Abstract
The WW domain-containing PQBP1 (polyglutamine tract-binding protein 1) protein regulates mRNA processing and gene transcription. Mutations in the PQBP1 gene were reported in several X chromosome-linked intellectual disability (XLID) disorders, including Golabi-Ito-Hall (GIH) syndrome. The missense mutation in the GIH syndrome maps within a functional region of the PQBP1 protein known as the WW domain. The causative mutation of PQBP1 replaces the conserved tyrosine (Y) at position 65 within the aromatic core of the WW domain to cysteine (C), which is a chemically significant change. In this short review, we analyze structural models of the Y65C mutated and wild type WW domains of PQBP1 in order to infer potential molecular mechanisms that render the mutated PQBP1 protein inactive in terms of ligand binding and its function as a regulator of mRNA splicing.
Keywords: WW domain, Intellectual disability, Cysteine oxidation, disulfide bridge, mRNA processing
1. Introduction
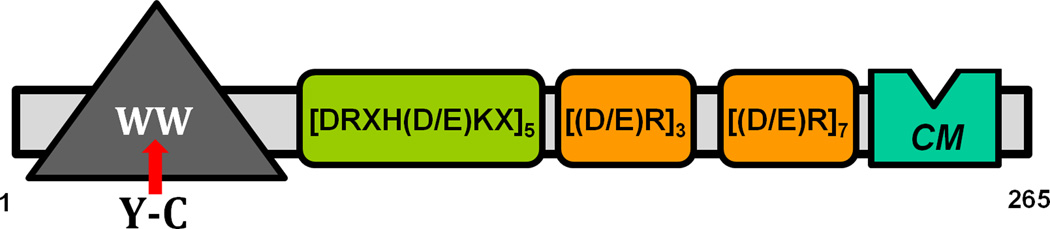
The WW domain-containing PQBP1 (polyglutamine tract-binding protein 1) gene encodes a protein that regulates mRNA processing and RNA Polymerase II-driven transcription [1–5]. The PQBP1 gene is highly conserved in evolution and its orthologs in plants, worms and mammals share a high degree of sequence similarity. In mammals, the PQBP1 gene is widely expressed, but its transcript is enriched in the brain [6]. Originally, the PQBP1 protein was identified as a protein partner of poly-glutamine-(poly-Q)-containing proteins, including huntingtin, ataxin-1 and neuronal transcription factor Brn-2. Mutations in the PQBP1 gene were reported in several X chromosome-linked intellectual disability (XLID) disorders, such as Renpenning, Sutherland-Haan, Hamel, Porteous, and Golabi-Ito-Hall (GIH) syndromes [7,8]. Interestingly, although caused by different mutations that affect the coding sequence of the PQBP1 gene - frame shifts that result in truncated protein products or a missense point mutation - these syndromes share similar clinical features [7]. In addition to severe intellectual disability, the patients also have a lean body, short stature, small head, and are frequently diagnosed with cardiac abnormalities [9,10]. The missense mutation in the GIH syndrome maps within a functional region of the PQBP1 protein known as the WW domain (Figure 1) [8,11–13]. In contrast to other mutations reported for PQBP1, the genetic lesion of GIH syndrome does not affect the size of the mutated protein, which normally migrates on SDS-PAGE as a singlet of 38 kilo-Daltons [14].
Figure 1.
Schematic of the organization of the PQBP1 protein. PQBP1 contains only one discernable modular domain, the WW domain located at the N-(amino)-terminus. The causative Y-C PQBP1 mutation of the GIH syndrome is located within the middle of the WW domain sequence and it is indicated with a red arrow. There are multiple copies of three distinct amino acid repeats found in PQBP1: five consecutive copies of DRXH(D/E)KX and ten copies of (D/E)R, which occur consecutively in repeats of three and seven separated by a small stretch of intervening amino acids. At the C-(carboxy)-terminal region there is a stretch of highly conserved amino acids –GPLFQQRPYPSPG-, which are identical among PQBP1 proteins of man (Homo sapiens), a sweet water polyp (Hydra magnipapilata) and a small flowering plant (Arabidopsis thaliana). We indicate the relative location of this sequence as a green box with CM, after “conserved motif”. An indentation on the box is to suggest a possibility that the CM motif interacts with the WW domain, shown as a gray triangle.
The WW domain is one of the smallest protein modules mediating specific protein-protein interactions with short proline-rich or proline-containing motifs [11–13]. The WW domain mutation implicated in the GIH syndrome replaces the conserved tyrosine (Y) at position 65 within the aromatic core of the domain to cysteine (C), which is a chemically significant change to the protein that we would like to discuss here in detail [14].
Recently, we have documented that the binding of the Y65C mutant of PQBP1 protein to its cognate ligand, WBP11 (also known as SIPP1), which regulates mRNA processing, was abrogated compared to that of the control wild-type protein [14]. More importantly, we have verified that in lymphoblasts derived from a GIH patient, the complex between the endogenous PQBP1 and endogenous WBP11 proteins was compromised compared with the intact complex between these two proteins found in lymphoblasts derived from a normal individual who was matched for race and sex with the GIH patient [14]. Using a cell culture model and a reporter gene construct that monitors efficiency of mRNA splicing, we have shown that the missense mutation in the PQBP1 WW domain resulted in a substantial decrease in pre-mRNA splicing [14].
2. Y65 of the WW domain of PQBP1 is conserved among the family of WW domains
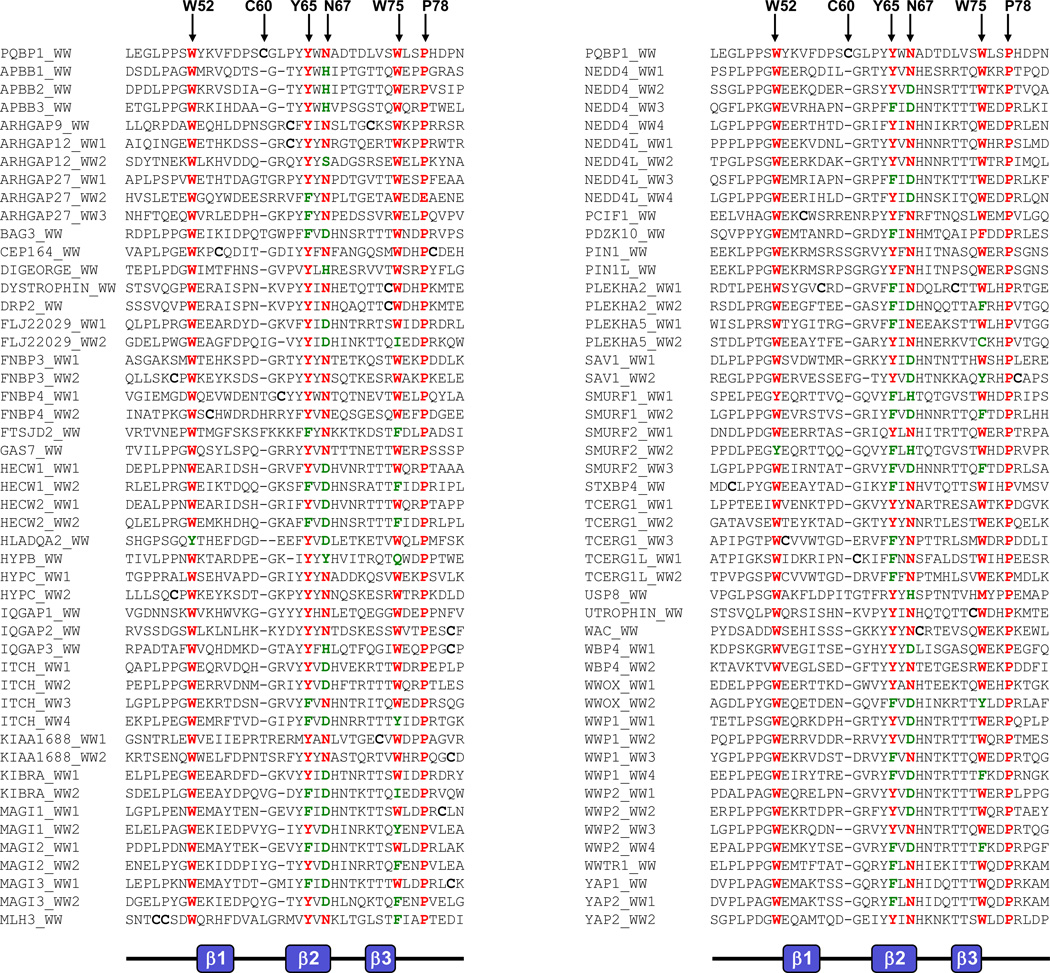
In an effort to understand the importance of tyrosine at position 65 (Y65) in the function of PQBP1, we performed amino acid sequence alignment of the entire human family of WW domains (Figure 2). Our analysis reveals that Y65 is predominantly conserved across all WW domains and that it is frequently substituted by its chemical analogue phenylalanine, further implicating its central role in the function of WW domains. In addition to Y65, a number of other residues in the WW domain of PQBP1 are also predominantly conserved across the entire human family of WW domains. These include the signature tryptophan residues at positions 52 and 75 (W52 and W75), an asparagine at position 67 (N67) and a proline at position 78 (P78). It is notable that while W52 is substituted by tyrosine in only a few WW domains, W75 displays more versatility and is replaced by amino acids as diverse as tyrosine, phenylalanine, isoleucine, glutamine and also cysteine. In a manner akin to the structural versatility of W75, the N67 residue can also be replaced by amino acids as diverse as aspartate, serine, tyrosine and histidine. In contrast, WW domains appear to have an obligate requirement for P78 as it is only absent from the WW2 domain of ARHGAP27 GTPase. The highly conserved nature of W52, Y65, N67, W75 and P78 residues in PQBP1 across the entire family of WW domains and their key role in the structural and functional integrity of WW domains is further corroborated by site-directed mutagenesis studies on the WW domain of Pin1 [15]. The mutagenesis of the WW domain of Pin1 revealed that W52, N67, Y65 and P78 form a hydrophobic core of the domain, which may be important in stabilizing the structure [15].
Figure 2.
Amino acid sequence alignment of the human family of WW domains. Alignment is divided into two columns for clarity and each column is headed by the WW domain of PQBP1. Highly conserved amino acid residues across the entire family of WW domains are colored red and green, and the equivalent residues in the WW domain of PQBP1 are indicated by vertical arrows. Cysteines are in bold font.
3. Y65C PQBP1 mutant and intramolecular disulfide bonds
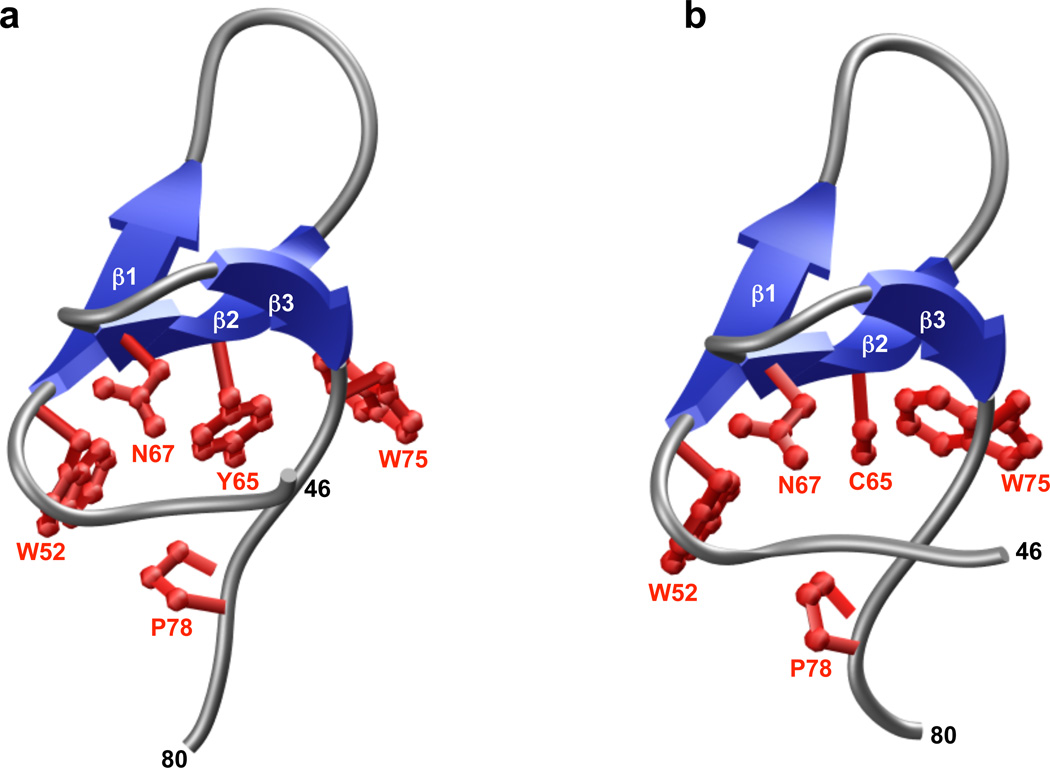
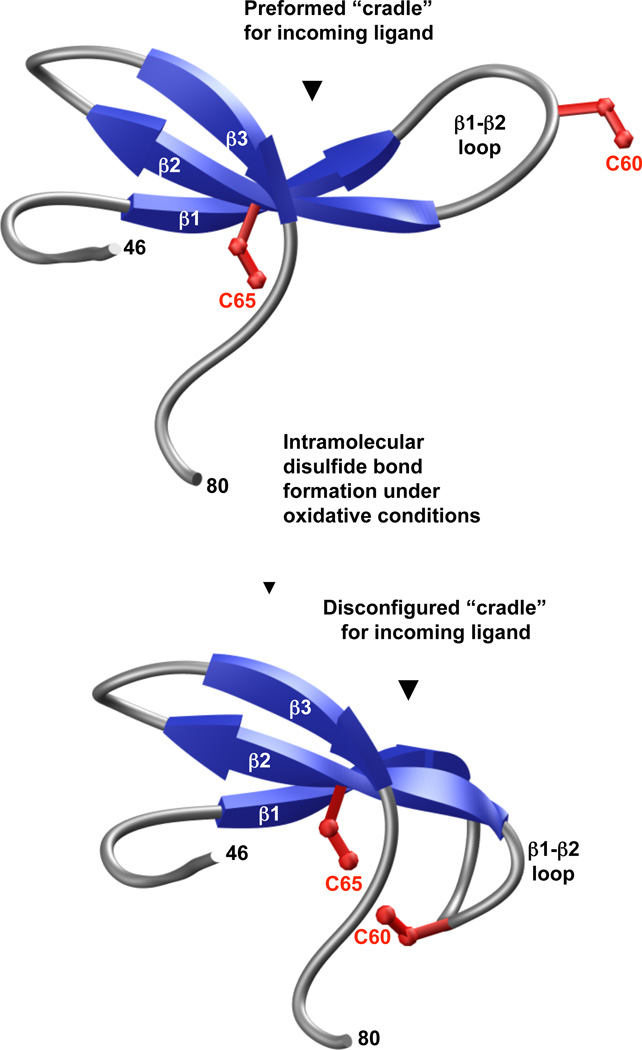
In order to gain better insight about the effect of Y65C mutation on the WW domain fold, we generated 3D atomic models of wt and Y65C-mutant WW domains of PQBP1 (Figure 3). As discussed in our recent study, the hydrophobic core that is located beneath the ligand-binding pocket of the wt WW domain is composed of a highly conserved quartet of hydrophobic residues W52, Y65, N67 and P78 (and perhaps to a lesser extent by W75). While Y65 constitutes a key component of the hydrophobic core within the wild type domain, the placement of cysteine at this position within the Y65C-mutant WW domain may not only result in the collapse of the triple-stranded β-sheet “roof” supporting the ligand-binding “cradle” [16,17], but under oxidative conditions, the formation of an intramolecular disulfide bond between C60 and C65 residues within the Y65C-mutant WW domain may also be favored (Figure 4). This scenario is conceivable due to the inherent flexibility of the β1-β2 loop allowing it to curve away from the ligand-binding “cradle” toward the hydrophobic core of the WW domain, so as to bring the C60 and C65 residues in close proximity and thereby drive their covalent linkage via a disulfide bond. As we have shown recently in an in vitro assay, the binding of Y65C-mutant WW domain to its cognate peptide ligands is indeed compromised, but perhaps not as dramatically as would be expected from the replacement of a conserved tyrosine with a cysteine within its hydrophobic core. This salient observation argues in favor of an alternative mechanism by which the Y65C mutation could lead to loss-of-function in PQBP1. Given that the in vitro binding studies, which we recently reported, were conducted under reducing conditions in the presence of a reducing agent, 5mM β-mercaptoethanol [14], it is thus conceivable that the Y65C-mutant WW domain may behave differently toward its cognate ligands within the milieu of the cell, than in an in vitro binding assay. Thus, under non-reducing conditions in response to cellular oxidative stress, for instance, it is conceivable that the Y65C-mutant WW domain may undergo intramolecular disulfide bond formation as proposed above. Accordingly, such intramolecular covalent modification would destroy the “cradle” optimized for ligand binding and the resulting Y65C-mutant domain would not be expected to bind to its cognate ligands in a physiologically-relevant manner, leading to loss-of-function in PQBP1, as we observed in co-immunoprecipitation assays performed on full length proteins expressed in cultured cells. It is also important to note that C60 occupies a unique position in the WW domain of PQBP1 and is largely absent from other WW domains. Moreover, it is intriguing that the position of C60 in the WW domain of PQBP1 is either absent in other WW domains or substituted for by other amino acids, with the exception of cysteine (Figure 2). Of particular interest is the observation that the occurrence of more than one cysteine in the WW domains is a rare event. Of ~100 human WW domains, only four contain more than one cysteine. These include the WW domains of ARHGAP9, CEP164, MLH3 and PLEKHA2 (Figure 2). Notably, these cysteine residues are located in the following areas: within the β2 and β3 strands (ARHGAP9), the β1 strand and the C-terminus following the β3 strand (CEP164), within the N-terminus preceding the β1 strand (MLH3), and within the β1-β2 loop and β3 strand (PLEKHA2). The markedly differential location of cysteine residues with the WW domain family suggests that they contribute to WW domain structure in distinct manners and may modulate their function via distinct mechanisms. Although the presence of more than one cysteine within a protein opens up the possibility of intramolecular disulfide bond formation, it should be noted that this remains an unlikely scenario in the case of WW domains of ARHGAP9, CEP164 and PLEKHA2 proteins on structural grounds. Thus, unlike the Y65C-mutant WW domain of PQBP1, the cysteine residues are either located within one of the three rigid β-strands or away from the flexible β1-β2 loop in the WW domains of ARHGAP9, CEP164 and PLEKHA2. Taken together, it seems that the Y65C mutation may predispose the WW domain of PQBP1 to loss-of-function via the formation of an intramolecular disulfide bond. However, it is important to mention that in some cases intra-molecular disulfide bond formation could lead to a gain of protein function, as exemplified by the bridge between cysteines 377 and 330 in Cdc25C phosphatase, which results in enhanced binding to 14-3-3 protein and the cytoplasmic sequestration of the protein [18]. Fittingly, in our in vitro binding studies, we did observe a small number of peptide ligands that bound more tightly to the Y65C PQBP1 WW domain compared to the wild type control. Therefore, it is possible, that in the network or systems view, the Y65C mutations may result in both loss and gain of function changes that in concert deregulate mRNA processing.
Figure 3.
3D atomic models of wild type (A) and Y65C-mutant (B) WW domains of human PQBP1. The triple-stranded β-sheet of the WW domains is shown in blue and the intervening loops in gray. The sidechains of residues W52, Y65/C65, N67, W75 and P78 that constitute the hydrophobic core of the domains are colored red.
Figure 4.
A hypothetical model for the loss-of-function in the Y65C mutant form of human PQBP1 in Golabi-Ito-Hall syndrome. Under oxidative conditions, the formation of an intra-molecular disulfide bond between C60 and C65 residues within the Y65C-mutant WW domain may be favoured. This scenario is conceivable due to the inherent flexibility of the β1-β2 loop, allowing it to curve away from the ligand-binding “cradle” toward the hydrophobic core of the WW domain, so as to bring the C60 and C65 residues in close proximity and thereby drive their covalent linkage via an S-S bond. Note that the S-S bond between the sidechain SG atoms of C60 and C65 residues is omitted for clarity.
4. Y65C PQBP1 regulation via oxidation
Although in general, proteins that signal in the cytoplasm are embedded in a reducing environment, the reversible oxidation of cysteines of cytoplasmic proteins is a well-established regulatory mechanism for various signaling proteins, including protein-tyrosine phosphatases [19]. The general cellular redox state and the extracellular-ligand stimulated production of reactive oxygen or reactive nitrogen species (ROS or RNS) affects oxidation of cysteines in proteins, in a reversible manner. Reaction of H2O2 oxidizes cysteine thiols at first to sulphenic acid (SOH), and at higher concentrations of the oxidant, to sulphinic (SO2H) or sulphonic acid (SO3H) derivatives [19]. The sulphenic acid (SOH) derivative of C65 of PQBP1 could form sulphenylamides by reaction with other amino acids of the PQBP1 that are located in the proximity of the WW domain, in addition to forming an intra-molecular disulfide bridge with C60. Interestingly, the wt PQBP1 protein does not contain any other cysteine residues except for the WW domain-located C60.
We cannot exclude other possibilities by which oxidation of C65 could regulate the function of PQBP1 protein. The sulphinic (SO2H) or sulphonic acid (SO3H) derivatives of C65 in PQBP1 may lead to conformational changes that also render the domain inactive in terms of ligand binding. Moreover, the cysteine thiols in a local environment with pKa below 6 could form a reactive thiolate anion that is permissive for posttranslational modifications such as phosphorylation, glutathionylation or even adduct formation with other endogenous electrophilic molecules [19]. Such modifications could also render the mutated WW domain inactive in terms of ligand binding and signaling of the PQBP1 complex in mRNA processing.
5. Concluding remarks
The atomic details of the Y65C lesion of the PQBP1 protein will hopefully come soon from crystal structures of the wild type and mutant proteins. Apart from the intrinsic changes that the mutation causes to the WW domain itself, it will be very important to see the relative location of the carboxy-terminal tail of the protein with its well-conserved motif (CM) (Figure 1). Since most of the PQBP1 mutations that result in XLID syndromes have the carboxy-terminal region of the protein missing, including the very CM sequence, we presume that somehow the WW domain and the CM region collaborate together to regulate mRNA splicing.
Apart from X-ray crystallography, two other approaches represent attractive ways to study the effect of the Y65C mutation on the molecular architecture of the PQBP1 protein. These are protein folding assays in the periplasm of bacteria [20] and atomic force microscopy [21]. Both of these approaches are able to interrogate the PQBP1-Y65C mutant and wild type proteins under reducing and non-reducing conditions. These two approaches could quickly validate several regulatory scenarios proposed here.
In sum, our recent studies on PQBP1 and various plausible mechanisms discussed here for the inactivation of PQBP1 provide the framework for conducting future studies to unlock the molecular basis of XILD disorders at atomic level with important consequences on the rationale design of therapeutic approaches.
ACKNOWLEDGMENTS
We thank Virginia Mazack for valuable comments on the first version of the manuscript.
This work was supported by the National Institutes of Health Grants R01-GM083897 and funds from the USylvester Braman Family Breast Cancer Institute (to AF), and by PA Breast Cancer Coalition Grants (#60707 an #920093) and by the Geisinger Clinic (to MS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Komuro A, Saeki M, Kato S. Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J Biol Chem. 1999;274:36513–36519. doi: 10.1074/jbc.274.51.36513. [DOI] [PubMed] [Google Scholar]
- 2.Komuro A, Saeki M, Kato S. Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res. 1999;27:1957–1965. doi: 10.1093/nar/27.9.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waragai M, Lammers CH, Takeuchi S, Imafuku I, Udagawa Y, Kanazawa I, Kawabata M, Mouradian MM, Okazawa H. PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet. 1999;8:977–987. doi: 10.1093/hmg/8.6.977. [DOI] [PubMed] [Google Scholar]
- 4.Okazawa H, Rich T, Chang A, Lin X, Waragai M, Kajikawa M, Enokido Y, Komuro A, Kato S, Shibata M, Hatanaka H, Mouradian MM, Sudol M, Kanazawa I. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron. 2002;34:701–713. doi: 10.1016/s0896-6273(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 5.Sudol M, Sliwa K, Russo T. Functions of WW domains in the nucleus. FEBS Lett. 2001;490:190–195. doi: 10.1016/s0014-5793(01)02122-6. [DOI] [PubMed] [Google Scholar]
- 6.Okazawa H, Sudol M, Rich T. PQBP-1 (Np/PQ): a polyglutamine tract-binding and nuclear inclusion-forming protein. Brain Res Bull. 2001;56:273–280. doi: 10.1016/s0361-9230(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 7.Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gecz J, Sefiani A, Hoffmann K, Moser B, Haas S, Gurok U, Haesler S, Aranda B, Nshedjan A, Tzschach A, Hartmann N, Roloff TC, Shoichet S, Hagens O, Tao J, Van Bokhoven H, Turner G, Chelly J, Moraine C, Fryns JP, Nuber U, Hoeltzenbein M, Scharff C, Scherthan H, Lenzner S, Hamel BC, Schweiger S, Ropers HH. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet. 2003;35:313–315. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- 8.Lubs H, Abidi FE, Echeverri R, Holloway L, Meindl A, Stevenson RE, Schwartz CE. Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J Med Genet. 2006;43:e30. doi: 10.1136/jmg.2005.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golabi M, Ito M, Hall BD. A new X-linked multiple congenital/mental retardation syndrome. Am J Med Genet. 1984;17:367–374. doi: 10.1002/ajmg.1320170130. [DOI] [PubMed] [Google Scholar]
- 10.Kunde SA, Musante L, Grimme A, Fischer U, Muller E, Wanker EE, Kalscheuer VM. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum Mol Genet. 2011;20:4916–4931. doi: 10.1093/hmg/ddr430. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 12.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 14.Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, Beullens M, Schwartz CE, Landgraf C, Volkmer R, Pastore A, Farooq A, Bollen M, Sudol M. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Structure-function-folding relationship in a WW domain. Proc Natl Acad Sci U S A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 18.Savitsky PA, Finkel T. Redox regulation of Cdc25C. J Biol Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 19.Ostman A, Frijhoff J, Sandin A, Bohmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem. 2011;150:345–356. doi: 10.1093/jb/mvr104. [DOI] [PubMed] [Google Scholar]
- 20.Mansell TJ, Linderman SW, Fisher AC, DeLisa MP. A rapid protein folding assay for the bacterial periplasm. Protein Sci. 2010;19:1079–1090. doi: 10.1002/pro.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baclayon M, Roos WH, Wuite GJ. Sampling protein form and function with the atomic force microscope. Mol Cell Proteomics. 2010;9:1678–1688. doi: 10.1074/mcp.R110.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]