Abstract
Breast cancer is one of the leading causes of cancer mortality in women. Recent advances in gene expression profiling have indicated that breast cancer is a heterogeneous disease and the current prognostication using clinico-pathological features is not sufficient to fully predict therapy response and disease outcome. In this retrospective study, we show that expression levels of BRE, which encodes a member of the BRCA1 DNA damage repair complex, predicted disease-free survival (DFS) in non-familial breast cancer patients. The predictive value of BRE expression depended on whether patients received radiotherapy as a part of their primary treatment. In radiotherapy-treated patients, high BRE expression predicted a favorable DFS (hazard ratio (HR) = 0.47, 95 % confidence interval (CI) = 0.28–0.78, p = 0.004), while in non-treated patients, high BRE expression predicted an adverse prognosis (HR = 2.59, 95 % CI = 1.00–6.75, p = 0.05). Among radiotherapy-treated patients, the prognostic impact of BRE expression was confined to patients with smaller tumors (HR = 0.23, 95 % CI = 0.068–0.75, p = 0.015) and it remained an independent factor after correction for the other prognostic factors age, tumor size, lymph node involvement, and histological grade (HR = 0.50, CI = 0.27–0.90, p = 0.021). In addition, high BRE expression predicted a favorable relapse-free survival in a publicly available dataset of 2,324 breast cancer patients (HR = 0.59, CI = 0.51–0.68, p < 0.001). These data indicate that BRE is an interesting candidate for future functional studies aimed at developing targeted therapies.
Keywords: BRE, Radiotherapy, DNA damage repair, BRCA1, Breast cancer
Introduction
Despite great improvements in diagnostic imaging techniques and treatment, breast cancer remains one of the leading causes of cancer mortality in women. Prognostication of breast cancer patients nowadays relies highly on classical clinico-pathological features, such as tumor size, histological grade, age, and lymph node metastases [1]. However, it remains a challenge to accurately predict disease outcome based on these parameters, which is necessary not to under or over treat the patients.
Over the last 20 years, there has been great interest in developing prognostic patient classification methods based on molecular screenings. Genome-wide gene expression screens have identified expression profiles that predict disease outcome and therapy response. For example, in several large patient studies, a 70-gene signature called the “MammaPrint” (Agendia, Amsterdam, the Netherlands) has been shown to outperform classical prognostication methods [2–4]. Together with other molecular classification methods [5, 6], these data indicate that the identification of differential gene expression has great potential for improved prediction of disease outcome and subsequent treatment decisions.
DNA double strand breaks (DSBs) are one of the most cytotoxic types of DNA damage. The importance of proper repair of these breaks to maintain genomic integrity is exemplified by recurrent mutations in genes involved in DSB repair in various cancers. For example, BRCA1 mutations occur in approximately 20 % of familial breast cancer cases [7–9]. The importance of the BRCA1 multi-protein complex has been exemplified by the identification of polymorphisms and haplotypes within other BRCA1 complex members, such as RAP80 and ABRAXAS, both in BRCA1/2 mutated and non-mutated familial breast cancer patients. However, the clinical impact of these polymorphisms remains to be confirmed [10–15]. Furthermore, BRCA1 expression levels seem to predict breast cancer outcome in non-familial cases [16–19] although data are not consistent [20].
Recently, it has been shown that high expression of BRE (Brain and Reproductive organ-Expressed), another member of the BRCA1 complex [21–24], denotes a favorable prognosis in acute myeloid leukemia (AML) [25–27]. In this study, we demonstrate that BRE expression levels in breast cancer tumor tissue contained prognostic information in a cohort of 229 non-familial breast cancer patients, establishing the relevance of this DNA damage repair factor in breast cancer.
Materials and methods
Breast cancer samples
Frozen breast cancer tissue sections were available for two independent cohorts of 229 patients in total who had undergone resection of their primary tumor, as described before [28–30]. Patients underwent surgical resection of their primary tumor between November 1987 and December 1997 and were selected by the availability of RNA samples in the tumor bank of the Department of Chemical Endocrinology of the Radboud University Nijmegen Medical Centre (Nijmegen, The Netherlands). This bank contains tumor material from five different hospitals of the Comprehensive Cancer Centre East in the Netherlands. Patients had no previous diagnosis of carcinoma, no distant metastases at time of diagnosis, and no evidence of disease within 1 month after primary surgery. Patients that received neoadjuvant therapy or were diagnosed with carcinoma in situ were excluded from this study. Patients were treated with protocols established at that time. 60 % of the patients underwent mastectomy (137/229) and the remaining patients underwent lumpectomy. 74 % of the patients (169/229) received radiotherapy following surgery and 39 % (90/229) received systemic adjuvant treatment, in combination with radiotherapy or not. Adjuvant treatment consisted of endocrine treatment with tamoxifen and/or chemotherapy. Detailed patient characteristics can be found in Table 1. The median follow-up period of censored patients was 107.5 months. This study was performed according to REMARK guidelines [31].
Table 1.
Clinico-pathological characteristics of 229 non-familial breast cancer patients
| Total cohort | Non-radiotherapy-treated | Radiotherapy-treated | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low BRE a (N = 115) | High BRE a (N = 114b) | p | Low BRE a (N = 32) | High BRE a (N = 27b) | p | Low BRE a (N = 83) | High BRE a (N = 86b) | p | |||||||
| Age (N = 229), mean (range) | 59.9 | (31–88) | 59.2 | (32–86) | 0.775d | 63.0 | (33–88) | 61.5 | (35–86) | 0.784d | 58.7 | (31–85) | 58.3 | (32–83) | 0.816d |
| Menopausal status (N = 229) | 1.000e | 1.000e | 1.000e | ||||||||||||
| Premenopausal, no. (%) | 29 | (25.2) | 28 | (24.6) | 6 | (18.8) | 5 | (18.5) | 23 | (27.7) | 23 | (26.7) | |||
| Postmenopausal, no. (%) | 86 | (74.8) | 86 | (75.4) | 26 | (81.3) | 22 | (81.5) | 60 | (72.3) | 63 | (73.3) | |||
| Nodal category (N = 208) | 0.923e | 0.310e | 0.806e | ||||||||||||
| Negative, no. (%) | 61 | (58.1) | 59 | (57.3) | 25 | (86.2) | 18 | (72.0) | 36 | (47.4) | 41 | (52.6) | |||
| 1–3 involved lymph nodes, no. (%) | 29 | (27.6) | 31 | (30.1) | 4 | (13.8) | 7 | (28.0) | 25 | (32.9) | 24 | (30.8) | |||
| ≥4 involved lymph nodes, no. (%) | 15 | (14.3) | 13 | (12.6) | 0 | (0) | 0 | (0) | 15 | (19.7) | 13 | (16.7) | |||
| Radiotherapy (N = 228) | 0.547e | NA | NA | ||||||||||||
| Treated, no. (%) | 83 | (72.2) | 86 | (76.1) | 0 | (0) | 0 | (0) | 83 | (100) | 86 | (100) | |||
| Non-treated, no. (%) | 32 | (27.8) | 27 | (23.9) | 32 | (100) | 27 | (100) | 0 | (0) | 0 | (0) | |||
| Surgery (N = 229) | 0.282e | 0.495f | 0.219f | ||||||||||||
| Mastectomy, no. (%) | 73 | (63.5) | 64 | (56.1) | 30 | (93.8) | 27 | (100) | 43 | (51.8) | 36 | (41.9) | |||
| Lumpectomy, no. (%) | 42 | (36.5) | 50 | (43.9) | 2 | (6.3) | 0 | (0) | 40 | (48.2) | 50 | (58.1) | |||
| Adjuvant systemic therapy (N = 228) | 0.230f | 0.537f | 0.269f | ||||||||||||
| None, no. (%) | 70 | (60.9) | 68 | (60.2) | 26 | (81.3) | 18 | (66.7) | 44 | (53.0) | 50 | (58.1) | |||
| Endocrine therapy, no. (%) | 30 | (26.1) | 28 | (24.8) | 3 | (9.4) | 4 | (14.8) | 27 | (32.5) | 24 | (27.9) | |||
| Chemotherapy, no. (%) | 13 | (11.3) | 9 | (8.0) | 2 | (6.3) | 2 | (7.4) | 11 | (13.3) | 7 | (8.1) | |||
| Endocrine + Chemotherapy, no. (%) | 2 | (1.7) | 8 | (7.1) | 1 | (3.1) | 3 | (11.1) | 1 | (1.2) | 5 | (5.8) | |||
| Histology grade (N = 168) | 0.406e | 0.151f | 0.123e | ||||||||||||
| I, no. (%) | 9 | (10.5) | 4 | (4.9) | 1 | (4.3) | 1 | (5.3) | 8 | (12.7) | 3 | (4.8) | |||
| II, no. (%) | 36 | (41.9) | 35 | (42.7) | 6 | (26.1) | 10 | (52.6) | 30 | (47.6) | 25 | (39.7) | |||
| III, no. (%) | 41 | (47.7) | 43 | (52.4) | 16 | (69.6) | 8 | (42.1) | 25 | (39.7) | 35 | (55.6) | |||
| Tumor type (N = 193) | 0.744e | 0.721f | 1.000e | ||||||||||||
| Ductal, no. (%) | 73 | (73.0) | 70 | (75.3) | 22 | (71.0) | 18 | (81.8) | 51 | (73.9) | 52 | (73.2) | |||
| Lubular, no. (%) | 15 | (15.0) | 15 | (16.1) | 3 | (9.7) | 2 | (9.1) | 12 | (17.4) | 13 | (18.3) | |||
| Other (mixed/unknown), no. (%) | 12 | (12.0) | 8 | (8.6) | 6 | (19.3) | 2 | (9.1) | 6 | (8.7) | 6 | (8.5) | |||
| Tumor size (N = 227)c | 0.014f | 0.853f | 0.005e | ||||||||||||
| pT1, no. (%) | 33 | (28.7) | 50 | (44.6) | 11 | (34.4) | 9 | (33.3) | 22 | (26.5) | 41 | (48.2) | |||
| pT2, no. (%) | 66 | (57.4) | 43 | (38.4) | 19 | (59.4) | 15 | (55.6) | 47 | (56.6) | 28 | (32.9) | |||
| pT3/4, no. (%) | 16 | (13.9) | 19 | (17.0) | 2 | (6.3) | 3 | (11.1) | 14 | (16.9) | 16 | (18.8) | |||
| Estrogen receptor status (N = 196) | 0.769e | 0.265e | 0.228e | ||||||||||||
| Positive, no. (%) | 61 | (61.0) | 61 | (63.5) | 18 | (66.7) | 12 | (50) | 43 | (58.9) | 49 | (69.0) | |||
| Negative, no. (%) | 39 | (39.0) | 35 | (36.5) | 9 | (33.3) | 12 | (50) | 30 | (41.1) | 22 | (31.0) | |||
| Progesterone receptor status (N = 197) | 0.776e | 0.579e | 1.000e | ||||||||||||
| Positive, no. (%) | 51 | (50.5) | 51 | (53.1) | 12 | (44.4) | 13 | (54.2) | 39 | (52.7) | 38 | (53.5) | |||
| Negative, no. (%) | 50 | (49.5) | 45 | (46.9) | 15 | (55.6) | 11 | (45.8) | 35 | (47.3) | 33 | (46.5) | |||
aHigh and low BRE expression is defined as expression above or below the median expression of the total cohort, respectively
bAs data on radiotherapy treatment was lacking for one patient (showing high BRE expression), the patient numbers in the radiotherapy-treated and non-treated groups do not add up to the total cohort
cpT1: tumor size ≤2 cm, pT2: tumor size of 2–5 cm, pT3/4 tumor size >5 cm and/or direct extension to chest wall or skin
d p-value is based on Mann–Whitney U test
e p-value is based on χ2 test
f p-value is based on Fisher Exact test
NA not applicable
BRE QPCR
Tissue collection, mRNA isolation, and cDNA preparation have been described before [29]. BRE expression was measured in both cohorts by QPCR using a commercially available primer/probe set (Hs01046283_m1, Life Technologies, Carlsbad, CA, USA) and normalized to expression of the housekeeping gene PBGD, as described in [26]. Normalized QPCR data were mean centered per analyzed cohort and afterward the data of the cohorts were combined to increase patient numbers for further analyses.
Statistical analyses
To statistically test the correlation of BRE expression with clinical parameters, the complete cohort was subdivided into two equally sized groups based on BRE expression. Differences in patient characteristics were tested by χ2, Fisher exact, or Mann–Whitney U tests, as indicated. Disease-free survival (DFS; defined as time between surgery and diagnosis of recurrent or metastatic disease) and overall survival (OS; defined as time between surgery and death by any cause) were used as feature for disease outcome. The prognostic impact of BRE expression was visualized by Kaplan–Meier plots and statistically tested via the logrank method and univariate or multivariate Cox regression analyses. Statistical analyses were carried out by means of Graphpad (La Jolla, CA, USA) or SPSS (IBM Corporation, Armonk, NY, USA) software.
Results
BRE expression correlates with tumor size
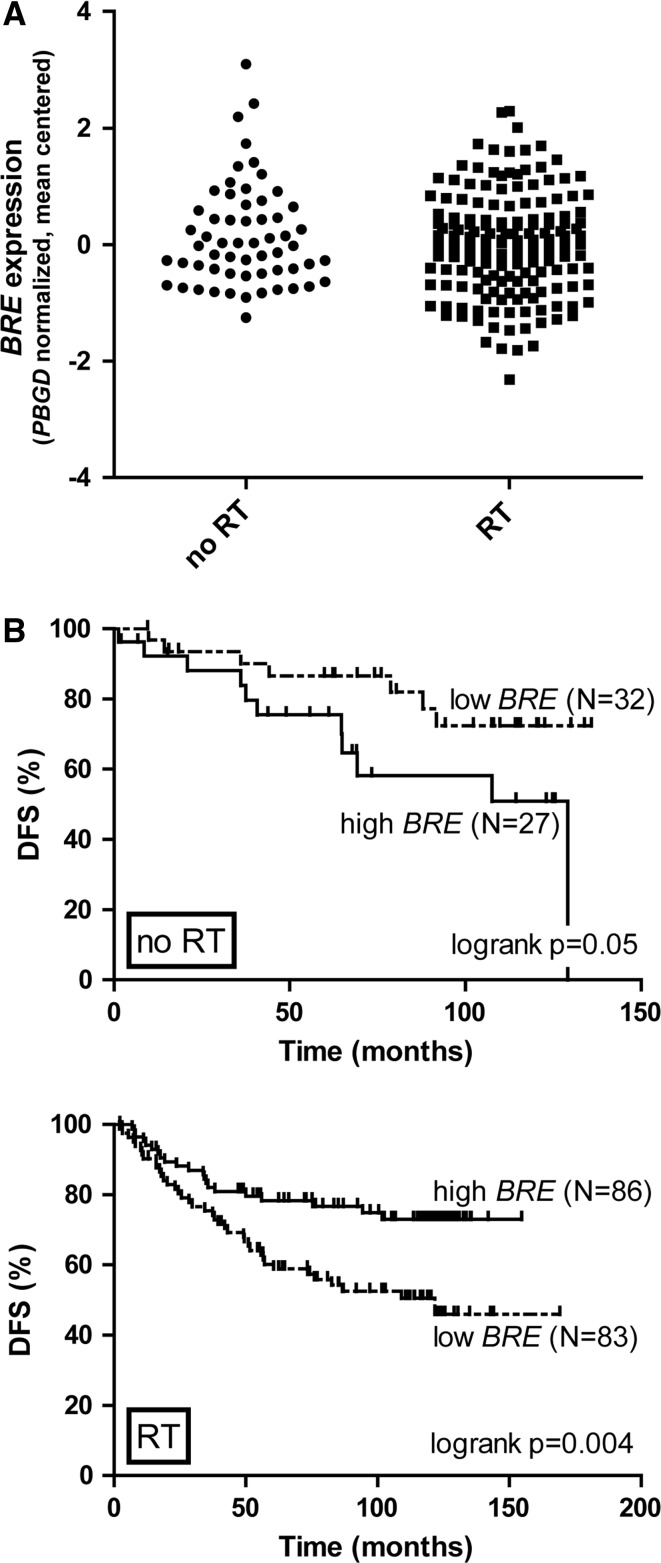
To study the prognostic effect of BRE expression in breast cancer, BRE mRNA levels were measured in tumor tissues collected at diagnosis for a cohort of 229 breast cancer patients by QPCR. Given the association of BRE with DNA damage repair, we subdivided the patient cohort a priori in two groups based on whether they had received radiotherapy as a part of their primary treatment or not. BRE levels were gradually distributed and no difference was observed between radiotherapy-treated or non-treated patients (p = 0.25). The dynamic range of expression was less than 50-fold (5.4 Ct) and levels were normally distributed (based on a Kolmogorov–Smirnov test) (see Fig. 1a). This is in contrast to AML in which BRE is highly expressed in a distinctive subset of patients, while the remaining patients show little variation [26].
Fig. 1.
BRE expression predicts DFS in breast cancer. a BRE expression was gradually distributed among 229 breast cancer patients. No significant differences were observed between radiotherapy- and non-radiotherapy-treated patients. BRE expression was measured by QPCR and normalized with the housekeeping gene PBGD by calculating the ΔCt. Data shown are mean centered. Expression levels between radiotherapy-treated and non-treated patients did not differ significantly (p = 0.25 based on student’s t test). b For Kaplan–Meier analyses, the total cohort was divided into two equally sized groups based on BRE expression (high: solid line; low: dashed line, as indicated). BRE expression has opposing prognostic impact in non-radiotherapy-treated (no RT: upper panel) and radiotherapy-treated (RT: lower panel) patients. In non-radiotherapy-treated patients, the 5-year DFS was 86.6 ± 6.2 % and 75.5 ± 8.7 % for low and high BRE expression, respectively (HR = 2.59, CI = 1.00–6.75, p = 0.05). In radiotherapy-treated patients, the 5-year DFS was 60.2 ± 5.5 % and 78.3 ± 4.5 % for low and high BRE expression, respectively (HR = 0.47, CI = 0.28–0.78, p = 0.004). Patient numbers included in the analyses are indicated in brackets. p values, HR’s and CI’s were calculated by the logrank method. Subdividing the cohort into three groups based on BRE expression obtained comparable results (data not shown)
Comparisons of BRE expression with known clinico-pathological factors showed that BRE expression correlated with tumor size (p = 0.014), but not with any of the other parameters (Table 1). The correlation of BRE expression with tumor size was only observed in radiotherapy-treated patients in which high BRE expression was more often found in smaller tumors (p = 0.005, Table 1).
BRE expression predicts DFS in breast cancer
Gradual differences in BRE expression (using continuous QPCR data) did not correlate with DFS or overall survival (OS) in the total cohort, as tested by univariate Cox regression analysis (DFS: Table 2, OS: data not shown). However, when the cohort was subdivided into radiotherapy-treated and non-treated patients, BRE expression (tested as continuous variable) had prognostic impact on DFS within both groups (Table 2). Remarkably, BRE expression showed opposite effects on prognosis. In the radiotherapy-treated group (N = 169), which accounted for the majority of the patients, high BRE expression correlated with a favorable DFS (Hazard ratio (HR) = 0.72, 95 % confidence interval (CI) = 0.53–0.97, p = 0.030), while in the non-radiotherapy-treated group (N = 59), high BRE expression correlated with a poor prognosis (HR = 1.79, CI = 1.11–2.87, p = 0.016). Similar results were obtained when subdividing patients into two or three groups based on BRE expression, instead of using gradual QPCR data (Table 3, and data not shown).
Table 2.
Univariate analysis of BRE expression in correlation with DFS
| Total cohort | Non-radiotherapy-treated patients | Radiotherapy-treated patients | ||||
|---|---|---|---|---|---|---|
| p | HR (95 % CI) | p | HR (95 % CI) | p | HR (95 % CI) | |
| BRE expression (QPCR data) | 0.342 | 0.877 (0.67–1.15) | 0.016 | 1.79 (1.11–2.87) | 0.030 | 0.72 (0.53–0.97) |
HR hazard ratio; CI confidence interval
Table 3.
Multivariate Cox regression analysis of BRE expression correlation with DFS
| Non-radiotherapy-treated patients | Radiotherapy-treated patients | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariatea | Univariate | Multivariatea | |||||
| p | HR (95 % CI) | p | HR (95 % CI) | p | HR (95 % CI) | p | HR (95 % CI) | |
| BRE | 0.059e | 2.51 | 0.083e | 2.38 | 0.004 | 0.46 | 0.021 | 0.50 |
| (2 groupsb) | (0.97–6.53) | (0.89–6.35) | (0.27–0.79) | (0.27–0.90) | ||||
| Age | 0.349 | 0.98 | 0.616 | 0.99 | 0.112 | 0.98 | 0.020 | 0.97 |
| (continuous) | (0.95–1.02) | (0.95–1.03) | (0.96–1.00) | (0.95–1.00) | ||||
| Menopausal status | 0.838 | 1.07 | 0.140 | 0.81 | ||||
| (post- vs. premenopausal) | (0.57–1.99) | (0.62–1.07) | ||||||
| Tumor sizec | 0.422 | 1.39 | 0.465 | 1.56 | <0.001 | 2.01 | 0.014 | 1.70 |
| (pT1 vs. pT2 vs. pT3/4) | (0.62–3.09) | (0.47–5.15) | (1.42–2.84) | (1.11–2.59) | ||||
| Histological grade | 0.941 | 0.97 | 0.895 | 1.01 | 0.032 | 1.70 | 0.313 | 0.95 |
| (I vs. II vs. III vs. NDd) | (0.39–2.42) | (0.86–1.19) | (1.05–2.74) | (0.86–1.05) | ||||
| Involved lymph nodes | 0.002 | 5.63 | 0.034 | 3.92 | 0.001 | 1.87 | 0.013 | 1.66 |
| (0 vs. 1–3 vs. ≥ 4) | (1.84–17.3) | (1.11–13.8) | (1.30–2.68) | (1.11–2.48) | ||||
| Estrogen receptor status | 0.680 | 0.82 | 0.362 | 0.78 | ||||
| (positive vs. negative) | (0.31–2.15) | (0.45–1.34) | ||||||
| Progesterone receptor status | 0.866 | 0.92 | 0.839 | 0.95 | ||||
| (positive vs. negative) | (0.36–2.39) | (0.55–1.62) | ||||||
aFactors included in multivariate analysis: BRE expression, age, tumor size, histological grade, and involved lymph nodes
bThe two groups are defined as BRE expression above or below the median expression of the total cohort, respectively
cpT1: tumor size ≤2 cm, pT2: tumor size of 2–5 cm, pT3/4: tumor size >5 cm and/or direct extension to chest wall or skin
dAs data on histological grading were missing for a substantial number of patients, this group (ND not done) was included in the multivariate analyses as separate group next to histological grade I, II, or III
eIn non-radiotherapy-treated patients, BRE expression lost its significance when the median expression was used to divide patients based on BRE expression. When subdividing patients into three groups based on BRE expression, BRE was a significant predictor for DFS in both univariate and multivariate models
HR hazard ratio; CI confidence interval
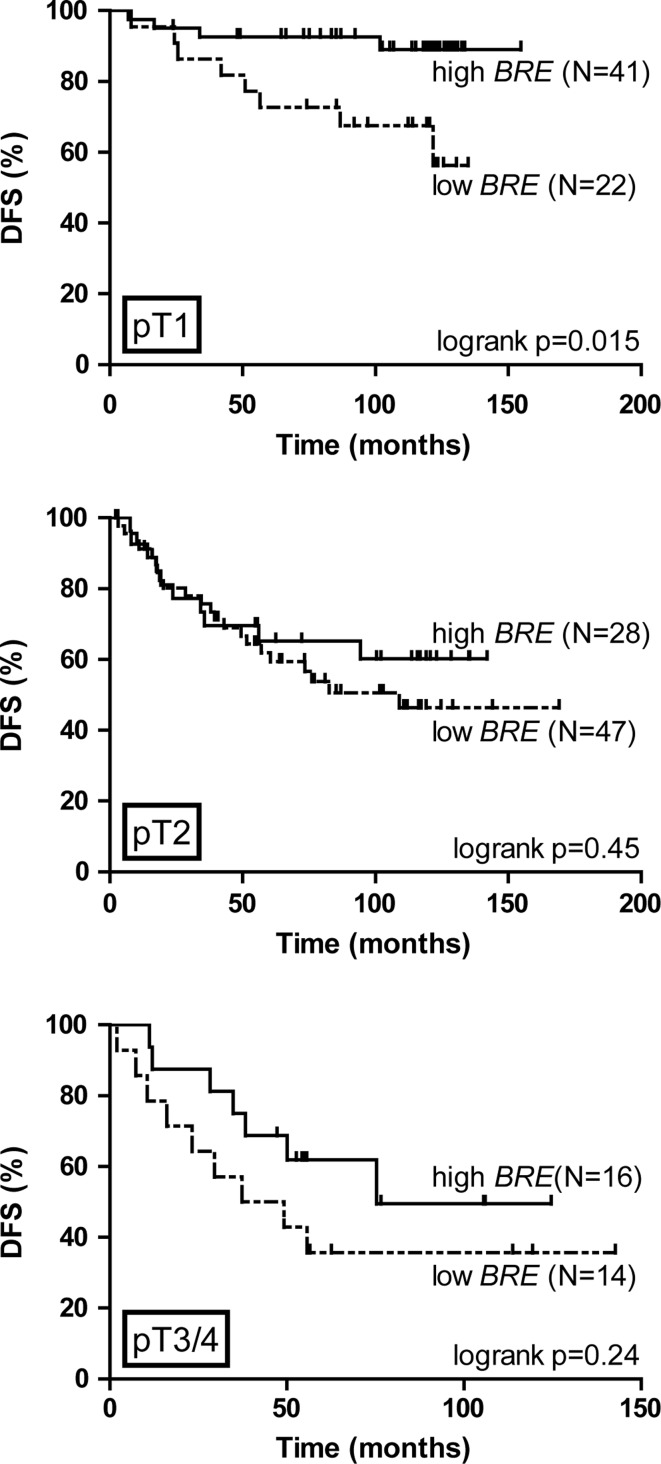
The effect of BRE expression on DFS was visualized by Kaplan–Meier plots by subdividing the total cohort into two groups using the median of BRE expression as cut-off. Among the patients who did not receive radiotherapy, high BRE expression predicted an adverse prognosis validating the Cox regression analysis (HR = 2.59, CI = 1.00–6.75, p = 0.05). High BRE expression predicted a favorable prognosis among the patients who received radiotherapy (HR = 0.47, CI = 0.28–0.78, p = 0.004) (Fig. 1b). Interestingly, within the radiotherapy-treated patients, a significant correlation between BRE expression and DFS was only observed for the group of patients with smaller tumors (HR = 0.23, CI = 0.068–0.75, p = 0. 015), which contained relatively more high BRE expressing patients (Table 1; Fig. 2). No significant prognostic impact was observed in patients with larger tumors (Fig. 2). Radiotherapy was combined with adjuvant systemic treatment for a part of the cohort (see Table 1). To exclude the possibility that the effect of BRE expression on prognosis depended on the combination of radiotherapy and adjuvant treatment, we calculated the effect of BRE expression on DFS for patients treated by radiotherapy only (94 of the 169 patients that received radiotherapy). This analysis showed that also within this subcohort, high BRE expression predicted favorable disease outcome (HR = 0.38, CI = 0.18–0.78, p = 0.009, data not shown). Within the group of patients who did not receive radiotherapy, 75 % did not receive adjuvant treatment either (44 of the 59 patients). Within this group of patients, the impact of BRE expression on DFS lost its significance (data not shown). This might indicate that in non-radiotherapy treated patients, the effect of BRE expression on DFS is dependent on adjuvant treatment. However, as the number of patients receiving adjuvant treatment without radiotherapy was too small, we were not able to test this hypothesis.
Fig. 2.
BRE expression predicts favorable DFS in radiotherapy-treated patients with small tumors. In radiotherapy-treated patients, BRE expression predicts DFS in patients with small tumors (pT1, upper panel). The 5-year DFS was 72.7 ± 9.5 % and 92.6 ± 4.1 % for low and high BRE expression, respectively (HR = 0.23, CI = 0.068–0.75, p = 0.015). For patients with larger tumors, no statistically significant prognostic effect of BRE expression was observed. For this analysis, patients were subdivided into two groups based on BRE expression, as explained in Fig. 1. p-values, HR’s, and CI’s were calculated by the logrank method
BRE expression is an independent prognostic factor in radiotherapy-treated patients
To determine whether BRE expression was an independent prognostic factor for DFS in breast cancer, multivariate Cox regression analyses were performed. These analyses showed that BRE expression was a prognostic factor within the group of radiotherapy-treated patients, independent of other tested prognostic factors such as age, tumor size, lymph node involvement, and histological grade (HR = 0.50, CI = 0.27–0.90, p = 0.021, shown in Table 3). Of note, also age, tumor size, and the number of involved lymph nodes were independent prognostic factors in this group of patients. For non-radiotherapy-treated patients, BRE expression did not correlate significantly with DFS in the multivariate analysis.
BRE expression predicts outcome in a large independent breast cancer cohort
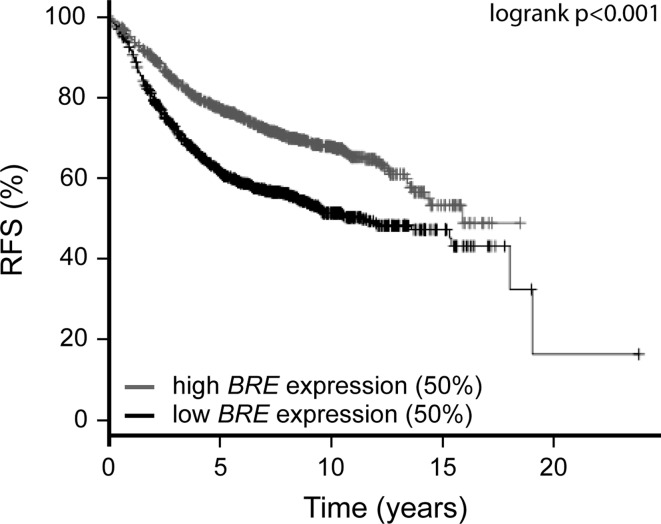
To determine whether BRE expression has an impact on survival in other patient cohorts, we extended our studies to a large independent, publicly available micro-array dataset of 2,324 patients (see Fig. 3, Kaplan–Meier Plotter [32] (www.kmplot.com)). We observed a favorable prognosis for patients with high BRE expression (upper 50 % of the patients) and an adverse survival for patients with low BRE expression (lower 50 %) (HR = 0.59, CI = 0.51–0.68, p < 0.001 after correction for multiple testing). The data of this cohort resembled the data of the first cohort of radiotherapy-treated patients (Fig. 1b). However, as no data were available on the number of patients who received radiotherapy within this publically available cohort, we were unable to test whether the prognostic effect of BRE expression was influenced by radiotherapy treatment.
Fig. 3.
BRE expression predicts relapse-free survival in a cohort of 2,324 breast cancer patients. A publicly available database (Kaplan–Meier Plotter [32]) was used to investigate the effect of BRE expression on relapse-free survival (RFS) in a cohort of 2,324 breast cancer patients. Array data (probe set 211566_s_at) of these patients were used to divide patients into two equally sized groups. High BRE expression predicts a favorable prognosis (HR = 0.51, CI = 0.51–0.68, p < 0.001). p-value, HR, and CI were calculated by the logrank method
Discussion
High expression of the BRCA1 complex member BRE has recently been identified in a subgroup of AML patients in whom it defines favorable prognosis [25, 26]. Here, we show that the expression of this gene also predicted disease outcome in a cohort of 229 non-familial breast cancer patients. Interestingly, the predictive value of BRE expression at diagnosis on DFS depended on whether the patient received subsequent radiotherapy treatment or not. In radiotherapy-treated patients, high BRE expression predicted a favorable disease outcome, whereas in non-radiotherapy-treated patients, it correlated with an adverse outcome (see Fig. 1; Table 3). To extend our studies, BRE expression was evaluated in a publicly available dataset of 2,324 breast cancer patients [32]. In this large cohort, high BRE expression predicted a favorable relapse-free survival, resembling the data of radiotherapy-treated patients within the cohort studied in this manuscript (Fig. 1b). The large cohort of 2,324 patients represents a collection of previously published gene expression datasets, for which integral data on clinico-pathological factors are unavailable. Therefore, we were unable to determine the impact of radiotherapy on the effect of BRE expression on disease outcome in this cohort. The identification of prognostic impact of BRE expression in two independent cohorts warrants further studies in large cohorts to validate the effects found in radiotherapy-treated and non-treated patients.
The fact that BRE expression predicted opposing effects on disease outcomes depending on radiotherapy treatment might imply that there are intrinsic differences in breast cancer patients who are treated or not treated with radiotherapy. Alternatively, there might be a direct effect of high BRE expression on radiotherapy response. The effect of BRE expression on disease outcome was not due to co-treatment with adjuvant therapy within the radiotherapy-treated group of patients as the effect of BRE expression on DFS was also present in the subgroup of radiotherapy-treated patients who did not receive adjuvant treatment. The decision for radiotherapy treatment is closely related to surgical treatment and depends on multiple factors like tumor size and the involvement of axillary lymph nodes. As the patients were consequently not randomly assigned for treatment, it was not possible to explain the opposing effect of BRE expression on the prognosis of radiotherapy-treated versus non-treated patients in this cohort. Therefore, it would be of particular interest to test BRE expression in a cohort of patients who received radiotherapy in a randomized fashion to evaluate a direct effect of BRE expression on therapy outcome.
BRE is a member of the BRCA1 complex involved in DNA double strand break repair [21–24]. This complex is recruited to DNA damaged sites via binding of the complex member Rap80 to ubiquitin chains, which are generated upon DNA damage [33–35]. Mutations in DNA damage repair factors are closely linked to familial breast cancer as 25 % of these cases is characterized by mutations in factors involved in the DNA damage repair pathway, like BRCA1, BRCA2, PTEN, p53, CHEK2, and ATM [7–9, 36–39]. However, in non-familial breast cancer, these mutations are rare. In non-familial cases, associations between low BRCA1 expressions with poor prognosis have been identified [16–19] resembling the observations we made for BRE expression in radiotherapy-treated patients.
Depletion of BRE abrogates BRCA1 foci formation, indicating that BRE is needed for complex formation and downstream DNA repair [22–24, 40]. Several studies have described an increased radiosensitivity of cells after BRE depletion [21, 22]. Next to a role in the BRCA1 complex, BRE is also involved in death receptor-mediated apoptosis as it binds TNFα and FAS receptors, and overexpression of BRE caused resistance to apoptosis induction by various stress-related stimuli [41]. This indicates that BRE serves an anti-apoptotic role following different types of stress. It was therefore unexpected to find a positive correlation between high BRE expression and breast cancer outcome in relation to radiotherapy. High expression would enhance DNA repair and hence would render cells resistant to radiotherapy. Indeed, this reasoning seems to be true for BRCA1 as radiotherapy has been shown to be especially beneficial for patients with low BRCA1 levels, whereas there was no benefit for patients with high BRCA1 levels [42]. On the other hand, high expression of the Mre11/Rad50/Nbs1 complex, also involved in DNA damage repair, predicts a good response to radiotherapy [43], indicating that DNA repair proteins can contribute differentially to radiotherapy response. In this case, high BRE expression might attenuate the DNA damage repair pathway following radiotherapy. Potentially, high BRE expression causes a misbalance in the BRCA1 multi-protein complex formation, thereby reducing the functionality of the complex and rendering cells more sensitive to radiation-induced DNA damage. It would be of particular interest to study the subcellular localization of BRE in these tumors to determine whether responses can be attributed to the DNA damage response or death receptor signaling. The data described in this study indicate that BRE is an interesting candidate for further functional studies in breast cancer to test its effect on radiotherapy responses.
Acknowledgments
This study was financially supported by the Vanderes foundation.
Conflict of interest
The authors declare no conflict of interest in this study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 2.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Buyse M, Loi S, van‘t Veer LJ, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 8.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 9.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 10.Akbari MR, Ghadirian P, Robidoux A, et al. Germline RAP80 mutations and susceptibility to breast cancer. Breast Cancer Res Treat. 2009;113:377–381. doi: 10.1007/s10549-008-9938-z. [DOI] [PubMed] [Google Scholar]
- 11.Nikkila J, Coleman KA, Morrissey D, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28:1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak DJ, Sabbaghian N, Maillet P, Chappuis PO, Foulkes WD, Tischkowitz M. Analysis of the genes coding for the BRCA1-interacting proteins, RAP80 and Abraxas (CCDC98), in high-risk, non-BRCA1/2, multiethnic breast cancer cases. Breast Cancer Res Treat. 2009;117:453–459. doi: 10.1007/s10549-008-0134-y. [DOI] [PubMed] [Google Scholar]
- 13.Osorio A, Barroso A, Garcia MJ, Martinez-Delgado B, Urioste M, Benitez J. Evaluation of the BRCA1 interacting genes RAP80 and CCDC98 in familial breast cancer susceptibility. Breast Cancer Res Treat. 2009;113:371–376. doi: 10.1007/s10549-008-9933-4. [DOI] [PubMed] [Google Scholar]
- 14.Rebbeck TR, Mitra N, Domchek SM, et al. Modification of BRCA1-associated breast and ovarian cancer risk by BRCA1-interacting genes. Cancer Res. 2011;71:5792–5805. doi: 10.1158/0008-5472.CAN-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solyom S, Patterson-Fortin J, Pylkas K, Greenberg RA, Winqvist R. Mutation screening of the MERIT40 gene encoding a novel BRCA1 and RAP80 interacting protein in breast cancer families. Breast Cancer Res Treat. 2010;120:165–168. doi: 10.1007/s10549-009-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008;39:857–865. doi: 10.1016/j.humpath.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Seery LT, Knowlden JM, Gee JM, et al. BRCA1 expression levels predict distant metastasis of sporadic breast cancers. Int J Cancer. 1999;84:258–262. doi: 10.1002/(SICI)1097-0215(19990621)84:3<258::AID-IJC10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 18.Taylor J, Lymboura M, Pace PE, et al. An important role for BRCA1 in breast cancer progression is indicated by its loss in a large proportion of non-familial breast cancers. Int J Cancer. 1998;79:334–342. doi: 10.1002/(SICI)1097-0215(19980821)79:4<334::AID-IJC5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Sakurai T, Mori I, et al. Prognostic significance of BRCA1 expression in Japanese sporadic breast carcinomas. Cancer. 2001;92:54–60. doi: 10.1002/1097-0142(20010701)92:1<54::AID-CNCR1291>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Lambie H, Miremadi A, Pinder SE, et al. Prognostic significance of BRCA1 expression in sporadic breast carcinomas. J Pathol. 2003;200:207–213. doi: 10.1002/path.1348. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y, Hakimi MA, Chen X, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–1099. doi: 10.1016/S1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 22.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao G, Patterson-Fortin J, Messick TE, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balgobind BV, Zwaan CM, Reinhardt D, et al. High BRE expression in pediatric MLL-rearranged AML is associated with favorable outcome. Leukemia. 2010;24:2048–2055. doi: 10.1038/leu.2010.211. [DOI] [PubMed] [Google Scholar]
- 26.Noordermeer SM, Sanders MA, Gilissen C, et al. High BRE expression predicts favorable outcome in adult acute myeloid leukemia, in particular among MLL-AF9-positive patients. Blood. 2011;118:5613–5621. doi: 10.1182/blood-2011-06-359182. [DOI] [PubMed] [Google Scholar]
- 27.Noordermeer SM, Monteferrario D, Sanders MA, Bullinger L, Jansen JH, van der Reijden BA. Improved classification of MLL-AF9-positive acute myeloid leukemia patients based on BRE and EVI1 expression. Blood. 2012;119:4335–4337. doi: 10.1182/blood-2012-02-405019. [DOI] [PubMed] [Google Scholar]
- 28.Wennemers M, Bussink J, Grebenchtchikov N, Sweep FC, Span PN. TRIB3 protein denotes a good prognosis in breast cancer patients and is associated with hypoxia sensitivity. Radiother Oncol. 2011;101:198–202. doi: 10.1016/j.radonc.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 29.Wennemers M, Bussink J, Scheijen B, et al. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 2011;13:R82. doi: 10.1186/bcr2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Span PN, Waanders E, Manders P, et al. Mammaglobin is associated with low-grade, steroid receptor-positive breast tumors from postmenopausal patients, and has independent prognostic value for relapse-free survival time. J Clin Oncol. 2004;22:691–698. doi: 10.1200/JCO.2004.01.072. [DOI] [PubMed] [Google Scholar]
- 31.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 32.Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 33.Sobhian B, Shao G, Lilli DR, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.FitzGerald MG, Marsh DJ, Wahrer D, et al. Germline mutations in PTEN are an infrequent cause of genetic predisposition to breast cancer. Oncogene. 1998;17:727–731. doi: 10.1038/sj.onc.1201984. [DOI] [PubMed] [Google Scholar]
- 37.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 38.Sidransky D, Tokino T, Helzlsouer K, et al. Inherited p53 gene mutations in breast cancer. Cancer Res. 1992;52:2984–2986. [PubMed] [Google Scholar]
- 39.Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97:813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, Kim JA, Castillo A, Huang M, Liu J, Wang B. NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes. J Biol Chem. 2011;286:11734–11745. doi: 10.1074/jbc.M110.200857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Ching AK, Chan BC, et al. A death receptor-associated anti-apoptotic protein, BRE, inhibits mitochondrial apoptotic pathway. J Biol Chem. 2004;279:52106–52116. doi: 10.1074/jbc.M408678200. [DOI] [PubMed] [Google Scholar]
- 42.Soderlund K, Skoog L, Fornander T, Askmalm MS. The BRCA1/BRCA2/Rad51 complex is a prognostic and predictive factor in early breast cancer. Radiother Oncol. 2007;84:242–251. doi: 10.1016/j.radonc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Soderlund K, Stal O, Skoog L, Rutqvist LE, Nordenskjold B, Askmalm MS. Intact Mre11/Rad50/Nbs1 complex predicts good response to radiotherapy in early breast cancer. Int J Radiat Oncol Biol Phys. 2007;68:50–58. doi: 10.1016/j.ijrobp.2006.12.005. [DOI] [PubMed] [Google Scholar]