Abstract
A novel recombinant hirudin, RGD-hirudin, inhibits the activity of thrombin and the aggregation of platelets. Here, we successfully expressed 15N, 13C-labeled RGD-hirudin in Pichia pastoris in a fermenter. The protein was subsequently purified to yield sufficient quantities for structural and functional studies. The purified protein was characterized by HPLC and MALDI-TOF mass spectroscopy. Analysis revealed that the protein was pure and uniformly labeled with 15N and 13C. A bioassay showed that the anti-thrombin activity and the anti-platelet aggregation ability of the labeled protein were the same as those of unlabeled RGD-hirudin. Multidimensional heteronuclear NMR spectroscopy has been used to determine almost complete backbone 15N, 13C and 1H resonance assignments of the r-RGD-Hirudin. The 15N-1H HSQC spectrum of uniformly 15N, 13C-labeled RGD-hirudin allowed successful assignment of the signals. Examples of the quality of the data are provided for the 15N-lH correlation spectrum, and by selected planes of the CBCA(CO)NH, CBCANH, and HNCO experiments. These results provide a basis for further studies on the structure-function relationship of RGD-hirudin with thrombin and platelets.
Introduction
Hirudin, an antithrombotic substance produced by the salivary glands of the medicinal leech (Hirudo medicinalis) [1], [2], is the most potent and specific thrombin inhibitor currently known. It blocks the thrombin-mediated conversion of fibrinogen to fibrin in clot formation, and, unlike heparin, it is a direct thrombin inhibitor (DTI) [3], which is not inactivated by platelet factor 4 (PF4) [4], [5].
Today, two preparations of recombinant hirudin are marketed: lepirudin (Refludan, Pharmion, UK and Berlex Laboratories, USA) and desirudin (Revasc, Novartis) [6]. Although there are minor differences in the amino-terminal composition of the two, the mechanism of interaction between hirudin and thrombin is identical: the carboxyl-terminal tail of hirudin binds to the substrate-binding site on thrombin and the globular amino-terminal domain then interacts with the active site of thrombin [7].
RGD-hirudin is a novel bi-functional molecule that contains the Arg-Gly-Asp (RGD) adhesion site recognition sequence. The abilities of this protein to inhibit thrombin and the aggregation of platelets were confirmed in our previous study [8]. More recent studies have focused on the structure-function relationship of RGD-hirudin. NMR was used to study its solution structure [9]. Purified 15N-labeled RGD-hirudin was prepared in our lab [10]. However, the interaction sites and precise mechanism between RGD-hirudin and its substrate are still unclear.
In this study, we describe the successful expression of 15N, 13C-labeled RGD-hirudin in yeast, Pichia pastoris (GS115). In total, 600 mg of 15N, 13C-labeled RGD-hirudin was generated through this, and we obtained a sufficient amount of purified and uniformly labeled RGD-hirudin for solution structure studies by NMR. Two- and three-dimensional double and triple resonance NMR techniques have been successfully applied to obtain most backbone 1H 15N, 13C and 13CO assignments of r-RGD-Hirudin(1–66).
Materials and Methods
Pichia pastoris cells carrying the RGD-hirudin gene (Mut+) were obtained from our lab. Briefly, the RGD-hirudin gene was synthesized in the Key Laboratory of Molecular Medicine at Fudan University. The cDNA encoding RGD-hirudin was cloned into the plasmid pPIC9K. The resulting expression vector was transformed into Pichia pastoris GS115. Vector integration into the Pichia pastoris chromosome was confirmed by PCR analyses [8].
Yeast nitrogen base with (or without) ammonium sulfate or amino acids was obtained from Sigma Aldrich. Isotope-enriched (98%) 15N ammonium sulfate, isotope-enriched (99%) 13C-glycerol and isotope-enriched (99%) 13C-methanol were obtained from Cambridge Isotope Lab. Blood plasma was from the Shanghai Blood Center. D2O and DCl for NMR experiments were from Sichuan Torch Chemistry Engineering Cooperation. Other reagents were of analytical purity. Sephacryl S-100 HR, Sephadex-G50, and Q-Sepharose-FF were purchased from GE.
Protein Expression
The production of the unlabeled RGD-hirudin was carried out in a fermenter (NBS Bioflow 3000) [11]. The production phase lasted 20 h at 30°C with a gradual increase in the methanol feeding rate, from 0.8 to 11.2 mL/L·h, allowing the culture to adapt to methanol consumption. After 6 h, the methanol feed rate was maintained at 11.2 mL/L·h for an additional 14 h.
The production of the 15N, 13C-labeled RGD-hirudin was carried out in the same fermenter vessel. BMD medium contained 100 mmol/L potassium phosphate (pH 6.0), 0.34% yeast nitrogen base without ammonium sulfate or amino acids, 1% 15N ammonium sulfate, 1% 13C glycerol, and 4×10−5% biotin [12], [13]. Fermentation medium (1.5 L) contained 30 g (15NH4)2SO4, 50 g 13C glycerol, 46.3 mL H3PO4 (85%), 1.61 g CaSO4, 10.8 g MgSO4, 7.16 g KOH, 27.3 g K2SO4, and 3 mL PTM1 solution. PTM1 solution (1 L) contained 6 g CuSO4·5H2O, 3 g MnSO4·H2O, 0.2 g H3BO4, 20 g ZnCl2, 0.8 g KI, 0.2 g Na2MoO4·2H2O, 0.5 g CoCl2, 65 g FeSO4·7H2O, 5 mL H2SO4, and 0.5 g CaSO4·2H2O.
For the expression of uniformly 15N, 13C-labeled RGD-hirudin, a single colony was picked and grown in 5 mL of BMD medium at 30°C overnight. This culture was diluted (1∶40) into 195 mL BMD medium and grown at 30°C until OD600 reached 4.0. The culture was transferred into 1.5 L medium in the fermenter and grown in batch mode for 20 h. A sharp increase in dissolved oxygen (DO) occurred when the OD600 reached 60, triggering a program for limited glycerol feed. 13C-glycerol (50%, v/v) was added from 4 mL/L·h to 40 mL/L·h for 3 h. In total, 120 mL 13C-glycerol (50%, v/v) was used until the OD600 reached 125; 50 g (15NH4)2SO4 was dissolved in 120 mL 13C-glycerol. The methanol-fed phase began once all the glycerol had been consumed. During 13C-methanol feeding, 22 g (15NH4)2SO4 dissolved in 50 mL H2O was added over 20 h. The fermenter was programmed to maintain the DO at 35% saturation and to maintain the pH at 5 by automatic addition of 4.0 mol/L KOH and 7.4 mol/L NaOH [14].
Protein Purification
The culture was centrifuged and the supernatant was ultra-filtered, followed by gel filtration and anion exchange chromatography. The concentrated supernatant was loaded onto a Sephacryl-S100 column (9.5 cm ×100 cm), pre-equilibrated with 20 mMol/L phosphate buffer (PB, pH 7.4). A volume of 1000 mL collected sample, which was eluted from the gel filtration, was loaded onto a Q-Sepharose FF column (2.6 cm ×20 cm), also pre-equilibrated with 20 mmol/L PB (pH 7.4). It was washed with 20 mmol/L PB (pH 7.4), followed by a single linear gradient of 0–1.0 mol/L NaCl-PB buffer. RGD-hirudin was eluted at 0.25 mol/L NaCl-PB. The sample that contained anti-thrombin activity was collected and desalted with Sephadex-G50 (1.6 cm ×20 cm). The loading sample volume was 5 mL each time. Protein concentration was measured by the Bradford assay. The desalted sample was lyophilized and stored in −80°C.
Protein Identification
Protein samples (both fermenter supernatant and purified protein) were analyzed by 15% SDS-PAGE.
The anti-thrombin activity of RGD-hirudin was tested by the Titration Testing Method, according to Markwardt [15]. Briefly, 200 µL of fresh plasma was added to a 1.5-mL tube; a 5-µL sample of RGD-hirudin was added to the plasma and mixed by vortexing. Thrombin (5 µL of 100 NIH units) was added to the above mixture and allowed to stand for 1 min at 37°C: if the plasma did not clot, the RGD-hirudin had 100 anti-thrombin units. Thus, consumption of each 1 NIH unit of thrombin is equivalent to 1 anti-thrombin unit.
A classic turbidity assay was used to measure the anti-platelet aggregation activity of RGD-hirudin [8].
HPLC-MS (Waters) was used to assess the purity and labeling efficiency of RGD-hirudin. Homogeneity of the purified protein was further confirmed using a Bruker Autoflex II. Both 15N, 13C-labeled and unlabeled RGD-hirudin were examined.
NMR Spectroscopy
Samples for NMR contained 2 mmol/L I5N/l3C-labeled RGD-hirudin, which was dissolved in 90% H2O/10% D2O, and the pH was adjusted with 1 mol/L DCl to 4.2–7.4. All NMR spectra were recorded on a Bruker DMX600 spectrometer equipped with a pulsed field gradient at 25°C. The following 3D spectra were recorded: CBCA(CO)NH [16], CBCANH [17], and HNCO [18]. The chemical shift of 15N was referenced indirectly [19]. The 2D 15N-1H HSQC spectra were recorded using the sensitivity-enhanced protocol with gradient echo-antiecho coherence selection. A time-proportional phase increment was used to discriminate between the positive and negative (15N) frequencies. Trim pulses were used in INEPT transfers [20]–[22]. NMR data processing and analysis were carried out using a Bruker XWIN-NMR and the Sparky software.
Results
Production of Unlabeled and 15N, 13C-labeled RGD-hirudin
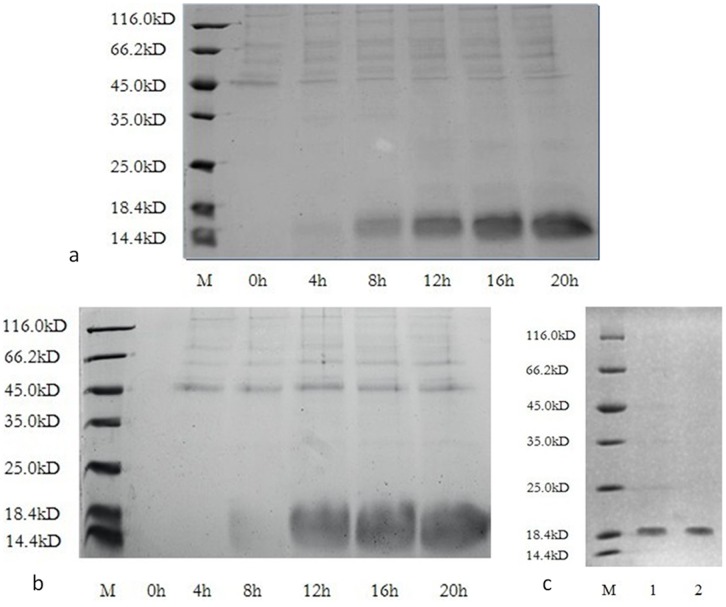
After 20 h of methanol induction, the anti-thrombin activity of the expressed product in the culture was up to 12,500 ATU/mL, as determined by fibrinogen solidification assay. SDS-PAGE showed that the expression level of unlabeled RGD-hirudin was as high as 85% of the culture (Fig. 1a). The expression level of 15N, 13C-labeled RGD-hirudin was the same as that of the unlabeled protein (Fig. 1b).
Figure 1. Expression and purification of unlabeled RGD-hirudin and labeled RGD-hirudin were analyzed by 15% SDS-PAGE.
Lane M, molecular mass standards (116 to 14.4 kDa ). a. Loading was 10 µL of culture supernatant of unlabeled RGD-hirudin for 0 to 20 h after methanol induction. b. Loading was 10 µL of culture supernatant of labeled RGD-hirudin for 0 to 20 h after methanol induction. c. Loading was 10 µL of the purified unlabeled RGD-hirudin (Lane 1) and labeled RGD-hirudin (Lane 2).
Purification and Characterization of Unlabeled and 15N, 13C-labeled RGD-hirudin
The protein was purified by gel filtration and anion exchange chromatography, as described in the Materials and Methods. Finally, 300 mg unlabeled RGD-hirudin (Fig. 1c, lane 1) and 250 mg 15N, 13C-labeled RGD-hirudin were obtained (Fig. 1c, lane 2; Table 1).
Table 1. Results of 15N, 13C-labeled RGD-Hirudin Purification.
| Step | Volume(mL) | Total ProteinConcentration(mg/mL) | Total Protein(mg) | Total Anti-ThrombinActivity (ATU) | Specific Activity(ATU/mg) | ActivityRecovery (%) |
| Supernatant | 1600 | 0.75 | 1200 | 9,000,000 | 7500 | 100 |
| After ultrafiltration | 400 | 2.6 | 1040 | 7,800,000 | 7500 | 86.7 |
| After Gel filtration | 1000 | 0.5 | 500 | 4,000,000 | 8000 | 44.4 |
| After anion exchange | 200 | 1.25 | 250 | 2,500,000 | 10000 | 27.8 |
The anti-thrombin specific activity of the purified 15N, 13C-labeled RGD-hirudin was more than 10,000 ATU/mg, the same as that of unlabeled RGD-hirudin (Table 2).
Table 2. The anti-thrombin activity of purified unlabeled and 15N, 13C-labeled RGD-hirudin.
| anti-thrombin activity (ATU/mg) Mean ± SD | |
| Unlabeled RGD-hirudin | 10280.0±672.3 |
| Labeled RGD-hirudin | 10460.0±1028.6 |
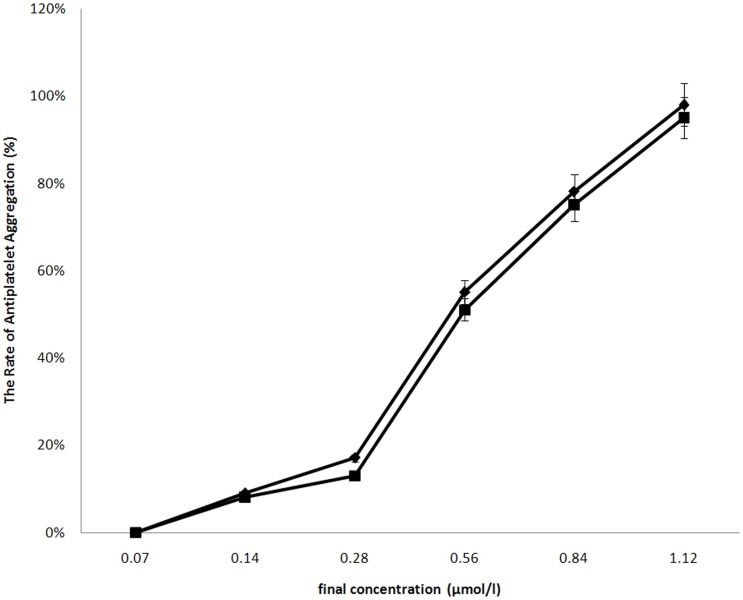
Anti-platelet aggregation activities of the proteins were determined by a classic turbidity assay. The results showed there was no the significant difference between unlabeled RGD-hirudin and 15N, 13C-labeled RGD-hirudin (Fig. 2).
Figure 2. Unlabeled RGD-hirudin and 15N, 13C-labeled RGD-hirudin contains the similar effects on rabbit platelet-aggregation induced by ADP.
(♦) unlabeled RGD-hirudin inhibited platelet aggregation induced by ADP in a dose-dependent manner; (▪) 15N, 13C-labeled RGD-hirudin was same capable of inhibiting platelet aggregation induced by ADP; two samples 0.14 µmol/l could inhibit the platelet aggregation, and their inhibitory effect on the platelet aggregation were dose-response.
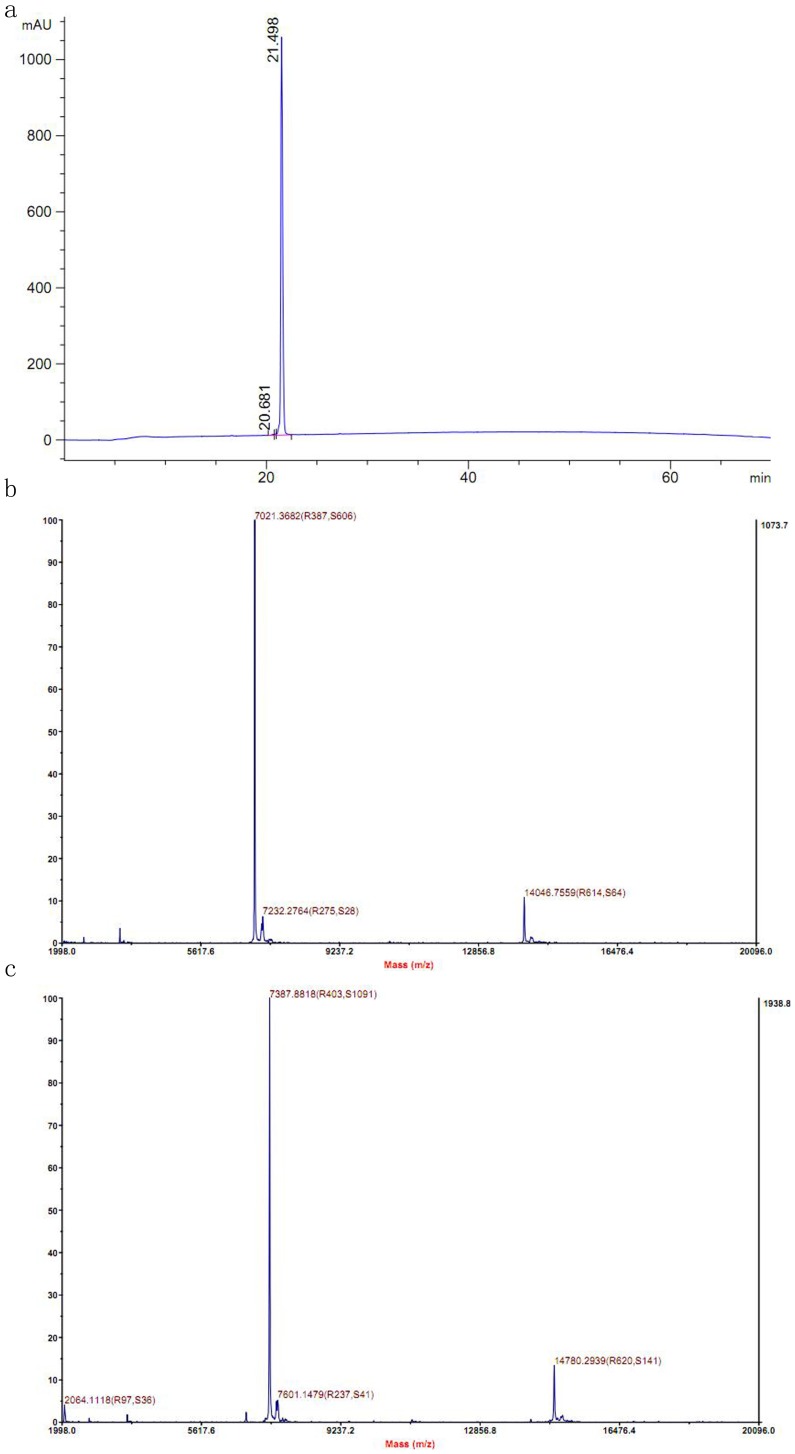
HPLC analyses revealed that the purity of the unlabeled RGD-hirudin (data not shown) and labeled RGD-hirudin (Fig. 3a) was up to 98%.
Figure 3. Purity and molecular weight of 15N, 13C-labeled RGD-hirudin were determined by HPLC-MS.
a. HPLC analysis of purified 15N, 13C-labeled RGD-hirudin showed the purity was up to 98%. b. Mass spectrometry of the purified proteins for unlabeled RGD-hirudin showed its molecular weight was 7021 Da c. The molecular weight of 15N, 13C-labeled RGD-hirudin was 7387 Da The total labeling ratio is more than 98%.
A mass spectrum of the unlabeled sample in Figure 3b showed its molecular weight was 7021 Da; the molecular weight of 15N, 13C-labeled RGD-hirudin was 7387 Da (Fig. 3c). The isotope labeling ratio calculated from MS was more than 98%, providing a basis for further studies of the structure-function relationship of RGD-hirudin by NMR and other methods.
NMR Spectroscopy of 15N, 13C-labeled RGD-hirudin
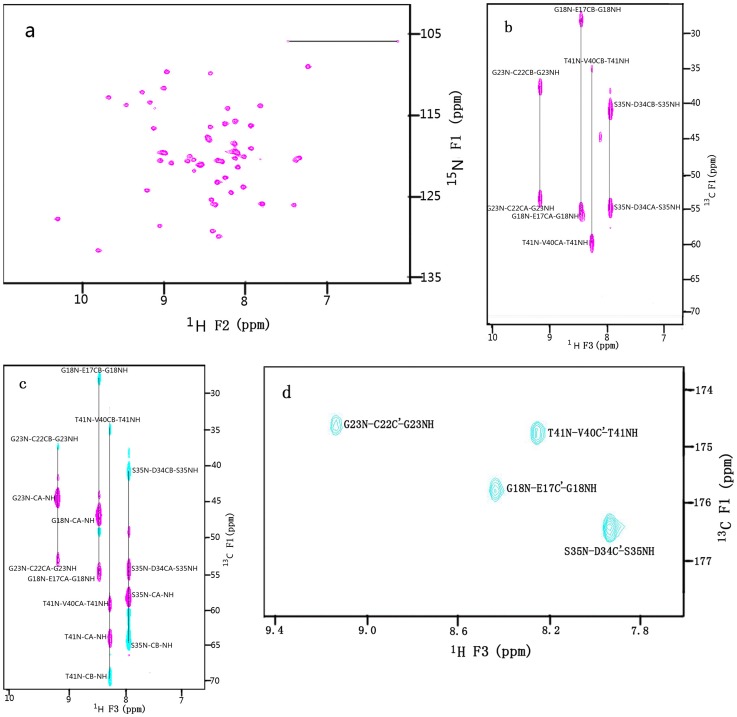
To further examine whether the protein had folded properly, a sensitivity-enhanced 2D 15N-1H HSQC [23] spectrum of uniformly 15N, 13C-labeled RGD-hirudin was recorded and is shown in Figure 4a. The 15N-1H HSQC analysis of a labeled protein displays one peak for each nitrogen-bound hydrogen atom (one peak per amino acid in the polypeptide backbone except proline) The position of these peaks in the spectrum is very sensitive to the structural environment of the resonating nuclei and thus to the global folding of the protein. The good dispersion of 15N-1H cross-peaks indicated the presence of a well-ordered structure and proper folding of the purified RGD-hirudin.
Figure 4. The 600MHz NMR spectra of 2.0 mM 15N, 13C-labeled RGD-hirudin (1–66) in 20 mM sodium phosphate buffer, pH 7.4.
a. 15N(F1 axis)-NH(F2 axis) region of the 2D 15N-1H Overbodenhausen correlation spectrum, b, c & d are selected 13C(F1)-1H(F3) planes at δ[15N(F2)] = 116.4 ppm of the 3D CBCA(CO)NH, CBCANH and HNCO spectra of RGD-hirudin (1–66) respectively. Sequential connectivities for four residues are indicated.
The 3D CBCANH experiment correlates the N and HN resonance of each amino acid with aliphatic resonances of both the same amino acid and the preceding amino acid. Thus, four cross peaks are obtained for each amino acid, which connect the chemical shifts of HN(i), N(i) with the chemical shifts of Cα(i), Cβ(i) and Cα(i-1), Cβ(i-1). The two cross peaks of each amino acid in 3D CBCA(CO)NH experiment only correlate the chemical shifts of HN(i), N(i), Cα(i-1), and Cβ(i-1). For example, in Figure 4b, HN of residue of T41 is correlated with Cα and Cβ of V40 and V41, respectively, in the 13C(F1)-1H(F3) planes at δ[15N(F3)] = 116.4 ppm of the 3D CBCANH, while in Figure 4c, HN of residue of T41 is connected to Cα and Cβ of V40 only, in the 13C(F1)-1H(F3) planes at δ[15N(F3)] = 116.4 ppm of the 3D CBCA(CO)NH of RGD hirudin (1–66). The HNCO experiment correlates the amide HN and 15N chemical shift of one amino acid with the carbonyl (C’) shift of the preceding residue by using the one-bond 15N-13CO J-coupling to establish the sequential correlation. As shown in Figure 4d, HN of residue of T41 is connected to C’ of V40, in the 13C(F1)-1H(F3) plane at δ[15N(F3)] = 116.4 ppm of the 3D HNCO of RGD hirudin (1–66) From the combination of CBCA(CO)NH and CBCANH experiments, the backbone resonance assignments and the sequential connectivities can be obtained, which can help to resolve a number of uncertainties in assignments due to degeneracy or the close similarity of backbone chemical shifts [24]. Almost complete 15N, 13 C and 1H resonance assignments of the backbone in r-RGD-Hirudin are reported (Table 3).
Table 3. 15N, 13C and 1H Resonance Assignments for r-RGD-Hirudin at pH 7.4 and 25°C.
| Residue | Chemical shift (ppm) | |||||
| 15N | C | Cβ | C’ | NH | Hα | |
| V1 | 60.0 | 33.1 | 176.3 | 3.53 | ||
| V2 | 125.8 | 61.6 | 32.6 | 175.3 | 8.32 | 4.03 |
| Y3 | 125.9 | 57.1 | 40.0 | 175.2 | 8.33 | 5.00 |
| T4 | 111.9 | 59.4 | 71.9 | 176,2 | 9.05 | 4.67 |
| D5 | 121.1 | 55.8 | 40.3 | 176.9 | 8.54 | 4.86 |
| C6 | 121.1 | 55.3 | 39.9 | 175.0 | 8.95 | 4.63 |
| T7 | 113.8 | 61.4 | 70.5 | 174.4 | 9.50 | 4.40 |
| E8 | 120.3 | 54.6 | 33.2 | 174.9 | 7.39 | 4.57 |
| S9 | 119.7 | 59.6 | 63.3 | 175.9 | 9.01 | 4.86 |
| G10 | 112.8 | 44.2 | 173.3 | 9.72 | 4.59, 3.34 | |
| Q11 | 120.6 | 54.8 | 32.7 | 175.6 | 7.42 | 5.30 |
| N12 | 118.7 | 51.9 | 39.9 | 174.5 | 8.21 | 4.54 |
| L13 | 113.6 | 55.3 | 37.8 | 175.2 | 9.23 | 3.25 |
| C14 | 109.2 | 53.1 | 43.6 | 172.0 | 7.30 | 4.98 |
| L15 | 119.8 | 54.9 | 39.2 | 174.0 | 9.08 | 4.26 |
| C16 | 126.1 | 56.6 | 40.6 | 172.3 | 7.46 | 4.75 |
| E17 | 120.7 | 54.8 | 27.8 | 175.8 | 8.67 | 4.46 |
| G18 | 116.5 | 46.3 | 174.5 | 8.48 | 3.95, 3.61 | |
| S19 | 120.1 | 57.6 | 62.9 | 173.4 | 8.70 | 4.39 |
| N20 | 119.2 | 52.7 | 39.2 | 176.4 | 7.97 | 4.81 |
| V21 | 128.7 | 63.3 | 31.7 | 174.4 | 9.07 | 3.40 |
| C22 | 129.3 | 53.0 | 37.3 | 174.6 | 8.47 | 5.12 |
| G23 | 116.7 | 44.0 | 172.2 | 9.16 | 4.03, 3.98 | |
| Q24 | 117.8 | 58.0 | 28.3 | 177.1 | 8.50 | 4.03 |
| G25 | 114.2 | 44.3 | 172.7 | 9.12 | 4.24, 3.66 | |
| N26 | 120.3 | 51.8 | 44.0 | 172.3 | 8.07 | 5.51 |
| K27 | 112.2 | 54.6 | 35.5 | 173.1 | 9.32 | 4.45 |
| C28 | 120.8 | 53.9 | 42.5 | 174.1 | 9.11 | 5.38 |
| I29 | 131.8 | 59.2 | 36.7 | 176.6 | 9.86 | 4.28 |
| L30 | 130.0 | 54.9 | 41.0 | 178.3 | 8.36 | 4.13 |
| G31 | 109.8 | 44.4 | 173.1 | 9.01 | 3.82 3.42, | |
| R32 | 121.5 | 54.9 | 31.4 | 177.1 | 8.11 | 4.51 |
| G33 | 113.0 | 46.5 | 174.9 | 8.87 | 4.69 3.81 | |
| D34 | 125.3 | 54.3 | 40.6 | 176.4 | 8.69 | 4.62 |
| S35 | 116.2 | 58.1 | 64.6 | 173.4 | 7.98 | 4.52 |
| K36 | 120.7 | 55.0 | 33.6 | 176.9 | 8.36 | 4.42 |
| N37 | 120.8 | 52.7 | 38.8 | 175.1 | 8.70 | 4.76 |
| Q38 | 118.0 | 54.6 | 33.5 | 173.1 | 8.45 | 4.73 |
| C39 | 127.9 | 54.8 | 38.8 | 174.6 | 10.3 | 5.42 |
| V40 | 124.4 | 59.2 | 34.7 | 174.8 | 9.27 | 4.88 |
| T41 | 116.1 | 63.7 | 69.7 | 174.7 | 8.28 | 4.46 |
| G42 | 114.2 | 44.1 | 171.3 | 8.27 | 4.12, 3.61 | |
| E43 | 119.6 | 56.8 | 29.2 | 176.4 | 8.09 | 4.18 |
| G44 | 113.8 | 44.3 | 172.7 | 7.85 | 4.58, 3.55 | |
| T45 | 115.8 | 57.8 | 70.6 | 180.8 | 8.19 | 4.85 |
| P46 | 62.4 | 31.9 | 175.1 | 4.62 | ||
| K47 | 125.4 | 55.0 | 32.9 | 174.9 | 8.42 | 4.38 |
| P48 | 63.7 | 31.9 | 177.0 | 4.46 | ||
| Q49 | 121.9 | 55.5 | 29.5 | 175.4 | 8.63 | 4.32 |
| S50 | 121.7 | 8.29 | 4.37 | |||
| H51 | 112.9 | 8.17 | 4.72 | |||
| N52 | 122.8 | 53.4 | 38.7 | 175.0 | 8.38 | 4.64 |
| Q53 | 121.3 | 56.6 | 28.9 | 176.4 | 8.54 | 4.76 |
| G54 | 109.9 | 44.4 | 173.8 | 8.44 | 3.90, 3.84 | |
| D55 | 120.4 | 54.5 | 40.9 | 175.7 | 8.17 | 4.12 |
| F56 | 119.4 | 57.3 | 39.6 | 175.1 | 8.04 | 4.61 |
| E57 | 124.5 | 53.7 | 29.9 | 173.4 | 8.18 | 4.41 |
| P58 | 62.4 | 31.7 | 176.6 | 4.38 | ||
| I59 | 123.3 | 58.6 | 38.4 | 175.0 | 8.25 | 4.41 |
| P60 | 63.4 | 31.9 | 177.0 | 4.38 | ||
| E61 | 121.2 | 56.4 | 30.0 | 176.3 | 8.58 | 4.09 |
| D62 | 120.8 | 54.3 | 40.9 | 175.7 | 8.29 | 4.53 |
| A63 | 123.9 | 52.3 | 19.0 | 177.2 | 8.05 | 4.24 |
| Y64 | 119.6 | 57.7 | 38.7 | 175.4 | 8.13 | 4. 57 |
| D65 | 122.7 | 54.0 | 41.2 | 174.9 | 8.25 | 4.61 |
| E66 | 126.0 | 58.0 | 31.1 | 176.6 | 7.83 | 3.96 |
Discussion
Two cultures were used under the same conditions, except for the nitrogen source and carbon source. In the first culture, NH4OH was used as the nitrogen source and for controlling the pH; unlabeled glycerol and methanol were used as the carbon source. In the second culture, (15NH4)2SO4, 13C-labeled glycerol, and 13C-labeled methanol were used and a mixture of 4 mol/L KOH and 7.4 mol/L NaOH was used for pH control, as described in the Materials and Methods. (NH4)2SO4 is known to inhibit cell growth, because it generates K2SO4 and increases the ionic strength of the culture medium [12], so (15NH4)2SO4 was added sequentially and the total amount was optimized, as described in the Materials and Methods. Both unlabeled (data not shown) and labeled RGD-hirudin were then expressed with about 20 h of methanol induction and were analyzed by SDS-PAGE. The final expression level was approximately 0.75 mg/mL.
The goal of this study was to obtain abundant 15N, 13C-labeled RGD-hirudin. Our results showed that high-density fermentation was well-suited for the production of 15N, 13C-labeled RGD-hirudin. During fermentation, KOH and NaOH were used to control pH, instead of NH4OH. (15NH4)2SO4 was used as the sole nitrogen source. Wood et al. reported that as the salt concentration was increased, a medium exchange was recommended for a MutS strain [25]. In our experiment, no medium exchange was carried out, and we suggest that salt concentration may not affect a Mut+ strain. After a three-step purification, we obtained sufficient 15N, 13C-labeled RGD-hirudin for a NMR study.
In NMR experiments, the spectra were affected by the concentration, pH, temperature, and salts. Previously, 1H NMR spectra of unlabeled RGD-hirudin were obtained at pH 7.4, and a concentration of 5 mmol/L [11]. For the 15N-1H HSQC spectrum, the signals were largely improved. When the sample concentration was 5 mmol/L, some cross peaks overlapped in the spectrum. The HSQC spectra of RGD-hirudin at different concentrations (2.0, 1.0, and 0.6 mmol/L) showed that even at 0.6 mmol/L, the peaks could be clearly separated. However, at 0.6 mmol/L, most peaks disappeared after injecting thrombin into RGD-hirudin in the initial NMR titration experiments (data not shown). Here, we chose the concentration of 2 mmol/L for the HSQC study. Because RGD-hirudin reacts with thrombin and GPIIa/IIIb in human blood, the 15N-1H HSQC spectrum was collected at the biological pH of 7.4.
Conclusions
In a previous study, we confirmed the function of RGD-hirudin, inhibiting thrombin and platelet aggregation, which may indicate RGD-hirudin will be useful in clinical applications [8]. RGD-hirudin needed to be modified, based on structure-function relationships, for advanced clinical use. Our lab has previously conducted a study on the solution structure of RGD-hirudin (1–49) [9]. Although 15N-labeled RGD-hirudin from Pichia pastoris was obtained, the structure of the C-terminal domain (from Ser50 to Glu66) has not yet been worked out. To study the relationship between the molecular structure in solution and ligand-binding properties, we have now prepared 15N/13C double-labeled RGD-hirudin in sufficient quantities to permit a full determination of the structure and dynamics of RGD-hirudin (1–66) using heteronuclear NMR spectroscopy. Almost complete backbone 1H 15N, 13C and 13CO assignments of RGD-Hirudin have been obtained. Complete side-chain assignment is in progress by using the combination of HCCH-COSY and HCCH-TOCSY now.
Acknowledgments
We thank Chunyang Cao and Wenxian Lan, of the State Key Laboratory of Bio-organic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, for providing us with instrument time on the Varian INOVA 600 NMR spectrometer and helpful discussions regarding our NMR experiments.
Funding Statement
This study was supported by the Foundation of Ministry of Science and Technology of the People’s Republic of China (2009ZX09503-006). The funders had no role in study design, data collection and analysis, decision to publish,or praparation of the manuscript.
References
- 1. Markwardt F (1992) Hirudin: the promising antithrombotic. Cardiovasc Drug Rev 10: 211–232. [Google Scholar]
- 2. Markwardt F (1994) The development of hirudin as an antithrombotic drug. Thromb Res 74: 1–23. [DOI] [PubMed] [Google Scholar]
- 3. Sun Z, Zhao Z, Zhao S, Sheng Y, Gao C, et al. (2009) Recombinant hirudin treatment modulates aquaporin-4 and aquaporin-9 expression after intracerebral hemorrhage in vivo. Mol Biol Rep 36: 1119–1127. [DOI] [PubMed] [Google Scholar]
- 4. Amiral J, Bridey F, Dreyfus M, Vissoc AM, Fressinaud E, et al. (1992) Platelet factor 4 complexed to heparin is the target for antibodies generated in heparin-induced thrombocytopenia. Thromb Haemost 68: 95–96. [PubMed] [Google Scholar]
- 5. Greinacher A, Potzsch B, Amiral J, Dummel V, Eichner A, et al. (1994) Heparin-associated thrombocytopenia: isolation of the antibody and characterization of a multimolecular PF4-heparin complex as the major antigen. Thromb Haemost 71: 247–251. [PubMed] [Google Scholar]
- 6. Warkentin TE (2004) Bivalent direct thrombin inhibitors: hirudin and bivalirudin. Best Pract Res Clin Haematol 17: 105–125. [DOI] [PubMed] [Google Scholar]
- 7. Wallis RB (1996) Hirudins: from leeches to man. Semin Thromb Hemost 22: 185–196. [DOI] [PubMed] [Google Scholar]
- 8. Mo W, Zhang YL, Chen HS, Wang LS, Song HY (2009) A novel hirudin derivative characterized with anti-platelet aggregations and thrombin inhibition. J Thromb Thrombolysis 28: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song X, Mo W, Liu X, Zhu L, Yan X, et al. (2007) The NMR solution structure of recombinant RGD-hirudin. Biochem Biophys Res Commun 360: 103–108. [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Zhang Y, Li S, Liu X, Yan X, et al. (2010) Overproduction of 15N-labeled r-RGD-hirudin in pichia pastoris for NMR studies. Protein Pept Lett 17: 1228–1233. [DOI] [PubMed] [Google Scholar]
- 11. Mo W, Zhang YL, Wang LS, Yang XY, Song HY (2004) [Fermentation, purification and identification of recombinant RGD-Hirudin]. Sheng Wu Gong Cheng Xue Bao 20: 126–129. [PubMed] [Google Scholar]
- 12. Wood MJ, Komives EA (1999) Production of large quantities of isotopically labeled protein in Pichia pastoris by fermentation. J Biomol NMR 13: 149–159. [DOI] [PubMed] [Google Scholar]
- 13. Denton H, Smith M, Husi H, Uhrin D, Barlow PN, et al. (1998) Isotopically labeled bovine beta-lactoglobulin for NMR studies expressed in Pichia pastoris. Protein Expr Purif 14: 97–103. [DOI] [PubMed] [Google Scholar]
- 14. Wendeler M, Hoernschemeyer J, John M, Werth N, Schoeniger M, et al. (2004) Expression of the GM2-activator protein in the methylotrophic yeast Pichia pastoris, purification, isotopic labeling, and biophysical characterization. Protein Expr Purif 34: 147–157. [DOI] [PubMed] [Google Scholar]
- 15. Markwardt F, Walsmann P (1967) Purification and analysis of the thrombin inhibitor hirudin. Hoppe Seylers Z Physiol Chem 348: 1381–1386. [PubMed] [Google Scholar]
- 16. Grzesiek S, Bax A (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc 114: 6291–6293. [Google Scholar]
- 17. Grzesiek S, Bax A (1992) An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins J Magn Reson. 96: 201–207. [Google Scholar]
- 18. Kay LE, Ikura M, Tschudin R, Bax A (1990) Three-dimensional triple-resonance NMR Spectroscopy of isotopically enriched proteins. J Magn Reson 213: 423–441. [DOI] [PubMed] [Google Scholar]
- 19. Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, et al. (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6: 135–140. [DOI] [PubMed] [Google Scholar]
- 20. Palmer AG, Cavanagh J, Wright PE, Rance M (1991) Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. Journal of Magnetic Resonance (1969) 93: 151–170. [Google Scholar]
- 21. Kay LE, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114: 10663–10665. [Google Scholar]
- 22. Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, et al. (1994) A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR 4: 301–306. [DOI] [PubMed] [Google Scholar]
- 23. Ernst JF (1988) Codon usage and gene expression. Trends Biotechnol 6: 196–199. [Google Scholar]
- 24. Garrett DS, Lodi PJ, Shamoo Y, Williams KR, Clore GM, et al. (1994) Determination of the secondary structure and folding topology of an RNA binding domain of mammalian hnRNP A1 protein using three-dimensional heteronuclear magnetic resonance spectroscopy. Biochemistry 33: 2852–2858. [DOI] [PubMed] [Google Scholar]
- 25. Cubeddu L, Moss CX, Swarbrick JD, Gooley AA, Williams KL, et al. (2000) Dictyostelium discoideum as expression host: isotopic labeling of a recombinant glycoprotein for NMR studies. Protein Expr Purif 19: 335–342. [DOI] [PubMed] [Google Scholar]