The ubiquitin ligases CUL-3 and SLMB collaborate to regulate the Drosophila circadian clock by controlling TIMELESS oscillations.
Abstract
Eukaryotic circadian clocks rely on transcriptional feedback loops. In Drosophila, the PERIOD (PER) and TIMELESS (TIM) proteins accumulate during the night, inhibit the activity of the CLOCK (CLK)/CYCLE (CYC) transcriptional complex, and are degraded in the early morning. The control of PER and TIM oscillations largely depends on post-translational mechanisms. They involve both light-dependent and light-independent pathways that rely on the phosphorylation, ubiquitination, and proteasomal degradation of the clock proteins. SLMB, which is part of a CULLIN-1-based E3 ubiquitin ligase complex, is required for the circadian degradation of phosphorylated PER. We show here that CULLIN-3 (CUL-3) is required for the circadian control of PER and TIM oscillations. Expression of either Cul-3 RNAi or dominant negative forms of CUL-3 in the clock neurons alters locomotor behavior and dampens PER and TIM oscillations in light-dark cycles. In constant conditions, CUL-3 deregulation induces behavioral arrhythmicity and rapidly abolishes TIM cycling, with slower effects on PER. CUL-3 affects TIM accumulation more strongly in the absence of PER and forms protein complexes with hypo-phosphorylated TIM. In contrast, SLMB affects TIM more strongly in the presence of PER and preferentially associates with phosphorylated TIM. CUL-3 and SLMB show additive effects on TIM and PER, suggesting different roles for the two ubiquitination complexes on PER and TIM cycling. This work thus shows that CUL-3 is a new component of the Drosophila clock, which plays an important role in the control of TIM oscillations.
Author Summary
Circadian clocks adjust the physiology and behavior of organisms to the day/night cycle and rely on molecular feedback loops that generate daily oscillations of transcription. In the Drosophila fruit fly, the PERIOD (PER) and TIMELESS (TIM) proteins coordinate the clock–they accumulate during the night, form a complex, and repress their own gene expression in the early morning. The temporal control of this oscillation involves the phosphorylation, ubiquitination, and proteasomal degradation of the PER and TIM proteins. The SUPERNUMERARY LIMBS (SLMB) ubiquitin ligase is known to play a key role in controlling the degradation of phosphorylated PER and TIM. In this study we investigated the role of another ubiquitin ligase, CULLIN-3 (CUL-3). We found that inhibition of CUL-3 activity results in the abolition of rest/activity rhythms in flies and flattens the PER and TIM oscillations. CUL-3 physically interacts and forms a complex with a lowphosphorylated version of TIM in the absence of PER, thereby allowing its accumulation during the night. In contrast, when PER is present SLMB preferentially interacts with phosphorylated TIM, favoring its degradation. The results suggest that CUL-3 and SLMB share the work to control the oscillations of the PER and TIM proteins during the day/night cycle.
Introduction
Circadian clocks are present in most living organisms and control a variety of physiological and behavioral functions. Eukaryotic clocks stem from a transcriptional negative feedback loop where activators induce the expression of repressors, which then inhibit the activators [1]. The accumulation of the repressor proteins is delayed by post-translational mechanisms, allowing us to define active and inactive phases of transcription, hence an oscillation. In Drosophila, the two basic helix-loop-helix PER-ARNT-SIM (bHLH PAS) proteins CLOCK (CLK) and CYCLE (CYC) activate the transcription of their targets in the evening [2],[3]. The target genes period (per) and timeless (tim) encode two proteins that associate in a complex, progressively accumulate, and become phosphorylated to repress CLK/CYC-dependent transcription in the late night. The delayed accumulation, nuclear entry, and transcriptional activity of PER and TIM involve their phosphorylation by several kinases such as DOUBLE TIME (DBT) CK1ε [4]–[10] and NEMO [11] that target PER. The CK2 casein kinase [12]–[14] and SHAGGY (SGG) GSK3 [15] rather target TIM. The phosphorylation of PER and TIM are also regulated by the phosphatases PP2A [16] and PP1 [17], respectively. Nuclear phosphorylated PER induces CLK phosphorylation and removal from the chromatin, then PER degradation in the morning allows CLK-CYC-dependent transcription to resume [18]–[21].
The pace of the oscillation depends largely on the speed of PER/TIM accumulation during the early night and degradation in the late night. The stability of phosphorylated PER and TIM is controlled by the SUPERNUMERARY LIMBS (SLMB) E3 ubiquitin ligase [22],[23]. For PER, the phosphorylation of a few serine residues around S47 by DBT cooperatively increases SLMB binding to PER [24] and is negatively regulated by NEMO-controlled phosphorylation in the PERS region [11] to finely tune PER stability. Both S47 phosphorylation and direct SLMB-binding occur at the end of the night, suggesting that SLMB controls nuclear PER at the end of the cycle [24]. DBT has been proposed to also induce cytoplasmic PER degradation in the absence of TIM and thus control the delayed accumulation of PER [4],[6]. This model predicts that the control of cytoplasmic PER stability depends on TIM accumulation. CK2 has been suggested to destabilize TIM [14], whereas PP1 appears to favor TIM stabilization [17], but whether they affect SLMB function is unknown.
In addition to SLMB that controls the circadian oscillations of TIM, the JETLAG (JET) E3 ligase targets TIM and CRYPTOCHROME (CRY) for the light-induced proteasome-dependent protein degradation that participates to the resetting of the clock [25]–[27]. The JET-dependent degradation, but not the SLMB-dependent one, appears to be regulated by the COP9 signalosome [28]. SLMB and JET are parts of CULLIN-1-based SCF complexes that belong to the RING family of E3 ubiquitin ligases [29]. SLMB plays a major role in CUBITUS INTERRUPTUS (CI) proteolysis, which generates the repressor form of the CI transcription factor in the absence of HEDGEHOG (HH) signaling [30]. In addition to SLMB, other components of the HH pathway such as CK1, CK2, and SGG are shared with the circadian oscillator [31]. Another ubiquitin complex, based on CULLIN-3, has been shown to participate in the proteolytic regulation of CI [32]–[35]. Since SLMB alone unlikely regulates all aspects of the circadian control of PER and TIM stability, we asked whether CULLIN-3 may participate in the control of PER and/or TIM oscillations. We show that CUL-3 downregulation induces strong defects of rest-activity rhythms, which mainly result from the loss of TIM cycling. Indeed, CUL-3 forms protein complexes with hypo-phosphorylated TIM, which are favored in the absence of PER, suggesting that CUL-3 participates in the control of the night accumulation of TIM. In contrast, SLMB is present in complexes that contain more phosphorylated TIM and are favored by the presence of PER, suggesting a rather later role in the TIM cycle.
Results
Deregulation of CUL-3 in the Clock Neurons Induces Behavioral Arrhythmicity
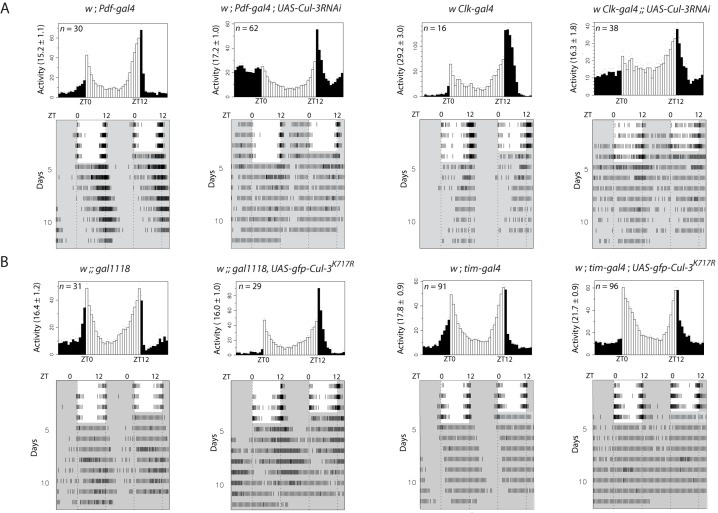
Since Cul-3 mutants are homozygous lethal, we first used targeted expression of Cul-3 RNAi to test the possible role of CUL-3 in the control of behavioral rhythms. About 150 neurons express PER and TIM in the brain, and various studies have assigned specific behavioral contributions to defined neuronal subsets, depending on the environmental conditions [36]. In particular, PER cycling in the lateral neurons (LNs) that express the Pigment-Dispersing Factor (PDF) generates morning activity in light-dark (LD) cycles and free running rhythms in constant darkness (DD), whereas PER cycling in the PDF-negative LNs generates LD evening activity [37],[38]. Expressing Cul-3 RNAi in all clock cells under tim-gal4 control induced 100% lethality, but a limited number of adult flies could be obtained with the Clk-gal4 driver, whose expression is less broad than tim-gal4 [39],[40], and no lethality was observed with the more restricted cry-gal4 and Pdf-gal4 drivers. In LD cycles, flies expressing Cul-3 RNAi under Pdf-gal4 or Clk-gal4 control displayed no or reduced morning anticipatory activity (Figure 1A and Table S1). The two genotypes also showed reduced lights-ON startle response. After transfer to DD, RNAi genotypes either showed weak rhythms or became arrhythmic (Figure 1A and Table 1). We also used the gal1118 driver, which is mostly expressed in the PDF cells [41]. Reduced morning anticipation in LD and strongly altered rhythms in DD were observed when Cul-3 RNAi expression was driven by gal1118 (Figure S1). No disappearance of the lights-ON startle response was observed, supporting a genetic background effect in the previous genotypes. Importantly, the effect of Cul-3 RNAi on Cul-3 mRNA levels was tested by quantitative RT-PCR (Figure S2). Cul-3RNAi induced a 2-fold decrease of Cul-3 mRNAs in head extracts when driven by Clk-gal4 or 5-fold in larvae when using the ubiquitous da-gal4 (daughterless), supporting decreased CUL-3 activity in the RNAi flies. For RNAi genotypes, two copies of both the gal4 and UAS transgenes were required for generating arrhythmic behavior (Figures 1A and S1, and Table 1).
Figure 1. Locomotor activity of Cul-3 downregulated flies in LD and DD.
Control flies and flies expressing either a Cul-3 RNAi (A) or a CUL-3K717R protein (B) were entrained for 4 d in LD 12∶12 and transferred to DD. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5-h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated three to four times with very similar results.
Table 1. Locomotor activity rhythms in constant darkness of flies with altered CUL-3 activity.
| Genotype | Total Flies | Rhythmic Flies (%) | Period (h) | Power |
| w ;; UAS-Cul-3RNAi | 27 | 93 | 24.1±0.1 | 154±14 |
| w ; Pdf-gal4 ; UAS-Cul-3RNAi | 47 | 55 | 25.1±1.2 | 57±5 |
| w ; Pdf-gal4 | 28 | 100 | 25.0±0.2 | 145±10 |
| w ; cry-gal4-39 ; UAS-Cul-3RNAi | 28 | 30 | 26.5±2.0 | 43±5 |
| w ; cry-gal4-39 | 20 | 90 | 24.6±0.1 | 159±7 |
| w Clk-gal4 ;; UAS-Cul-3RNAi | 26 | 61 | 24.1±0.7 | 55±7 |
| w Clk-gal4 | 16 | 94 | 23.7±0.1 | 117±15 |
| w ;; gal1118, UAS-Cul-3RNAi | 25 | 44 | 23.3±0.4 | 74±9 |
| w ;; gal1118 | 29 | 100 | 24.5±0.1 | 118±10 |
| w ;; UAS-gfp-Cul-3K717R | 27 | 100 | 23.7±0.1 | 173±12 |
| w ;; gal1118, UAS-gfp-Cul-3K717R | 29 | 72 | 27.1±1.0 | 75±9 |
| w ; tim-gal4 ; UAS-gfp-Cul-3K717R | 96 | 40 | 26.8±0.9 | 56±7 |
| w ; tim-gal4 ; cry-gal80, UAS-gfp-Cul-3K717R/UAS-gfp-Cul-3K717R | 37 | 97 | 24.1±0.1 | 123±8 |
| w ;; UAS-flag-Cul-3K717R/+ | 32 | 100 | 23.3±0.1 | 169±12 |
| w ; tim-gal4 ; UAS-flag-Cul-3K717R/+ | 23 | 17 | 27.4±2.9 | 31±5 |
| w ;; UAS-Cul-3ΔC | 30 | 93 | 24.3±0.1 | 174±11 |
| w ; tim-gal4 ; UAS-Cul-3ΔC | 48 | 29 | 25.4±0.7 | 73±12 |
| w ;; UAS-gfp-Cul-3K717R/UAS-Cul-3ΔC | 32 | 97 | 24.0±0.2 | 113±11 |
| w ; tim-gal4 ; UAS-gfp-Cul-3K717R/UAS-Cul-3ΔC | 54 | 9 | 28.3±3.8 | 49±20 |
| w ;; UAS-gfp-Cul-3 | 40 | 100 | 24.3±0.1 | 178±10 |
| w ; tim-gal4 ; UAS-gfp-Cul-3 | 25 | 36 | 25.4±1.6 | 53±10 |
| w ; tim-gal4 | 96 | 89 | 25.3±0.2 | 113±7 |
| w ; Pdf-gal4 ; UAS-Cul-3RNAi (20°C) | 61 | 82 | 23.9±0.4 | 91±8 |
| w ; Pdf-gal4 (20°C) | 30 | 100 | 24.6±0.2 | 131±9 |
The mean values of period and associated power (see Materials and Methods) are given ± s.e.m. In genotypes with altered CUL-3 activity, rhythmic flies display weak rhythms as indicated by the large s.e.m. of the period and the low associated power.
Two inactive forms of CUL-3 were then expressed in the clock neurons. Both forms lack the site for the addition of Nedd8, a CULLIN modification that is required for ubiquitin transfer from E2 to the substrate [42]. CUL-3K717R bears a mutation in the neddylation site [43], whereas CUL-3ΔC lacks the conserved C-terminal domain that includes this site [44]. Expression of one or the other inactive protein under the control of a Pdf-gal4, cry-gal4, or Clk-gal4 driver did not significantly affect behavior (unpublished data), suggesting that they were less efficient than RNAi for decreasing CUL-3 activity in the clock neurons. Driving their expression with tim-gal4 did not induce lethality, also supporting weaker effect compared to Cul-3 RNAi. We thus used gal1118 and tim-gal4 to test the effect of CUL3K717R on behavioral rhythms. LD morning anticipation was reduced and DD free running rhythms were lost or altered in flies expressing either one of two different CUL3K717R-encoding constructs (Figures 1B and S3, Tables 1 and S1), indicating that the modified CUL-3 protein was acting as a dominant negative form. Rhythms were restored when GAL4 activity was inhibited by a cry-gal80 transgene, confirming that CUL-3 was required in clock cells for controlling behavior (Table 1). Altered morning anticipation in LD (Figure S3, Table S1, and unpublished data) and behavioral arrhythmicity in DD (Figure S3 and Table 1) were also observed in flies expressing CUL-3ΔC, both CUL3K717R and CUL3ΔC, or wild-type CUL-3, under tim-gal4 control. The latter genotype suggested that supernumerary CUL-3 molecules might interfere with the function of the protein, as reported for SLMB [22]. Targeted expression of RNAi or mutant protein as well as overexpression of the wild type protein thus revealed that CUL-3 activity was required in the clock neurons for strong morning anticipation in LD and rhythmic behavior in DD. Although tim-gal4 and Clk-gal4 are expressed in all clock neurons, evening anticipation could still be observed in flies with altered CUL-3 activity.
Since Cul-3 mutants affect the axonal growth of the mushroom body neurons [43], we tested the possibility that developmental effects of CUL3 deregulation would be responsible for the behavioral defects. We took advantage of the fact that GAL4-driven expression is temperature-dependent [45] and compared the behavior of RNAi flies grown at 25°C and tested at either 25°C or 20°C. Adult flies tested at 20°C showed significantly weaker defects than those tested at 25°C (Figure S4 and Table 1), indicating that the adult stage was determinant for CUL-3 function in activity rhythms. We also checked the morphology of the PDF cells expressing Cul-3 RNAi or inactive CUL-3 proteins (Figure S5). Anti-PDF labeling did not show any detectable morphological defects of the PDF-positive ventral lateral neurons in flies expressing Cul-3 RNAi. Slightly more defasciculated projections and reduced arborization in the medulla were observed for flies expressing CUL-3ΔC but not in the other genotypes. One or two additional PDF-positive cells were sometimes seen in flies expressing either CUL-3ΔC or CUL-3K717R (unpublished data). Altogether, the data indicated that the behavioral phenotype induced by CUL-3 downregulation was, at least for a large part, not a consequence of a developmental defect.
CUL-3 Controls PER and TIM Oscillations in the Clock Neurons
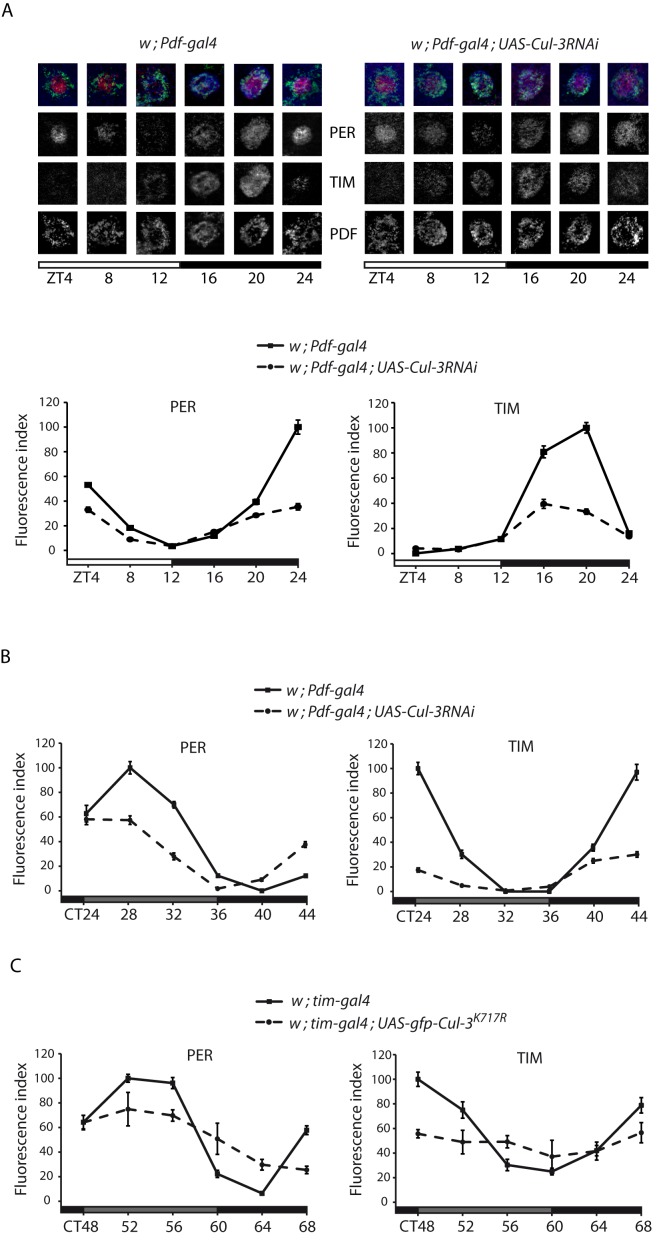
To understand the molecular bases of the behavioral alterations displayed by the flies with deregulated CUL-3, PER and TIM oscillations were analyzed in their PDF-expressing small ventral lateral neurons (s-LNvs). In LD conditions, w ; Pdf-gal4; UAS-Cul-3RNAi (hereafter Pdf>RNAi) flies showed dampened PER and TIM oscillations (Figure 2A). The two proteins failed to reach wild-type peak levels during the night, indicating that their nighttime accumulation was affected by Cul-3 downregulation. Interestingly, PER and TIM appeared slightly more evenly distributed between nuclear and cytoplasmic compartments in the RNAi flies (see ZT24 in particular). Similarly dampened oscillations of the two proteins were also observed in the second day after transfer in DD (Figure 2B), providing a molecular basis for the behavioral arrhythmicity of these flies. Arrhythmic w ; tim-gal4 ; UAS-gfp-Cul-3K717R (tim>Cul-3K717R) flies also showed altered PER and TIM oscillations, with a complete loss of TIM cycling on the third day of DD (Figure 2C). Although these flies should express inactive CUL-3 in all clock cells, they still displayed evening activity in LD conditions (see above). We thus suspected that PER and TIM oscillations might not be affected in their “evening” CRY-expressing dorsal lateral neurons (LNds). Indeed, a 30%–40% decrease of TIM immunoreactivity was observed at peak time (ZT16-20) in the s-LNvs of tim>Cul-3K717R flies, whereas mutants and controls showed similar TIM labeling in the CRY-positive LNds (Figure S6).
Figure 2. PER and TIM in the s-LNvs of flies expressing Cul-3 RNAi.
Flies were entrained for 3 d in LD and collected every 4 h on the fourth day of LD (A) or transferred to DD and collected on the second (B) or third (C) day of DD. Images are optical sections of individual PDF-expressing s-LNvs from Pdf>RNAi flies and controls in LD. Merged images: red is PER, blue is TIM, and green is PDF. Graphs represent quantifications of PER and TIM immunolabeling (see Materials and Methods) in the PDF-positive s-LNvs of Pdf>RNAi or tim>Cul-3K717R flies and relevant controls. White and gray bars indicate lights-ON and lights-OFF, respectively. Gray and black bars indicate subjective day and subjective night, respectively. Time (h) is indicated as ZT or CT (Circadian Time) where CT0 is 12 h after lights-OFF of the last LD day. Error bars indicate s.e.m. Two independent experiments were done for each genotype/condition with very similar results.
CUL-3 Acts on PER and TIM Post-Transcriptionally
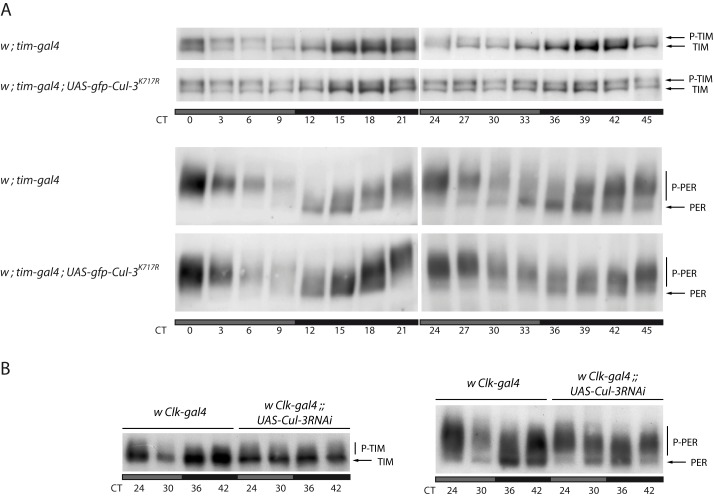
PER and TIM oscillations were analyzed in head extracts of tim>Cul-3K717R flies. In LD conditions, mutant flies still showed robust PER (unpublished data) and TIM (Figure S7) cycling. Since a strong dampening of PER and TIM oscillations was observed in the PDF cells under the same conditions, it suggested CUL-3 might have a less crucial role in the eye, which is the major source of clock proteins in the head [46], at least in LD cycles. Alternatively, the lower expression of the tim-gal4 driver in the eye (unpublished data) might not produce sufficiently high levels of CUL-3K717R protein to affect LD oscillations, which are strongly driven by light. We then looked at protein oscillations in DD (Figure 3A and Figure S8). At DD1, only subtle defects could be observed on PER and TIM cycling, with slightly more phosphorylated proteins at night in the mutant. At DD2, TIM cycling was strongly altered in the mutant with similar levels of phosphorylated TIM at all time points and weak cycling of unphosphorylated TIM, which stayed at relatively low levels. A very similar phenotype was observed in flies with two copies of tim-gal4 driving a single copy of UAS-flag-Cul-3K717R (Figure S9) or a single copy of both UAS-gfp-Cul-3K717R and UAS-Cul-3ΔC (unpublished data). Phosphatase treatment abolished the slow migrating TIM forms of tim>Cul-3K717R flies (Figure S10), demonstrating that they were indeed phosphorylated TIM. The constitutive presence of both phosphorylated and unphosphorylated forms of TIM supported a post-translational effect of CUL-3. Similarly altered TIM cycling was observed in flies with overexpressing wild type CUL-3 (Figure S11), suggesting that excess of wild type protein might interfere with the formation of functional complexes with the proper stoichiometry. Since tim>Cul-3RNAi were lethal, we analyzed TIM in head extracts of Clk>Cul-3RNAi flies, although it is less efficient than tim-gal4 to affect molecular oscillations in head extracts when used to drive RNAi targeted against various clock genes (unpublished data). At DD2, TIM cycling was strongly reduced, but the accumulation of phosphorylated TIM was less prominent than in flies expressing CUL-3K717R (Figure 3B). The data indicated that deregulating CUL-3 strongly reduces TIM cycling, with mostly hypo-phosphorylated TIM in Clk>Cul-3RNAi flies and similar amounts of phosphorylated TIM and hypo-phosphorylated TIM in tim>Cul-3K717R flies.
Figure 3. PER and TIM in head extracts of flies expressing CUL-3K717R or Cul-3 RNAi.
Flies were entrained for 3 d in LD, then transferred to DD. Gray and black bars indicate subjective day and subjective night, respectively. (A) TIM (top, Tris-acetate gel) and PER (bottom, Tris-glycine gel) Western blot analysis of head extracts from flies collected the first and second days of DD. Phosphorylated (P-) and hypo-phosphorylated forms of PER and TIM are indicated. (B) TIM (left) and PER (right) Western blots (both Tris-glycine gels) of head extracts from flies collected the second day of DD. Two (Cul-3RNAi) or three (Cul-3K717R) independent Western blots were done for each condition with very similar results.
In both tim>Cul-3K717R and Clk>Cul-3RNAi flies, PER cycling was still observed at day 2, but it was dampened. More phosphorylated and less unphosphorylated protein was observed at the beginning of the night (CT36), with a stronger phenotype in the tim>Cul-3K717R flies (Figure 3A,B). Since PER oscillations were less rapidly affected than TIM oscillations, it suggested that CUL-3 might control TIM more directly than PER.
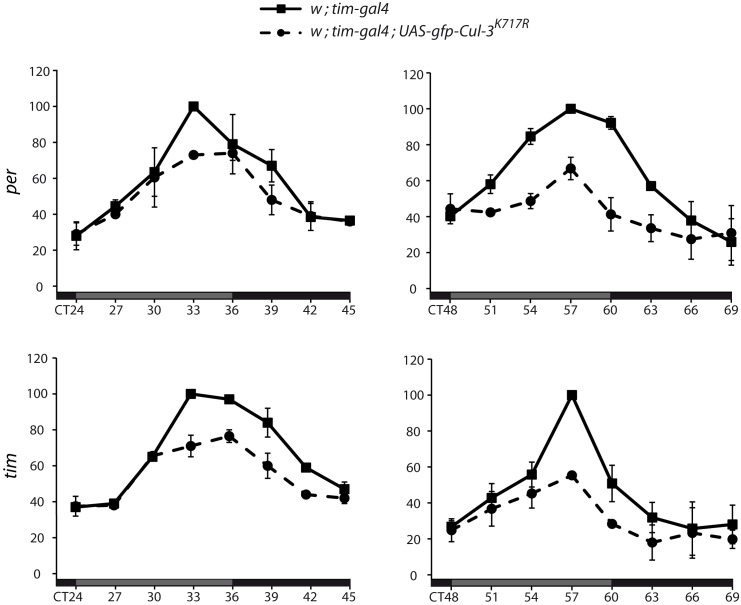
We then asked how CUL-3 deregulation affected per and tim mRNA levels. mRNA were quantified in head extracts of tim>Cul-3K717R and control flies (Figure 4). For both per and tim, mRNA levels still cycled at day 2 (left panels) in the mutant, but peak levels were about 25% lower. At day 3 (right panels), per and tim mRNA oscillations were more severely dampened, with peak levels lowered by about 40%. Decreased mRNA thus likely contributed to the lower levels of unphosphorylated PER and TIM at the beginning of the night (see Figure 3A). Since tim mRNA levels still showed oscillations at DD2 while phosphorylated TIM was already flat, it was unlikely that the protein defect was a consequence of altered mRNA cycling. PER protein and per mRNA showed dampened oscillations at DD2 in the flies expressing CUL-3K717R. However, the mRNA increase began to slow down at CT30 when more phosphorylated PER was observed (see Figure 3A), suggesting that higher phosphorylated PER levels were responsible for lower mRNA accumulation and subsequent lower PER and TIM synthesis. The data thus supported CUL3 acting at the protein level to control PER and TIM oscillations.
Figure 4. per and tim mRNA oscillations in flies expressing CUL-3K717R.
Flies were entrained for 3 d in LD and transferred to DD for collection every 3 h in the second (left) and third (right) days of DD. Relative per and tim mRNA levels in head extracts were determined by quantitative RT-PCR (see Materials and Methods). For each experiment, the averaged normalized values of both mutant and control genotypes were expressed as a percentage of the maximum value set to 100. Means of two independent experiments are reported in the graphs. Error bars indicate the range between the two experimental values.
The Control of Phosphorylated TIM by CUL-3 Does Not Require PER
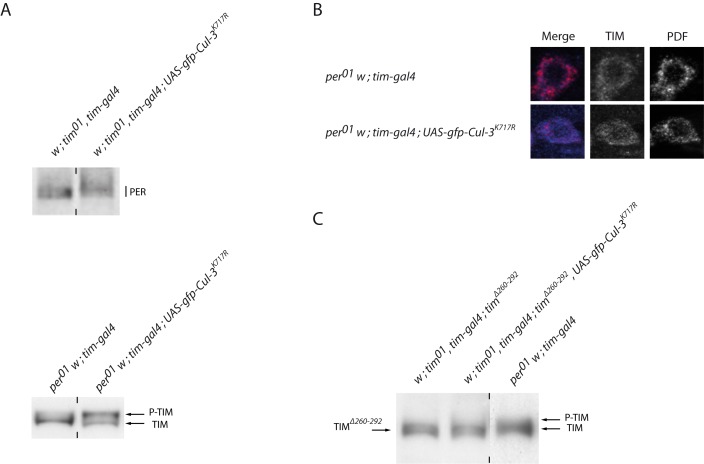
To understand how CUL-3 controls PER and TIM proteins, we analyzed PER in tim0 tim>Cul-3K717R flies and TIM in per0 tim>Cul-3K717R flies (Figure 5). TIM is not required for PER phosphorylation, but highly phosphorylated PER does not accumulate in tim0 mutants [47]. We observed a small increase of PER molecular weight in tim0 tim>Cul-3K717R flies compared to tim0 controls, indicating that CUL-3 could affect PER in the absence of TIM (Figure 5A, top). The increase in PER molecular weight was slightly higher in a tim+ background, when PER is hypophosphorylated in the control (CT12 and CT36 in Figure 3A and CT12 in Figure 6A). This suggested that TIM could favor the accumulation of phosphorylated PER in Cul-3 mutants.
Figure 5. PER and TIM proteins in per or tim mutants expressing CUL-3K717R.
Flies were entrained for 3 d in LD, transferred to DD, and collected at CT27. (A) anti-PER (top) and anti-TIM (bottom) Western blots of head extracts in a tim0 and per0 backgrounds, respectively. (B) Optical section of an individual s-LNv immunolabeled for TIM (blue) and PDF (red) in a per0 background. (C) Anti-TIM Western blot of head extract in a tim0 timΔ260–292 background. Vertical dashes indicate that the two surrounding slots were not contiguous on the gel (A) or that a different exposure was used (C) (shorter exposure for per0 extracts, which have more TIM). At least two independent experiments were done for each assay with very similar results.
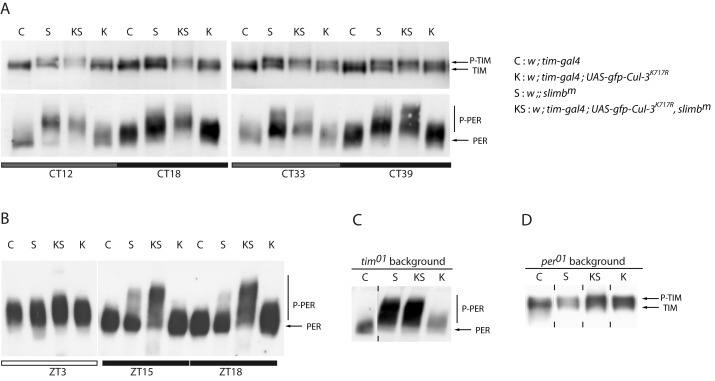
Figure 6. PER and TIM proteins in flies Cul-3K717R and slmbm flies.
(A) Anti-TIM (top) and anti-PER (bottom) Western blots of head extracts. Flies were entrained 3 d in LD and transferred to DD for collection during the first and second day of DD. Gray and black bars indicate subjective day and subjective night, respectively. (B) Anti-PER Western blots of head extracts. Flies were entrained 3 d in LD and collected the fourth day. White and black bars indicate day and night, respectively. (C, D) Anti-PER (C) and anti-TIM (D) Western blots of head extracts tim0 and per0 backgrounds, respectively. Flies were entrained for 3 d in LD and transferred to DD for collection at CT27. Vertical dashes indicate that the two surrounding slots were not contiguous on the gel. Two to three independent Western blots were done for each condition with very similar results.
As described previously [48], TIM remained mostly unphosphorylated in per0 flies (Figure 5A, bottom). In contrast, per0 tim>Cul-3K717R flies showed accumulation of phosphorylated TIM, thus indicating that CUL-3 does not require PER to induce destabilization of phosphorylated TIM. This result thus supports TIM as being phosphorylated and degraded in per0 flies, in the presence of CUL-3. Interestingly, per0 tim>Cul-3K717R flies had a higher phosphorylated/unphosphorylated TIM ratio than per+ tim>Cul-3K717R flies (see CT 12 in Figures 3A and 6A), suggesting that the presence of PER partly inhibits CUL-3 action on TIM. It has been reported that hypo-phosphorylated TIM is mostly cytoplasmic in per0 flies [49]. Since CUL-3 downregulation affected TIM in the absence of PER, CUL-3 could act in the cytoplasmic compartment. Indeed, TIM immunolabeling was mainly cytoplasmic in both per0 and per0 tim>Cul-3K717R flies, although some faint nuclear labeling was observed in the mutant (Figure 5B). This supported the hypothesis that CUL-3 is able to control the accumulation of phosphorylated TIM in the cytoplasm.
We then asked whether a tim deletion affecting TIM phosphorylation and stability would affect CUL-3-dependent stabilization. tim0 timΔ260–292 flies carry a tim transgene with a 32 aa deletion in a conserved N-terminal region [50]. This deletion removes a serine-rich region that contains putative phosphorylation sites and TIMΔ260–292 migrates faster than TIM in a per0 background on gel electrophoresis [14]. No effect of CUL-3 downregulation could be observed in tim0 timΔ260–292 flies, indicating that the TIMΔ260–292 protein is insensitive to CUL-3 control as opposed to the unphosphorylated TIM that accumulates in per0 flies (Figure 5C).
CUL-3 and SLMB Stabilize Phosphorylated TIM through Different Pathways
In DD, both PER and TIM are stabilized in slmbm mutants, which do not have detectable SLMB protein [22]. We thus asked whether decreasing CUL-3 activity in slmbm mutants would show additional effects on TIM stability. In the presence of both PER and TIM (Figure 6A), slmbm and tim>Cul-3K717R flies showed a progressive increase of phosphorylated TIM in DD, leading to an equivalent amount of hypo-phosphorylated and phosphorylated protein at day 2 (CT 33 and 39). However, slmbm mutants showed a slightly higher TIM molecular weight than tim>Cul-3K717R flies. In the double mutants, a progressive loss of unphosphorylated TIM was observed, leading to a large excess of phosphorylated protein at day 2, migrating similarly to the high molecular weight TIM of slmbm mutants. Moderately phosphorylated PER accumulated in the tim>Cul-3K717R flies, whereas hyper-phosphorylated protein was observed in both slmbm and double mutants, with even higher forms progressively accumulating in the double mutants. To confirm this interaction, PER was analyzed in LD (Figure 6B), where SLMB has limited effects on PER cycling [22]. At ZT3, only a small increase of PER levels was observed in slmbm and tim>Cul-3K717R flies, confirming that the downregulation of the two ubiquitin ligases was for a large part compensated by light, although PER was a bit more phosphorylated in the double mutant. At night, moderately phosphorylated PER accumulated in tim>Cul-3K717R flies, higher forms of phosphorylated PER were observed in slmbm, and very highly phosphorylated protein accumulated in the double mutant. Our data thus indicated that SLMB and CUL-3 had additive or synergistic effects on PER and TIM phosphorylation. Furthermore, slmbm mutants appeared to stabilize higher forms of both PER and TIM compared to tim>Cul-3K717R flies.
To better understand the respective roles of SLMB and CUL-3, we compared the effects of the two mutants on PER and TIM in tim0 and per0 backgrounds, respectively. Whereas tim0 tim>Cul-3K717R flies showed a mild increase of PER phosphorylation, slmbm (see also [22]) and double mutants accumulated large amounts of similar highly phosphorylated PER when associated with a tim0 mutation (Figure 6C). The absence of TIM thus enhanced the effect of slmbm mutants on PER, revealing that SLMB-dependent PER degradation is decreased by TIM. Since no difference was observed between slmbm and the double mutant in a tim0 background, it indicated that CUL-3 requires TIM to show synergistic effects with SLMB on the PER protein. In the absence of PER (Figure 6D), slmbm mutants accumulated less phosphorylated TIM than in a per+ background, revealing that SLMB-dependent TIM degradation is increased by PER. Since similarly high levels of phosphorylated TIM accumulated in both per0 tim>Cul-3K717R and per0 slmbm tim>Cul-3K717R mutants, it indicated that SLMB requires PER to show additive or synergistic effects with CUL-3 on the TIM protein.
Different Forms of TIM Make Complexes with CUL-3 and SLMB
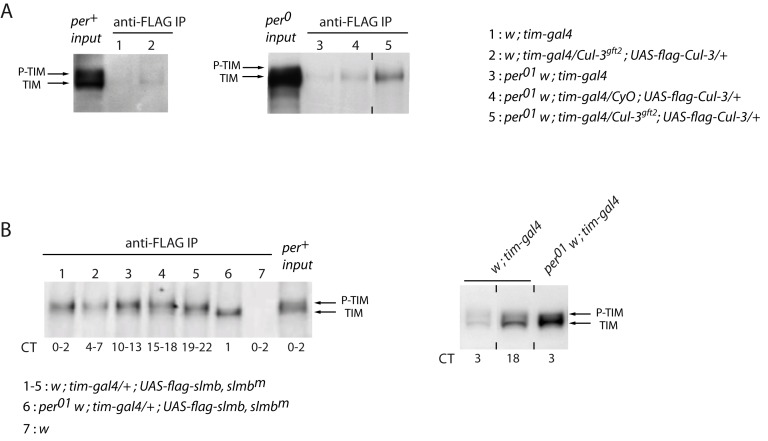
Since TIM appears to be the main target of CUL-3, we asked whether the two proteins could be co-immunoprecipitated from head extracts of flies expressing a FLAG-tagged version of CUL-3. A BTB-domain protein is likely to provide the substrate-binding component of the CUL-3 complex and indirect interactions would be predicted. We generated tim>flag-Cul-3 flies in a heterozygous Cul-3 mutant background to reduce competition between endogenous and tagged CUL-3 proteins for immunoprecipitation assays. Anti-FLAG immunoprecipitated extracts from such flies showed a faint TIM band that corresponded to hypo-phosphorylated TIM (Figure 7A, left). Since CUL-3 effect on TIM was higher in the absence of PER, a per0 background would be expected to favor TIM-CUL-3 interactions. Indeed, a stronger hypo-phosphorylated TIM band was detected in per0 flies, with TIM amount being further increased in heterozygous Cul-3gft2 mutants (Figure 7A, right). The results thus indicated that CUL-3 and hypo-phosphorylated TIM associate in complexes that are more abundant in the absence of PER. We then compared the properties of TIM/CUL-3 complexes and TIM/SLMB complexes. Immunoprecipitation of tagged SLMB was done in slmbm flies where tagged SLMB was expressed under tim-gal4 control to reduce competition between FLAG-SLMB and the endogenous protein. SLMB co-immunoprecipitated more phosphorylated TIM than hypo-phosphorylated TIM in per+ flies, but also co-immunoprecipitated hypo-phosphorylated TIM in a per0 background (Figure 7B, left). However, the amount of TIM/SLMB complexes did not increase in per0 flies, where much higher levels of TIM are observed (Figure 7B, right). In contrast to TIM/CUL-3 complexes, TIM/SLMB complexes thus mostly involve phosphorylated TIM. These complexes are favored in the presence of PER, suggesting that they involve PER-bound TIM.
Figure 7. TIM interactions with CUL-3 and SLMB.
(A) Anti-CUL-3-FLAG immunoprecipitation from head extracts followed by anti-TIM immunoblotting in per+ (left) and per0 (right) backgrounds. Slots 1 and 3 are negative controls (faint band reveals low non-specific anti-FLAG background), and 50 µg of total proteins were used for per+ and per0 input extracts. Flies were entrained 3 d in LD and transferred to DD for collection at CT15. (B) Left: anti-FLAG-SLMB immunoprecipitation from head extracts collected at different time points, followed by anti-TIM immunoblotting. Two consecutive time points were pooled for IPs in a per+ background. Slot 7 is the negative control, and 50 µg of extracts were used for per+ input. Right: anti-TIM Western blot of head extracts of w; tim-gal4 (50 µg) and per0 w; tim-gal4 (25 µg) indicate that TIM levels are much higher in a per0 background. At least two independent experiments were done for each assay with very similar results.
Discussion
The control of PER and TIM oscillations in the Drosophila clock relies for a large part on post-translational modifications of the two proteins. PER stability depends on the SLMB-containing SCF ubiquitin ligase complex and requires PER phosphorylation by DBT [11],[22]–[24]. Although TIM stops cycling in slmbm mutants [22], the circadian control of TIM stability is poorly understood. We have shown here that in addition to SLMB, CUL-3-based ubiquitin ligase complexes play a major role in the control of PER and TIM cycling, with TIM appearing to be the major target of CUL-3 effects. The results indicate that SLMB and CUL-3 differently control the stability of phosphorylated TIM and support opposite effects of PER on the two TIM degradation pathways.
In both LD and DD conditions, Pdf>Cul-3RNAi and tim>Cul-3K717R flies show a nice correlation between dampened PER/TIM oscillations in the PDF neurons and the loss or reduction of morning activity in LD as well as free running rhythms in DD. Since light strongly induces TIM degradation, it likely counteracts the effect of CUL-3 downregulation on TIM cycling and makes difficult the analysis of CUL3 function in LD. The apparently normal TIM oscillations observed in the LNds in LD correlates with the presence of evening activity but contrasts with the dampened oscillations of the sLNvs. It suggests that light-induced TIM degradation is more efficient in the LNds than in the sLNvs, as recently reported in an early night short light pulse paradigm [51], although we cannot exclude that CUL-3 function is more important in the PDF cells. Head extracts of tim>Cul-3K717R flies in LD also show robust PER and TIM oscillations, but the lower expression of tim-gal in the eye likely explains the weaker effects in head extracts compared to PDF cells.
In DD, PER and TIM oscillations are both affected in flies with downregulated CUL-3. However, the dampening of TIM oscillations occurs faster (Figure 3A,B). In addition, limited effects of CUL-3 donwregulation were observed on PER, especially in tim0 flies, whereas a strong effect was observed on TIM and it was increased in a per0 background. Finally, TIM-CUL-3 complexes could be isolated and were more abundant in per0 extracts. These results support TIM as the main target of CUL-3 circadian function, with a major part of the effects on PER being a consequence of TIM modifications. The presence of phosphorylated TIM at all time points in flies expressing dominant negative CUL-3 suggests a post-translational effect of CUL-3 on TIM. tim>Cul-3K717R flies also show tim mRNA changes, but the kinetics of CUL-3 effects on tim mRNA levels and TIM protein suggests that the protein is first affected. The strong effect of CUL-3 downregulation on TIM protein in the absence of PER also makes unlikely the possibility that CUL-3 acts on the circadian control of CLK-CYC-dependent transcription. Finally, a decrease of per and tim mRNA peak levels is the expected consequence of more phosphorylated PER at the end of the day.
Since phosphorylated TIM accumulates in tim>Cul-3K717R flies, CUL-3 downregulation could either enhance TIM phosphorylation or stabilize phosphorylated TIM. The putative ubiquitin ligase function of CUL-3 rather favors a direct role in the control of TIM levels in the mutants. This would be in agreement with the presence of TIM and CUL-3 in protein complexes, but only the identification of the substrate-recognition protein of the complex will allow us to test for direct interactions. The present data do not exclude that CUL-3 destabilizes an associated TIM kinase. Since no changes in SGG or CK2β levels were observed in tim>Cul-3K717R flies (unpublished data), more subtle modifications would have to be involved or a yet uncharacterized TIM kinase would have to be destabilized by CUL-3. We thus believe that the simplest interpretation is a direct control of TIM by CUL-3. In this hypothesis, CUL-3 could either control the stability of phosphorylated TIM or control the stability of hypo-phosphorylated TIM, which would then be stabilized and subsequently phosphorylated more extensively. For example, accumulation of phosphorylated PER has been reported in long period dbt mutants [5],[52], where defective phosphorylation of the SLMB-binding site induces the subsequent accumulation of normally unstable phosphorylated forms [24]. Why does phosphorylated TIM not accumulate in Clk>Cul-3RNAi flies? Since they only show a 2-fold decrease of Cul-3 mRNA in head extracts, they are likely to display a weak phenotype compared to tim>Cul-3K717R flies. The absence of phosphorylated TIM in the RNAi flies would thus support CUL-3 acting on several forms of TIM, with phosphorylated TIM being more sensitive to degradation than hypo-phosphorylated TIM.
Even if TIM phosphorylation facilitates CUL-3-controlled degradation, our experiments suggest that CUL-3 preferentially targets TIM proteins that are less phosphorylated than those targeted by SLMB. First, the Co-IP experiments in per+ flies indicate that CUL-3 preferentially binds to hypo-phosphorylated TIM (Figure 7A), whereas SLMB preferentially interacts with phosphorylated TIM (Figure 7B). In addition, TIM-CUL-3 complexes are more abundant in per0 flies, where TIM is hypo-phosphorylated and cytoplasmic, whereas TIM-SLMB complexes are similarly abundant. Second, the absence of PER enhances the effects of CUL-3 on TIM (Figures 5C and 6D), suggesting that CUL-3 preferentially targets unbound TIM protein, which is mostly present in the evening when TIM is less phosphorylated [48]. In contrast, the absence of PER reduced the accumulation of phosphorylated TIM in slmbm mutants (Figure 6D), suggesting that SLMB preferentially targets PER-bound TIM protein. Most importantly, higher forms of phosphorylated TIM accumulate in slmbm and double mutants compared to Cul-3K717R flies (Figure 6A). This suggests that highly phosphorylated TIM is still degraded when CUL-3 is downregulated, while it is not degraded in the absence of SLMB. The results thus favor a model where CUL-3 plays a major role in the control of free less phosphorylated TIM, whereas SLMB appears more important to control PER-bound highly phosphorylated TIM. The accumulation of PER also depends on both TIM-dependent and TIM-independent effects. Both CUL-3 and SLMB play a role in PER degradation in the absence of TIM, but TIM increases CUL-3-controlled degradation, whereas it decreases SLMB-controlled degradation.
CUL-3 complexes recognize substrates through a BTB-domain [53], and the ROADKILL (RDX) protein has been shown to target the CI transcription factor [34],[35]. Expression of several rdx RNAi constructs in the clock cells did not show any behavioral phenotype (unpublished data), suggesting that TIM is targeted by another member of the large BTB-domain protein family. A recent study has revealed that the BTB domain protein encoded by the insomniac gene is involved in the control of sleep by specifically affecting sleep bout duration, but insomniac does not appear to have a circadian function [54]. The importance of serine-rich regions as putative degrons has been revealed for the degradation of CI by the CUL-3/RDX complex [55]. Several serine-rich regions are present in TIM [14] and might thus contain target sites for CUL-3 ubiquitin ligase complexes. The fast migration of TIMΔ260–292 in both Cul-3+ and Cul-3K717R flies suggests that no phosphorylation can occur on this form of TIM or that phosphorylated TIMΔ260–292 cannot accumulate even with downregulated CUL-3. The absence of putative phosphorylation sites in TIMΔ260–292 [14] may suggest that such sites are required to generate CUL-3-sensitive TIM proteins. At the end of the cycle, TIM is degraded before PER [56], suggesting that TIM degradation may occur within the nuclear PER/TIM complex. Our results rather support SLMB as the major player of this late night nuclear degradation, as it is for PER. SLMB acts on PER through an atypical SLMB-binding site whose progressive phosphorylation during the night increases its affinity for the ubiquitin ligase [24]. No canonical SLMB recognition site was observed in TIM, but the loose conservation of such motifs [24],[57] leaves room for the occurrence of non-canonical sites in the TIM protein. A detailed analysis of TIM phosphorylation sites will be required to understand how the phosphorylation of the protein determines its destabilization by CUL-1 and CUL-3 complexes.
Although both CUL-1 and CUL-3 complexes might control TIM indirectly, the possibility that they share the work to control TIM ubiquitination is interesting. For example, CUL-1/SLMB controls the proteolysis of highly phosphorylated CI, whereas CUL-3/RDX appears to control the degradation of a less phosphorylated form of the protein (see [35]). The preference of CUL-3 for complexing with hypo-phosphorylated TIM and the one of SLMB for targeting more phosphorylated forms suggests that similar mechanisms may apply to TIM. In the mammalian clock, CRY has a function similar to TIM by acting within PER/CRY complexes to repress CLK/BMAL1-dependent transcription [58]. It has been shown that the SLMB ortholog βTrCP controls PER1/2 stability [59]–[61], whereas another F-box ubiquitin ligase, FBXL3, controls CRY stability [62]–[64]. It will be interesting to see whether the two ubiquitin ligases also show preferences for different phosphorylation levels of the clock components.
Materials and Methods
Fly Strains and Constructs
The UAS-Cul-3RNAi line carries two independent insertions (CG11861-R1 and -R2) of the same RNAi construct, which are described at http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp. The Cul-3gft2 loss-of-function allele has been described in [32], UAS-Cul-3ΔC in [44], and UAS-flag-Cul-3 and UAS-flag-Cul-3K717R in [65]. The UAS-gfp-Cul-3K717R and UAS-gfp-Cul-3 have been described in [43], with UAS-gfp-Cul-3 expression shown to rescue Cul-3 mutant cellular clones [43]. Pdf-gal4 [66], gal1118 [41], cry-gal4-39 [67], Clk-gal4-6/1 [40], tim-gal4-62 [39], cry-gal80 [38], and timΔ260–292 [50] flies have been previously reported. slmbm (slmb8 hs-slmb) adults were produced by providing hs-slmb expression with daily heat-shocks during development, as described in [22]. tim-gal4 and tim-gal4 UAS-Cul-3K717R flies were in an s-tim background (see [26]). For generating w;;UAS-flag-slmb flies, the slmb ORF was amplified from cDNA LD08669 (http://flybase.org/reports/FBcl0155568.html) and cloned into the pENTR/D-TOPO vector (Invitrogen). The ORF was recombined into pTFMW (https://dgrc.cgb.indiana.edu/vectors/store/vectors.html) containing a UASt promoter and N-terminal 3xFLAG and 6xMyc tags using LR clonase (Invitrogen). The construct was transformed into w1118 embryos using standard procedures and inserts on chromosomes 2 and 3 were selected. Line 23 was used for the anti-FLAG IP experiments.
Behavioral Analysis
Experiments were carried out with 1- to 7-d-old males at 25°C, except otherwise indicated, in Drosophila activity monitors (TriKinetics). Light was provided by standard, white, fluorescent, low-energy bulbs. Flies were entrained in 12 h∶12 h LD cycles for 4 d and then transferred to DD. Actograms are double-plotted graphs representing the absolute activity levels for each 0.5-h interval, averaged over the total number of flies of a given genotype. LD analysis was done on days 2–4 and DD analysis on days 6–12. Data analysis was done with FaasX software 1.16, which is derived from the Brandeis Rhythm Package (see [68]) and is freely available upon request (Apple Mac OSX only). For LD data, histograms represent the distribution of the activity through 24 h, in 0.5-h intervals, averaged for the total number of flies over three LD cycles. The anticipation phase score is defined as the percentage of averaged activity in the 6 h before the lights-on (or lights-off) transition that occurs in the 3 h before the transition [69]. For DD data, rhythmic flies were defined by χ2 periodogram analysis of a 7 d dataset with the following criteria (filter ON): power ≥20, width ≥2 h, with no selection on period value. Power and width are the height and width of the periodogram peak, respectively, and give the significance of the calculated period. Experiments were reproduced two or three times with very similar results.
Immunolabelings
Experiments were done on whole-mounted brains as previously described [41]. Primary antibodies were rabbit anti-PER [70] at 1∶15,000, rat anti-TIM [22] at 1∶10,000, and mouse anti-PDF (Developmental Studies Hybridoma Bank) at 1∶50,000. Secondary goat antibodies were FP546-conjugated anti-rabbit (Interchim) at 1∶2,000, Alexa 647–conjugated anti-rat (Molecular Probes) at 1∶5,000, and Alexa 488–conjugated anti-mouse (Molecular Probes) at 1∶2,000. Fluorescence signals were analyzed with a Zeiss AxioImager microscope with an ApoTome structured illumination module. Fluorescence intensity of individual cells was quantified from digital images of single confocal plans with the NIH ImageJ software. We calculated a fluorescence index I = 100n(S−B)/4B, which gives the fluorescence percentage above background (where n is the number of labeled cells among the four PDF-expressing s-LNvs, or the three CRY-expressing LNds, S is fluorescence intensity, and B is average intensity of the region adjacent to the positive cell). Index values were then averaged over 12–20 brain hemispheres for each time point.
Quantitative RT-PCR
Total RNA were prepared from adult heads (about 35) or larvae (3 to 5) using the Promega SV total RNA isolation system. They were quantified using the Nanodrop ND-1000 spectrophotometer and the integrity of the RNA was verified using the Agilent 2100 bioanalyser with the eukaryote total RNA Nano assay. 1 µg of total RNA was reverse-transcribed in a 50 µl final reaction in presence of 0.4 µm oligodT(15), 8 mM dNTP, 40 units of RNasine, and 400 units of M-MLV RTase H-minus (Promega), during 3 h at 37°C. Quantitative PCR was performed with a Roche LightCycler using the SYBR green detection protocol of the manufacturer. 5 µl of a 25 times diluted cDNA were mixed with FastStart DNA masterPLUS SYBR green I mix with 500 nM of each primer, the reaction mix was loaded on the capillaries and submitted to 40 cycles of PCR (95°C, 15 s; 60°C, 10 s; 72°C, 20 s), followed by a fusion cycle in order to analyze the melting curve of the PCR products. Negative control without the RTase was introduced to verify the absence of genomic DNA contaminants. Primers (Table S2) were defined within exons using the PrimerSelect program of the Lasergene software (DNAstar). BLAST searches were performed to confirm gene specificity and the absence of multi-locus matching at the primer site. The amplification efficiencies of each pair of primers were generated using the slopes of the standard curves obtained by a four 10-fold dilution series for tubulin or an eight 2-fold dilution series for the other genes. All experimental points fell within the linear portion of the curves. The efficiency of the q-PCR amplifications for all pairs of primers is indicated in the table. Amplification specificity for each q-PCR reaction was confirmed by the dissociation curve analysis. Determined Ct values (Table S2) were then used for quantification, with the tubulin gene as reference. For each biological replicate (at least two independent samples), measurement was made at least in duplicate (technical replicate).
Western Blots and Immunoprecipitations
Protein extracts were made from frozen heads homogenized in ice-cold HE extraction buffer (20 mM hepes pH 7.5, 0.1 M KCl, 10 mM EDTA), supplemented with 20 mM glycerophosphate, 0.1 mM DTT, phosphatase inhibitor cocktails 1 (0.5%) and 2 (1%) (Sigma), and protease inhibitor tablets (Roche) according to the manufacturer's instructions. Anti-FLAG immunoprecipitations were performed using EZview Red anti-FLAG affinity gel (Sigma) according to the manufacturer's instructions. For immunoprecipitations, 1 mg of total head proteins were extracted in supplemented HE buffer, and DTT was replaced by 0.1% NP40. For SDS-PAGE, 50 µg of extracts (or immunoprecipitated proteins from 1 mg of extracts) were separated on 3%–8% Tris-acetate (TIM) or 4% Tris-glycine (PER and TIM in Figure 3B) gels (Invitrogen) and transferred to PVDF membranes. Immunoblotting was performed as described previously [22]. Primary antibodies were rabbit anti-PER [70] at 1∶10,000 and rat anti-TIM [22] at 1∶2,000. HRP-conjugated secondary antibodies (Santa Cruz biotechnology) were anti-rabbit goat antibody (1∶40,000) and anti-rat goat antibody (1∶50,000).
Supporting Information
Locomotor activity of flies expressing Cul-3RNAi under gal1118 control. Flies were entrained for 5 d in LD 12∶12 and then transferred to DD. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5 h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated two times with very similar results.
(PDF)
Cul-3 mRNA levels in flies expressing Cul-3RNAi. Quantitative RT-PCR was done as described in Materials and Methods, from either adult heads (top) or L3 larvae (bottom) cDNA with Cul-3 primers E7-E9. da-gal4 (daughterless) is a ubiquitously expressed gal4 driver, and da-gal4/+ ; UAS-Cul-3RNAi/+ flies were lethal after the larval stages. For each experiment, averaged normalized values of the mutant and control genotypes were expressed as a percentage of the maximum value set to 100. Means of three (top) or five (bottom) independent experiments are reported in the graphs. Error bars indicate s.e.m. Two other sets of Cul-3 primers were tested and gave similar results: Cul-3 mRNA levels were decreased to 46% (E7-E8 primers) or 45% (E3-E4 primers) of the control levels in Clk-gal4 ;; UAS-Cul-3-RNAi heads, and to 28% (E7-E8 primers) or 22% (E3-E4 primers) of the control levels in da-gal4/+ ; UAS-Cul-3-RNAi/+ larvae.
(PDF)
Locomotor activity of flies expressing flag-Cul-3K717R, Cul-3ΔC, or gfp-Cul-3 transgenes. Flies were entrained for 4 d in LD 12∶12 and then transferred to DD. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5 h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated two or three times with very similar results.
(PDF)
Locomotor activity of Cul-3 downregulated flies at different temperatures. Flies expressing Cul-3 RNAi and controls were grown at 25°C, and the adults were then either transferred at 20°C or kept at 25°C for 4 d in LD 12∶12 followed by DD. 25°C data are those already shown in Figure 1A. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5-h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated twice with very similar results.
(PDF)
Morphology of PDF-positive s-LNvs in Cul-3 mutants and controls. Stacks of optical sections from adult brains immunolabeled with anti-PDF. Short arrow indicates slightly more defasciculated projections, and long arrow indicates reduced arborization in the medulla. Flies with a homozygous tim-gal4 insertion very often show some additional PDF-positive fibers that appear to derive from the Posterior Optic Tract (arrowheads). Scale bars, 50 µM.
(PDF)
TIM immunoreactivity in the LNs of tim>Cul-3K717R flies. Flies were entrained for 3 d and collected the fourth day of LD at ZT12, 16, or 20. Graphs represent quantifications of TIM immunolabeling in the four “morning” PDF-positive s-LNvs and the three “evening” CRY-positive LNds of tim>Cul-3K717R flies and controls. Error bars indicate s.e.m. Experiments were repeated twice with very similar results.
(PDF)
TIM Western blot of head extracts of flies expressing CUL-3K717R and controls. Flies were entrained for 3 d in LD and collected every 3 h the fourth day of LD. White and black bars indicate day and night, respectively. ZT is Zeitgeber time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated. Two independent Western blots were done for each genotype with very similar results.
(PDF)
Quantification of PER and TIM in head extracts of flies expressing CUL-3K717R. Phosphorylated (P-) and hypo-phosphorylated forms of PER and TIM were quantified with the Gel Analyzing Tool of Image J software (NIH), which compares the signal density and background of each track. Average values of at least three independent Western blots were used for each genotype/condition. The results are normalized to the maximum value of each blot, set to 100. Error bars indicate s.e.m. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time.
(PDF)
TIM protein in head extracts of flies expressing another Cul-3K717R transgene and controls. Flies were entrained for 3 d in LD and transferred to DD for collection in the second day of DD. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated.
(PDF)
Low mobility TIM observed in tim>Cul-3K717R flies disappears after phosphatase treatment. Flies were entrained for 3 d in LD, transferred to DD, and collected the second day at CT39. Head extracts were treated (+) or not (−) with L-Phosphatase for 30 mn at 30°C and then subjected to electrophoresis and Western Blot analysis with anti-TIM antibodies. Untreated samples stored at 4°C were used to estimate endogenous phosphatase activities, which are not inhibited in the extraction buffer. Phosphatase treatment: protein extracts were made in Lysis Phosphatase buffer (10 mM hepes pH 7.5, 0.1 M KCl, 0.1 mM EDTA, 5% glycerol, 0.1% Triton X-100, 5 mM DTT, 1 mM Mncl2, protease inhibitor tablets) and incubated with 1,000 units of L-phosphatase (New England Biolabs) for 30 mn at 30°C. The experiment was repeated twice with very similar results.
(PDF)
TIM protein in head extracts of flies overexpressing CUL-3 or CUL-3K717R protein and controls. Flies were entrained for 3 d in LD and transferred to DD for collection in the second day of DD. Top: anti-TIM Western blot. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated. Two independent experiments were done with similar results for CUL-3 overexpression. A CUL-3K717R blot is shown here for comparison (see Figure 3 for another blot and Figure S8 for quantification). Bottom: quantification of Phosphorylated (P-) and hypo-phosphorylated forms of TIM in flies overexpressing CUL-3 and controls. Bars represent the higher and lower value for each time point in the two independent experiments.
(PDF)
Anticipation phase score of fly group activity preceding Lights-ON/Lights-OFF transitions in flies with altered CUL-3 activity. Corresponding activity graphs with activity levels and number of flies are shown in Figures 1, S1, S3, and S4. Anticipation phase score is defined in the Materials and Methods section and is given ± s.e.m. Genotypes with altered CUL-3 activity show no (50%) or low (<60%) morning anticipation compared to controls.
(DOCX)
Primers specifications (see Materials and Methods). Efficiency, DNA concentration ratio between cycles n+1 and n (1<E<2); R 2, coefficient of determination of the calibration curve; Ct, Cycle threshold. The Ct values interval defines the linear range of the standard curve for each pair of primers.
(DOCX)
Acknowledgments
We thank C. T. Chien, H. Mistry, A. Sehgal, and the Bloomington stock center for fly strains, as well as R. Stanewsky for anti-PER antibody. We address special thanks to M. Boudinot for the FaasX software, to T. Legnaioli for his participation in preliminary experiments, and to R. Ueda and T. D. Murphy for the generous gift of unpublished cul-3 RNAi flies and flag-slmb flies, respectively. We also thank E. Jaquet (IMAGIF) for expert advice on quantitative PCR, J-P. Bouillot and L. Collet for help with digital imaging, and S. Bouleau, A. Klarsfeld, A. Lamaze, and A. Szabo for their critical reading of the manuscript.
Abbreviations
- CI
CUBITUS INTERRUPTUS
- CLK
CLOCK
- CRY
CRYPTOCHROME
- CT
circadian time
- CUL-3
CULLIN-3
- CYC
CYCLE
- DBT
DOUBLE TIME
- DD
constant darkness
- HH
HEDGEHOG
- JET
JETLAG
- LD
light-dark
- LNds
dorsal lateral neurons
- LNs
lateral neurons
- PER
PERIOD
- RDX
ROADKILL
- SGG
SHAGGY
- SLMB
SUPERNUMERARY LIMBS
- s-LNvs
small ventral lateral neurons
- TIM
TIMELESS
- ZT
Zeitgeber time
Funding Statement
This work was supported by Agence Nationale de la Recherche “DrosoClock” and “ClockGene”, “Equipe FRM” program of Fondation pour la Recherche Médicale, European Union 6th Framework Programme “EUCLOCK”. AD was supported by Ministère de l'Enseignement Supérieur et de la Recherche and FR by Institut National de la Santé et des Etudes et Recherches Médicales. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allada R, Chung BY (2010) Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hardin PE (2011) Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet 74: 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, et al. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- 5. Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, et al. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- 6. Kloss B, Rothenfluh A, Young MW, Saez L (2001) Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the drosophila clock. Neuron 30: 699–706. [DOI] [PubMed] [Google Scholar]
- 7. Nawathean P, Rosbash M (2004) The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13: 213–223. [DOI] [PubMed] [Google Scholar]
- 8. Kim EY, Ko HW, Yu W, Hardin PE, Edery I (2007) A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27: 5014–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nawathean P, Stoleru D, Rosbash M (2007) A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol 27: 5002–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL (2007) Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27: 8049–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiu JC, Ko HW, Edery I (2011) NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akten B, Jauch E, Genova GK, Kim EY, Edery I, et al. (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 16: 251–257. [DOI] [PubMed] [Google Scholar]
- 13. Lin J-M, Kilman V, Keegan K, Paddock B, Emery-Le M, et al. (2002) A role for casein kinase 2a in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- 14. Meissner RA, Kilman VL, Lin JM, Allada R (2008) TIMELESS is an important mediator of CK2 effects on circadian clock function in vivo. J Neurosci 28: 9732–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinek S, Inonog S, Manoukian AS, Young MW (2001) A role for the segment polarity gene shaggy/GSK-3 in the drosophila circadian clock. Cell 105: 769–779. [DOI] [PubMed] [Google Scholar]
- 16. Sathyanarayanan S, Zheng X, Xiao R, Sehgal A (2004) Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116: 603–615. [DOI] [PubMed] [Google Scholar]
- 17. Fang Y, Sathyanarayanan S, Sehgal A (2007) Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev 21: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim EY, Edery I (2006) Balance between DBT/CKI{varepsilon} kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A 103: 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE (2006) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu W, Zheng H, Price JL, Hardin PE (2009) DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol 29: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M (2010) Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 24: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, et al. (2002) The F-box protein SLIMB controls the levels of clock proteins PERIOD and TIMELESS. Nature 429: 178–182. [DOI] [PubMed] [Google Scholar]
- 23. Ko HW, Jiang J, Edery I (2002) A role for Slimb in the degradation of Drosophila PERIOD protein phosphorylated by DOUBLETIME. Nature 420: 673–678. [DOI] [PubMed] [Google Scholar]
- 24. Chiu JC, Vanselow JT, Kramer A, Edery I (2008) The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22: 1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koh K, Zheng X, Sehgal A (2006) JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312: 1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peschel N, Veleri S, Stanewsky R (2006) Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila's circadian clock. Proc Natl Acad Sci U S A 103: 17313–17318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peschel N, Chen KF, Szabo G, Stanewsky R (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol 19: 241–247. [DOI] [PubMed] [Google Scholar]
- 28. Knowles A, Koh K, Wu JT, Chien CT, Chamovitz DA, et al. (2009) The COP9 signalosome is required for light-dependent timeless degradation and Drosophila clock resetting. J Neurosci 29: 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakayama KI, Nakayama K (2005) Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol 16: 323–333. [DOI] [PubMed] [Google Scholar]
- 30. Jiang J, Struhl G (1998) Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature 391: 493–496. [DOI] [PubMed] [Google Scholar]
- 31. Hooper JE, Scott MP (2005) Communicating with Hedgehogs. Nat Rev Mol Cell Biol 6: 306–317. [DOI] [PubMed] [Google Scholar]
- 32. Ou CY, Lin YF, Chen YJ, Chien CT (2002) Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 16: 2403–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang J (2006) Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 5: 2457. [DOI] [PubMed] [Google Scholar]
- 34. Kent D, Bush EW, Hooper JE (2006) Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133: 2001–2010. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, et al. (2006) A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell 10: 719–729. [DOI] [PubMed] [Google Scholar]
- 36. Dubruille R, Emery P (2008) A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol 38: 129–145. [DOI] [PubMed] [Google Scholar]
- 37. Grima B, Chélot E, Xia R, Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431: 869–873. [DOI] [PubMed] [Google Scholar]
- 38. Stoleru D, Peng P, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behavior of Drosophila . Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- 39. Kaneko M, Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422: 66–94. [DOI] [PubMed] [Google Scholar]
- 40. Gummadova JO, Coutts GA, Glossop NR (2009) Analysis of the Drosophila clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J Biol Rhythms 24: 353–367. [DOI] [PubMed] [Google Scholar]
- 41. Blanchardon E, Grima B, Klarsfeld A, Chélot E, Hardin PE, et al. (2001) Defining the role of Drosophila lateral neurons in the control of circadian activity and eclosion rhythms by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci 13: 871–888. [DOI] [PubMed] [Google Scholar]
- 42. Merlet J, Burger J, Gomes JE, Pintard L (2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66: 1924–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu S, Perez R, Pan M, Lee T (2005) Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J Neurosci 25: 4189–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mistry H, Wilson BA, Roberts IJ, O'Kane CJ, Skeath JB (2004) Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech Dev 121: 1495–1507. [DOI] [PubMed] [Google Scholar]
- 45. Duffy JB (2002) GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- 46. Zeng H, Hardin PE, Rosbash M (1994) Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J 13: 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Price JL, Dembinska ME, Young MW, Rosbash M (1995) Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless . EMBO J 14: 4044–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeng HK, Qian ZW, Myers MP, Rosbash M (1996) A light-entrainment mechanism for the Drosophila circadian clock. Nature 380: 129–135. [DOI] [PubMed] [Google Scholar]
- 49. Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW (1996) Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271: 1736–1740. [DOI] [PubMed] [Google Scholar]
- 50. Ousley A, Zafarullah K, Chen YF, Emerson M, Hickman L, et al. (1998) Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics 148: 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang CH, Hinteregger E, Shang Y, Rosbash M (2010) Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron 66: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suri V, Hall JC, Rosbash M (2000) Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in drosophila. J Neurosci 20: 7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pintard L, Willems A, Peter M (2004) Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. Embo J 23: 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stavropoulos N, Young MW (2011) insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72: 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Q, Shi Q, Chen Y, Yue T, Li S, et al. (2009) Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci U S A 106: 21191–21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rothenfluh A, Young MW, Saez L (2000) A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26: 505–514. [DOI] [PubMed] [Google Scholar]
- 57. Smelkinson MG, Zhou Q, Kalderon D (2007) Regulation of Ci-SCF(Slimb) binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell 13: 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Takahashi JS, Hong HK, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shirogane T, Jin J, Ang XL, Harper JW (2005) SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem 280: 26863–26872. [DOI] [PubMed] [Google Scholar]
- 60. Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, et al. (2005) Control of mammalian circadian rhythm by CKI{varepsilon}-regulated proteasome-mediated PER2 degradation. Mol Cell Biol 25: 2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, et al. (2007) {beta}-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms 22: 375–386. [DOI] [PubMed] [Google Scholar]
- 62. Siepka SM, Yoo SH, Park J, Song W, Kumar V, et al. (2007) Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, et al. (2007) The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316: 897–900. [DOI] [PubMed] [Google Scholar]
- 64. Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, et al. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316: 900–904. [DOI] [PubMed] [Google Scholar]
- 65. Wu JT, Lin HC, Hu YC, Chien CT (2005) Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol 7: 1014–1020. [DOI] [PubMed] [Google Scholar]
- 66. Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- 67. Klarsfeld A, Malpel S, Michard-Vanhée C, Picot M, Chélot E, et al. (2004) Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci 24: 1468–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klarsfeld A, Leloup JC, Rouyer F (2003) Circadian rhythms of locomotor activity in Drosophila . Behav Processes 64: 161–175. [DOI] [PubMed] [Google Scholar]
- 69. Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN (2007) Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci 27: 12489–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, et al. (1997) Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci 17: 676–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Locomotor activity of flies expressing Cul-3RNAi under gal1118 control. Flies were entrained for 5 d in LD 12∶12 and then transferred to DD. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5 h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated two times with very similar results.
(PDF)
Cul-3 mRNA levels in flies expressing Cul-3RNAi. Quantitative RT-PCR was done as described in Materials and Methods, from either adult heads (top) or L3 larvae (bottom) cDNA with Cul-3 primers E7-E9. da-gal4 (daughterless) is a ubiquitously expressed gal4 driver, and da-gal4/+ ; UAS-Cul-3RNAi/+ flies were lethal after the larval stages. For each experiment, averaged normalized values of the mutant and control genotypes were expressed as a percentage of the maximum value set to 100. Means of three (top) or five (bottom) independent experiments are reported in the graphs. Error bars indicate s.e.m. Two other sets of Cul-3 primers were tested and gave similar results: Cul-3 mRNA levels were decreased to 46% (E7-E8 primers) or 45% (E3-E4 primers) of the control levels in Clk-gal4 ;; UAS-Cul-3-RNAi heads, and to 28% (E7-E8 primers) or 22% (E3-E4 primers) of the control levels in da-gal4/+ ; UAS-Cul-3-RNAi/+ larvae.
(PDF)
Locomotor activity of flies expressing flag-Cul-3K717R, Cul-3ΔC, or gfp-Cul-3 transgenes. Flies were entrained for 4 d in LD 12∶12 and then transferred to DD. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5 h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated two or three times with very similar results.
(PDF)
Locomotor activity of Cul-3 downregulated flies at different temperatures. Flies expressing Cul-3 RNAi and controls were grown at 25°C, and the adults were then either transferred at 20°C or kept at 25°C for 4 d in LD 12∶12 followed by DD. 25°C data are those already shown in Figure 1A. White and black/gray indicate lights-ON and lights-OFF, respectively. ZT is Zeitgeber Time (ZT0 corresponds to lights-ON). Top panels: averaged activity distribution of n flies in LD (see Materials and Methods). Dots indicate the s.e.m. of the activity for each 0.5-h interval. Average activity per 0.5-h is indicated in parentheses on the left. Bottom panels: averaged actograms during both LD and DD conditions (see Materials and Methods). Behavioral analyses were repeated twice with very similar results.
(PDF)
Morphology of PDF-positive s-LNvs in Cul-3 mutants and controls. Stacks of optical sections from adult brains immunolabeled with anti-PDF. Short arrow indicates slightly more defasciculated projections, and long arrow indicates reduced arborization in the medulla. Flies with a homozygous tim-gal4 insertion very often show some additional PDF-positive fibers that appear to derive from the Posterior Optic Tract (arrowheads). Scale bars, 50 µM.
(PDF)
TIM immunoreactivity in the LNs of tim>Cul-3K717R flies. Flies were entrained for 3 d and collected the fourth day of LD at ZT12, 16, or 20. Graphs represent quantifications of TIM immunolabeling in the four “morning” PDF-positive s-LNvs and the three “evening” CRY-positive LNds of tim>Cul-3K717R flies and controls. Error bars indicate s.e.m. Experiments were repeated twice with very similar results.
(PDF)
TIM Western blot of head extracts of flies expressing CUL-3K717R and controls. Flies were entrained for 3 d in LD and collected every 3 h the fourth day of LD. White and black bars indicate day and night, respectively. ZT is Zeitgeber time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated. Two independent Western blots were done for each genotype with very similar results.
(PDF)
Quantification of PER and TIM in head extracts of flies expressing CUL-3K717R. Phosphorylated (P-) and hypo-phosphorylated forms of PER and TIM were quantified with the Gel Analyzing Tool of Image J software (NIH), which compares the signal density and background of each track. Average values of at least three independent Western blots were used for each genotype/condition. The results are normalized to the maximum value of each blot, set to 100. Error bars indicate s.e.m. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time.
(PDF)
TIM protein in head extracts of flies expressing another Cul-3K717R transgene and controls. Flies were entrained for 3 d in LD and transferred to DD for collection in the second day of DD. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated.
(PDF)
Low mobility TIM observed in tim>Cul-3K717R flies disappears after phosphatase treatment. Flies were entrained for 3 d in LD, transferred to DD, and collected the second day at CT39. Head extracts were treated (+) or not (−) with L-Phosphatase for 30 mn at 30°C and then subjected to electrophoresis and Western Blot analysis with anti-TIM antibodies. Untreated samples stored at 4°C were used to estimate endogenous phosphatase activities, which are not inhibited in the extraction buffer. Phosphatase treatment: protein extracts were made in Lysis Phosphatase buffer (10 mM hepes pH 7.5, 0.1 M KCl, 0.1 mM EDTA, 5% glycerol, 0.1% Triton X-100, 5 mM DTT, 1 mM Mncl2, protease inhibitor tablets) and incubated with 1,000 units of L-phosphatase (New England Biolabs) for 30 mn at 30°C. The experiment was repeated twice with very similar results.
(PDF)
TIM protein in head extracts of flies overexpressing CUL-3 or CUL-3K717R protein and controls. Flies were entrained for 3 d in LD and transferred to DD for collection in the second day of DD. Top: anti-TIM Western blot. Gray and black bars indicate subjective day and subjective night, respectively. CT is circadian time. Phosphorylated (P-) and hypo-phosphorylated forms of TIM are indicated. Two independent experiments were done with similar results for CUL-3 overexpression. A CUL-3K717R blot is shown here for comparison (see Figure 3 for another blot and Figure S8 for quantification). Bottom: quantification of Phosphorylated (P-) and hypo-phosphorylated forms of TIM in flies overexpressing CUL-3 and controls. Bars represent the higher and lower value for each time point in the two independent experiments.
(PDF)
Anticipation phase score of fly group activity preceding Lights-ON/Lights-OFF transitions in flies with altered CUL-3 activity. Corresponding activity graphs with activity levels and number of flies are shown in Figures 1, S1, S3, and S4. Anticipation phase score is defined in the Materials and Methods section and is given ± s.e.m. Genotypes with altered CUL-3 activity show no (50%) or low (<60%) morning anticipation compared to controls.
(DOCX)
Primers specifications (see Materials and Methods). Efficiency, DNA concentration ratio between cycles n+1 and n (1<E<2); R 2, coefficient of determination of the calibration curve; Ct, Cycle threshold. The Ct values interval defines the linear range of the standard curve for each pair of primers.
(DOCX)