Abstract
Textbook descriptions of signal transduction complexes provide a static snapshot view of highly dynamic events. Despite enormous strides in identifying the key components of signaling complexes and the underlying mechanisms of signal transduction, our understanding of the dynamic behavior of these complexes has lagged behind. Using the example of receptor tyrosine kinases, this perspective takes a fresh look at the dynamics of the system and their potential impact on signal processing.
It is now well accepted that signal transduction depends on the specific, regulated binding of proteins to other proteins and to other cellular structures such as membrane lipids. From the pioneering work of Tony Pawson and others, we know that such interactions are often mediated by modular protein domains [1,2]. With the availability of complete genome sequences and the advent of high-throughput methods to analyze protein interactions, this insight has led to a wealth of information on various signaling pathways. From “hairball” network diagrams to detailed pathway cartoons, it would seem that we have more than enough information to understand both the general design principles and specific mechanistic details of most signaling pathways. Despite this progress, however, the dynamic behavior of individual signaling complexes remains something of a mystery. In this brief perspective, I will focus on emerging ideas on the dynamics of one of the most intensively studied classes of signal transduction complexes, those involving receptor tyrosine kinases (RTKs).
Binding of specific ligands to RTKs leads to increased activity of their cytosolic tyrosine kinase catalytic domains [3]. Similarly, nonreceptor tyrosine kinases can be activated by ligand binding to transmembrane receptors lacking intrinsic kinase activity, such as cytokine and T cell receptors and integrins. For downstream signaling, the most critical substrates of the activated kinases are generally the receptor itself, along with receptor-associated scaffold proteins. This is because the resulting phosphorylation sites serve to recruit effector and adaptor proteins that contain phosphotyrosine (pTyr) specific binding domains, predominantly Src homology 2 (SH2) domains [4,5]. The net result is the relocalization of a set of effector proteins to the receptor on the membrane, and the assembly of multi-component signaling complexes that set in motion downstream signaling cascades.
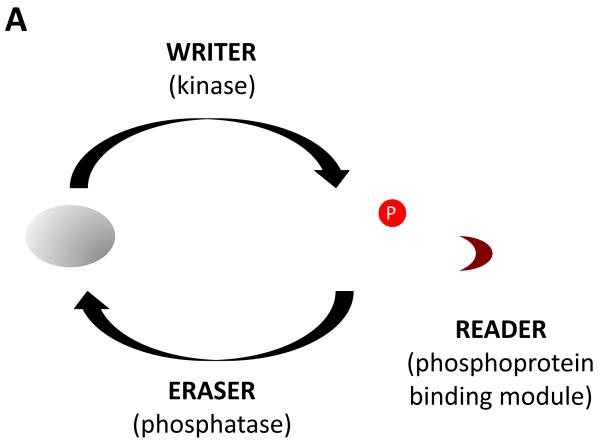
This mechanism is but one example of the many signaling systems that follow the “reader-writer-eraser” paradigm [6,7]. In these systems, signal output depends on post-translational marks laid down by writer enzymes; these marks can be removed by eraser enzymes, and are read by proteins that bind specifically to the marked sites. In the case of tyrosine kinase signaling, the writers are protein tyrosine kinases, the erasers are protein-tyrosine phosphatases, and the readers are modular pTyr binding domains such as SH2 and PTB domains [6] (Figure 1a).
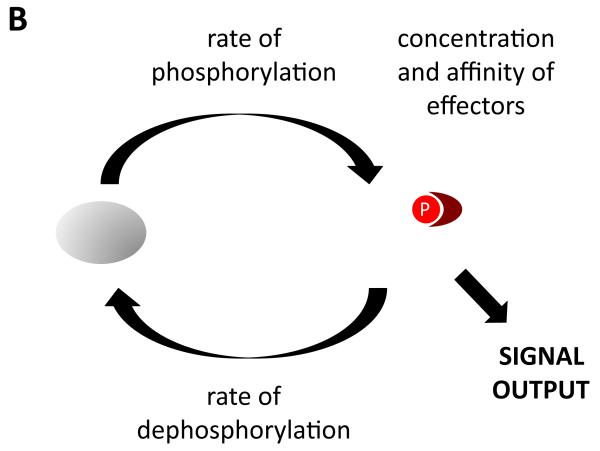
Figure 1. Regulation of signal output from receptor tyrosine kinases.
(a) Signaling is controlled by the activities of tyrosine kinases, tyrosine phosphatases, and effector proteins that can bind to tyrosine-phosphorylated sites. (b) The amount of signal output is proportional to the amount of effector complexes, which in turn depends on the rates of phosphorylation and dephosphorylation, and the local concentration and affinities of effectors.
Let’s consider the RTK system a bit more closely. Downstream signaling (signal output) depends on the amount of effectors recruited to the activated receptor and the identity of those effectors. What then actually controls output levels at any point in time? Of course the rate of tyrosine phosphorylation is important, but so too is the rate of dephosphorylation, as well as the concentration and affinity of effectors (since the levels of a bimolecular complex depend on the concentrations of the two partners and the dissociation constant, Kd, of the interaction) (Figure 1b). Typically, descriptions of RTK signaling focus on kinase activity, because this is what is known to change upon ligand binding (phosphatase activity and the concentrations of effectors are usually considered to be relatively constant, at least over the rather short time scales relevant to signaling). I will argue, however, that the actual dynamic behavior of such complexes can be understood fully only when all three factors are considered.
Cellular phosphatase activities are high
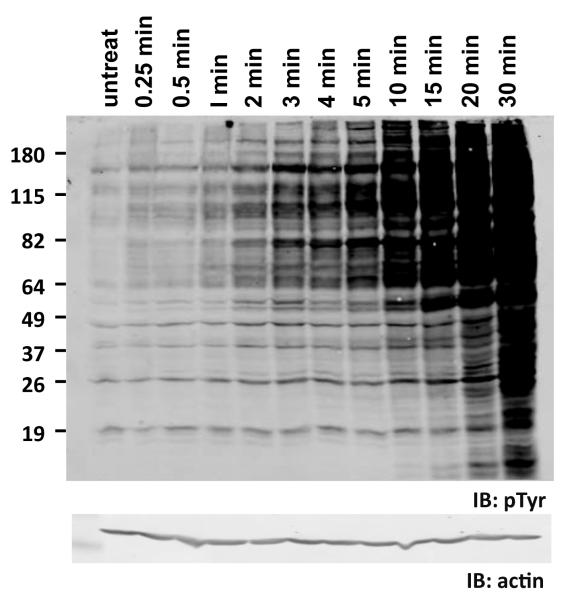
There is evidence that the activity of endogenous tyrosine phosphatases is surprisingly high. Anyone who has ever treated cells in culture with the potent tyrosine phosphatase inhibitor vanadate has seen that tyrosine phosphorylation levels increase enormously, suggesting that phosphatase activity normally holds the steady-state level of pTyr in check [8]. These effects are also quite rapid (Figure 2), suggesting that kinases must continuously fight against a strong and steady headwind of phosphatase activity. The same conclusion can be drawn from recent studies looking at the rate of dephosphorylation of an RTK, the epidermal growth factor receptor (EGFR), upon treatment of cells with EGFR-specific kinase inhibitors [9]. In these experiments the half-life of phosphorylation on receptors was found to be very short, ~15 sec. The implication of both types of studies is that steady-state rates of phosphorylation and dephosphorylation are both very high, and that sites may be phosphorylated and then dephosphorylated within a matter of seconds.
Figure 2. Constitutive rate of tyrosine dephosphorylation in cells is high.
Human 293T cells were treated with 50 micromolar pervanadate for the indicated times to inhibit cellular tyrosine phosphatases. Cell lysates were then immunoblotted with anti-phosphotyrosine antibody (pTyr). Levels of tyrosine phosphorylation rise rapidly in pervanadate-treated cells. Lysates were also immunoblotted with anti-actin as a loading control (bottom). Positions of molecular weight markers are indicated on left (in kDa).
Conceptually, this insight fundamentally alters how we view changes in tyrosine phosphorylation upon RTK activation. Currently, we tend to think that little is going on in unstimulated cells, and that ligand stimulation dramatically changes the situation by introducing an activated kinase into this quiet scene. Instead, it seems there is considerable flux through the system even in the unstimulated cell: a constant battle between spontaneously activated kinases and the phosphatases that remove their marks. Intuitively this makes little sense, because each round of phosphorylation and dephosphorylation costs an ATP molecule; high flux in the absence of specific signals would seem to be enormously wasteful. Of course, we are willing to accept other seemingly wasteful processes, such as the transcription of lengthy introns in most eukaryotic genes, so such apparent profligacy cannot be a disqualifying feature in biological systems.
Presumably, however, there must be advantages to high flux that more than make up for the waste of energy. What might these advantages be? A number of plausible scenarios can be proposed. For example, high flux is likely to make the system more dependent on dimerization and higher-order clustering, presumably helping eliminate background “noise” from spontaneously activated kinases. In the presence of high constitutive phosphatase levels, a kinase that spontaneously adopts an activated conformation in the absence of ligand would be rapidly dephosphorylated, as would any other substrates it might phosphorylate during its brief period of activity. Genuine ligand-induced signals, on the other hand, induce or stabilize receptor dimerization and often lead to higher-order clustering [3]. Furthermore, the active conformation of the catalytic domain is generally stabilized by phosphorylation in trans on the “activation loop” [10]. The proximity of multiple kinase molecules allows each activated kinase to repeatedly phosphorylate and trans-activate other kinases, and also repeatedly phosphorylate substrates in the immediate vicinity. Any kinase that happens to be dephosphorylated can be rapidly re-phosphorylated by other nearby kinases. Thus the equilibrium shifts dramatically from dephosphorylation to phosphorylation in the vicinity of the cluster. In the absence of high rates of dephosphorylation, spontaneous activity would likely lead to much higher basal (unstimulated) levels of substrate phosphorylation.
High phosphatase activity may also present advantages in terms of protein engineering. The protein tyrosine kinases evolved relatively recently, around the time when metazoans emerged [6,11,12], and may not have had the evolutionary time to “perfect” themselves, in contrast to more ancient enzymes such as those involved in metabolism. High constitutive phosphatase activity may allow tyrosine kinases to function in signaling despite imperfect control over their activity and substrate specificity. First, if constitutive phosphatase activity is high, a fairly high level of leaky activity can be tolerated; that is, the difference in average catalytic activity between unliganded and liganded RTKs can be modest. As outlined above, spontaneous activity (in absence of specific ligand) is unlikely to have serious consequences if the kinase is not clustered with other kinases. Second, substrate specificity is not absolutely crucial, since any “mistakes”—proteins phosphorylated on sites that are not useful or may even be deleterious—are quickly erased by phosphatases. Of course, this raises the question of how tyrosine-phosphorylated sites important for signaling can selectively survive the high phosphatase activity— the subject of the next section.
Effectors actively edit the cellular pTyr profile
When phosphatase rates are high in cells, the half-life of phosphorylated sites must be relatively short. It follows from this that proteins that bind particular phosphorylated sites are likely to affect their levels, by shielding them from phosphatases and thus delaying their dephosphorylation. While it has long been speculated that this might be the case, there has been remarkably little direct experimentation to confirm it. It has been shown, not surprisingly, that SH2 binding protects phosphorylated binding sites from dephosphorylation in vitro [13]. The oncogenic Crk SH2/SH3 adaptor protein, which when highly expressed leads to increased steady-state levels of its SH2 binding partners such as p130Cas and paxillin, also supports this idea [14,15]; however since Crk also binds to tyrosine kinases, a role for those kinases in increased pTyr cannot be ruled out [16-18]. Consistent with expectations, more recent experiments in my group have confirmed that overexpression of an SH2 domain such as that of Grb2 can dramatically increase the amount of phosphorylation of SH2-binding sites in cells both before and after stimulation by EGF (J. Jadwin, K. Machida and BJM, unpublished observations).
Thus is seems that effectors are not just passive readers of the post-translational marks laid down by tyrosine kinases, but actually play a much more active role in determining which sites will predominate. By the very act of reading the signal, effectors prevent the sites being read from being erased. Which readers are present (and more quantitatively, their local concentrations and on- and off-rates) determines not only the ultimate output of the signal, but also the pattern of phosphorylation that is observed.
When dephosphorylation rates are high, effector recruitment (and ultimately signal output) becomes a race between the rate of dephosphorylation and the rate of effector binding, which in turn is dependent on the local concentration of those effectors. The popular arcade game “Whac-a-Mole” provides an apt analogy (Figure 3). In this game, mechanical moles periodically pop up from one of a number of holes, then almost immediately pop back down. In order to score points, the player needs to whack the mole with a padded mallet before it disappears. The more moles whacked, the higher the score. Similarly, SH2-containing effectors in the cytosol must recognize and bind to tyrosine-phosphorylated receptors before they are dephosphorylated. The strength of the signal output (the score) depends on the number of sites that can be bound per unit time.
Figure 3.
The Whac-a-Mole game (used with permission of Bob’s Space Racers, Inc.)
Ptyr-effector complexes are highly transient
SH2 domains and other pTyr binding modules have moderate affinity and specificity; typical Kd’s for preferred phosphopeptides are in the ~100 nM range. Furthermore, when on-rates and off-rates have been examined, for example by surface plasmon resonance (SPR), the off-rates have been found to be remarkably rapid. In the case of Grb2 SH2 domain examined by SPR, the off-rate is so fast as to be unmeasurable, and could only be estimated from the Kd and on-rate to be >10 sec−1 [19,20]. In other words, the average half-life of an SH2-pTyr peptide complex in vitro is less than 100 milliseconds. Thus clearly, if all of the binding energy of RTK-effector binding were provided by the SH2-pTyr interaction, then these signaling complexes must be extremely short-lived. The highly transient nature of such complexes has been confirmed by single molecule imaging, both in membrane preparations of EGF-treated cells [21] and in live EGF-treated cells (Oh DM, Ogiue-Ikeda M, Jadwin JA, Machida M, Mayer BJ, Yu J, manuscript submitted).
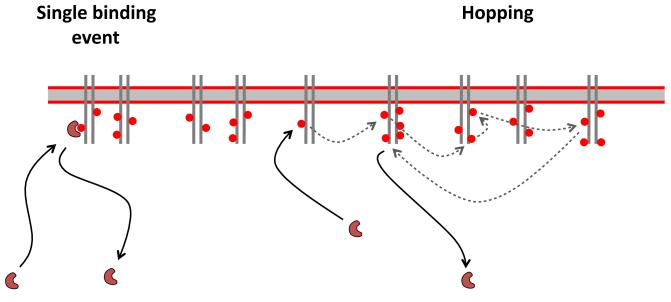
In vivo experiments done by Ji Yu’s group in collaboration with my own have also revealed a number of other interesting properties of SH2-RTK complexes, all of which are consistent with a model in which each SH2 domain “hops” from site to site multiple times before finally escaping the membrane into solution (Oh et al., manuscript submitted) (Figure 4). Such hopping is consistent with theoretical work suggesting that a high local concentration of binding sites on a surface, coupled with a high on-rate for association of a ligand, can generate slower apparent off-rates due to repeated rebinding to the surface [22-24]. In the case of individual SH2 domains examined in vivo by single-molecule imaging, even with repeated rebinding the apparent off-rate for SH2 domains is quite fast, with half-times of a second or two.
Figure 4. Surface rebinding affects apparent off-rates.
Binding is illustrated for an SH2 domain to phosphorylated sites (red circles) on an RTK on the membrane. For simple chemical kinetics, the amount of binding of the SH2 domain is determined by the on-rate and off-rate for the bimolecular interaction (left). If density of binding sites and on-rate are high, however, the domain may “hop” from site to site on the membrane (dotted arrows) before finally escaping. The dwell-time on the membrane is much longer than predicted by the simple off-rate.
Now, a major caveat of these studies is that the isolated SH2 domain may behave very differently from full-length effector or adaptor proteins, all of which contain other interaction domains that could work cooperatively to assemble larger and more stable signaling complexes. Recent data have demonstrated secondary interactions beyond the pTyr-SH2 interface providing additional affinity and specificity to interactions [25], and cooperative binding is a ubiquitous and under-appreciated aspect of biological signaling mechanisms [26]. In the specific case of Grb2, in vitro studies showed that full-length Grb2, complexed with its primary effector Sos, had a somewhat higher affinity for pTyr peptides than the SH2 domain alone [19]. Consistent with this, in vivo single molecule imaging experiments showed that binding of full-length Grb2 was moderately more stable than the SH2 alone, but still was highly transient and was dominated by the SH2 interaction; no evidence of cooperative interactions via other binding proteins was found (Oh et al., manuscript submitted).
The apparent transience of SH2-pTyr interactions in vivo raises an interesting technical question: why do the workhorse methods for detecting protein interactions, co-immunoprecipitation and pulldown assays, work at all? Such methods absolutely depend on relatively stable protein interactions lasting many minutes. If the actual half-time of SH2-mediated associations is on the order of seconds, then binding partners should be lost almost immediately in the washing steps. Of course, the relatively high density of an antibody or fusion protein on a bead might suppress escape from the surface in the same way that is seen in vivo; however protein densities on beads are unlikely to be significantly higher than those seen in living cells, where binding appears to be highly transient. Another possibility is that cooperative, multivalent, multi-protein complexes are assembled on beads, but again it is curious that this should happen in cell lysates but not in living cells. We also need to consider possible experimental artifacts, such as non-physiological disulfide bond formation, or irreversible denaturation or precipitation of proteins onto the surface of beads.
What are some of the implications if the hopping mechanism proves to be an accurate representation of RTK signaling complexes? First, such a mechanism would make signaling even more dependent on the local density of pTyr sites. The number of hops an SH2-containing effector makes before it can escape depends on the density of binding sites, so membrane regions with higher pTyr density will promote longer average dwell-times on the membrane for their binding partners. Indeed, in single-molecule experiments the dwell-time of Grb2 SH2 domain increased substantially with increasing time after EGF stimulation, correlating with increased clustering of activated receptors (Oh et al., manuscript submitted).
In terms of the bigger picture, a hopping mechanism provokes us think more carefully about what we actually mean when we say that effectors are “recruited” to the phosphorylated receptor on the membrane. The static cartoon is easy to understand: effectors are affixed to the membrane-bound receptor, so they have ready access to their substrates (lipids, membrane-associated proteins). But what does it mean if the effector is hopping from site to site, spending much of its time in solution “in flight” between different binding sites? Perhaps what is most important is increasing the likelihood of collision of effectors with their substrates on the membrane [27,28].
Finally, such a mechanism might lessen the importance for downstream signaling of individual high-affinity sites for a particular effector. The Grb2 single-molecule data are consistent with each SH2 domain transiently binding tô20 different sites before finally escaping from the membrane (Oh et al., manuscript submitted). Most RTKs lead to phosphorylation of many different sites upon activation. When an effector samples multiple sites with varying affinities during one encounter with the membrane, these different affinities are effectively averaged together. Loss of a particularly high affinity site will lower the average dwell-time to some extent, but is unlikely to make a binary (binds/does not bind) difference. This averaging effect due to hopping might help explain some earlier studies in which individual RTK phosphorylation sites were mutated. Contrary to expectation, in many cases loss of the preferred binding site for a particular SH2-containing effector had little effect on downstream signaling. It was only in a background where all major phosphorylation sites were eliminated and single high-affinity sites were added back that signfiicant effects could be seen [29,30].
A more practical effect of binding site hopping might be to simplify the quantitative modeling of RTK signaling. Because each RTK has many different possible phosphorylation sites, and because receptors dimerize upon activation, the number of possible discrete phosphorylated receptor species can become enormous. This combinatorial explosion of sites is a major potential problem for quantitative modeling, because in principle each different species must be explicitly accounted for in any mathematical model [31,32]. Most current models get around this problem by making simplifying assumptions, such as defining the receptor as existing in only two states, unphosphorylated or phosphorylated. If binding of effectors to the receptor is not really a discrete event, but instead is averaged over many different transient interactions, then such simplifications are much more likely to be valid representation of actual signaling behavior.
Conclusion
In this speculative look at the dynamic behavior of RTK signaling complexes, I have argued that the constitutive tyrosine phosphatase activity in cells is quite high, and that this has significant implications for downstream signaling. It allows tyrosine kinases to be rather sloppy enzymes, and it makes signal output highly dependent on receptor clustering. Furthermore, it ensures that pTyr-binding effectors play an active role in determining the phosphorylation pattern in cells. I also argue that effector binding is relatively transient in vivo, and may involve sampling multiple binding sites during each membrane encounter. What emerges is a highly dynamic motion picture of RTK signaling, one that is quite different from static textbook cartoons of receptors in their inactive and active states.
Acknowledgments
I would like to thank colleagues in my group and in the Richard D. Berlin Center for Cell Analysis and Modeling, especially Joshua Jadwin, Kazuya Machida, Dongmyung Oh, and Ji Yu, for many interesting discussions relevant to this article. I would also like to thank Lin Jia for providing Fig. 2. I am particularly grateful to Wendell Lim for sharing his ideas about flux and its possible roles in tyrosine kinase signaling, and to Tony Pawson for his unparalleled generosity throughout the years. Unpublished studies discussed here were funded by grant CA146843 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- [1].Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- [2].Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- [3].Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- [5].Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003 doi: 10.1126/stke.2003.191.re12. 2003, RE12. [DOI] [PubMed] [Google Scholar]
- [6].Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- [8].Ruff SJ, Chen, Cohen S. Peroxovanadate induces tyrosine phosphorylation of multiple signaling proteins in mouse liver and kidney. J Biol Chem. 1997;272:1263–1267. doi: 10.1074/jbc.272.2.1263. [DOI] [PubMed] [Google Scholar]
- [9].Kleiman LB, Maiwald T, Conzelman H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- [11].Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc. Natl Acad. Sci. USA. 2008 doi: 10.1073/pnas.0801314105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in pre-metazoan lineages. Proc. Natl Acad. Sci. USA. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rotin D, et al. SH2 Domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase Cγ. EMBO J. 1992;11:559–567. doi: 10.1002/j.1460-2075.1992.tb05087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- [15].Mayer BJ, Hanafusa H. Mutagenic analysis of the v-crk oncogene: requirement for SH2 and SH3 domains and correlation between increased cellular phosphotyrosine and transformation. J. Virol. 1990;64:3581–3589. doi: 10.1128/jvi.64.8.3581-3589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mayer BJ, Hanafusa H. Association of the v-crk oncogene product with phosphotyrosine-containing proteins and protein kinase activity. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2638–2642. doi: 10.1073/pnas.87.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ren R, Ye Z-S, Baltimore D. Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes & Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- [19].Chook YM, Gish GD, Kay CM, Pai EF, Pawson T. The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. J Biol Chem. 1996;271:30472–30478. doi: 10.1074/jbc.271.48.30472. [DOI] [PubMed] [Google Scholar]
- [20].de Mol NJ, Dekker FJ, Broutin I, Fischer MJE, Liskamp RMJ. Surface plasmon resonance thermodynamic and kinetic analysis as a strategic tool in drug design. Distinct ways for phosphopeptides to plug into Src- and Grb2 SH2 domains. J Med Chem. 2005;48:753–763. doi: 10.1021/jm049359e. [DOI] [PubMed] [Google Scholar]
- [21].Morimatsu M, Takagi H, Ota KG, Iwamoto R, Yanagida T, Y S. Multiple-state reactions between the epidermal growth factor receptor and Grb2 as observed by using single-molecule analysis. Proc Natl Acad Sci U S A. 2007;104:18013–18018. doi: 10.1073/pnas.0701330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- [24].Gopalakrishnan M, Forsten-Williams K, Nugent MA, Täuber UC. Effects of receptor clustering on ligand dissociation kinetics: theory and simulations. Biophys J. 2005;89:3686–3700. doi: 10.1529/biophysj.105.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bae JH, Lew ED, Yuzawa S, Tome F, Lax I, Schlessinger J. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell. 2009;138:514–524. doi: 10.1016/j.cell.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem. Sci. 2009;34:471–482. doi: 10.1016/j.tibs.2009.06.007. [DOI] [PubMed] [Google Scholar]
- [27].Kholodenko BN, Hoek JB, Westerhoff HV. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol. 2000;10:173–178. doi: 10.1016/s0962-8924(00)01741-4. [DOI] [PubMed] [Google Scholar]
- [28].Mugler A, Gotway Bailey A, Takahashi K, Rein ten Wolde P. Membrane clustering and the role of rebinding in biochemical signaling. Biophys J. 2012;102:1069–1078. doi: 10.1016/j.bpj.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dankort DL, Wang Z, Blackmore V, Moran MF, Muller WJ. Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol Cell Biol. 1997;17:5410–5425. doi: 10.1128/mcb.17.9.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Valius M, Kazlauskas A. Phospholipase C-γ1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell. 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- [31].Hlavacek WS, Faeder JR, Blinov ML, Perelson AS, Goldstein B. The complexity of complexes in signal transduction. Biotechnol Bioeng. 2003;84:783–794. doi: 10.1002/bit.10842. [DOI] [PubMed] [Google Scholar]
- [32].Mayer BJ, Blinov ML, Loew LM. Molecular machines or pleiomorphic ensembles: signaling complexes revisited. J Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]