Abstract
Metaproteomics and its potential applications are very promising to study microbial activity in environmental samples and to obtain a deeper understanding of microbial interactions. However, due to the complexity of soil samples the exhaustive extraction of proteins is a major challenge. We compared soil protein extraction protocols in terms of their protein extraction efficiency for two different soil types. Four different protein extraction procedures were applied based on (a) SDS extraction without phenol, (b) NaOH and subsequent phenol extraction, (c) SDS–phenol extraction and (d) SDS–phenol extraction with prior washing steps. To assess the suitability of these methods for the functional analysis of the soil metaproteome, they were applied to a potting soil high in organic matter and a forest soil. Proteins were analyzed by two-dimensional liquid chromatography/tandem mass spectrometry (2D-LC–MS/MS) and the number of unique spectra as well as the number of assigned proteins for each of the respective protocols was compared. In both soil types, extraction with SDS–phenol (c) resulted in “high” numbers of proteins. Moreover, a spiking experiment was conducted to evaluate protein recovery. To this end sterilized forest soil was amended with proteins from pure cultures of Pectobacterium carotovorum and Aspergillus nidulans. The protein recovery in the spiking experiment was almost 50%. Our study demonstrates that a critical evaluation of the extraction protocol is crucial for the quality of the metaproteomics data, especially in highly complex samples like natural soils.
Keywords: Soil metaproteomics, Soil protein extraction, Microbial communities, Mass spectrometry, Microbiology, Microbial ecology
Highlights
► Comparison of protein recovery from natural soil samples with different protocols. ► Direct soil protein extraction with gel-free separation techniques (2D-LC–MS/MS). ► Unique sets of proteins were observed for individual extraction approaches. ► Only small overlap of unique spectra between four different extraction protocols.
1. Introduction
Out of all natural environments, soils probably contain the greatest microbial community in terms of biomass and diversity, which classifies them as one of the most challenging habitats for microbiologists (Mocali and Benedetti, 2010). Recently, several novel molecular techniques have been employed to soil samples, for example metagenomics, metatranscriptomics and – so far to a smaller degree – metaproteomics (Bastida et al., 2009). While metagenomics and -transcriptomics allow to assess “biodiversity” in depth, metaproteome analyses provide a direct measure of proteins present in an environmental sample, offering functional information especially at intracellular level (Bastida et al., 2012). Metaproteomics have been used to analyse the community function and structure in more detail, as demonstrated in samples with limited diversity by VerBerkmoes et al. (2009). This “new approaches” will reach its greatest accomplishments through integration with other approaches such as process measurements, e.g. soil respiration and enzyme activities, as partly demonstrated by Schneider et al. (2012). Metaproteomics in environmental samples is an ambitious task regarding resolution and yields of proteins. The preparation of samples is a crucial step in proteome analysis to obtain high-quality resolution (Wang et al., 2006). There are several methodological challenges for the application of proteomics to soil samples due to their complexity (Bastida et al., 2009; Nannipieri, 2006) as (i) the abundance of proteins in soil is sometimes low, (ii) samples feature spatial distribution, heterogeneity, high microbial diversity and dynamics of soil microbial communities and (iii) extracellular enzymes often strongly adhere or adsorb onto soil minerals or entrapment by humic colloids (Nannipieri, 2006). The adsorption of proteins has several implications on soil proteomics, for it stabilizes secreted enzymes and protects them against proteolysis (Nannipieri, 2006), and can lead to reduced although not eliminated catalytic activity after clay adsorption (Nielsen et al., 2006). Furthermore this may constitute a strong background against protein expression profiles, and hinder detection of metabolic pathways in a soil microbial community at a given time because stabilized proteins are not related to actual microbial activity (Giagnoni et al., 2012; Nannipieri, 2006). Correct application of soil metaproteomics needs to consider that intracellular N represents on average only 4% of total organic N in soils (Nannipieri, 2006), which means that most organic N (30–45%) is present in form of stabilized extracellular amino acids. Recent soil metaproteomic studies based their investigations mostly on indirect extractions of proteins, were microbial cells are extracted from the soil matrix, including an enrichment of these cells prior to protein extraction (Williams and Taylor, 2010). However, approaches that directly extract proteins from soil could be advantageous in terms of completeness of the extracted proteome and they minimize changes in proteome composition during sample preparation (Benndorf et al., 2007; Nannipieri, 2006; Schulze et al., 2005). A critical step of direct extraction is cell lysis within the soil sample, which was tackled with different approaches, namely (i) sonification of the extracts, (ii) boiling and (iii) use of SDS or NaOH in the extraction buffer. Soils are chemically complex environments with a variety of adsorbing surfaces such as clays and humic acids (Nielsen et al., 2006). Proteins are generally rapidly adsorbed onto clays, a process which is reversible to a limited extent (Nielsen et al., 2006) but is suggested to obstruct protein extraction and purification (Giagnoni et al., 2012) as well as quantification, separation and identification (Benndorf et al., 2007). The extraction step of proteins is specifically critical due to interfering humic acids which are usually co-extracted (Siggins et al., 2012). To target the problem of co-extraction of contaminants three out of four of the applied protocols in this study contain a phenol extraction step in order to remove interfering substances.
The aim of the present study was to evaluate different extraction protocols with respect to protein extraction efficiency. To this end we applied four recently described protocols to extract proteins from two different soils, a forest soil and a potting soil and analysed them by a state-of-the art proteomics approach (2D-LC–MS/MS). Furthermore, we used two protocols in a spiking experiment were a protein mixture from pure cultures were added to a forest soil sample to evaluate protein recovery rates.
2. Materials and methods
2.1. Soil samples
The forest soil was sampled in February 2010 from the Ah layer (0–10 cm) of the soil profile in Schottenwald (48°14′N 16°15′E) in the Vienna Woods. Soil type is a Dystric Cambisol over sandstone, soil texture is silty loam with a pH of 4.4 and a C:N ratio of 16, Corg is 37.7 mg g−1 and Ntot is 2.38 mg g−1 (Kitzler et al., 2006) with high clay fraction (0.19) in comparison to other forest soil types (Stange et al., 2000). For comparison a customary potting soil bought from “https://einheitserde.de”, consisting of 50% white peat, 25% clay and 25% pumice, which is fertilized with nitrogen (340 mg l−1), phosphorus (260 mg l−1) and potassium (330 mg l−1), was used to evaluate differences in extraction protocols and the resulting microbial community composition. One replicate per extraction protocol and soil sample was processed, in order to evaluate the different extraction protocols on the two different soil samples.
2.2. Metaproteome analysis
2.2.1. Soil protein extraction protocols
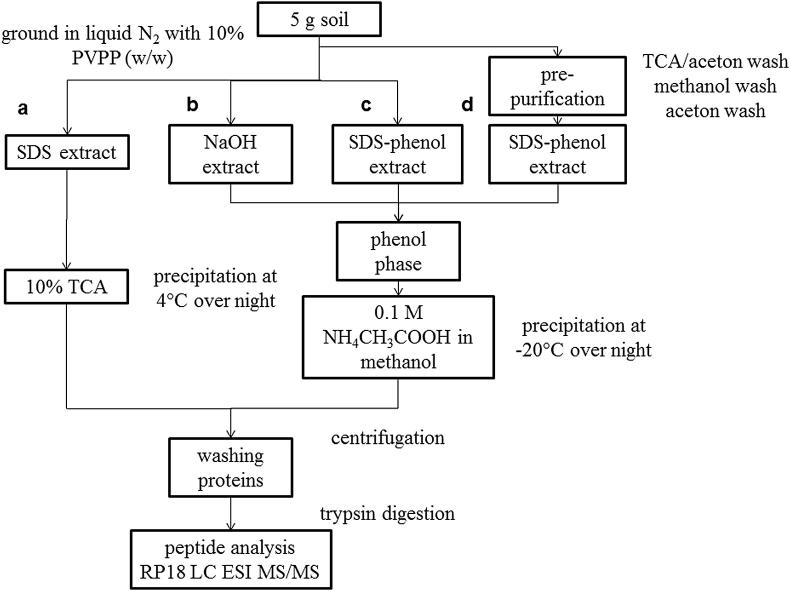
Four different extraction protocols were tested in the present study (the schematic flow of all four extraction procedures is given in Fig. 1). Five gram of fresh sieved (<2 mm) soil was amended with 10% (w/w) polyvinylpolypyrrolidone (PVPP) to clean samples from interfering substances (e.g. humic acids) and ground with a pestle in liquid nitrogen to minimize proteolysis and protein degradation. Extraction buffer was added in a 1:2 to 1:3 (w/v) ratio depending on the water content of the respective soil (dry soil 1:3, wet soil 1:2; potting soil was drier than forest soil). The extraction buffer varied according to the respective protocol (see description of protocols in the next sections: a, b, c, d). The suspension was sonicated with an ultrasonic homogenizer (SONOPLUS, Bandelin, Berlin, Germany) on ice at 90% pulsing and a maximum of 40% energy twice for 1 min to break soil aggregates and to lyse the cells, followed by shaking at 150 rpm and 20 °C (30 min). A second extraction step (sequential extraction) was performed as described in the respective protocols with shaking in fresh extraction buffer for 1 h instead of 30 min. After both steps the suspension was centrifuged at 3220 g 20 min at 4 °C and the resulting supernatants were pooled and precipitated using either 10% tri-chloric acid (TCA) overnight at 4 °C (TCA precipitation) or a 5-fold amount of 0.1 M ammonium acetate in methanol (p.a) over night at −20 °C (ammonium acetate precipitation). The rationale behind precipitation is to concentrate the samples and to remove substances which interfere with further processing (protein digestion, peptide separation and MS analysis). The protocols including a phenol extraction step were precipitated using ammonium acetate in methanol, whereas in the extraction protocol using only SDS proteins were precipitated with TCA.
-
a)
SDS (sodiumdodecylsulfate)
Fig. 1.
Schematic workflow for protein extraction protocols. Four different protocols were used for the extraction of proteins from a forest soil and a potting soil, while for the described spiking experiment procedures b) and c) were used. Abbreviations: TCA (Trichloric acid), ESI (Electron spray ionisation), MS (mass spectroscopy), RP (reversed phase), SDS (sodium dodecyl sulfate), PVPP (polyvinylpolypyrrolidone).
SDS is an effective solubilizing detergent and is therefore used to maximize protein extraction of proteins by breaking existing intra-protein interactions and preventing protein aggregation. Soil samples amended with the SDS extraction buffer (50 mM Tris, 1% SDS pH 7.5) were vortexed and sonicated as described above. To lyse cells, the suspension was boiled for 20 min and vortexing and sonication was repeated prior to centrifugation (20 min and 4 °C at 3220 g). The supernatants were combined and proteins were precipitated with TCA.
-
b)
NaOH (Benndorf et al., 2007)
Benndorf et al. (2007) described that incubation with NaOH releases humic acids and proteins from soil minerals, and simultaneously disrupt microbes. To separate humic acids and proteins a subsequent phenol extraction was used. Soil samples in extraction buffer (0.1 M NaOH) were vortexed and sonicated as described above. The supernatants were amended with a mixture of phenol and milliQ water (8:5). This suspension was shaken vigorously for 1 h at 20 °C, followed by centrifugation (20 min and 4 °C at 3220 g). The water was removed and the lower phenol phase was washed with the same amount of milliQ water by gentle vortexing for 5 min and centrifugation (20 min and 4 °C at 3220 g). The lower phenol phases were transferred to a new tube and precipitated with 0.1 M ammonium acetate.
-
c)
SP (SDS–phenol)
SDS–phenol extraction was applied because SDS is used to improve solvent action for effective and efficient extraction of proteins from soil, soil samples in extraction buffer (1:1 (v:v) SDS–phenol buffer – 50 mM Tris, 1% SDS pH 7.5 + phenol (pH 8.0)) were vortexed and sonicated. The suspension was shaken for 1 h and vortexing and sonication was repeated prior to centrifugation (20 min and 4 °C at 3220 g). The water was removed and the lower phenol phase was washed with the same amount of milliQ water by gentle vortexing for 5 min and centrifugation (20 min and 4 °C at 3220 g). The purified phenol phases were combined and proteins were precipitated with ammonium acetate.
-
d)
WSP (SDS–phenol with prior washing steps modified from Wang et al. (2006))
We modified a protocol used by Wang et al. (2006) to extract proteins from leaf tissue. The procedure includes pre-washing steps to remove contaminants affecting the separation (TCA/acetone wash) and to additionally remove (poly)phenolic compounds (methanol wash) (Wang et al., 2006). Ground soil samples were first washed with 10% TCA in acetone, followed by a methanol washing step and a final acetone washing prior to extraction with 1:1 (v/v) (SDS–phenol buffer (50 mM Tris, 1% SDS pH 7.5) and phenol (pH 8.0)) by vortexing and sonication as described above. Then the suspension was shaken for 1 h and vortexing and sonication was repeated prior to centrifugation (20 min and 4 °C at 3220 g). The water was removed and the phenol phase was washed with the same amount of milliQ water by gentle vortexing for 5 min and centrifugation (20 min and 4 °C at 3220 g). The purified phenol phases were combined and proteins were precipitated with ammonium acetate.
2.2.2. Processing of precipitates
Precipitated proteins obtained from the different extraction procedures either with ammonium acetate in methanol or TCA were collected by centrifugation at 10640 g for 20 min at 4 °C. Supernatants were discarded. The pellets were washed with 100% pre-chilled acetone by gentle vortexing and a further centrifugation step, as described above. The supernatant was discarded and the pellets were dried. Pellets were resuspended in a maximum of 1 ml 0.5 M TEAB buffer containing 10 mM dithiothreiol (DTT), 6 M urea and 1 M thiourea by vortexing and gentle shaking over night at 4 °C. After centrifugation for 3 min at 17960 g, the resulting supernatant was used for further processing. Extracted proteins were separated by 1D-SDS–PAGE (Laemmli, 1970) in a 12% polyacrylamide gel to evaluate the protein separation pattern (Supplementary Fig. 1).
2.2.3. Protein digestion
Protein solutions (containing ∼0.1–0.6 μg μl−1 proteins) were diluted 1:7 (in 0.5 M triethylammonium bicarbonate (TEAB) buffer) to lower urea concentration to suitable amounts for trypsin digestion. An amount of 500 μl of protein solution was pre-treated with 10 mM DTT for 30 min at 60 °C and with 25 mM iodoacetamide for 1 h at room temperature. Finally, trypsin (sequencing grade modified trypsin, Promega, reference V5111) was added to a final concentration of 2 μg ml−1 and the sample was incubated overnight at 30 °C.
The digested solutions were dried with vacuum-centrifugation using a Concentrator plus (Eppendorf AG, Hamburg, Germany) at 30 °C. Pellets were resuspended in 1 ml 5% acetonitrile (ACN) 25 mM KH2PO4 pH 8.5 and pre-purified on a C18 column (SepPak, Waters). Eluted samples (elution buffer – 70% ACN, 0.1% formic acid) were dried with vacuum-centrifugation and resuspended in 500 μl of LC -buffer (25 mM KH2PO4, 5% ACN, pH 8.5).
2.2.4. Separation via reverse HPLC
To reduce sample complexity, peptides were fractionated using two-dimensional liquid chromatography separation. The first dimension was reversed phase chromatography at alkaline pH conditions on a C18-column (YMC Pack PRO C18 RS, 150 × 2.1 mm). Samples were put in the ultrasonic bath at 30 °C for 10 min and centrifuged for 3 min at room temperature. Buffers for LC were, buffer A (25 mM KH2PO4, 5% ACN, pH 8.5) and buffer B (25 mM KH2PO4, 50% ACN, pH 8.5). The flow rate was adjusted to 200 μl min−1 with a linear gradient from 5 to 30% acetonitrile in 60 min. Twenty-six fractions were collected per sample: two fractions from retention time of 8–25 min and further 24 fractions from 25 to 90 min. Three of the obtained fractions were pooled together respectively and peptide mixtures were analysed on a hybrid LTQ-Orbitrap mass spectrometer (ThermoFischer Scientific) interfaced with a nano-electrospray ion source. Second-dimension peptide fractionation was achieved by low pH reversed phase LC. Chromatographic separation of peptides was achieved on an Eksigent nano LC system (Eksigent Technologies, Dublin, CA, USA), equipped with a 11 cm fused silica emitter, 75 μm inner diameter (BGB Analytik, Böckten, Switzerland), packed inhouse with a Magic C18 AQ 3 μm resin (Michrom BioResources, Auburn, CA, USA). Peptides were loaded from a cooled (4 °C) Spark Holland auto sampler and separated using an acetonitrile/water solvent system containing 0.1% formic acid at a flow rate of 200 μl min−1 with a linear gradient from 3 to 35% acetonitrile in 60 min. Up to 6 data-dependent MS/MS spectra were acquired in the linear ion trap for each Fourier-transform (FT)-MS spectral acquisition range. The latter was acquired at 60,000 full-width half-maximum (FWHM) nominal resolution settings with an overall cycle time of approximately 1 s. Charge state screening was employed to select for ions with two charges and rejecting ions in single-charge state. The automatic gain control (AGC) was set at 5e5 for ion injection control and at 1e4 for full FT-MS and linear ion trap MS/MS. The instrument was calibrated externally according to the manufacturer's instructions. All samples were acquired using internal lock mass calibration on m/z 429.088735 and 445.120025.
2.2.5. Database searches
The MASCOT Search Engine (version no.2.2.04) was used for protein database searches. Data were searched against a database containing all proteins from UniRef100 (9808438 entries, downloaded from the European Bioinformatics Institute webpage http://www.ebi.ac.uk/uniref/ at the 26st January 2010), protein sequence information derived from the metagenome of a Minnesota farm silage soil microbial community (Tringe et al., 2005) (184,374 entries, downloaded from http://img.jgi.doe.gov at the 15th of October 2009) as well as common contaminants like keratin and trypsin (total no. of entries 9,993,117). The following search parameters were applied: (i) trypsin was chosen as protein-digesting enzyme and up to two missed cleavages were tolerated, (ii) carbamidomethylation of cysteine was chosen as fixed modification, and (iii) oxidation of methionine was chosen as variable modification. Searches were performed with a parent-ion mass tolerance of ±5 ppm and a fragment-ion mass tolerance of ±0.8 Da.
2.2.6. Data processing
Scaffold (version Scaffold 3.00, Proteome Software, Portland, OR, USA) was used to validate and quantify MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at greater than 90% probability and one peptide was assigned to a respective protein in one of our samples. Protein probability was assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that were identified with the same set of peptides and could not be differentiated based on the MS/MS analysis alone were grouped to satisfy the principles of parsimony. Protein false-discovery rate was calculated by the Scaffold software. At the 90% protein identification probability level false-discovery rate was 4.9%.
2.2.7. Data validation and assignment of data to phylogenetic and functional groups
Protein hits obtained in the database searches were assigned to phylogenetic and functional groups and assignments were validated by our newly developed perl-script based PROteomics result Pruning & Homology group Annotation Engine (PROPHANE) (Schneider et al., 2011). The normalized spectral abundance factor (NSAF) was calculated as a marker for protein abundance (Florens et al., 2006; Zybailov et al., 2006). The approach is based on the empirical observation that the more protein is present in a sample the more tandem MS spectra are collected for peptides of that protein (Bantscheff et al., 2007). To calculate the NSAF, the number of unique spectra assigned to each protein was divided by the number of the amino acid chain length of the longest candidate in the protein cluster giving the spectral abundance factor (SAF). The SAF allows the comparison of protein abundances in one sample with taking into account protein molecular weight. Afterwards each SAF is divided by the sum of all SAFs in the respective sample to allow cross-sample abundance comparison.
2.2.8. Spiking of soil with bacterial and fungal proteins
To reveal if binding of proteins to the soil matrix is a problem for the extraction procedure, a spiking experiment was conducted. Autoclaved forest soil was spiked with whole-cell proteins from Pectobacterium carotovorum and secreted proteins of Aspergillus nidulans obtained from pure cultures of the two organisms. To this end, A. nidulans (Austrian Center of Biological Resources and Applied Mycology no. MA5366) and P. carotovorum (American Type Culture Collection no. 39048) cultures were grown in 1 l Erlenmeyer flasks containing 200 ml AB minimal medium (Clark and Maaloe, 1967) supplemented with 0.2% sterile filtered glucose (Sigma–Aldrich) or 1% sterile beech leaf litter, for the bacterium and the fungus respectively as described in Schneider et al. (2010). A. nidulans proteins in the culture supernatant were precipitated and used for the spiking experiment. P. carotovorum cultures were grown over night for the inoculation. The main culture was inoculated with the over-night culture to a final OD600 of 0.05 and grown on minimal medium supplemented with 0.2% glucose at 28 °C with vigorous agitation. P. carotovorum culture was grown until an OD600 of 1.087 was reached. Cell lysis of was obtained with a French press (Hypramag AG, Zürich, Switzerland), with “Pressure Cell” (Aminco, Maryland, USA) and hydraulic motor (Ruetschi AG, Suhr, Switzerland). Proteins in the lysate were precipitated with 10% TCA, and centrifuged at 5213 g for 15 min. The resulting pellet was resuspended in 0.5 M TEAB buffer pH 8.5 (Sigma–Aldrich). These precipitated und resuspended proteins from A. nidulans supernatant and P. carotovorum lysate (∼20 mg protein in total) were added to 20 g of autoclaved sterile soil in 0.1 M Na3PO4 buffer pH 6.0 and vigorously shaken over night at 4 °C. Subsequently, in the soil-protein suspensions the supernatant was reduced using a rotary vacuum evaporator (Heidolph, USA), in order to ensure sample conditions similar to in situ soil extractions. Extractions of the spiked soil were conducted using the NaOH extraction procedure and the SDS–phenol protocol (as described previously in 2.2.1 sections b) and c)). Extractions of the fungal and bacterial protein solutions were combined and amended with the respective extraction buffer and extracted with the same protocols as the soil samples (see description of protocols b) and c) in section 2.2.1), the only deviance being that those samples were not ground in liquid nitrogen. Subsequently, the number of extracted proteins from the spiked soil was compared with those identified from P. carotovorum lysates and A. nidulans supernatants to evaluate the protein recovery ratio using.
3. Results
3.1. Comparison of protein extraction protocols from natural soil samples
As a first step, numbers of unique spectra were obtained as quantification is based on these data (Fig. 2) while the other results and figures derive from protein assignments. Generally, more unique spectra (i.e. peptide matching spectra) could be assigned to proteins extracted from the forest soil than from the potting soil sample (Figs. 2 and 3). A list of all assigned proteins and the number of proteins in each sample are provided in Supplementary Table S2. For the forest soil the sum of proteins assigned with a protein probability of 90% of the four protocols was 881, while for the potting soil only a total number of 498 proteins could be assigned (Supplementary Table S2).
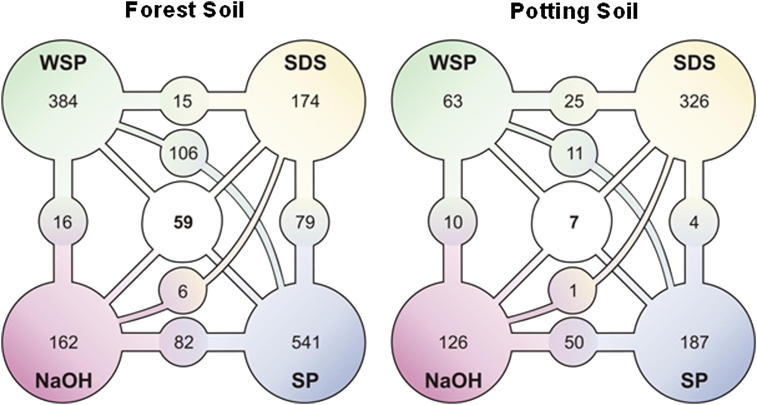
Fig. 2.
Venn diagram of the number of unique spectra (cutoff: protein probability 90%, minimum peptides 1, minimum peptide probability 95%) observed with the four different extraction protocols. The four main circles contain the numbers of unique spectra that were found using the respective protocol, while smaller circles display the set of spectra that were also detected with another protocol. The set of spectra that could be identified with all four approaches is given in the middle circle. Numbers derive from Venn-diagrams calculated with Scaffold (Scaffold 3.00, Proteome Software, Portland, OR, USA) (SDS, extraction with sodium dodecyl sulfate; WSP, extraction with SDS and Phenol with pre-purification according to Wang et al.; NaOH, extraction based on sodium hydroxide with further phenol purification; SP, extraction with SDS and phenol).
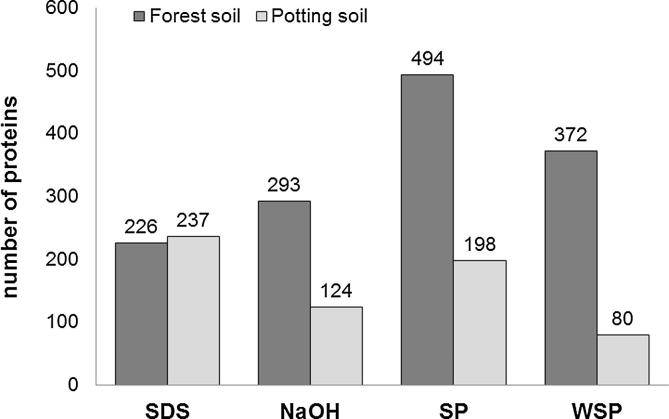
Fig. 3.
Bar chart of number of proteins for the four different protocols (SDS, extraction with sodium dodecyl sulfate; WSP, extraction with SDS and phenol with pre-purification according to Wang et al. (2006); NaOH, extraction based on sodium hydroxide with further phenol purification; SP extraction with SDS and phenol) which were used for the extraction of proteins and the two different soil types (dark grey forest soil; light grey potting soil), to compare the total number of proteins obtained from the respective protocols with special emphasis on precipitation. (cutoff: 90% protein probability). Protein identifications were accepted when one peptide with 95% peptide probability was assigned to the respective protein.
When comparing the number of proteins assigned for each protocol in the different soils we observed a similar number of proteins for both samples when using the SDS protocol (Fig. 3), while the number of proteins in the forest soil was always higher when using the protocols based on phenol extraction and ammonium acetate precipitation (NaOH, SP, WSP) (Fig. 3).
For the potting soil the extraction procedure using SDS showed the highest number of spectra, followed by the SP (SDS–phenol) protocol (Fig. 2). The pre-purification using the WSP protocol (Wang pre-purification, SDS–phenol) resulted in the least number of unique spectra in the potting soil. For the forest soil the SP (SDS–phenol) extraction protocol resulted in the highest number of proteins (Fig. 3). The pre-purification used in the WSP protocol (Wang pre-purification, SDS–phenol) proved to be advantageous for the forest soil and resulted in the 2nd highest number of proteins. The SDS protocol revealed the lowest number of proteins (Fig. 3), in the forest soil. However, the NaOH protocol resulted in the 2nd lowest number of proteins in both soils. In conclusion, our results show that “highest” numbers of proteins could be gained with the SP (SDS–phenol) protocol independent of soil type. The set of unique spectra that could be identified with all four protocols within a sample was remarkably low (Fig. 2). Although a low protein probability level of 90% was used for the calculation, only 2.9% (59 out of 1982) and 0.8% (7 out of 856) of the spectra were found with all four methods in the forest soil and in the potting soil sample, respectively (Fig. 2).
Comparing total number of spectra obtained and those assigned to peptide sequences by protein database searches (Supplementary Table S1), we observed that generally the number of assigned spectra was lower in the potting soil and the SDS protocol resulted in the lowest amount of total spectra acquired. Nevertheless, SDS protocol applied to the potting soil showed a percentage of 24% of total acquired spectra assigned to peptides which was highest of all methods in this soil type (Supplementary Table S1). Whereas SP, WSP and NaOH protocol resulted in an assignment of close to 6%, although strongly variable numbers of total acquired spectra were obtained. In the forest soil the SP protocol and to a minor extent also the WSP protocol were favourable in terms of percent of spectra assigned to peptides in the database, as finally observed in the number of proteins as well (Fig. 3; Supplementary Table S1). When comparing our results of the two different soil types with the spiking experiment, a generally higher percentage (32–41%) of assigned spectra was observed in our spiking experiment (Supplementary Table S1). This is the result of using sequenced microorganisms (A. nidulans and P. carotovorum) in this experiment.
3.1.1. Comparison of protein phylogeny
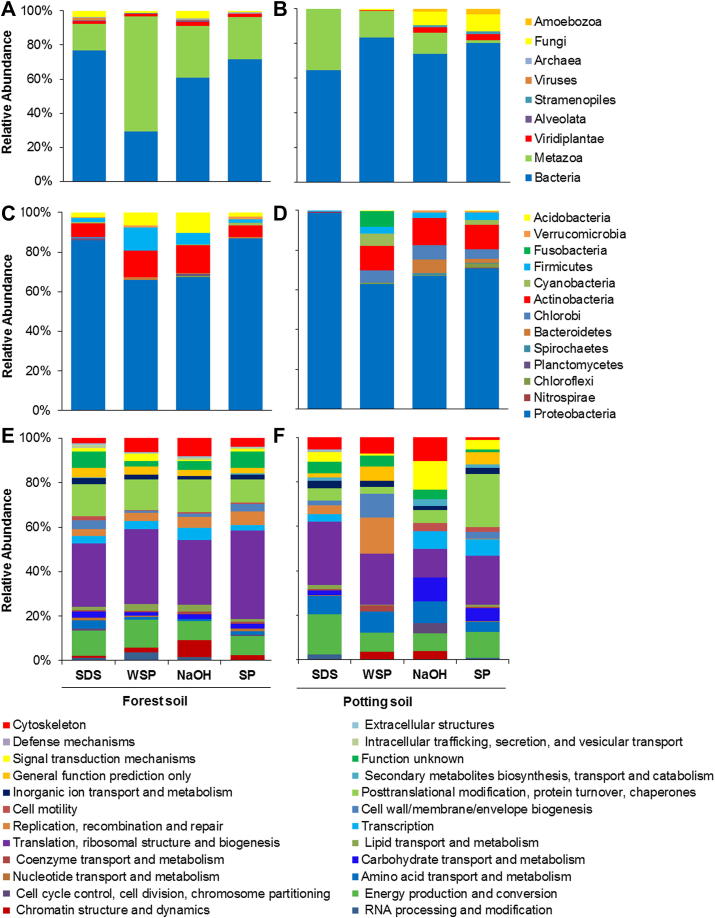
To evaluate extraction protocols we compared the relative abundance of spectra at different taxonomic levels. Six general groups (among them Bacteria, Metazoa, Fungi, Viridiplantae, Archaea and Alveolata) were found in the forest soil with each applied protocol, whereas in the potting soil the number of general groups represented by the extracted proteome ranged from two (SDS) to seven (SP).
Both soils were dominated by Bacteria followed by Metazoa (Fig. 4A, B). The only exception in that respect was the result from the WSP protocol in the forest soil sample, where the community was dominated by Metazoa. Fungi seemed to be of minor importance within both soil samples (Fig. 4A, B) and could not be detected with the SDS approach in the potting soil sample. Additionally, proteins were assigned to taxa of Archaea, Stramenopiles, Viridiplantae and Alveolata (Fig. 4A, B). Alveolata proteins were extracted with all four protocols from the forest soil but could not be found in the potting soil.
Fig. 4.
Community structure and functional groups. Relative abundance of proteins in % A and B assigned to general groups, C and D assigned to bacterial proteins, and E and F assigned to functional categories of the bacterial proteome based on cluster of orthologous groups (COG) classification; all based on NSAFs (normalized spectral abundance factors) for two different soil types, the left column shows the forest soil and the right column the potting soil and four different extraction protocols (SDS, extraction with sodium dodecyl sulfate; WSP, extraction with SDS and phenol with pre-purification according to Wang et al. (2006); NaOH, extraction based on sodium hydroxide, with further phenol purification; SP extraction with SDS and phenol).
A more detailed itemization of the bacterial community indicated a dominance of Proteobacteria (averaged 76%, n = 8, Fig. 4C, D), followed by Actinobacteria (averaged 10%, n = 8, Fig. 4C, D). Due to the fact that we did not analyze replicate samples in our methodological approach we decided against analyzing the phylogeny in more detail, because the results showed strong variation. As an example, for the phylum of Proteobacteria we obtained 15–95% for γ-Proteobacteria and 2–38% for α-Proteobacteria with the different protocols in the potting soil sample.
3.1.2. Comparison of observed functional groups of proteins
In order to separate proteins by their function, the obtained proteins were classified into cluster of orthologous groups (COG; prokaryotic proteins) categories based on their protein assignment (Fig. 4E, F). Generally, there were differences between soil types in functional groups assignments. Differences between the extraction protocols were more pronounced in the potting soil than in the forest soil (Fig. 4E, F). Most of the acquired proteins could be assigned to “Translation” processes, followed by “Post translational modifications, protein turnover and chaperones” as well as “Energy production and conversion”. When comparing the assignment of proteins to functional groups gained with the respective protocols, we observed that the SP protocol revealed more assignments to the functional groups of “Transcription” and “Post translational modifications, Protein turnover and chaperones” (Fig. 4E, F). The protocol using NaOH revealed a huge proportion of assignments to “Carbohydrate transport and metabolism” and “Amino acid transport and metabolism” (Fig. 4E, F), but revealed lower proportion of proteins in the categories of “Energy production and conversion”, “Translation, ribosomal structure and biogenesis”, “Inorganic ion transport” and “General function, prediction only” (Fig. 4E, F). There were no proteins assigned to the “Nucleotide transport and metabolism” extracted with the NaOH procedure in both soil types. Using the SDS protocol fewer proteins could be assigned to the category “Chromatin structure and dynamics” (Fig. 4E, F). It was not possible to assign proteins to the functional group “Secondary metabolite synthesis, transport and catabolism” with the WSP extraction procedure.
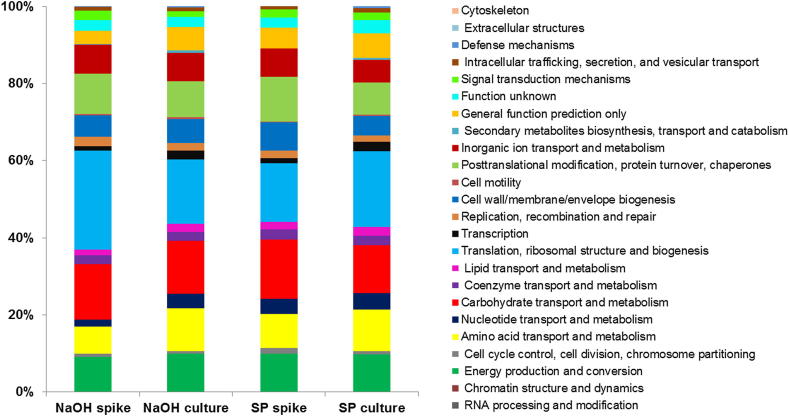
The functional categorization of the proteins identified in the spiking experiment is given in Fig. 5. Most of the acquired proteins could be assigned to “Translation” processes, followed by “Carbohydrate transport and metabolism” and in a similar portions of “Post translational modifications, Protein turnover and chaperones”, Energy production and conversion” as well as “Amino acid transport and metabolism” (Fig. 5). When comparing the assignment of proteins to functional groups gained with the respective protocols (only NaOH and SP used for the spiking experiment), we observed almost no differences (Fig. 5) for the spiked soil and the pure culture extractions.
Fig. 5.
Relative abundance of proteins in % assigned to functional categories based on cluster of orthologous groups (COG) classification; all based on NSAFs (normalized spectral abundance factors) for two different protocols (NaOH, extraction based on sodium hydroxide, with further phenol purification; SP extraction with SDS and phenol) for the culture and the spiked soil.
3.2. Recovery of proteins from spiked soil
In our spiking experiment, the full protein complement of P. carotovorum (producing cellulases, pectinases and proteases (Schneider et al., 2010) together with secreted extracellular proteins of A. nidulans was added to sterilized soil. To estimate the number of proteins that can be recovered from a soil sample two protocols, namely NaOH (b) and SDS–phenol (c), were used. Almost 50% of proteins could be recovered from the spiked soil with each of the protocols (Table 1). The number of total spectra obtained, and the number of spectra assigned and a percentage value of the assigned spectra is given in Supplementary Table S1. Recovery was calculated as the number of identified proteins extracted from the fungal and bacterial protein solutions compared to the number of assigned proteins extracted from the spiked soil. A list of all assigned proteins is provided in Supplementary Table S2. The spiking experiment revealed almost no differences in recovered functional groups between the two different protocols (Fig. 5). Comparison of the two extraction protocols in terms of recovery of secreted extracellular enzymes (i.e. cellulases, pectinases) showed that the NaOH based protocol reached a slightly higher recovery (46.5%) of pectinases than the SDS–phenol based protocol (42.6%). For secreted cellulases there was almost no difference in recovery (∼41% for both). Recovery of extracellular enzymes was slightly lower than general recovery.
Table 1.
Recovery of proteins from spiked soil samples. Number of proteins identified from spiked soil and mixed extracellular fungal and intracellular bacterial protein solutions (protein identification cutoff: 90% protein probability. Protein identifications were accepted when one peptide with 95% peptide probability was assigned to the respective protein. (NaOH, extraction based on sodium hydroxide with further phenol purification; SP, extraction with SDS and phenol).
| NaOH | SDS–phenol | |
|---|---|---|
| Protein solution | 820 | 1019 |
| Spiked soil | 400 | 483 |
| Recovery [%] | 48.8 | 47.4 |
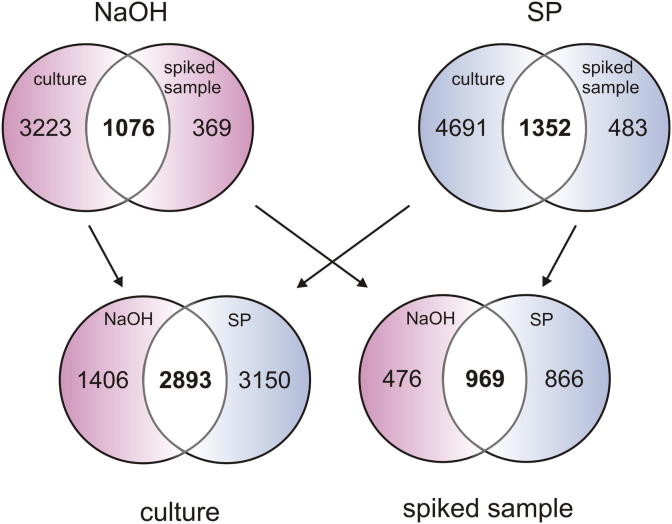
The Venn diagrams of the spiking experiment show that for both, the pure culture and the spiked soil, higher numbers of unique spectra could be obtained with the SP (SDS–phenol) protocol compared to the NaOH protocol (Fig. 6). The set of unique spectra that could be identified with both protocols within a sample was higher than for the comparison of the four extraction procedures in a natural sample (Figs. 2 and 6). Approximately one quarter of the unique spectra assigned in the culture could also be found in the spiked soil (25% for NaOH and 22% for SP). On the other hand 74% of the unique spectra assigned in the spiked sample were also found in the respective culture for both protocols (1076 out of 1445 for NaOH and 1352 out of 1835 for SP). Finally we compared the overlap of unique spectra found with the two different protocols for the forest soil, the potting soil, the culture and the spiked soil. Extraction in the natural samples resulted in an overlap of 10.4% (82 out of 785) and 13.8% (50 out of 363) for the forest soil and the potting soil respectively. Comparing these data with a less complex system we found an overlap of 38.8% (2893 out of 7449) for the pure culture and 42% (969 out of 2311) for the spiked soil sample.
Fig. 6.
Venn diagram of the number of unique spectra (cutoff: protein probability 90%, minimum peptides 1, minimum peptide probability 95%) obtained in the spiking experiment. The upper part shows the experimental bias by comparing the numbers of unique spectra for the pure culture and the subsequent recovery from the spiked soil sample for both protocols (NaOH, extraction based on sodium hydroxide with further phenol purification; SP extraction with SDS and phenol). The lower venn diagrams depict the methodological bias by showing the number of unique spectra that could be found with either one or both protocols.
4. Discussion
So far there is little knowledge about gene expression and microbial identification in the soil metaproteome (Bastida et al., 2009) as well as about the distinct role of soil microorganisms in biogeochemical nutrient cycling (Hettich et al., 2010). In order to shed light on these fundamental questions, metaproteomics has to overcome several obstacles like heterogeneity and hydrophobic nature of the soil matrix (Hettich et al., 2010), low protein extraction yields, high microbial diversity (most of it not jet covered by metagenome data), soil components that interfere with soil protein extraction and purification (Giagnoni et al., 2012) and therefore the inability to reliably extract the complete proteome from a random soil sample (Williams and Taylor, 2010). However, resolving these problems is crucial to successfully study the microbial proteome and possible changes within, as response to environmental stress (Nannipieri, 2006).
Although environmental applications of metaproteomics are still in their infancy, the present study takes advantage of a gel-free metaproteomics approach (2D-LC-MS), while most of earlier studies employed a combination of 1D-SDS–PAGE and liquid chromatography to separate proteins/peptides prior to the MS analysis (Renella et al., 2011; Wang et al., 2009; Williams and Taylor, 2010). Gel-free separation is commonly used to analyse highly complex protein mixtures as it provides (compared to gel-based approaches) a greater reduction in sample complexity without loss of information (Gevaert et al., 2003), larger scale quantitative analysis and the possibility of high throughput analysis (Griffin and Roe, 2006).
We conducted a comparison of four different soil extraction protocols, which were applied to two different soil types. Both soil types, a forest soil and a potting soil, were rich in clay and humus content and therefore probably had higher sorption ability compared to low organic matter soils (Williams and Taylor, 2010). As this experiment was a methodological approach focussing on the issue of extracting proteins out of soils containing clay and humics the experiments did not include a soil matrix without these components. Sodium hydroxide and SDS both result in bacterial lysis, which improves direct protein extraction (Benndorf et al., 2007). In three of the four protocols (SP, WSP and NaOH) used in the present study proteins were precipitated with ammonium acetate in methanol after extraction and phenol purification, while precipitation was conducted with TCA when extraction was done with SDS only. A comparison of protocols used for precipitation of recalcitrant plant tissue revealed promising results using phenol extraction methanol/ammonium acetate precipitation for plant tissue (Carpentier et al., 2005). Generally, more unique spectra could be assigned to proteins extracted from the forest soil than from the potting soil sample. This trend can be attributed to the NaOH, SP and WSP protocols, which include phenol for purification and precipitation with ammonium acetate in methanol. Since SDS protocol showed slightly higher numbers of proteins in the potting soil, which might be an effect of the precipitation technique. In the present study only SDS extractions were precipitated with TCA instead of ammonium acetate in methanol. As Carpentier et al. (2005) showed that large proteins might be lost during TCA precipitation when extracting plant proteins, which lead to an enrichment in small proteins compared to phenol based extraction. This effect was explained by differences in proteolytic breakdown during the extraction procedure and an easier re-solubilisation of small proteins precipitated with TCA (Carpentier et al., 2005). While TCA is known to be an efficient precipitation agent, there is the disadvantage that proteins are difficult to redissolve after TCA precipitation. However, in terms of total number of assigned proteins the SDS protocol seemed to be the least effective protocol in forest soil, compared to protocols that include a phenol purification step. Nevertheless, the highest number of proteins assigned was obtained with the SDS protocol in the potting soil. The data presented show that efficacy of the extraction method varies depending on the soil matrix and the proteins present.
In three (SP, WSP and NaOH) out of four protocols, phenol was used for further purification in combination with ammonium acetate in methanol precipitation. Bastida et al. (2009) proposed that for soil samples phenol extraction is most effective to remove interfering substances (e.g. humic acids) and to overcome the problem of co-extraction of contaminants. The phase separation with phenol and water intends higher proteins yields in the phenol phase and an increase in humic acids (brown substances) in the water phase (Benndorf et al., 2007). This procedure might have caused losses of proteins as protein–humic complexes which will be removed through the aqueous phase (Giagnoni et al., 2012) and, thus, specific proteins will be lost (Giagnoni et al., 2012; Nannipieri, 2006). However, we obtained the second lowest number of proteins using the NaOH protocol, in both soil types. As sodium hydroxide is widely used to extract humic compounds like fulvic and humic acids from soil, our results probably can be assigned to co-extraction of humic compounds. The combination of NaOH with phenol might co-extract protein–humic complexes, which will be removed in the aqueous phase resulting in lower protein abundances (Benndorf et al., 2007). Polyphenol-protein interactions may shield proteins by incorporation into hydrophobic compartments of soil organic matter (Bastida et al., 2009) and thereby influence the extraction efficiency. Also, Renella et al. (2011) claimed that the purification of protein extracts form humic substances using the NaOH protocol and phenol can be insufficient.
It was suggested, that extraction using SDS and phenol resulted in a more efficient extraction than SDS and phenol alone for recalcitrant plant tissue (Wang et al., 2006). This is consistent with our findings where “high” numbers of proteins were gained with the SP protocol for both soil matrices (highest number of proteins in the forest soil and the second highest number in the potting soil). The pre-washing step which was applied in the WSP protocol probably removed substances that interfere with protein extraction, separation and quantification, and therefore results obtained by this protocol might strongly depend on the soil type and whether the contained interfering substances were extractable with the used pre-washing solvents. While the WSP protocol obtained the second highest number of proteins in the forest soil, the lowest numbers of proteins were observed in the potting soil, where the pre-washing step maybe resulted in a loss of proteins.
Generally, the use of phenol during extraction and the subsequent precipitation with ammonium acetate seemed to have a favourable effect on the resulting diversity patterns (i.e., increasing the number of recovered taxonomic groups); however, taxonomic groups were only increased in the potting soil, but not in the forest soil samples (Figs. 2 and 4A–D). Extraction with SDS alone yielded the lowest number of microbial taxonomic groups for the potting soil (Fig. 4B, D) although highest number of proteins were observed.
These disparities in numbers of microbial taxonomic groups suggest that the extraction protocol used most likely biased the information about microbial community composition in different soil types. In addition, the high complexity of soils would result in a bias towards the most abundant phylogenetic groups due to blocked spectra of low abundant organisms. Apart from the fact that incomplete information about soil genomes and proteomes still aggravates the assignment of peptides to their phylogenetic origins, it is nearly impossible to favour one diversity pattern over another on the basis of profound knowledge. An additional metagenomic approach might have improved this result by assignment of spectra to related genome sequences from the respective soil samples.
While metagenomics and -transcriptomics allows assessing “biodiversity” in depth, metaproteome analyses enables the measurement of protein presence and abundance. Metatranscriptomics is closer to identify active metabolic pathways, but a lack of correlation between mRNA and protein levels has been reported (Siggins et al., 2012). The obtained phylogeny data should be considered with respect to the fact that protein identification relies on availability of relevant metagenome data when searching against databases with the generated spectra. As metaproteomics cannot be seen as an isolated method it benefits greatly from available sequencing data (Siggins et al., 2012). Although there is not always the possibility to afford a corresponding metagenome analysis this would be favourable in future studies. Still the database situation has already improved strongly by an increasing number of soil pyro-sequencing studies and continuing sequencing effort of soil samples will result in additional database information which will be available in the future.
Our results show that the choice of the extraction protocol used for a certain soil sample can affect yields of proteins and to some extend influence the obtained diversity pattern. The observed overlap of unique spectra for all four different protocols was very low with 0.9% and 2.9% for potting soil and forest soil respectively. There were no clear trends whether SDS-based protocols or the approach using NaOH gave more reliable results. In the end, relatively low numbers of proteins (ranging from 80 to 494) were extracted from the soil matrix. Therefore we assume that every protocol has unique biases towards certain sets of proteins. These results suggest that studies dealing with proteins in complex environmental samples might consider the application of different protocols in order to get a representative profile of a given microbial community and its functional capacity. This can be done by either pooling the different extracts before performing mass spectrometry or, as done in the present study, by analyzing them separately and combining the data in the end. Further research is required to remove interfering substances like humics and clays.
The forest soil rich in humic substances and clay fraction was used for the spiking experiment, which represented a challenging task compared to soils with less interfering substances but on the other hand allowed a critical evaluation of protein recovery. Moreover, spiking the soil with extracellular fungal proteins in addition to the intracellular bacterial proteins enabled us to compare the soil-binding capacity of secreted and intracellular proteins, which has never been investigated before.
The protein recovery rate obtained in our spiking experiment was between 47 and 49%, which was slightly lower compared to former investigations. For example a microcosm based approach with artificial soil mixtures by Renella et al. (2011) showed that the protein recovery using a 2D-PAGE approach was lower – (53.6% – 317 proteins identified out of 591) in the mix of sand + kaolinite + montmorillonite + goethite + humic acids (78:18:2:1:1) than in pure sand (close to 100%) or kaolinite (76% – 449 proteins identified out of 591), which was likely due to protein sorptive capacity of clay minerals and humic substances (Renella et al., 2011). A spiking experiment by Hettich et al. (2010) using a pure culture of Pseudomonas putida observed extraction efficiency close to 69% (925 proteins out of 1343) from a sandy soil, a soil-type which is usually low in interfering substances. Spiking of soil with bovin serum albumin (BSA) and employing a sequential extraction protocol resulted in an estimated recovery of 76% as determined by relative intensity of BSA bands in a gel based approach (Wang et al., 2009). Comparing all three studies with the results obtained in our approach (protein recovery about 50%) suggests that clay and organic matter content strongly influence extraction of proteins from soils due to protein sorption processes. Clay minerals have large reactive surfaces that are covered by Fe–Al oxyhydroxides and/or humic substances (Renella et al., 2011). Possible encapsulation effects of proteins with humic substances can lead to decreased extraction efficiency and might lower enzyme activity due to conformational changes by covalent coupling, and/or blocking of the active site (Sander et al., 2011). Since in our study no changes in the distribution of proteins into functional groups were observed, between the datasets derived from protein solutions and spiked soil, intracellular proteins seemed to be equally well extracted by our protocols. In contrast, there might be differences in the sorption capacity of extracellular enzymes to soil minerals and humic colloids. Protein extraction methods are of crucial interest in terms of optimizing selective recovery of active proteins because efficiency can be low due to physic-chemical factors such as irreversible adsorption of extracellular enzymes to the soil surface, protein occlusion or enzymatic hydrolysis (Giagnoni et al., 2012; Ogunseitan, 2006). Often, special emphasis was given on secreted extracellular enzymes in relation to nutrient cycling processes within terrestrial habitats (Murase et al., 2003). In our study, protein recovery of extracellular enzymes was slightly lower (41–46%) than for intracellular proteins, probably again due to protein binding to clay particles or to their substrates. Comparing functional categorization of the added proteins, in the spiked soil and the cultures themselves suggests representative protein extraction from samples with low complexity. These similar results from the spiking experiment using two different protocols, underpins the reproducibility of the extraction procedures especially in terms of protein functionality. The percentage of spectra assigned by protein database searches is almost the same for both protocols. Although the proteins used for the spiking experiment were derived from fully sequenced microbial organisms (A. nidulans and P. carotovorum) only ∼40% of the spectra matched to database entries (Supplementary Table S1) which could be explained by co-extracted substances such as contaminants.
5. Conclusion
Fully aware that there is still room for improvement regarding the protein recovery rate or the quality of databases applied, this study demonstrates that a critical evaluation of extraction methods applied to the soils investigated is of major importance to study the metaproteome of soil samples. The protein extraction efficiency from soil samples is severely hampered by the complex matrix and needs to be optimized for particular soil types and/or research questions. Moreover, the tested SDS–phenol protocol seems to be most promising to obtain “high” numbers of unique spectra from different soil samples.
Acknowledgements
The Austrian Academy of Sciences (ÖAW) granted Katharina Keiblinger with a DOC-fFORTE fellowship and the European Science Foundation (ESF) for an exchange grant entitled “Climatic Change – Manipulation Experiments in Terrestrial Ecosystems”. This research was performed within the National Research Network MICDIF (S100) of the Austrian Science Fund FWF (Project numbers S10001, 2, 3, 4, 6, 7-B17). Thanks to Alexander Grunau for lab assistance. We gratefully acknowledge many helpful suggestions that we have received from two anonymous reviewers and from the editor of Soil Biology and Biochemistry, this manuscript has benefited substantially from their comments.
Footnotes
Supplementary data related to this article can be found online at doi:10.1016/j.soilbio.2012.05.014.
Supporting information
The complete MASCOT search data set is provided on the PRIDE database website (http://www.ebi.ac.uk/pride/; the accession number is 18968).
Reviewer account details:
Username: review64083
Password: %P*9XmtH
Appendix A. Supplementary material
The following are the Supplementary data related to this article:
References
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and Bioanalytical Chemistry. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Bastida F., Moreno J.L., Nicolas C., Hernandez T., Garcia C. Soil metaproteomics: a review of an emerging environmental science. Significance, methodology and perspectives. European Journal of Soil Science. 2009;60:845–859. [Google Scholar]
- Bastida F., Algora C., Hernandez T., Garcia C. Feasibility of a cell separation-proteomic based method for soils with different edaphic properties and microbial biomass. Soil Biology & Biochemistry. 2012;45:136–138. [Google Scholar]
- Benndorf D., Balcke G.U., Harms H., von Bergen M. Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME Journal. 2007;1:224–234. doi: 10.1038/ismej.2007.39. [DOI] [PubMed] [Google Scholar]
- Carpentier S.C., Witters E., Laukens K., Deckers P., Swennen R., Panis B. Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics. 2005;5:2497–2507. doi: 10.1002/pmic.200401222. [DOI] [PubMed] [Google Scholar]
- Clark D.J., Maaløe O. DNA replication and the division cycle in Escherichia coli. Journal of Molecular Biology. 1967;23:99–112. [Google Scholar]
- Florens L., Carozza M.J., Swanson S.K., Fournier M., Coleman M.K., Workman J.L., Washburn M.P. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G.R., Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nature Biotechnology. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- Giagnoni L., Magherini F., Landi L., Taghavi S., van der Lelie D., Puglia M., Bianchi L., Bini L., Nannipieri P., Renella G., Modesti A. Soil solid phases effects on the proteomic analysis of Cupriavidus metallidurans CH34. Biology and Fertility of Soils. 2012;48(4):425–433. [Google Scholar]
- Griffin T.J., Roe M.R. Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–4687. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- Hettich R.L., Chourey K., Jansson J., VerBerkmoes N., Shah M., Chavarria K.L., Tom L.M., Brodie E.L. Direct cellular Lysis/protein extraction protocol for soil metaproteomics. Journal of Proteome Research. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kitzler B., Zechmeister-Boltenstern S., Holtermann C., Skiba U., Butterbach-Bahl K. Nitrogen oxides emission from two beech forests subjected to different nitrogen loads. Biogeosciences. 2006;3:293–310. [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mocali S., Benedetti A. Exploring research frontiers in microbiology: the challenge of metagenomics in soil microbiology. Research in Microbiology. 2010;161:497–505. doi: 10.1016/j.resmic.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Murase A., Yoneda M., Ueno R., Yonebayashi K. Isolation of extracellular protein from greenhouse soil. Soil Biology & Biochemistry. 2003;35:733–736. [Google Scholar]
- Nannipieri P. In: Role of Stabilised Enzymes in Microbial Ecology and Enzyme Extraction from Soil with Potential Applications in Soil Proteomics Nucleic Acids and Proteins in Soil. Nannipieri P., Smalla K., editors. Springer; Berlin Heidelberg: 2006. pp. 75–94. [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nielsen K.M., Calamai L., Pietramellara G. In: Stabilization of Extracellular DNA and Proteins by Transient Binding to Various Soil Components Nucleic Acids and Proteins in Soil. Nannipieri P., Smalla K., editors. Springer; Berlin Heidelberg: 2006. pp. 141–157. [Google Scholar]
- Ogunseitan O. In: Soil Proteomics: Extraction and Analysis of Proteins from Soils Nucleic Acids and Proteins in Soil. Nannipieri P., Smalla K., editors. Springer; Berlin Heidelberg: 2006. pp. 95–115. [Google Scholar]
- Renella G., Giagnoni L., Magherini F., Landi L., Taghavi S., Modesti A., Bini L., Nannipieri P., Van der Lelie D. Extraction of microbial proteome from soil: potential and limitations assessed through a model study. European Journal of Soil Science. 2011;62:74–81. [Google Scholar]
- Sander M., Tomaszewski J.E., Schwarzenbach R.P. Protein encapsulation by humic substances. Environmental Science & Technology. 2011;45:6003–6010. doi: 10.1021/es200663h. [DOI] [PubMed] [Google Scholar]
- Schneider T., Gerrits B., Gassmann R., Schmid E., Gessner M.O., Richter A., Battin T., Eberl L., Riedel K. Proteome analysis of fungal and bacterial involvement in leaf litter decomposition. Proteomics. 2010;10:1819–1830. doi: 10.1002/pmic.200900691. [DOI] [PubMed] [Google Scholar]
- Schneider T., Schmid E., de Castro J.V., Cardinale M., Eberl L., Grube M., Berg G., Riedel K. Structure and function of the symbiosis partners of the lung lichen (Lobaria pulmonaria L. Hoffm.) analyzed by metaproteomics. Proteomics. 2011;11:2752–2756. doi: 10.1002/pmic.201000679. [DOI] [PubMed] [Google Scholar]
- Schneider T., Keiblinger K.M., Schmid E., Sterflinger-Gleixner K., Ellersdorfer G., Roschitzki B., Richter A., Eberl L., Zechmeister-Boltenstern S., Riedel K. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME Journal. 2012 doi: 10.1038/ismej.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze W.X., Gleixner G., Kaiser K., Guggenberger G., Mann M., Schulze E.D. A proteomic fingerprint of dissolved organic carbon and of soil particles. Oecologia. 2005;142:335–343. doi: 10.1007/s00442-004-1698-9. [DOI] [PubMed] [Google Scholar]
- Siggins A., Gunnigle E., Abram F. Exploring mixed microbial community functioning: recent advances in metaproteomics. Fems Microbiology Ecology. 2012 doi: 10.1111/j.1574-6941.2011.01284.x. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange F., Butterbach-Bahl K., Papen H., Zechmeister-Boltenstern S., Li C.S., Aber J. A process-oriented model of N(2)O and NO emissions from forest soils 2. Sensitivity analysis and validation. Journal of Geophysical Research-Atmospheres. 2000;105:4385–4398. [Google Scholar]
- Tringe S.G., von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., Bork P., Hugenholtz P., Rubin E.M. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- VerBerkmoes N.C., Denef V.J., Hettich R.L., Banfield J.F. SYSTEMS BIOLOGY Functional analysis of natural microbial consortia using community proteomics. Nature Reviews Microbiology. 2009;7:196–205. doi: 10.1038/nrmicro2080. [DOI] [PubMed] [Google Scholar]
- Wang W., Vignani R., Scali M., Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27:2782–2786. doi: 10.1002/elps.200500722. [DOI] [PubMed] [Google Scholar]
- Wang W., Chen S.N., Rillig M.C. Improving soil protein extraction for metaproteome analysis and glomalin-related soil protein detection. Proteomics. 2009;9:4970–4973. doi: 10.1002/pmic.200900251. [DOI] [PubMed] [Google Scholar]
- Williams M.A., Taylor E.B. Microbial protein in soil: influence of extraction method and C amendment on extraction and recovery. Microbial Ecology. 2010;59:390–399. doi: 10.1007/s00248-009-9593-x. [DOI] [PubMed] [Google Scholar]
- Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. Journal of Proteome Research. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.