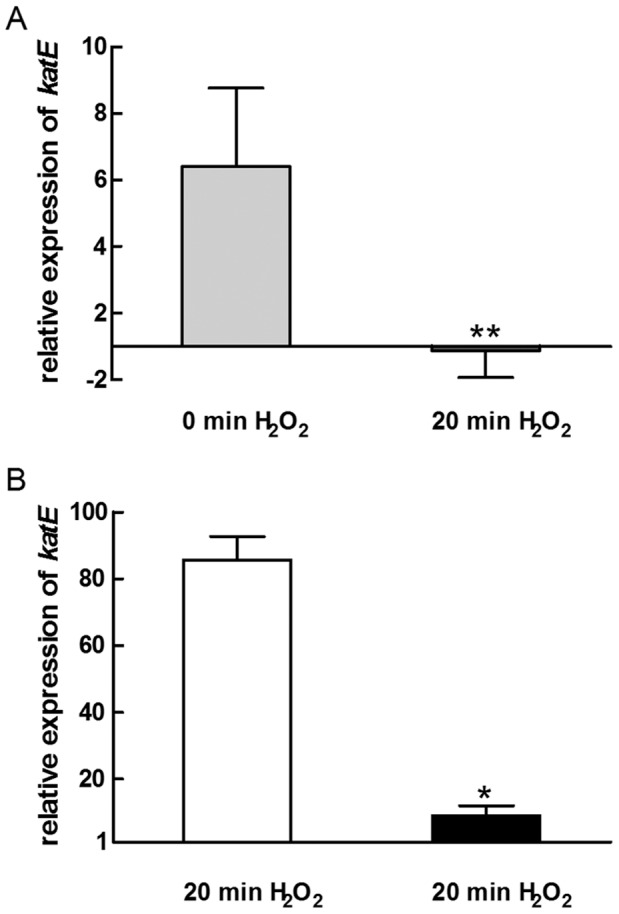
(A) Real-time RT-PCR was used to investigate the relative expression of katE in 2.4.1Δirr mutant 0 min (light gray bar) and 20 min (dark gray bar) after exposure to 1 mM H2O2 compared to the wild type. (B) Relative katE expression 20 min of 1 mM H2O2 in the R. sphaeroides wild type (white bar) and the irr deletion mutant (black bar). Values were normalized to rpoZ and to the control at time point 0. The data represent the mean of three independent experiments and error bars indicate standard deviation. Levels of significance are indicated as follows: *P≤0.01; **P≤0.05.
Response of the Protein-coding Transcriptome of the 2.4.1Δirr Mutant to Iron Limitation
We have previously characterized the response of the wild type transcriptome to iron limitation applying a high-density oligonucleotide microarray [12]. In the study presented here the effect of Irr on the transcriptome of R. sphaeroides was analyzed.
Under normal iron conditions, the abundance of 0.2% of the genes with a reliable A value (≥12) was changed by ≥1.75-fold and of 44.6% by ≤0.57-fold in the 2.4.1Δirr mutant compared to wild type cells under the same conditions (Table S1). Under iron limitation, the abundance of 2.6% of the genes with a reliable A value was changed by ≥1.75-fold and of 14.6% by ≤0.57-fold in the 2.4.1Δirr mutant compared to wild type cells under the same conditions (Table S1).
Under iron limitation, the abundance of 44.1% of those genes with a reliable A value was changed by ≥1.75-fold and of 0.6% by ≤0.57-fold in the 2.4.1Δirr mutant compared to mutant cells under normal iron conditions (Table S1). Table 1 gives an overview on those genes, grouped to functional categories, that are differently expressed (≥1.75 or ≤0.57) in the mutant strain and also lists the changes previously determined for the wild type under iron limitation [12].
Table 1. Selection of iron-responsive genes in R. sphaeroides.
| Ratio | |||||||
| Category and RSP no. | Gene | Δirr +Fe vs. wild type +Fea | Δirr –Fe vs. wild type –Fea | Δirr –Fe vs. Δirr +Fea | wild type –Fe vs. wild type +Feb | Description | |
| Iron uptake | |||||||
| RSP_0920 | exbB | (0.66) | 1.09 | 14.82 | 4.29 | Biopolymer transport protein | |
| RSP_0921 | exbD | (0.56) | 0.92 | 14.98 | 3.49 | Biopolymer transport protein | |
| RSP_0922 | 0.84 | 0.99 | 3.70 | 2.36 | Putative TonB protein | ||
| RSP_1440 | (0.75) | 0.73 | 7.19 | 3.92 | TonB dependent ferrisiderophore | ||
| RSP_1548 | irpA | (0.56) | 1.25 | 19.66 | 4.05 | Iron-regulated protein | |
| RSP_1818 | feoB | 0.46 | 1.01 | 3.65 | 1.36 | Fe2+ transport system protein | |
| RSP_1819 | feoA1 | 0.49 | 1.11 | 3.72 | 1.55 | Fe2+ transport system protein | |
| RSP_2913 | (0.46) | 0.96 | 13.21 | 3.22 | ABC Fe3+ siderophore transporter | ||
| RSP_3220 | (0.50) | 0.70 | (7.35) | 1.91 | ABC ferric siderophore transporter | ||
| RSP_6006 | hemP | (0.64) | 1.27 | 34.65 | 5.18 | Hemin uptake protein | |
| RSP_6020 | feoA2 | 0.43 | 0.86 | 4.03 | 1.28 | Fe2+ transport system protein | |
| RSP_7397 | (0.31) | 0.81 | (4.85) | 1.79 | ABC Fe3+ siderophore transporter | ||
| Iron storage | |||||||
| RSP_0352 | 0.46 | 0.67 | 2.05 | 1.15 | Probable ferredoxin | ||
| RSP_0850 | mbfA | 3.65 | 8.04 | 1.75 | (1.47) | Membrane-bound ferritin | |
| RSP_1546 | bfr | (0.55) | 1.27 | 4.52 | 1.99 | Bacterioferritin | |
| RSP_1547 | bfd | (0.49) | 1.32 | 12.31 | 2.71 | Bacterioferritin-associated ferredoxin | |
| RSP_2424 | 0.67 | 0.77 | 1.76 | 1.51 | Ferredoxin II | ||
| RSP_3342 | bfr | 0.54 | 0.70 | 0.90 | 1.01 | Bacterioferritin | |
| Iron utilization | |||||||
| RSP_0434 | sufD | 1.48 | 2.60 | 4.32 | 2.46 | Fe-S cluster assembly/repair | |
| RSP_0437 | sufC | 1.46 | 2.42 | 4.22 | 1.93 | Fe-S cluster assembly/repair | |
| RSP_0439 | 1.50 | 2.86 | 3.42 | 1.81 | Hypothetical protein | ||
| RSP_0440 | sufB | 1.72 | 2.74 | 3.69 | 1.63 | Fe-S cluster assembly/repair | |
| RSP_0442 | (0.74) | 1.39 | (3.92) | 1.56 | Putative aminotransferase | ||
| RSP_0443 | (0.62) | 1.34 | (4.67) | 1.77 | Rrf2 family transcriptional regulator | ||
| RSP_2395 | ccpA | 0.90 | 1.80 | 2.32 | 0.78 | BCCP, cytochrome c peroxidase | |
| Stress response | |||||||
| RSP_0166 | dksA | 0.51 | 0.99 | 1.63 | 1.12 | DnaK suppressor protein | |
| RSP_0697 | 0.43 | 1.17 | 1.81 | 0.75 | Universal stress protein | ||
| RSP_1172 | dnaJ | 0.50 | 0.68 | 1.40 | 1.09 | Chaperone | |
| RSP_1194 | grxC | 0.54 | 0.96 | 1.55 | 1.05 | Glutaredoxin | |
| RSP_1219 | grpE | 0.57 | 0.77 | 1.74 | 1.11 | Putative chaperone protein GrpE | |
| RSP_1529 | trxA | 0.56 | 0.82 | 1.70 | 1.07 | Thioredoxin | |
| RSP_1572 | 0.55 | 1.47 | 1.52 | 0.73 | Heat shock protein. Hsp20 family | ||
| RSP_2310 | groES | 0.72 | 0.82 | 2.31 | 1.49 | Chaperonin Cpn10 (GroES) (protein folding) | |
| RSP_2311 | groEL | 0.60 | 0.66 | 2.41 | 1.21 | Chaperonin GroEL | |
| RSP_2654 | 0.58 | 0.68 | 1.90 | 1.59 | DnaK suppressor protein | ||
| RSP_2693 | (0.42) | 0.67 | 2.45 | 1.62 | Superoxide dismutase (Fe-Mn) | ||
| RSP_2843 | hfq | 0.50 | 0.81 | 1.77 | 1.22 | RNA-binding protein Hfq | |
| RSP_4203 | 0.39 | 1.03 | 2.31 | 1.05 | putative glutaredoxin family protein/Thio-disulfide isomerase | ||
| Oxidative phosphorylation | |||||||
| RSP_0100–0104 | nuo | (0.44)–0.56 | 0.83–1.15 | 1.21–1.86 | 0.90–1.05 | Putative NADH dehydrogenase | |
| RSP_1035–39 | atp | 0.45–0.54 | 0.43–0.62 | 1.89–2.41 | 1.23–1.75 | F0F1 ATP synthase | |
| RSP_2296–2300 | atp | 0.48–0.56 | 0.56–0.66 | 1.90–2.25 | 1.35–1.65 | ATP synthase | |
| RSP_2512–30 | nuo | 0.44–0.57 | 0.58–0.76 | 1.87–2.35 | 1.30–1.60 | NADH dehydrogenase | |
| Transporter | |||||||
| RSP_0371 | 0.50 | 0.88 | 1.84 | 1.10 | ABC basic amino acid transporter | ||
| RSP_0372 | 0.55 | 0.75 | 1.55 | 1.18 | ABC basic amino acid transporter | ||
| RSP_0910–12 | dct | 0.52–0.62 | 0.41–0.51 | 1.85–2.23 | 1.71–1.90 | TRAP-T family transporter | |
| RSP_1747 | bztA | 0.47 | 0.83 | 2.02 | 1.03 | ABC glutamate/glutamine/aspartate/asparagines transporter | |
| RSP_1804 | ccmD | 0.53 | 1.00 | 1.72 | 1.08 | Heme exporter protein D | |
| RSP_2399 | 0.51 | 0.71 | 1.73 | 1.23 | ABC putrescine transporter | ||
| RSP_2400 | 0.49 | 0.64 | 1.74 | 1.36 | ABC putrescine transporter | ||
| RSP_3571 | znuA | (2.98) | 0.93 | (0.26) | (1.62) | ABC zinc transporter | |
| Photo-synthesis | |||||||
| RSP_0261–63 | bch | (1.17)–1.38 | 2.35–2.73 | 0.48–0.53 | 0.67–0.85 | Chlorophyllide reductase | |
| RSP_0277 | bchP | 0.95 | 1.81 | 0.88 | 0.95 | Geranylgeranyl hydrogenase | |
| RSP_0279 | bchG | 0.71 | 1.81 | 0.95 | 0.65 | bacteriochlorophyll a synthase | |
| RSP_0314 | pucB | 1.30 | 3.16 | 0.51 | 0.57 | LHII beta, light-harvesting B800/850 protein | |
| RSP_0315 | pucC | 0.97 | 2.57 | 0.60 | (0.89) | Light-harvesting 1 (B870) complex assembly | |
| RSP_0317 | hemN | 0.38 | 0.83 | 2.10 | 0.94 | Coproporphyrinogen III oxidase | |
| RSP_6256 | pucA | 1.14 | 3.12 | 0.64 | 0.45 | LHII alpha, light-harvesting B800/850 protein | |
| RSP_0679 | hemC | 0.57 | 1.07 | 1.37 | 0.92 | Porphobilinogen deaminase | |
| RSP_0680 | hemE | 0.57 | 0.93 | 1.48 | 1.05 | Uroporphyrinogen decarboxylase | |
| RSP_0693–96 | cco | 0.44–0.47 | 0.63–0.66 | 1.53–2.21 | 1.03–1.22 | Cbb 3-type cytochrome c oxidase | |
| RSP_0699 | hemZ | 0.56 | 0.75 | 1.84 | 1.64 | Coproporphyrinogen III oxidase | |
| RSP_1556 | puc2B | 1.23 | 2.75 | 0.60 | 0.68 | Light-harvesting complex, beta subunit | |
| RSP_6158 | puc2A | 1.08 | 2.34 | 0.70 | 0.56 | Light-harvesting complex, alpha subunit | |
Significant changes are in bold. Numbers in parentheses failed to meet the set A value criteria, while plain numbers show a lower fold change than ≥1.75 or ≤0.57. Selected genes that missed the cut-offs are included in this table to fully represent functional groups discussed.
Values are taken from Peuser and colleagues (2011).
When comparing expression in the wild type and the mutant and the response to iron limitation three types of expression patterns can be discriminated. Genes of group I show lower expression in the mutant than in the wild type under normal growth conditions. Iron limitation results in stronger induction in the mutant and consequently expression levels are similar in both strains under iron limitation. For group II genes expression in the mutant is higher than in the wild type in presence or absence of iron and group III genes behave oppositely (lower expression in mutant under both conditions).
Most genes with predicted function in iron uptake or iron storage fall into group I. Genes for systems involved in the uptake of iron include e.g. exbBD and tonB (RSP_0920–22, 3.7–15.0), hemP (RSP_6006, 34.7), a gene encoding an iron-regulated protein (RSP_1548, 19.7), and genes for Fe3+-siderophore transporters (RSP_2913, 13.2; RSP_1440, 7.2). The genes RSP_3220 (7.4) and RSP_7397 (4.9) encoding subunits of siderophore transporter were also up-regulated in response to iron limitation, but did not pass our filtering criteria. The expression of iron storage genes encoding bacterioferritin bfr (RSP_1546, 4.5), bacterioferritin-associated ferredoxin bfd (RSP_1547, 12.3) and ferredoxin (RSP_0352, 2.1; RSP_2424, 1.8) was also increased under iron limitation.
Genes involved in stress responses or proteolysis also showed the group I expression pattern. They were weakly induced in the mutant upon iron limitation, while induction in the wild type was even less or not observed. As a consequence expression levels in both strains were similar under iron limitation. Genes with function in stress responses include e.g. genes encoding the DnaK suppressor protein (RSP_0166, RSP_2654), a universal stress protein (RSP_0697), chaperones like grpE, groES, groEL and hfq (RSP_1219, RSP_2310-11, RSP_2843), the superoxide dismutase encoding gene RSP_2693, and glutaredoxin and thioredoxin encoding genes.
In contrast to other genes of iron storage, mbfA showed a different expression pattern, that it shares with the suf genes (RSP_0434–0443) which are involved in de novo assembly and/or repair of iron-sulfur clusters (group II): expression levels in the mutant are higher than in the wild type in presence or absence of iron. mbfA (RSP_0850) encodes a membrane-bound ferritin and showed about 8 fold higher expression in the mutant compared to the wild type under iron limiting conditions. This results from 3 fold higher expression levels under normal growth and stronger induction in the mutant strain in response to iron limitation (factor 1.8). Some suf genes showed slightly higher expression in the mutant compared to wild type under normal growth and more than two-fold higher expression under iron limitation due to the stronger induction.
Most of the selected iron-responsive genes follow the group III expression pattern. Genes for energy metabolism (glycolysis, citric acid cycle, oxidative phosphorylation), fatty acid metabolism, some genes for transporters, genes involved in transcription, RNA processing, amino acid metabolism and translation or chemotaxis showed lower expression levels in the mutant strain compared to the wild type in the presence and absence of iron and showed slightly increased expression in response to iron limitation in the mutant (Table 1 and Table S2).
Interestingly, the ABC zinc transporter gene znuA (RSP_3571) showed a different expression pattern. Its expression was about three times higher in the irr mutant compared to the wild type under normal iron conditions and did not change in the presence of iron in the mutant strain compared to the wild type. All three types of expression patterns (group I, II or III) can be observed among the genes with a role in photosynthesis (Table 1 and Table S2).
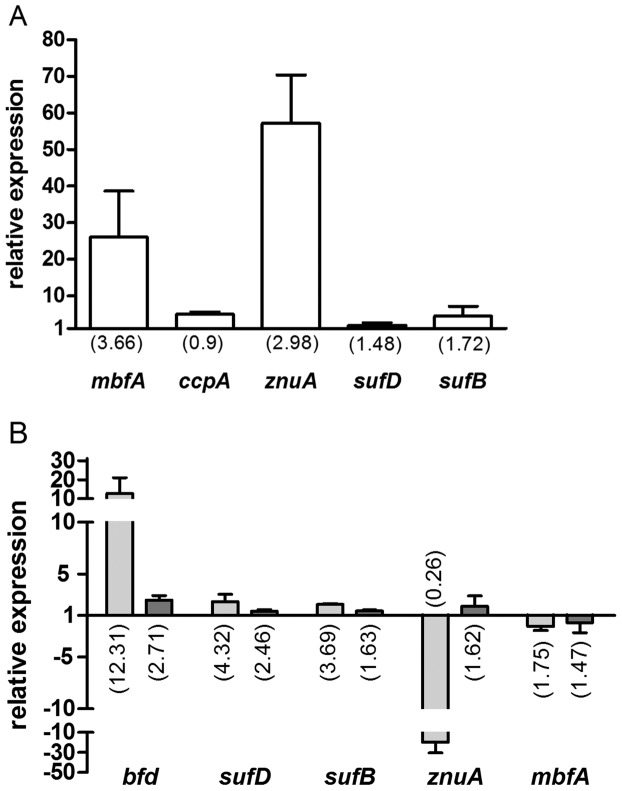
To validate the microarray data real-time RT-PCR was used for the quantification of mRNAs transcribed from some selected genes. On the one hand the ratio of the expression in the 2.4.1Δirr mutant was compared to that of the wild type strain under normal iron conditions (Fig. 4A), on the other hand the ratios of mRNA levels for the mutant strain grown in normal medium or under iron limitation were determined (Fig. 4B). Fig. 4B also includes the respective relative expression values of the wild type. Increase or decrease of expression levels as revealed by microarrays were confirmed by real-time RT-PCR, the extent of change was however different for some genes, mostly larger in the real-time RT-PCR data set.
Figure 4. Validation of microarray data by real-time RT-PCR.
Relative gene expression (A) in 2.4.1Δirr under normal iron conditions compared to the wild type under normal iron conditions and (B) in 2.4.1Δirr under iron limitation compared to normal iron conditions (light gray bars) and in wild type under iron limitation compared to normal iron conditions (dark gray bars). Values were normalized to rpoZ and to the respective control treatment. The data represent the mean of at least three independent experiments and error bars indicate standard deviation. Numbers in parentheses show the fold change of the respective genes as determined by microarray analysis.
In addition to protein-coding genes, the expression of intergenic regions (IGR) and small non-coding RNAs (sRNA) was analyzed as they can contribute to iron metabolism regulation as well. Northern Blot analysis was used to validate the expression levels as determined by the microarray approach. A selection of five sRNAs that showed altered expression in the microarray data in 2.4.1Δirr mutant under iron limitation was used for Northern hybridization (Fig. S3). The abundance of RSs0827 and RSs0680a were increased in the wild type and in the mutant under iron limitation (2.4 fold and 2.5 fold, respectively) as confirmed by Northern blot analysis (2.9 fold and 2.1 fold, respectively). Northern blots revealed that the abundance of RSs0682 and RSs2978 was not altered in the irr deletion strain under iron starvation (0.9 fold and 1.4 fold, respectively), although microarray analysis gave a ratio of about 4 fold and 2 fold, respectively. RSs2430 showed a higher expression in the mutant under iron-limiting conditions in the microarray analysis (about 3 fold), which could not be confirmed by Northern Blot analysis (0.4 fold). All five sRNAs showed a similar pattern in the wild type under iron limitation compared to the mutant (Fig. S3). Thus, it was only possible to confirm the microarray data of sRNAs in parts. It is conceivable, that the discrepancy between both techniques is due to mis-hybridization on the chip. In general, sRNAs have a size between 50 and 250 nt. Consequently, in many cases only one or two 60nt-oligonucleotides per sRNA or IGR could be designed for microarray analysis.
Irr Binds to Target Sequences in the Promoter Regions of the mbfA and ccpA Genes
Rodionov et al. (2006) predicted binding sites for the Irr protein in the upstream regions of the R. sphaeroides genes mbfA (RSP_0850) and ccpA (RSP_2395). MbfA encodes a membrane- bound ferritin, ccpA encodes a cytochrome c peroxidase. Iron limitation resulted in weak up-regulation of mbfA (about 1.5 fold) and weak down-regulation (factor 0.8) of ccpA in the wild type [12]. MbfA was strongly up-regulated (about 9 fold) by hydrogen peroxide stress, while ccpA was significantly down-regulated (about 5 fold) [30]. In the irr deletion mutant mbfA shows about 8 fold higher expression in comparison to the wild type under iron limitation and it is up-regulated by iron limitation (factor 1.75). The expression of ccpA is increased in 2.4.1Δirr under iron limitation (about twofold) and its expression level is also higher in the mutant compared to the wild type under iron limitation (factor 1.8) (Table 1).
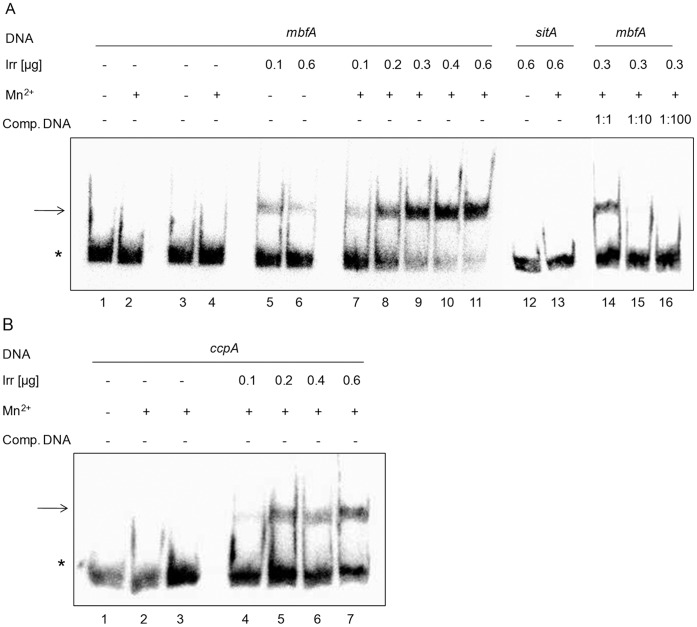
To verify the binding of Irr to the upstream regions of mbfA and ccpA Irr was purified after heterologous overexpression and suitable DNA fragments were amplified for gel retardation analyses. For both fragments the formation of a retarded DNA protein complex was observed (Fig. 5 A+B). The presence of manganese favored formation of the Irr complexes. Molar excess of unlabeled DNA fragment decreased the amount of labeled DNA in the complex (Fig. 5A lanes 14–16 and data not shown), while the presence of molar excess of unspecific DNA (salmon sperm DNA) did not compete with complex formation. From the binding curves dissociation constants of 0.8±0.15 µM for mbfA and 0.9±0.1 µM for ccpA, respectively, were determined (Fig. S4). The Irr protein did not bind to a DNA fragment from the upstream region of sitA (Fig. 5 A lanes 12 and 13), which was predicted as a target for the Mur protein in R. sphaeroides. Furthermore, Figure S5 shows that the Irr protein neither binds to the upstream region of katE nor to the upstream region of iscR (RSP_0443). Taken together, these analyses confirmed a specific interaction of Irr and the mbfA and ccpA upstream regions.
Figure 5. Binding of purified Irr to the promoter of mbfA and ccpA as determined by Electrophoretic Mobility Shift Assays.
(A) Binding of Irr to the promoter region of mbfA (180 bp). All reactions contain the same amount of 32P end-labeled DNA fragment (3.08 fmol/lane) comprising the promoter sequence. Lanes 1–4 contain no Irr; lanes 3 and 4 contain 0.6 µg BSA; lanes 5 and 7 contain 0.1 µg Irr; lane 6 and 11–13 contain 0.6 µg Irr; lane 8 contains 0.2 µg Irr; lanes 9 and 14–16 contain 0.3 µg Irr; lane 10 contains 0.4 µg. Reactions contain 1 mM MnCl2 as indicated. Lanes 14–16 contain non-labeled DNA fragment mbfA in excess amount as cold competitor. Lanes 12 and 13 contain radioactively labeled sitA DNA fragment (180 bp) as unspecific DNA. (B) Binding of Irr to the promoter region of ccpA (168 bp). All reactions contain the same amount of 32P end-labeled DNA fragment (3.68 fmol/lane) comprising the promoter sequence. Lanes 1–3 contain no Irr; lane 3 contains 0.6 µg BSA; lane 4 contains 0.1 µg Irr; lane 5 contains 0.2 µg Irr; lane 6 contains 0.4 µg Irr; lane 7 contains 0.6 µg Irr. Reactions contain 1 mM MnCl2 as indicated. All reactions contain 1 µg of salmon sperm DNA as unspecific competitor. The asterisks and arrows show the location of free and Irr-bound 32P end-labeled DNA fragments, respectively.
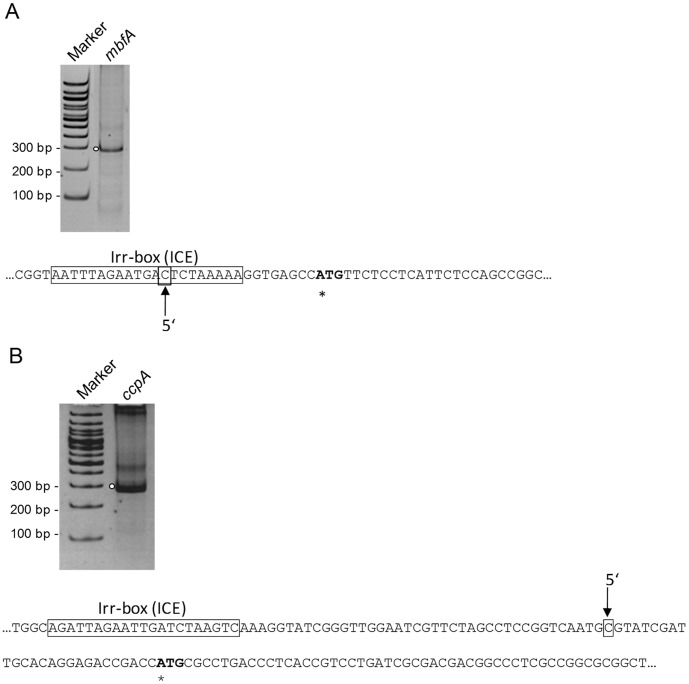
To verify that the Irr binding sites are located in an appropriate distance from the transcriptional start, we performed 5′ RACE to determine 5′ends of the mbfA and ccpA mRNAs. As indicated in Figure 6 5′ends were identified downstream of the Irr-box for ccpA (Fig. 6 B), while for mbfA the 5′end mapped within the Irr-box (Fig. 6 A).
Figure 6. Determination of 5′ ends of mbfA (RSP_0850) (A) and ccpA (RSP_2395) (B) mRNA by 5′ rapid amplification of cDNA ends (RACE).
Separation of 5′-RACE products mbfA and ccpA obtained from RNA extracts of the wild type strain under normal iron conditions. PCR products obtained after second PCR (nested) were separated on a 10% polyacrylamid gel and stained with ethidium bromide. Determined 5′ ends are indicated by an arrow. The putative translational start is indicated by an asterisk. The Irr-box (ICE, iron control element) is marked by a frame.
Discussion
The Irr protein was identified as an important regulator of iron metabolism in several alpha-proteobacteria. A mutant of R. sphaeroides lacking the predicted Irr homolog has a similar growth phenotype like the wild type under normal iron and iron limitation conditions. This suggests that Irr has no major function in the adaptation to growth under iron-limiting conditions. This was also observed for a mutant of Bru. abortus lacking Irr [32], while deletion of the irr gene in Agrobacterium tumefaciens diminished growth under iron limitation [33]. Furthermore, irr expression was not affected by the presence of iron in the R. sphaeroides wild type. This was also reported for irr expression of Bru. abortus [32]. Irr mutants of Brad. japonicum, Rhizobium leguminosarum and Bru. abortus accumulate protoporphyrin IX under iron limitation [11], [19], [32]. However, no protoporphyrin accumulation was observed in the R. sphaeroides irr deletion strain under iron-limiting conditions. Unlike the Rhizobiales species, R. sphaeroides requires protoporphyrin also as precursor for bacteriochlorophyll synthesis. The Brad. japonicum and the Bru. abortus Irr bind heme, although the latter does not contain the heme regulatory motif (HRM) and conserves only two histidines (HXH) of the second heme binding motif [34]. The HXH motif is also found in R. sphaeroides Irr and heme bound to the recombinant protein. In R. sphaeroides Irr and Irr proteins classified to the same branch [13] a proline is positioned between the two histidines, whereas in Bru. abortus Irr glutamine is the middle amino acid of the heme binding motif. No other Irr proten with a HPH motif has been analyzed in regard to heme binding up to now. Irr was identified in Brad. japonicum as a transcriptional repressor of hemB (5-aminolevulinate dehydratase) in the heme biosynthetic pathway [19]. However, hemB expression was not changed in 2.4.1Δirr under iron limitation.
The view that Irr has no major function in iron regulation in R. sphaeroides is supported by a transcriptome analysis of the mutant strain. Many genes in strain 2.4.1Δirr showed a lower expression level compared to the wild type in presence and absence of iron. The difference was mostly in the range of a factor of two and the affected genes fall into several different functional categories. It is highly unlikely that Irr would directly target all these genes, especially since no target sites are predicted [13]. Expression of katE and other OxyR-dependent genes was higher in the mutant compared to the wild type under non-stress conditions. Since activation of OxyR depends on oxidative stress this is indicative of a higher level of oxidative stress in the mutant. We hypothesize that the lower expression level of many genes in the mutant is most likely due to this increased oxidative stress in strain 2.4.1. Δirr. The expression pattern of genes in the Irr mutant under non-stress conditions is however partly different from the effect of hydrogen peroxide on gene expression in R. sphaeroides [30]. The higher resistance of the Irr mutant to oxidative stress is also in agreement with the higher expression levels of genes for the oxidative stress response. Thus, the cells are prepared to counteract oxidative stress better than cells, in which the oxidative stress response still needs to be mounted.
All genes for iron metabolism that we reported before to be induced in response to iron limitation in R. sphaeroides wild type [12] are also induced in strain 2.4.1Δirr. Indeed the induction in the Irr mutant was mostly stronger than in the wild type. Thus, Irr prevents even stronger induction of genes in response to iron starvation in the wild type.
Our study also identified two sRNAs, which are induced upon iron limitation. Irr had no influence on the expression of RSs0827, but expression levels of RSs0680a were higher in the absence of Irr. While the function of RSs0827 is unknown, RSs0680a is induced in response to singlet oxygen and even more in response to superoxide [25]. Thus, increased expression of RSs0680a in the mutant strain is another hint to increased levels of oxidative stress in the absence of Irr. Whether these sRNAs have a role in regulation of iron metabolisms as reported for sRNAs in other species [35], [36], [37] needs to be analyzed in the future.
Among the genes with reliable A value only mbfA (RSP_0850) encoding a membrane-bound ferritin showed significantly higher expression in the irr mutant compared to the wild type under normal iron and iron limitation conditions. In addition the znuA gene (RSP_3571) for an ABC zinc transporter showed higher expression in the mutant under normal iron conditions compared to the wild type. Expression levels where just below the A value but real-time RT-PCR confirmed the higher expression level in strain 2.4.1.Δirr (Fig. 4 A). However, no Irr-box (also named ICE for iron control element) was predicted in the promoter region of znuA [13]. MbfA is one of the two genes of R. sphaeroides for which an Irr-box was predicted [13]. A putative transcriptional start site for the mbfA gene was determined within the Irr-box motif. This position of the Irr binding site in relation to the transcriptional start is in agreement with a repressor function of Irr as also demonstrated for e.g. the bll6680 (bfr, bacterioferritin) and blr7895 (rubrerythrin-like protein) genes in Brad. japonicum [14]. The affinity of Irr to these ICE motifs in Brad. japonicum was very high with KDs of 7 to 19 nM [15]. The KD of the R. sphaeroides Irr protein to mbfA as determined by electrophoretic mobility shifts was about 800 nM, indicating a much lower in vitro affinity. The mbfA gene was only weakly induced by iron starvation in the Irr mutant (1.8 fold), similarly as in the wild type (about 1.5 fold). Thus, Irr has no major role in regulation of iron-dependent expression of mbfA.
The only other gene of R. sphaeroides for which an Irr-box in the promoter region was predicted [13] is ccpA (RSP_2395) encoding a cytochrome c peroxidase. This heme-iron protein reduces peroxides, which are generated by oxidative stress. Its expression level was similar in the wild type and the mutant under normal iron conditions but was slightly increased in the mutant compared to the wild type under iron limitation. Iron limitation resulted in weak induction of ccpA in the irr mutant, while it had no significant effect on ccpA expression in the wild type under the same conditions. We demonstrated in vitro binding of Irr to the ccpA promoter region with similar affinity as to the mbfA promoter region. The putative transcriptional start site mapped around 40 nt downstream of the Irr-box. In Brad. japonicum 17 iron-regulated genes with a putative Irr-box and a total of 172 ICE-like motifs were identified [14]. This search applied a more variable motif than the search by Rodionov et al. (2006), which predicted 23 Irr binding sites for Brad. japonicum and only 2 for R. sphaeroides. in vitro binding of Irr to these motifs was demonstrated for blr7895 (rubrerythrin-like protein) and for bll6680 (bfr) [14]. Only for few of the Brad. japonicum iron-regulated genes with a putative Irr binding site homologs with good similarity are found in R. sphaeroides (e.g. acnA, leuC, lguL, fumC). None of these genes showed a significant response to iron in R. sphaeroides. Our data imply different roles of Irr in gene regulation in individual alpha-proteobacteria. Table S3 summarizes the different features of Irr and Irr mutants in R. sphaeroides and the Rhizobiales species, which have been investigated in this regard.
Like the Irr mutants of Bru. abortus [38] and A. tumefaciens [33] the R. sphaeroides mutant showed higher resistance to oxidative stress, which is in agreement with increased katE expression under normal growth conditions. Interestingly both genes with Irr-boxes in R. sphaeroides, mbfA and ccpA have a function in oxidative stress defense. MbfA (RSP_0850) shares good homology with blr7895 (rubrerythrin-like protein) and showed an increased expression in the irr deletion strain compared to the wild type under non-stress conditions. Rubrerythrin is a structurally and biophysically well-characterized non-heme iron protein [39], [40]. It is hypothesized that rubrerythrin provides oxidative stress protection via catalytic reduction of intracellular hydrogen peroxide [41], [42], [43], although this function was disputed [44]. Since iron limitation causes oxidative stress due to decreased levels of iron-sulfur proteins with important roles in oxidative stress defense increased katE and maybe also mbfA and ccpA expression levels help to counteract this oxidative stress. A direct effect of Irr on katE expression is unlikely since no Irr-box is present in the katE promoter region and no in vitro binding was observed by gel shift experiments. Alternatively, the lack of Irr, which causes an enhanced induction of genes for iron uptake in response to iron starvation may cause the generation of ROS, which consequently activate katE expression and cause lower expression of many other genes. It is conceivable that under microaerobic growth a limitation of the up-regulation of genes in response to iron starvation is the biological function of Irr in R. sphaeroides. It would thus contribute to a balance between increase of iron-uptake systems, which counteracts iron limitation and the formation of ROS by too much iron import.
Materials and Methods
Bacterial Strains, Growth Conditions and Iron Limitation
Strains and plasmids used in this study are listed in Table S4.
R. sphaeroides strains were cultivated at 32°C in 50 ml Erlenmeyer flasks containing 40 ml malate minimal medium [45] with continuous shaking at 140 rpm. This growth is designated as microaerobic growth. At the chosen growth conditions the cultures contain about 2% oxygen (approx. 30 µM oxygen).
Conditions of iron limitation were achieved by cultivating R. sphaeroides without adding external Fe(III)citrate to the growth medium but with the iron chelator 2,2′-dipyridyl (30 µM; Merck). A small aliquot of cells were transferred from normal cultivation medium to iron-limited medium. The cells were grown overnight and then transferred two times more into iron-limited medium. Inductively coupled plasma mass spectrometry (ICP-MS) using Agilent 7500ce confirmed that the iron content was drastically reduced in iron-limited medium (from 140 µg/l to 16 µg/l) [46]. For gene expression studies cells were harvested at an OD660 of 0.4.
When required antibiotics were added to the liquid or solid growth medium at the following concentrations: kanamycin, 25 µg ml−1; tetracycline 2 µg ml−1 (for R. sphaeroides); and ampicillin, 200 µg ml−1; kanamycin, 25 µg ml−1; tetracycline, 20 µg ml−1 (for E. coli).
Construction of an R. sphaeroides irr Deletion Mutant
R. sphaeroides strain 2.4.1Δirr was generated by transferring the suicide plasmid pPHU2.4.1Δirr::Km into R. sphaeroides 2.4.1, and screening for insertion of the kanamycin resistance cassette into the chromosome by homologous recombination. Briefly, parts of the irr gene (RSP_3179) of R. sphaeroides 2.4.1, together with upstream and downstream sequences, were amplified by PCR using oligonucleotides KO3179-Eco_A1 (5′-CGA AGC GAA TTC CCT GCC AGC C-3′), KO3179-Pst_A2 (5′-GAT TGC CGA TCG CTG CAG CAT TCC-3′) and KO3179-Pst_B1.3 (5′- CGA CAA CCA TCT GCA GTT CTA CTG GG -3′), KO3179-Pae_B2.3 (5′- GGC AGT TCC GCA TGC GGG ATC TCG -3′).
The amplified PCR fragments were cloned into the EcoRI-PstI and PstI-PaeI sites of the suicide plasmid pPHU281, generating the plasmid pPHU2.4.1Δirr. A 1.3 kb PstI fragment containing the kanamycin cassette from pUC4K [47] was inserted into the PstI site of pPHU2.4.1Δirr to generate pPHU2.4.1Δirr::Km. This plasmid was transferred into E. coli strain S17-1 and diparentally conjugated into R. sphaeroides 2.4.1 wild type strain. Conjugants were selected on malate minimal salt agar plates containing 25 µg kanamycin ml–1. By insertion of the kanamycin cassette, 285 bp of the 441 bp R. sphaeroides irr gene (RSP_3179) was deleted. PCR analysis of chromosomal DNA was carried out to confirm the double crossover event of the kanamycin cassette into the R. sphaeroides chromosome (Fig. S6).
Complementation of the R. sphaeroides Deletion Mutant 2.4.1Δirr
For complementation of the irr deletion mutant of R. sphaeroides a 539 bp PCR fragment containing the entire irr gene (RSP_3179) along with 57 bp of the upstream and 49 bp of the downstream sequence of the irr gene was amplified by using the oligonucleotides 3179compl_fwd (5′-GCC GTC TAG AAA ACA TGG GTC TTT C-3′) and 3179compl_rev (5′-CTG CCC GCA GAA TTC GCA GAC G- 3′). Following digestion with XbaI and EcoRI, the fragment was cloned into the corresponding sites of pRK415, resulting in plasmid pRK2.4.1irr. To complement the irr deletion in the wild type strain 2.4.1, the plasmid pRK2.4.1irr was transferred into E. coli S17-1 and conjugated into the 2.4.1Δirr strain by diparental conjugation.
Inhibition Zone Assays
For inhibition zone assays cultures were grown microaerobically overnight at 32°C and then diluted to an OD660 of 0.2. Cultures were grown to an OD660 of 0.4 and 200 µl of the culture were mixed with 5 ml prewarmed top agar (0.8% (w/v) agar) and layered onto malate minimal salt medium plates. A 0.55 cm filter disk, containing 5 µl of hydrogen peroxide (100, 200 and 500 mM), was placed on the hardened top agar. Zones of inhibition were measured after incubation for 72 h at 32°C in the dark. Inhibition zone assays were also performed under a fluorescent tube (model NL 36 W/860 daylight) with filter disks containing 5 µl of 10 and 50 mM methylene blue to generate singlet oxygen. The assays were performed at least three times.
Extraction of RNA and Quantitative Real-time RT-PCR
Cell samples from growth experiments (OD660 0.4) were rapidly cooled on ice and harvested by centrifugation at 10 000 g in a cooled centrifuge. Total RNA was isolated by the peqGOLD TriFast ™ Kit (Peqlab) as described by the manufactures protocol. Samples were treated with 1 unit of RNase-free DNase I (Invitrogen) per 1 µg RNA to remove contaminating DNA. After DNase I treatment, the RNA was purified by standard procedures using a mixture of phenol/chloroform/isoamyl alcohol and chloroform/isoamyl alcohol before precipitating with sodium acetate and isopropanol. Contamination with remaining DNA was checked by PCR amplification of RNA samples using primers targeting gloB (RSP_0799-A: 5′-GAA CAA TTA CGC CTT CTC-3′, RSP_0799-B: 5′-CAT CAG CTG GTA GCT CTC-3′) as described previously [26].
Oligonucleotides used for gene amplification are listed in Table S5. Conditions for real-time RT-PCR were described earlier in detail [26]. A final concentration of 4 ng µl−1 of total RNA was used in an one-step RT-PCR kit (Qiagen). For detection of double stranded DNA SYBR Green I (Invitrogen) was added in a final dilution of 1∶50 000 to the master mix. For normalization of mRNA levels the rpoZ gene was used, which encodes the ω-subunit of RNA-polymerase of R. sphaeroides [48]. Relative expression of target genes was calculated relative to the expression of untreated samples and relative to rpoZ [49]. PCR efficiencies were determined experimentally using serial dilutions of RNA between a final concentration of 8 and 0.5 ng µl−1 (Table S6).
Microarray Analysis
Microarray experiments were performed as described previously [12]. In brief, total RNA from iron-limited and control cultures grown under microaerobic conditions (OD660 0.4) was extracted by the hot phenol method as described earlier [50], [51]. Genomic DNA contamination from RNA samples was removed by DNase treatment (Invitrogen). After DNA digestion, RNA was purified on RNeasy® MinElute™ spin columns (Qiagen). All RNA preparations were tested for the lack of genomic DNA contamination by PCR amplification using primers targeting gloB (RSP_0799) as described previously [26].
High-density oligonucleotide R. sphaeroides microarrays (Agilent gene chips corresponding to the whole 4.6-Mb genome) were used for transcriptome profiling. The microarray contains probes against 4.304 protein coding genes, 79 rRNA and tRNA genes, and 144 intergenic regions; its construction and performance analysis was performed according to the instructions of Agilent (www.chem.agilent.com). Three antisense probes with a length of 60 nt were designed for hybridization to each gene. The ULS™ Fluorescent Labeling Kit for Agilent arrays (Kreatech) was used for RNA labeling and fragmentation. The RNA of three independent experiments of R. sphaeroides wild type under normal and iron limitation conditions and the irr deletion mutant under normal and iron limitation conditions was pooled and hybridized to one array. Transcriptome profiles were analyzed on two arrays (Δirr normal iron vs. wild type normal iron; Δirr iron limitation vs. wild type iron limitation; Δirr iron limitation vs. Δirr normal iron) including six biological replicates. Genechip hybridizations and scanning were performed according to the specifications from Agilent. Multiarray analysis was performed with the Bioconductor package Limma for R [52], [53]. Background correction and normalization (Lowess, locally weighted scatterplot smoothing) were performed as described previously [54], [55]. To filter out unreliably measured and unchanged genes, two criteria were used as described previously [12]. (i) Genes were considered reliable if the mean intensity (A value) was ≥12. (ii) A cut-off value was applied, i. e., those genes were retained whose average expression value (ratio) was either ≥1.75 or ≤0.57. The fold changes are shown in the text in parentheses preceded by the RSP numbers of their corresponding genes. When expression of several genes is discussed, the lower and upper fold changes are shown, e. g., a range of two- to fivefold increase is shown as “2.0–5.0”. The expression data obtained here were deposited in the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (www.ncbi.nih.gov/geo) under superseries GSE33535.
Expression and Isolation of the R. Sphaeroides Irr Protein
Oligonucleotides 3179-His_fwd (5′-GCGCCCGCAATGGGATGGATCCCATTTC-3′) and 3179-His_rev (5′-GCGGGAATAAGCTTTCAGGTACGCTT-3′) were used for amplifying the coding region of irr. The 474-bp PCR product was ligated into the pJET1.2/blunt cloning vector (Qiagen) which was transformed into E. coli JM109. Afterwards the plasmid containing irr was digested with BamHI and HindIII and the purified irr fragment was ligated into the overexpression vector pQE30 (Qiagen) to generate pQE2.4.1irr, which was transformed into E. coli JM109. The correct construct was transformed into E. coli M15 (pREP-4) for overexpression of His-tagged Irr. For this purpose M15 (pREP-4/pQE2.4.1irr) was grown in 50 ml of Luria-Bertani medium to an OD600 of 0.5 to 0.6 at 37°C. The cells were induced with 1 mM IPTG for 3 h at 37°C. Following harvest, cells were resuspended in ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and disrupted by brief sonication. The lysate was centrifuged at 13 000 rpm and 4°C for 15 min. The clear supernatant was loaded onto Ni-NTA agarose (Qiagen) and incubated at 4°C for 3 h. Proteins were washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Aliquots of the fractions were analyzed on 15% sodium dodecyl sulfate-polyacrylamide gels, and fractions containing Irr protein were used for the experiments described below.
Electrophoretic Mobility Shift Assays (EMSAs)
Binding of the recombinant Irr protein to the upstream regions of RSP_0850 (mbfA) and RSP_2395 (ccpA) was determined by an EMSA. As controls served DNA fragments containing the sitA (RSP_0904), the katE (RSP_2779) and the iscR (RSP_0443) promoter region, respectively. The following oligonucleotides were used to generate DNA fragments containing the respective promoter region by PCR. RSP_0850∶0850up_fwd (5′-GTC AAC TTG CCG CAG GCG CTC C-3′) and 0850up_rev (5′-GCC GGT TGA CAT AGG AGC GGT AG-3′); RSP_2395∶2395up_fwd (5′-CGG TCA ACC CTG GTC GCC GCC GAA-3′) and 2395up_rev (5′-GCC GCG TCG ACG AGG GCC GTC-3′); RSP_0904∶0904up_fwd (5′-CAG TTA ACT GCG AAC GGC TCG CAG A-3′) and 0904up_rev (5′-GAC CGT TAA CGT CGT GGC GAC CT-3′); RSP_0443∶0443up_fwd (5′-CGC GGC GTA ATG TTG ACA AAA ACG-3′) and 0443up_rev (5′-CGA CAC GTC GAC AAG CGA GAC AAG-3′). The PCR fragments with a length of 180, 168, 180 and 246 bp for mbfA, ccpA, sitA and iscR, respectively, were cloned into pDrive cloning vector (Qiagen), and isolated from the vector by using the restriction enzyme HincII. In the case of katE the plasmid pkatEup was used that contains a 352 bp fragment of the upstream region of katE [29]. The fragment was isolated from the pDrive cloning vector by using the restriction enzymes BamHI and PstI. The restricted DNA fragments were then radioactively end-labeled with γ32P ATP using the T4 polynucleotide kinase (Fermentas).
An appropriate amount of the purified Irr protein, ranging from 0.1 to 0.6 µg, was mixed with approx. 3 fmol γ32P ATP-labeled DNA probe (5000 c.p.m.) in a 15 µl reaction volume containing 20 mM TB (pH 7.8), 5% v/v glycerol, 1 mM DTT, salmon sperm DNA (1 µg), and 0.1 mg/ml BSA. Binding incubations were carried out for 30 min at 32°C before the samples were loaded onto a 6% polyacrylamide gel in 0.5x TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1.25 mM EDTA, pH 8.3) and run at 180 V for 3 h at 4°C.
Competitive assays were performed to determine the specificity of the protein for the putative target site (Irr-box). In this case, the γ32P ATP-labeled DNA probes were mixed with a 1 to 100 fold molar excess of the respective unlabeled DNA fragment before adding to the binding reaction.
5′ RACE
For the determination of 5′ mRNA ends using 5′ rapid amplification of cDNA ends (RACE), 3 µg of total RNA isolated from wild type cells cultivated under normal iron conditions were reverse transcribed into cDNA by using avian myeloblastosis virus reverse transcriptase (Promega) and gene-specific primers (0850_RACE1 and 2395_RACE1; see Table S5). The 5′RACE protocol was performed as described previously [56].
Heme-binding Experiments
The interaction of Irr with heme was studied through the spectral properties of heme. Hemin (C34H32ClFeN4O4; Sigma) was dissolved in 0.1 M NaOH and binding studies were carried out using an appropriate dilution in buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0). The absorption spectrum of 8 µM heme was recorded in the presence or absence of 8 µM Irr. As positive control the absorption spectrum of 5 µM heme was recorded in the presence and absence of 5 µM BSA. As negative control the absorption spectrum of 8 µM heme was recorded in presence and absence of 8 µM IscR.
Detection of Protoporphyrin IX in Supernatants of R. sphaeroides Cultures
40 ml R. sphaeroides cells grown to saturation under iron limiting conditions were harvested by centrifugation (4 600 g, 15 min, 4°C) and the porphyrins from the supernatants were extracted with 10 ml ethyl/glacial acetic acid (3∶1, v/v) at 18°C overnight with shaking. Then, the ethyl acetate layer was washed with pure water and concentrated at low temperature in a vacuum system. The absorption spectrum of this extract was recorded between 350 and 650 nm with a Specord 50 spectrophotometer (Analytik Jena).
Supporting Information
Relative gene expression under iron limitation comparing aerobic and microaerobic conditions. Real-time RT-PCR was used to investigate the relative expression of hemB (RSP_2848), hemH (RSP_1197), mbfA (RSP_0850) and sufD (RSP_0434) under iron limitation in R. sphaeroides 2.4.1Δirr (light gray bars) and wild type (dark gray bars) under microaerobic conditions (non-striped bars) and aerobic conditions (striped bars). Values were normalized to rpoZ and to the respective control treatment under normal iron conditions. The data represent the mean of three independent experiments and error bars indicate standard deviation.
(TIF)
Effect of Irr on the absorption spetrum of heme. (A) Absorption spectrum of 8 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 8 µM recombinant Irr. A scan of 8 µM Irr alone (dotted line) is also shown. (B) Absorption spectrum of 5 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 5 µM BSA as positive control. A scan of 5 µM BSA alone (dotted line) is also shown. (C) Absorption spectrum of 8 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 8 µM recombinant IscR. A scan of 8 µM IscR alone (dotted line) is also shown. Absorption peak wavelengths are indicated.
(TIF)
The abundance of small RNAs under iron limitation in the wild type and the 2.4.1Δirr mutant as determined by Northern Blot analysis. After hot phenol extraction RNA was separated on 10% polyacrylamide gels containing 7 M urea and then transferred onto nylon membranes by semidry electroblotting. 10 µg total RNA was loaded per sample. For detection of sRNAs radioactively-labeled oligodeoxynucleotides were used. Membranes were exposed on phosphoimaging screens and analyzed with the 1D-Quantity One software (Bio-Rad). 5 S rRNA served as loading control.
(TIF)
Determination of Irr affinity for Irr-box motif containing DNA. (A) Binding of Irr to the promoter region of mbfA. (B) Binding of Irr to the promoter region of ccpA. To determine the dissociation constant (KD) of Irr-DNA binding, the percentage of DNA bound to total labeled DNA was plotted against increasing Irr concentrations. The KD was defined as the protein concentration required to shift 50% of the probe.
(TIF)
Binding of purified Irr to the promoter region of katE and iscR as determined by Electrophoretic Mobility Shift Assays. All reactions contain the same amount of 32P end-labeled DNA fragment (∼ 3 fmol/lane) comprising the respective promoter sequence. (A) Binding of Irr to the promoter region of katE (352 bp). Lanes 1 and 4–6 contain no Irr; lane 6 contains 0.6 µg BSA; lanes 2 and 7 contain 0.1 µg Irr; lane 8 contains 0.2 µg Irr; lane 9 contains 0.3 µg Irr; lane 10 contains 0.4 µg Irr; lanes 11 and 3 contain 0.6 µg Irr. Reactions contain 1 mM MnCl2 as indicated. Lanes 1–3 contain radioactively labeled mbfA DNA fragment (180 bp) as positive control. (B) Binding of Irr to the promoter region of iscR (246 bp). Lanes 1 and 5 contain no Irr; lanes 2 and 6 contain 0.1 µg Irr; lanes 3 and 7 contain 0.3 µg Irr; lanes 4 and 8 contain 0.6 µg Irr. All reactions contain 1 mM MnCl2. Lanes 1–4 contain radioactively labeled mbfA DNA fragment as positive control. The asterisks and arrows show the location of free and Irr-bound 32P end-labeled DNA fragments, respectively.
(TIF)
Confirmation of the irr knock-out by PCR (A) using oligodeoxynucleotides KO3179_Test-A (5′-CCA CGC CGA GCG CGA AGC CC-3′) and KO3179_Test-B (5′-GCA CCT CGT CGG GCA GTT CCG-3′) to amplify the irr locus with its upstream and downstream regions (estimated product length: WT (− Kmr cassette): 1352 bp; Δirr (+ Kmr cassette): 2435 bp), (B) using oligodeoxynucleotides KanR2_fwd (5′-CAT GAA CAA TAA AAC TGT CTG C-3′) and KanR2_rev (5′-GAA GAT GCG TGA TCT GAT CC-3′) to amplify the kanamycin resistance cassette (estimated product length: 983 bp) and (C) using oligodeoxynucleotides KanR2_fwd and KO3179_Test-B (estimated product length: Δirr (+ Kmr cassette): ∼1800 bp). Used template for PCR: chromosomal DNA (wild type, WT; irr deletion mutant, Δirr) and H2O as negative control. PCR products were separated on an 1% agarose gel (1x TAE) and stained with ethidium bromide. (D) Construction of R. sphaeroides 2.4.1Δirr. Oligodeoxynucleotides used for cloning are indicated as black arrows (A1, A2, B1, B2), oligodeoxynucleotides used for testing knock-out candidates are indicated as red arrows (KO3179_Test-A, KO3179_Test-B) and oligodeoxynucleotides for amplifying the kanamycin resistance cassette are indicated as blue arrows (KanR2_fwd, KanR2_rev).
(TIF)
Gene expression changes in 2.4.1Δirr.
(XLS)
Selection of iron-responsive genes in R. sphaeroides grouped to functional categories.
(DOC)
Summary of Irr features in R. sphaeroides and Rhizobiales species.
(DOCX)
Bacterial strains and plasmids.
(DOC)
Oligodeoxynucleotides used for real-time RT-PCR and 5′RACE.
(DOCX)
Primer efficiencies for real-time RT-PCR.
(DOCX)
Acknowledgments
We thank Sebastian Metz for instructions on microarray analysis and Carmen Haas for construction of the irr deletion strain.
Funding Statement
This work was supported by Deutsche Forschungsgemeinschaft (Kl563/25) (http://www.dfg.de). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hantke K (2001) Iron and metal regulation in bacteria. Curr Opin Microbiol 4: 172–177. [DOI] [PubMed] [Google Scholar]
- 2. Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181: 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, et al. (2007) Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 20: 501–511. [DOI] [PubMed] [Google Scholar]
- 4. Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ (2006) IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli . Mol Microbiol 60: 1058–1075. [DOI] [PubMed] [Google Scholar]
- 5. Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, et al. (2001) IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A 98: 14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, et al. (2002) RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum . Microbiology 148: 4059–4071. [DOI] [PubMed] [Google Scholar]
- 7. Viguier C, P OC, Clarke P, O’Connell M (2005) RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol Lett 246: 235–242. [DOI] [PubMed] [Google Scholar]
- 8. Chao TC, Becker A, Buhrmester J, Puhler A, Weidner S (2004) The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol 186: 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, et al. (2004) The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150: 1447–1456. [DOI] [PubMed] [Google Scholar]
- 10. Platero R, Peixoto L, O’Brian MR, Fabiano E (2004) Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti . Appl Environ Microbiol 70: 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wexler M, Todd JD, Kolade O, Bellini D, Hemmings AM, et al. (2003) Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum . Microbiology 149: 1357–1365. [DOI] [PubMed] [Google Scholar]
- 12. Peuser V, Metz S, Klug G (2011) Response of the photosynthetic bacterium Rhodobacter sphaeroides to iron limitation and the role of a Fur orthologue in this response. Environ Microbiol Rep 3: 397–404. [DOI] [PubMed] [Google Scholar]
- 13. Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW (2006) Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput Biol 2: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, et al. (2006) The Iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol 188: 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sangwan I, Small SK, O’Brian MR (2008) The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J Bacteriol 190: 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Small SK, Puri S, Sangwan I, O’Brian MR (2009) Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum . J Bacteriol 191: 1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, et al. (2006) Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol 60: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, et al. (2010) Heme-responsive DNA Binding by the Global Iron Regulator Irr from Rhizobium leguminosarum . Journal of Biological Chemistry 285: 16023–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamza I, Chauhan S, Hassett R, O’Brian MR (1998) The bacterial irr protein is required for coordination of heme biosynthesis with iron availability. J Biol Chem 273: 21669–21674. [DOI] [PubMed] [Google Scholar]
- 20. Qi Z, O’Brian MR (2002) Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell 9: 155–162. [DOI] [PubMed] [Google Scholar]
- 21. Yang J, Panek HR, O’Brian MR (2006) Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum . Mol Microbiol 60: 209–218. [DOI] [PubMed] [Google Scholar]
- 22. Elsen S, Swem LR, Swem DL, Bauer CE (2004) RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev 68: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeilstra-Ryalls JH, Kaplan S (2004) Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol Life Sci 61: 417–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anthony JR, Warczak KL, Donohue TJ (2005) A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A 102: 6502–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G (2009) Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides . Mol Microbiol 74: 1497–1512. [DOI] [PubMed] [Google Scholar]
- 26. Glaeser J, Klug G (2005) Photo-oxidative stress in Rhodobacter sphaeroides: Protective role of carotenoids and expression of selected genes. Microbiology 151: 1927–1938. [DOI] [PubMed] [Google Scholar]
- 27. Glaeser J, Zobawa M, Lottspeich F, Klug G (2007) Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter . J Proteome Res 6: 2460–2471. [DOI] [PubMed] [Google Scholar]
- 28. Li K, Pasternak C, Klug G (2003) Expression of the trxA gene for thioredoxin 1 in Rhodobacter sphaeroides during oxidative stress. Arch Microbiol 180: 484–489. [DOI] [PubMed] [Google Scholar]
- 29. Zeller T, Klug G (2004) Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter . Microbiology 150: 3451–3462. [DOI] [PubMed] [Google Scholar]
- 30. Zeller T, Moskvin OV, Li K, Klug G, Gomelsky M (2005) Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides . J Bacteriol 187: 7232–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeller T, Mraheil MA, Moskvin OV, Li K, Gomelsky M, et al. (2007) Regulation of hydrogen peroxide-dependent gene expression in Rhodobacter sphaeroides: regulatory functions of OxyR. J Bacteriol 189: 3784–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martinez M, Ugalde RA, Almiron M (2005) Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 151: 3427–3433. [DOI] [PubMed] [Google Scholar]
- 33. Hibbing ME, Fuqua C (2011) Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens . J Bacteriol 193: 3461–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang J, Ishimori K, O’Brian MR (2005) Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J Biol Chem 280: 7671–7676. [DOI] [PubMed] [Google Scholar]
- 35. Dühring U, Axmann IM, Hess WR, Wilde A (2006) An internal antisense RNA regulates expression of the photosynthesis gene isiA . Proc Natl Acad Sci U S A 103: 7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Massé E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli . Proc Natl Acad Sci U S A 99: 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, et al. (2004) Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 101: 9792–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez M, Ugalde RA, Almiron M (2006) Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus . Microbiology 152: 2591–2598. [DOI] [PubMed] [Google Scholar]
- 39. LeGall J, Prickril BC, Moura I, Xavier AV, Moura JJ, et al. (1988) Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry 27: 1636–1642. [DOI] [PubMed] [Google Scholar]
- 40. Li M, Liu MY, LeGall J, Gui LL, Liao J, et al. (2003) Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement. J Biol Inorg Chem 8: 149–155. [DOI] [PubMed] [Google Scholar]
- 41. Lumppio HL, Shenvi NV, Summers AO, Voordouw G, Kurtz DM Jr (2001) Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol 183: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM Jr (2002) Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis . Mol Microbiol 44: 479–488. [DOI] [PubMed] [Google Scholar]
- 43. Weinberg MV, Jenney FE Jr, Cui X, Adams MW (2004) Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J Bacteriol 186: 7888–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jean D, Briolat V, Reysset G (2004) Oxidative stress response in Clostridium perfringens . Microbiology 150: 1649–1659. [DOI] [PubMed] [Google Scholar]
- 45. Drews G (1983) Mikrobiologisches Praktikum. Heidelberg: Springer Verlag. [Google Scholar]
- 46. Peuser V, Metz S, Klug G (2011) Response of the photosynthetic bacterium Rhodobacter sphaeroides to iron limitation and the role of a Fur orthologue in this response. Environmental Microbiology Reports 3: 397–404. [DOI] [PubMed] [Google Scholar]
- 47. Vieira J, Messing J (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19: 259–268. [DOI] [PubMed] [Google Scholar]
- 48. Gomelsky L, Sram J, Moskvin OV, Horne IM, Dodd HN, et al. (2003) Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides . Microbiology 149: 377–388. [DOI] [PubMed] [Google Scholar]
- 49. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Janzon L, Lofdahl S, Arvidson S (1986) Evidence for a coordinate transcriptional control of apha-toxin and protein-a synthesis in Staphylococcus aureus . FEMS Microbiol Lett 33: 193–198. [Google Scholar]
- 51. von Gabain A, Belasco JG, Schottel JL, Chang AC, Cohen SN (1983) Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A 80: 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 53. Smyth GK (2005) Limma: Linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. [Google Scholar]
- 54. Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, et al. (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707. [DOI] [PubMed] [Google Scholar]
- 55. Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- 56. Nuss AM, Glaeser J, Klug G (2009) RpoHII activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides . J Bacteriol 191: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative gene expression under iron limitation comparing aerobic and microaerobic conditions. Real-time RT-PCR was used to investigate the relative expression of hemB (RSP_2848), hemH (RSP_1197), mbfA (RSP_0850) and sufD (RSP_0434) under iron limitation in R. sphaeroides 2.4.1Δirr (light gray bars) and wild type (dark gray bars) under microaerobic conditions (non-striped bars) and aerobic conditions (striped bars). Values were normalized to rpoZ and to the respective control treatment under normal iron conditions. The data represent the mean of three independent experiments and error bars indicate standard deviation.
(TIF)
Effect of Irr on the absorption spetrum of heme. (A) Absorption spectrum of 8 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 8 µM recombinant Irr. A scan of 8 µM Irr alone (dotted line) is also shown. (B) Absorption spectrum of 5 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 5 µM BSA as positive control. A scan of 5 µM BSA alone (dotted line) is also shown. (C) Absorption spectrum of 8 µM heme was recorded in the absence (dashed line) and in the presence (continuous line) of 8 µM recombinant IscR. A scan of 8 µM IscR alone (dotted line) is also shown. Absorption peak wavelengths are indicated.
(TIF)
The abundance of small RNAs under iron limitation in the wild type and the 2.4.1Δirr mutant as determined by Northern Blot analysis. After hot phenol extraction RNA was separated on 10% polyacrylamide gels containing 7 M urea and then transferred onto nylon membranes by semidry electroblotting. 10 µg total RNA was loaded per sample. For detection of sRNAs radioactively-labeled oligodeoxynucleotides were used. Membranes were exposed on phosphoimaging screens and analyzed with the 1D-Quantity One software (Bio-Rad). 5 S rRNA served as loading control.
(TIF)
Determination of Irr affinity for Irr-box motif containing DNA. (A) Binding of Irr to the promoter region of mbfA. (B) Binding of Irr to the promoter region of ccpA. To determine the dissociation constant (KD) of Irr-DNA binding, the percentage of DNA bound to total labeled DNA was plotted against increasing Irr concentrations. The KD was defined as the protein concentration required to shift 50% of the probe.
(TIF)
Binding of purified Irr to the promoter region of katE and iscR as determined by Electrophoretic Mobility Shift Assays. All reactions contain the same amount of 32P end-labeled DNA fragment (∼ 3 fmol/lane) comprising the respective promoter sequence. (A) Binding of Irr to the promoter region of katE (352 bp). Lanes 1 and 4–6 contain no Irr; lane 6 contains 0.6 µg BSA; lanes 2 and 7 contain 0.1 µg Irr; lane 8 contains 0.2 µg Irr; lane 9 contains 0.3 µg Irr; lane 10 contains 0.4 µg Irr; lanes 11 and 3 contain 0.6 µg Irr. Reactions contain 1 mM MnCl2 as indicated. Lanes 1–3 contain radioactively labeled mbfA DNA fragment (180 bp) as positive control. (B) Binding of Irr to the promoter region of iscR (246 bp). Lanes 1 and 5 contain no Irr; lanes 2 and 6 contain 0.1 µg Irr; lanes 3 and 7 contain 0.3 µg Irr; lanes 4 and 8 contain 0.6 µg Irr. All reactions contain 1 mM MnCl2. Lanes 1–4 contain radioactively labeled mbfA DNA fragment as positive control. The asterisks and arrows show the location of free and Irr-bound 32P end-labeled DNA fragments, respectively.
(TIF)
Confirmation of the irr knock-out by PCR (A) using oligodeoxynucleotides KO3179_Test-A (5′-CCA CGC CGA GCG CGA AGC CC-3′) and KO3179_Test-B (5′-GCA CCT CGT CGG GCA GTT CCG-3′) to amplify the irr locus with its upstream and downstream regions (estimated product length: WT (− Kmr cassette): 1352 bp; Δirr (+ Kmr cassette): 2435 bp), (B) using oligodeoxynucleotides KanR2_fwd (5′-CAT GAA CAA TAA AAC TGT CTG C-3′) and KanR2_rev (5′-GAA GAT GCG TGA TCT GAT CC-3′) to amplify the kanamycin resistance cassette (estimated product length: 983 bp) and (C) using oligodeoxynucleotides KanR2_fwd and KO3179_Test-B (estimated product length: Δirr (+ Kmr cassette): ∼1800 bp). Used template for PCR: chromosomal DNA (wild type, WT; irr deletion mutant, Δirr) and H2O as negative control. PCR products were separated on an 1% agarose gel (1x TAE) and stained with ethidium bromide. (D) Construction of R. sphaeroides 2.4.1Δirr. Oligodeoxynucleotides used for cloning are indicated as black arrows (A1, A2, B1, B2), oligodeoxynucleotides used for testing knock-out candidates are indicated as red arrows (KO3179_Test-A, KO3179_Test-B) and oligodeoxynucleotides for amplifying the kanamycin resistance cassette are indicated as blue arrows (KanR2_fwd, KanR2_rev).
(TIF)
Gene expression changes in 2.4.1Δirr.
(XLS)
Selection of iron-responsive genes in R. sphaeroides grouped to functional categories.
(DOC)
Summary of Irr features in R. sphaeroides and Rhizobiales species.
(DOCX)
Bacterial strains and plasmids.
(DOC)
Oligodeoxynucleotides used for real-time RT-PCR and 5′RACE.
(DOCX)
Primer efficiencies for real-time RT-PCR.
(DOCX)