Abstract
It has recently been proposed that a three-gene model (SCMGENE) that measures ESR1, ERBB2, and AURKA identifies the major breast cancer intrinsic subtypes and provides robust discrimination for clinical use in a manner very similar to a 50-gene subtype predictor (PAM50). However, the clinical relevance of both predictors was not fully explored, which is needed given that a ~30 % discordance rate between these two predictors was observed. Using the same datasets and subtype calls provided by Haibe-Kains and colleagues, we compared the SCMGENE assignments and the research-based PAM50 assignments in terms of their ability to (1) predict patient outcome, (2) predict pathological complete response (pCR) after anthracycline/taxane-based chemotherapy, and (3) capture the main biological diversity displayed by all genes from a microarray. In terms of survival predictions, both assays provided independent prognostic information from each other and beyond the data provided by standard clinical–pathological variables; however, the amount of prognostic information was found to be significantly greater with the PAM50 assay than the SCMGENE assay. In terms of chemotherapy response, the PAM50 assay was the only assay to provide independent predictive information of pCR in multivariate models. Finally, compared to the SCMGENE predictor, the PAM50 assay explained a significantly greater amount of gene expression diversity as captured by the two main principal components of the breast cancer microarray data. Our results show that classification of the major and clinically relevant molecular subtypes of breast cancer are best captured using larger gene panels.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-012-2143-0) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Microarrays, PAM50, Prognosis, Gene expression
Introduction
Over the years, global gene expression analyses have identified at least four intrinsic subtypes of breast cancer (Luminal A, Luminal B, HER2-enriched, and Basal-like) and a normal-like group with significant differences in terms of their risk factors, incidence, baseline prognoses and responses to systemic therapies [1–4]. In 2009, we reported a clinically applicable gene expression-based predictor that robustly identifies these main intrinsic subtypes by quantitative measurement of 50 genes (i.e., PAM50) [1]. Identification of these molecular subtypes using pathology-based surrogate definitions based upon hormone receptors (HRs), HER2 and Ki-67 expressions has been adopted by the 2011 St. Gallen Consensus Conference for treatment decision-making in early breast cancer [5], however, controversy exists as to whether these complex molecular subtypes can be effectively captured using four or less biomarkers.
Recently, Haibe-Kains et al. [6] reported a mRNA expression predictor that classifies tumors into four molecular entities (ER+/HER2−/Low Proliferative, ER+/HER2−/High Proliferative, HER2+ and ER−/HER2−) by quantitative measurement of three genes (ESR1, ERBB2 and AURKA). Similar to the PAM50 subtype predictions, the molecular entities identified by the SCMGENE predictor were found significantly associated with survival outcome [6]. However, a direct head-to-head comparison between both predictors was not performed despite that fact that the concordance (i.e., κ score) between these two predictors was 0.59 (0.58–0.61), which is considered moderate agreement and similar to the κ scores obtained when histological grade is evaluated by two independent observers [7].
In this study, we compared the SCMGENE assignments and the research-based PAM50 assignments in terms of their ability to (1) predict patient outcome, (2) predict pathological complete response (pCR) after anthracycline/taxane-based chemotherapy, and (3) capture the main biological diversity displayed by all genes from a microarray.
Materials and methods
Clinical and gene expression data
We used the clinical (Supplemental file: jnci-JNCI-11-0924-s02.csv) and gene expression data (http://www.compbio.dfci.harvard.edu/pubs/sbtpaper/data.zip) as provided by Haibe-Kains et al. [6]. For survival predictions, we used distant metastasis-free survival as the endpoint since it provides the largest number of patients that can be evaluated across 13 datasets (CAL [8], EMC2 [9], DFHCC [10], MAINZ [11], MDA5 [12], MSK [13], NKI [14], TAM [15], TRANSBIG [16], UCSF [17], UNT [18], VDX [19] and VDX3 [20]). None of the datasets (or samples) used for survival (or response prediction) were used to derive the SCMGENE or the PAM50 subtype predictor.
To compare chemotherapy response data, we used the clinical data of one of the datasets (MAQC2 [GSE20194] [21]) evaluated by Haibe-Kains et al. [6], which is composed of 230 pre-treatment samples with annotated response data (pCR vs. residual disease [RD]) after neoadjuvant anthracycline/taxane-based chemotherapy. Samples that received trastuzumab were excluded.
Combined microarray dataset
Eighteen Affymetrix and Agilent-based datasets (CAL [8], DFHCC [10], DUKE [22], EORTC10994 [23], EXPO [24], KOO [25], MAINZ [11], MAQC2 [21], MDA4 [26], MSK [13], NKI [14], PNC [27], STK [28], TRANSBIG [16], UNC337 [29], UNT [18], UPP [30] and VDX [19]) as provided in Haibe-Kains et al. [6] and with an appropriate distribution of ER+ (50–90 %, as defined by IHC) versus ER− tumors were combined into a single gene expression matrix. Probes mapping to the same gene (Entrez ID as defined by the manufacturer) were averaged to generate independent expression estimates. In each cohort, genes were median centered and standardized to zero mean and unit variance.
Statistical analyses
Distant metastasis-free survival univariate and multivariate analysis were calculated using a Cox proportional regression model. Likelihood ratio statistics of subtypes defined by the PAM50 or the SCMGENE predictors were also evaluated after accounting for clinical–pathological variables (age at diagnosis, nodal status, and tumor size) and type of systemic adjuvant treatment (chemotherapy, endocrine, and none). Models were first conditioned on one predictor and the clinical–pathological variables, and then the significance of the other was tested. Chemotherapy response (pCR vs. RD) predictions of each variable were evaluated using univariate and multivariate logistic regression analyses. Finally, R 2 values of each predictor (SCMGENE or PAM50) for each principal component (PC) were calculated using a simple linear regression model. All statistical computations were performed in R v.2.8.1 (http://www.cran.r-project.org).
Results
Outcome prediction
To compare the ability of the SCMGENE and PAM50 assays to predict patient outcome, we performed Cox proportional hazard regression analyses using the entire combined dataset as provided by Haibe-Kains et al. [6]. In the multivariate model (MVA), both predictors were found significantly associated with distant metastasis-free survival (Table 1) and the Luminals A and B segregation of the PAM50 assay was found significantly associated with outcome, whereas the ER+/HER2−/Low Proliferative and ER+/HER2−/High Proliferative segregation of the SCMGENE predictor was not. Conversely, distant metastasis-free survival differences of the ER−/HER2− versus the ER+/HER2−/Low Proliferative groups were found significant, whereas the Basal-like versus Luminal A segregation was not.
Table 1.
Distant metastasis-free survival Cox proportional hazards models of primary breast cancer patients
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower 95 % | Upper 95 % | p Value | HR | Lower 95 % | Upper 95 % | p Value | |
| Age (cont. variable) | 0.989 | 0.983 | 0.996 | 0.003 | 0.996 | 0.988 | 1.003 | 0.257 |
| Node status | 1.176 | 0.851 | 0.992 | 0.063 | 1.695 | 1.315 | 2.184 | <0.001 |
| Tumor size T2–T4 versus T0–T1 | 1.305 | 1.104 | 1.541 | 0.002 | 1.242 | 1.042 | 1.480 | 0.015 |
| Treatment (yes vs. no) | 0.973 | 0.845 | 1.121 | 0.707 | 0.547 | 0.428 | 0.700 | <0.001 |
| PAM50 | ||||||||
| Luminal A | 1.0 | – | – | – | 1.0 | – | – | – |
| Luminal B | 1.797 | 1.503 | 2.149 | <0.001 | 2.041 | 1.578 | 2.641 | <0.001 |
| HER2-E | 2.677 | 2.120 | 3.380 | <0.001 | 1.648 | 1.073 | 2.530 | 0.023 |
| Basal-like | 2.144 | 1.737 | 2.647 | <0.001 | 1.312 | 0.812 | 2.121 | 0.268 |
| Normal-like | 1.073 | 0.670 | 1.718 | 0.769 | 1.024 | 0.572 | 1.835 | 0.936 |
| Three-gene signature | ||||||||
| ER+/HER2−/Low Prolif | 1.0 | – | – | – | 1.0 | – | – | – |
| ER+/HER2−/High Prolif | 1.852 | 1.531 | 2.241 | <0.001 | 1.153 | 0.882 | 1.508 | 0.297 |
| HER2+ | 2.785 | 2.196 | 3.533 | <0.001 | 1.588 | 1.053 | 2.395 | 0.028 |
| ER−/HER2− | 2.536 | 2.041 | 3.150 | <0.001 | 1.762 | 1.095 | 2.835 | 0.020 |
HER2-E HER2-enriched, Prolif proliferation, HR hazard ratio
To compare the amount of independent prognostic information provided by each predictor, we estimated the likelihood ratio statistic of each predictor in a model that already included clinical–pathological variables (age, tumor size, treatment and nodal status) and the other predictor. The results revealed that the PAM50 subtypes provide a larger amount of independent prognostic information than the SCMGENE subtypes when using the entire cohort of heterogeneously treated patients (Fig. 1A, B). Similar results were observed when using the subset of patients that did not receive adjuvant systemic therapy (Fig. 1C, D), and in the subset of patients with HR+ tumors that received adjuvant tamoxifen-only (Fig. 1E, F).
Fig. 1.
Distant metastasis-free survival likelihood ratio statistics of subtypes defined by the PAM50 or the SCMGENE predictors, after accounting for clinical–pathological variables (age at diagnosis, nodal status, treatment and tumor size). Models were first conditioned on one predictor and the clinical–pathological variables, and then the significance of the other was tested. (A – B) Entire combined dataset (n = 2,008), (C–D) subset of patients that did not receive adjuvant systemic therapy (n = 994), (E–F) subset of patients with HR+ tumors that received adjuvant tamoxifen-only (n = 491). Similar results are obtained if a term for dataset is included in the model
Chemotherapy response prediction
To compare the ability of the PAM50 and SCMGENE assays to predict response to chemotherapy, we evaluated the MAQC2 (GSE20194) [21] dataset included in Haibe-Kains et al. [6] analyses. This cohort is composed of 226 pre-treatment samples with annotated response data (pCR vs. RD) after neoadjuvant anthracycline/taxane-based chemotherapy (without trastuzumab for HER2+ disease). As shown in Table 2, although both assays predicted response in univariate analysis, the PAM50 assay was the only one to provide independent predictive information in the MVA model.
Table 2.
| Variables | N | pCR rate (%) | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | Lower 95 % | Upper 95 % | p Value | OR | Lower 95 % | Upper 95 % | p Value | |||
| Age (cont. variable) | – | – | 1.0 | 0.95 | 1.01 | 0.169 | – | – | – | – |
| Tumor size | ||||||||||
| T0–T1 | 23 | 35 | 1.0 | – | – | – | 1.0 | – | – | – |
| T2–T4 | 203 | 19 | 2.3 | 0.92 | 5.86 | 0.076 | 0.4 | 0.13 | 1.23 | 0.111 |
| PAM50 | ||||||||||
| Luminal A | 66 | 3 | 1.0 | – | – | – | 1.0 | – | – | – |
| Luminal B | 66 | 9 | 3.2 | 0.62 | 16.47 | 0.164 | 5.2 | 0.68 | 37.97 | 0.108 |
| HER2-E | 28 | 46 | 23.5 | 5.25 | 105.36 | <0.001 | 12.5 | 1.46 | 145.68 | 0.030 |
| Basal-like | 59 | 42 | 27.7 | 5.65 | 136.18 | <0.001 | 25.3 | 2.64 | 255.95 | 0.005 |
| Normal-like | 7 | 0 | 0.0 | 0.00 | – | 0.988 | 0.0 | 0.00 | – | 0.988 |
| Three-gene signature | ||||||||||
| ER+/HER2−/Low Prolif | 52 | 4 | 1.0 | – | – | – | 1.0 | – | – | – |
| ER+/HER2−/High Prolif | 85 | 8 | 2.2 | 0.45 | 11.23 | 0.325 | 0.6 | 0.08 | 4.62 | 0.633 |
| HER2+ | 24 | 50 | 25.0 | 4.93 | 126.80 | <0.001 | 3.9 | 0.34 | 46.46 | 0.275 |
| ER−/HER2− | 65 | 38 | 15.6 | 3.49 | 69.93 | <0.001 | 0.9 | 0.09 | 9.97 | 0.954 |
HER2-E HER2-enriched, Prolif proliferation, OR odds ratio
Of note, the association of the PAM50 subtype with response was strengthened when PAM50 subtyping of the MAQC2 dataset was performed after median centering the PAM50 genes/rows (Supplemental Table 1). In fact, we and others have previously proposed median gene centering to minimize technical bias and allow the correct identification of the PAM50 intrinsic subtypes when appropriate representation of ER−, ER+, and HER2+ samples is available [31, 32]. Median gene centering of the UNC337 dataset before PAM50 or SCMGENE predictions also improved the survival classifications (Supplemental Fig. 1).
Capturing the main biological diversity
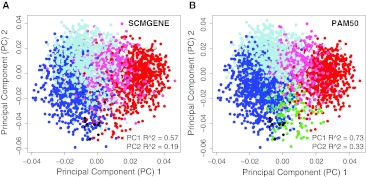
Finally, to compare both predictors in terms of their ability to capture the main biological diversity displayed by all genes in a breast cancer microarray, we first combined 18 datasets evaluated by Haibe-Kains et al. [6] and identified the two main principal components (PC1 and PC2). Compared to the SCMGENE subtypes, the PAM50 subtypes explained substantially more variation in gene expression for both PC1 and PC2 (Fig. 2a, b), with these components being especially prominent for the separation of the Luminal A (or ER+/HER2−/Low Proliferative) and Luminal B (or ER+/HER2−/High Proliferative) subtypes. To confirm these findings, we also evaluated all PCs in each normalized dataset provided by Haibe-Kains et al. [6] and observed that among 483 PCs significantly explained by either one of the predictors, the PAM50 explained 2.27 times more independent variation in expression than the SCMGENE assay.
Fig. 2.
PC1 and PC2 loading plots of 3,316 samples using 18 Affymetrix and Agilent-based datasets taken from Haibe-Kains et al. [6]. Samples colored based on the a SCMGENE calls, or b PAM50 subtype calls. PC1 and PC2 R 2 values obtained from simple linear regression models are shown. Only datasets with >50 % and <90 % ER+ tumors were included in this analysis. Blue Luminal A or ER+/HER2−/Low Proliferative, light blue Luminal B or ER+/HER2−/High Proliferative, pink HER2-enriched or HER2+, red Basal-like or ER−/HER2−, green normal-like, black normal breast samples (only present in the UNC337 dataset [29]). For the UNC337 dataset, we colored samples based on the subtype calls obtained after median centering as shown in Supplemental Fig. 1
Discussion
Our results presented here, using the same data provided by Haibe-Kains et al. [6], suggest that (1) the SCMGENE and the PAM50 predictors should not be considered the same in terms of outcome prediction; (2) both provide independent prognostic information; (3) the amount of prognostic information provided by the PAM50 predictor is greater than the information provided by the SCMGENE predictor; and (4) the PAM50 assay is the only independent predictor of neoadjuvant chemotherapy response.
A potential explanation of our findings is that the biological diversity of breast cancer is better captured using the quantitative measurement of the 50 PAM50 gene set compared to the 3 genes of the SCMGENE assay. This finding is further supported by our previous data during the PAM50 assay development, where the minimum number of genes required to identify the intrinsic molecular subtypes, as defined by subtype classifications based upon the ~1,900 intrinsic gene list with a 93 % accuracy, was the final selected 50 genes [1]. In fact, gene sets with less than 50 genes showed significantly worse accuracies, particularly for tumors of the Luminal B and HER2-enriched subtypes (Supplemental Fig. 2). Importantly, only 33.3 % (12/36) of all microarray datasets evaluated in Haibe-Kains et al. [6] had all the PAM50 genes available, whereas 100 % of the datasets had all three genes of the SCMGENE assay, thus highlighting another caveat of this study.
In total, these analyses show that a combination of ER, HER2, and a single proliferation biomarker (i.e., AURKA) is prognostic, but is suboptimal to capture the biological diversity of breast cancers, which has similar implications for the capture of this biological diversity using IHC-based methods. Although a head-to-head comparison of both assays in terms of their clinical utility might be warranted in the future, our results suggest that classification of the major and clinically relevant molecular subtypes is better achieved using larger gene sets that capture a greater proportion of the biological diversity of breast cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1. Logistic regression models of response in the MAQC2 neoadjuvant breast cancer dataset (n = 226) using the PAM50 subtype calls obtained after median centering the dataset as recommended in Perou et al. [31] and Lusa et al. [32]. Supplemental Fig. 1. PAM50 and SCMGENE subtype call differences obtained in the UNC337 dataset (GSE18229) with and without a platform normalization step. a Distribution of the SCMGENE and PAM50 subtype calls before and after median gene value centering of the dataset. Relapse free survival curves of the subtypes identified using the SCMGENE and PAM50 predictors obtained b before and c after median gene centering. Supplemental Fig. 2. Cross-validation performance on the PAM50 training dataset of different gene subsets of the starting ~1,900 genes, using the selected nearest centroid classification model. Note that the Luminal B, and HER2-enriched subtypes, are the most sensitive to the lower numbers of genes being used in the model, and thus if less than the 50 genes are used, these two subtypes accuracy will be the most compromised. (PDF 999 kb)
Acknowledgments
This study was supported by funds from the NCI Breast SPORE Program (P50-CA58223-09A1), by RO1-CA138255, by the Breast Cancer Research Foundation, and the Sociedad Española de Oncología Médica (SEOM). A. Prat is affiliated to the Medicine PhD Program of the Autonomous University of Barcelona (UAB), Spain.
Conflict of interest
C. M. P. is a stock holder of BioClassifier LLC. C. M. P. and J. S. P. have filed a patent on the PAM50 assay. A. P. and C. F. have declared no conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn H-J, Members P Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haibe-Kains B, Desmedt C, Loi S, Culhane AC, Bontempi G, Quackenbush J, Sotiriou C. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104:311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, Ellis M, Perou C. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2011;6:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin K, DeVries S, Fridlyand J, Spellman P, Roydasqupta R, Kuo W, Lapuk A, Neve R, Quian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Bos PD, Zhang XHF, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Eklund AC, Juul N, Haibe-Kains B, Workman CT, Richardson AL, Szallasi Z, Swanton C. Minimising immunohistochemical false negative ER classification using a complementary 23 gene expression signature of ER status. PLoS ONE. 2010;5(12):e15031. doi: 10.1371/journal.pone.0015031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr H-A, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 12.Symmans WF, Hatzis C, Sotiriou C, Andre F, Peintinger F, Regitnig P, Daxenbichler G, Desmedt C, Domont J, Marth C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minn A, Gupta G, Siegel P, Bos P, Shu W, Giri D, Viale A, Oshen A, Gerald W, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vijver MJ, He YD, van ‘t Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 15.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt A, Gillet C, Ellis P, Ryder K, Reid J, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9(1):239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 17.Korkola J, Blaveri E, DeVries S, Moore D, Hwang ES, Chen Y-Y, Estep A, Chew K, Jensen R, Waldman F. Identification of a robust gene signature that predicts breast cancer outcome in independent data sets. BMC Cancer. 2007;7(1):61. doi: 10.1186/1471-2407-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Sieuwerts A, McGreevy M, Casey G, Cufer T, Paradiso A, Harbeck N, Span P, Hicks D, Crowe J, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116(2):303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 21.Popovici V, Chen W, Gallas B, Hatzis C, Shi W, Samuelson F, Nikolsky Y, Tsyganova M, Ishkin A, Nikolskaya T. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 23.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, MacGrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24(29):4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 24.EXPO Project of the International Genomics Consortium (IGC). https://expo.intgen.org/geo/. Accessed 20 May 2012
- 25.Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 26.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 27.Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M, Deplus R, Luciani J, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3(12):726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perou C, Parker J, Prat A, Ellis M, Bernard P. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010;11(8):718–719. doi: 10.1016/S1470-2045(10)70176-5. [DOI] [PubMed] [Google Scholar]
- 32.Lusa L, McShane LM, Reid JF, De Cecco L, Ambrogi F, Biganzoli E, Gariboldi M, Pierotti MA. Challenges in projecting clustering results across gene expression profiling datasets. J Natl Cancer Inst. 2007;99(22):1715–1723. doi: 10.1093/jnci/djm216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Logistic regression models of response in the MAQC2 neoadjuvant breast cancer dataset (n = 226) using the PAM50 subtype calls obtained after median centering the dataset as recommended in Perou et al. [31] and Lusa et al. [32]. Supplemental Fig. 1. PAM50 and SCMGENE subtype call differences obtained in the UNC337 dataset (GSE18229) with and without a platform normalization step. a Distribution of the SCMGENE and PAM50 subtype calls before and after median gene value centering of the dataset. Relapse free survival curves of the subtypes identified using the SCMGENE and PAM50 predictors obtained b before and c after median gene centering. Supplemental Fig. 2. Cross-validation performance on the PAM50 training dataset of different gene subsets of the starting ~1,900 genes, using the selected nearest centroid classification model. Note that the Luminal B, and HER2-enriched subtypes, are the most sensitive to the lower numbers of genes being used in the model, and thus if less than the 50 genes are used, these two subtypes accuracy will be the most compromised. (PDF 999 kb)