Abstract
BACKGROUND
Capecitabine, an oral 5-fluorouracil (5-FU) prodrug, is widely used in the treatment of breast, colorectal, and gastric cancers. To guide selection of patients potentially at greatest benefit of experiencing antitumor efficacy, or, alternatively, of developing toxicities, identifying genomic predictors of capecitabine sensitivity could permit its more informed use.
METHODS
Our objective was to perform capecitabine sensitivity genome-wide association studies (GWAS) using 503 well-genotyped human cell lines from individuals representing multiple different world populations. Meta-analysis including all ethnic populations then enabled identification of novel germline determinants (SNPs) of capecitabine susceptibility.
RESULTS
First, intra-population GWAS of Caucasian individuals identified rs4702484 (within ADCY2) at a level reaching genome-wide significance (P=5.2×10−8). This SNP is located upstream of MTRR (5-methyltetrahydrofolate-homocysteine methyltransferase reductase), a gene whose enzyme is known to be involved in the methionine-folate biosynthesis and metabolism pathway that is the primary target of 5-FU-related compounds, although we were unable to find a direct relationship between rs4702484 and MTRR expression in a tested subset of cells. In the meta-analysis, 4 SNPs comprised the top hits including again rs4702484 and three additional SNPs (rs8101143, rs576523, rs361433) that approached genome-wide significance (P values 1.9×10−7—8.8×10−7). Meta-analysis also identified one missense variant (rs11722476; Ser to Asn) within SMARCAD1, a novel gene for association with capecitabine/5-FU susceptibility.
CONCLUSIONS
Toward the goal of individualizing cancer chemotherapy, our study identified novel SNPs and genes associated with capecitabine sensitivity that are potentially informative and testable in any patient, regardless of ethnicity.
Keywords: capecitabine, susceptibility, pharmacogenomics, genome-wide, meta-analysis
Introduction
Variation in drug response is both clinically expected and relatively poorly predicted1, 2. Chemotherapy, in particular, is plagued by highly variable response rates as well as significant toxicity1. Capecitabine is a chemotherapeutic agent widely used in the treatment of breast, colorectal, and gastric cancers3. It is an oral, 5-fluorouracil (5-FU) prodrug designed to have limited toxicity due to preferential activation in tumor cells4. However, toxicities can result including, in particular, gastrointestinal toxicity and hand-foot syndrome5. Identifying genetic predictors of capecitabine susceptibility could permit more informed use of this therapy by guiding selection of patients potentially most likely to experience antitumor efficacy, or alternatively to recognize patients who might be at particular risk of developing toxicities. Notably, capecitabine is particularly well suited for pharmacogenomic study because, compared to other oncologic drugs, it is often used as single-agent therapy6,7.
Toward discovery of genetic variants governing chemotherapeutic susceptibility in patients, we developed a human cell-based model8. The model utilizes lymphoblastoid cell lines (LCLs) collected from individuals across the globe as part of the International HapMap project, with genotype information publicly available for each individual9. Previous genome-wide discovery work has utilized populations of these cells for several pharmacologic agents including cytarabine10, daunorubicin11, etoposide12, cisplatin, and carboplatin13–15. LCLs offer a model for genome-wide discovery without the confounders of diet, co-medications, and comorbidities16. Genetic variants discovered using this model have been validated in clinical settings17.
Pharmacoethnicity18, the concept that different ethnic populations have different responses to the same drug, makes population-based studies particularly informative. Although genetics may not be the only factor contributing to different responses across different ethnic groups, it is likely an important component. While discovery of ethnic-specific polymorphisms is useful, the ultimate goal for clinical translation of pharmacogenomics remains the discovery of genetic polymorphisms (SNPs) that are informative and testable in any patient, regardless of ethnicity1, 13, 18. Therefore, to identify SNPs for testing in the clinical setting, our objective was to interrogate capecitabine susceptibility using genome-wide association in over 500 individuals’ samples and to perform cross-population meta-analysis to characterize genetic determinants of sensitivity in individuals representing diverse global backgrounds.
Materials and Methods
Phenotyping
HapMap cell lines from six different panels were purchased from Coriell Institute for Medical Research (www.coriell.org) and used for susceptibility phenotyping: 84 unrelated Asian (ASN) individuals’ LCLs from HapMap Phase I (individuals from Tokyo, Japan, and Beijing, China); 84 LCLs representing Caucasian individuals from Utah, U.S. with northern/western European ancestry (CEU) in trio structure (two parents plus their child) from HapMap I (CEU1); 80 LCLs from CEU in trio structure from HapMap Phase III (CEU3); 87 LCLs from the Yoruba individuals of Ibadan, Nigeria (YRI) in trio structure from HapMap I (YRI1); 86 LCLs from YRI in trio structure from HapMap III (YRI3); and 82 LCLs from an African-American population from the Southwest U.S. (ASW) in trio structure. LCLs were cultured in RPMI 1640 media (Cellgro, Herndon, VA) containing 15% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) and 20mM L-glutamine. Cell lines were diluted 3 times/week at a concentration of 300,000–350,000 cells/mL and maintained in a 37°C, 5% CO2 humidified incubator.
Using capecitabine in LCLs is hindered by the lack of expression of cytidine deaminase19 which is required for conversion of capecitabine to its active form. To circumvent this step in enzymatic activation, 5′-deoxy-5-fluorouridine (5′DFUR), a major metabolite of capecitabine, was used to evaluate capecitabine sensitivity using a short-term cellular growth inhibition assay20. LCLs in the exponential growth phase with >85% viability (Vi-Cell XR viability analyzer, Beckman Coulter, Fullerton, CA) were plated in triplicate at density=1×105 cells/mL in 96-well round-bottom plates (Corning, Corning, NY) 24 hours prior to drug treatment. Drug was added immediately after preparation of stock at the following concentrations: 2.5, 10, 20, and 40 μM and left on cells for 72 hours. AlamarBlue was added 24 hours before absorbance reading at wavelengths=570nm and 600nm (Synergy-HT multi-detection plate reader, BioTek, Winooski, VT). Percent survival was quantified relative to a control well without drug and, at each concentration, represents two separate experiments each performed in triplicate. Area under the survival curve (AUC), representing sensitivity to the drug, was calculated for each cell line using the trapezoidal rule and was log2-transformed for all data analysis. For comparisons between populations, AUC values were corrected for cellular growth rate20, 21 by subtracting each cell line’s AUC phenotype with the cellular growth rate multiplied by the linear regression coefficient for growth rate. For performing the genome-wide association studies (GWAS), uncorrected AUCs were utilized so that any growth rate-associated variants could also potentially be identified.
Genotyping
Genotypes were downloaded from the HapMap Consortium release. Fewer genotypes were available for LCLs from Phase III HapMap (ASW, YRI3, CEU3) when compared to Phase I samples (for which >2 million SNPs were available). To make these populations more comparable, imputation was performed for Phase III lines individually using BEAGLE22. For CEU3 and YRI3, CEU1 and YRI1 (HapMap r22) respectively were used as reference. To measure accuracy of imputation at each SNP, R2 was calculated as described following 100 imputations22. Imputed genotypes with R2>0.80, minor allele frequency (MAF)>0.05, no Mendelian errors, and in Hardy-Weinberg equilibrium (P>0.001) were carried through the rest of the analysis. For ASW, the same process was followed using both YRI1 and CEU1 as reference.
GWAS Analyses
Each of the six HapMap panels was analyzed by a GWAS independently. Since GWA studies assume normality in the data, we first ensured this for each population. For five of the six panels, log2-transformed AUC phenotypes achieved normal distributions. Because log2-transformation in the ASW did not yield a normal distribution (Shapiro-Wilk test), the ASW population required rank-normalization to achieve normality. Rank-normalization was performed using the rntransform function in the R GenABEL package.
For CEU1, CEU3, YRI1, and YRI3, >2 million SNPs (MAF>5% within the panel, no Mendelian errors, and in Hardy Weinberg equilibrium [P>0.001]) were tested for association using the quantitative trait disequilibrium test (QTDT)23. In ASW, local ancestry at each SNP was estimated using HAPMIX24. Phased genotypes from CEU1 and YRI1 were used as the ancestral populations to estimate ancestry. GWAS was performed using QTDT with local ancestry (a fractional predicted number of chromosomes) as covariate.
A genomic control value25 was calculated for each GWAS. Correction for residual inflation of the test statistic was done for studies with λ>1. Resulting P values (possibly adjusted) were carried forward to the meta-analysis.
Meta-analysis
To identify SNPs associated with capecitabine-induced cytotoxicity, we conducted a meta-analysis to assimilate the results of the GWA studies from the individual populations. We used METAL26, which combines P values across the studies for each SNP using a study-specific weight (sample size) and the direction of effect (β). At each SNP, the direction of effect and the P values from the individual studies were converted into signed Z-scores. Z-scores were combined with weights proportional to the square root of the sample size for each study.
Results
Phenotype Variation Across Ethnic Groups
Fig. 1 shows the susceptibility phenotype results, grouped by ethnic population, for all 503 included cell lines upon exposure to the capecitabine metabolite 5′DFUR. Inter-population comparisons show that YRI were most sensitive to growth inhibitory effects of capecitabine, with a median AUC significantly lower than CEU (P=8.5×10−8) and ASN (P=6×10−3) but not significantly different from ASW. The CEU population was the most resistant. Because previous work has shown significant growth rate differences across HapMap populations21 and that growth rate is a significant confounder of pharmacologic endpoints including for 5′DFUR20, the AUC measurement is corrected for growth rate to allow the most appropriate comparisons.
Figure 1. Celluar sensitivity to capecitabine in 503 HapMap lymphoblastoid cell lines.
The YRI population was the most sensitive to growth inhibitory effects of 5′DFUR (capecitabine) with a median AUC significantly lower than that of CEU (P=8.5×10−8) and ASN (P=6×10−3) but not significantly different from ASW.
Individual Population GWAS Reveal Top SNP Finding in Caucasians
Since some inter-population differences in sensitivity were observed, we first conducted individual, intra-population GWA studies for each ethnic population so that any potentially important population-restricted SNPs might be identified. Such SNPs may in fact be important toward explaining inter-ethnic susceptibility differences13, 27 like those seen in Fig. 1.
Manhattan plots showing GWAS results for each population are illustrated in Fig. 2A (CEU) and Fig. 3 (YRI, ASW, ASN). While there are interesting findings for each population, the most intriguing result was produced by the CEU GWAS. The top CEU signal—considerably stronger than any other signal in the entire CEU GWAS—identified rs4702484 at a level that approximated genome-wide significance (P=5.2×10−8). This SNP, located in an intronic region of ADCY2 (adenylate cyclase), has a CEU MAF=12%. Additionally, as can be seen in the chromosomal plot of this region (Fig. 2B), rs4702484 is located just upstream of MTRR (5-methyltetrahydrofolate-homocysteine methyltransferase reductase), a gene whose enzyme is known to be involved in the methionine-folate biosynthesis and metabolism pathway28. 5-FU-related compounds have been previously shown to target other enzymes in this pathway29, but a potential pharmacogenomic relationship with MTRR would, to our knowledge, be novel.
Figure 2. GWAS in Caucasian individuals identified a novel variant, rs4702484, associated with capecitabine sensitivity at a level reaching genome-wide significance (P=5.2×10−8).
(A) Manhattan plot showing capecitabine susceptibility GWAS results in CEU. (B) Zoom-in view of chromosome 5 region around top CEU GWAS SNP, rs4702484. The location within ADCY2 (adenylate cyclase) is shown, in addition to its close proximity (upstream) of MTRR (5-methyltetrahydrofolate-homocysteine methyltransferase reductase). r2 patterns designate LD probabilities with other SNPs in the region.
Figure 3. Manhattan plots of GWAS results for capecitabine sensitivity in different populations.

(A): YRI; (B): ASW; (C): ASN.
Using whole-genome mRNA expression data that we previously generated in our CEU1 LCLs using Affymetrix Exon Array 1.019, we investigated in an exploratory manner whether there was a statistical correlation between rs4702484 and MTRR mRNA expression levels in 30 CEU LCL trios (90 samples) using QTDT software, however we were unable to find a direct relationship in this limited subset.
Meta-Analysis Results of the Population-Based GWA Studies
A meta-analysis of the individual population data was next performed to identify the most significant SNPs incorporating all populations. Fig. 4 shows the full meta-analysis results in a Manhattan plot. While none of the top SNP associations reached the traditional cutoff for GWA statistical significance30, 4 SNPs approached this threshold (rs8101143, rs576523, rs4702484, rs361433; P values 1.9×10−7—8 8×10−7). An additional 23 top SNPs had P values <10−5 (Table 1). It is noteworthy that the previously identified top CEU SNP, rs4702484, remained highly significant (and ranked third overall) in the meta-analysis (meta P=6.4×10−7). This SNP did not have a MAF>0.05 in the YRI or ASW populations; in ASN, P=0.34.
Figure 4. Meta-anlaysis of individual LCLs from CEU, YRI, ASW and ASN populations.

Manhattan plot showing meta-analysis results of capecitabine susceptibility GWA studies including all of the utilized combined world populations (n=503 individuals).
Table 1. Top SNPs from the multi-population meta-analysis of genome-wide association findings for capecitabine susceptibility.
While 503 individuals’ samples were tested overall for capecitabine susceptibility, the number of samples analyzed for each SNP (far right column) was less if the SNP was monomorphic or rare (MAF<5%) in some ethnic groups, or, in infrequent cases, if HapMap or imputation genotypes were unavailable.
| Chromosome | Gene | SNP | Location | Meta-analysis P Value | Overall Rank (by P value) | Number of Individual Samples Tested |
|---|---|---|---|---|---|---|
| 1 | --- | rs576523 | --- | 2.3×10−07 | 2 | 172 |
| PDE4DIP | rs2863344 | intron | 1.9×10−06 | 9 | 156 | |
| SLC44A5 | rs1249675 | intron | 6.5×10−06 | 17 | 491 | |
| 2 | --- | rs4848143 | 9.3×10−06 | 27 | 329 | |
| 3 | --- | rs6771019 | --- | 2.1×10−06 | 10 | 83 |
| --- | rs9824150 | --- | 6.6×10−06 | 18 | 244 | |
| 4 | --- | rs11941399 | --- | 1.3×10−06 | 6 | 328 |
| SMARCAD1 | rs3106136 | intron | 7.2×10−06 | 23 | 484 | |
| SMARCAD1 | rs183993 | intron | 8.3×10−06 | 25 | 502 | |
| 5 | ADCY2 | rs4702484 | intron | 6.4×10−07 | 3 | 248 |
| 6 | --- | rs2524276 | --- | 6.3×10−06 | 16 | 246 |
| LOC643281 | rs12198063 | intron | 7.6×10−06 | 24 | 245 | |
| 7 | --- | rs361433 | --- | 8.8×10−07 | 4 | 251 |
| --- | rs2882834 | --- | 1.3×10−06 | 5 | 494 | |
| --- | rs6971109 | --- | 6.8×10−06 | 20 | 503 | |
| 10 | --- | rs705469 | --- | 6.8×10−06 | 19 | 495 |
| SH2D4B | rs6586111 | intron | 7.1×10−06 | 21 | 487 | |
| SH2D4B | rs7915642 | intron | 8.5×10−06 | 26 | 489 | |
| --- | rs705471 | --- | 7.2×10−06 | 22 | 493 | |
| 11 | SOX6 | rs16932455 | intron | 1.5×10−06 | 7 | 329 |
| SOX6 | rs12577378 | intron | 1.8×10−06 | 8 | 330 | |
| SOX6 | rs7947008 | intron | 2.6×10−06 | 11 | 336 | |
| SOX6 | rs12576205 | intron | 4.9×10−06 | 14 | 257 | |
| SOX6 | rs16932445 | intron | 4.9×10−06 | 15 | 257 | |
| 13 | --- | rs6490525 | --- | 3.9×10−06 | 13 | 246 |
| 18 | --- | rs9953852 | --- | 3.1×10−06 | 12 | 252 |
| 19 | --- | rs8101143 | --- | 1.9×10−07 | 1 | 474 |
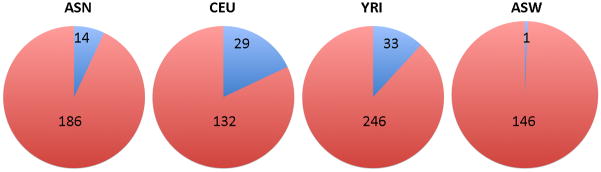
We undertook an analysis to determine the proportion of SNPs identified from each intra-population GWAS that remained strongly significant in the meta-analysis (Fig. 5). Using an arbitrary cutoff of significance of P<10−4, the ASN-only GWAS identified 200 top SNPs, 14 (7%) of which remained significant in the across-population meta-analysis. For CEU, 161 SNPs were identified by CEU-only GWAS, of which 29 (18%) were also significant in the meta-analysis. For YRI the proportion was 12% (33 of 279 SNPs). For ASW, 1 SNP remained significant in the meta-analysis out of 147 that had been identified by ASW-only GWAS.
Figure 5. Graphical depiction of the proportion of SNPs identified from each intra-population GWAS that remained strongly significant in the meta-analysis.
For example, the ASN-only GWAS identified 200 top SNPs (the sum of 14 + 186), 14 (7%) of which remained significant in the meta-analysis. An arbitrary cutoff of significance of P<10−4 was used. None of the SNPs from an individual population that remained strongly significantly in the meta-analysis were the same in multiple populations.
Top Meta-Analysis SNPs Considering Directionality in All Populations
For analysis of the top meta-analysis SNPs having potential importance, we more closely interrogated all SNPs having P<10−4, consistent with many of our previous cell-based analyses8, 11, 13, 31. In the meta-analysis, 321 SNPs met this threshold. Upon inspection, it became apparent that these SNPs generally fell into one of three categories: 1) Directional agreement across all populations regarding the demonstrated association independent of significance of the association; 2) The meta-analysis P value was entirely driven by the association within a single population; 3) The direction of the genotype-phenotype association for the SNP was opposite for one or more of the ethnic populations.
One-third of the top SNPs (108 of 321 SNPs) fit the description of consistent genotype-phenotype association direction across all six individually evaluated populations (ASN, CEU1, CEU3, ASW, YRI1, YRI3). A representative example of this is shown for rs8101143 (P=1.9×10−7; top-ranked in the meta-analysis; Fig. 6A). This SNP was not identified by any of our single population GWAS since the strength of the association was low in any single ethnic population (P>10−4), but strong when considering multiple populations in the meta-analysis, apparently as a result of reproducible, consistent directional effects in all of the included populations. Many additional SNPs (193 SNPs) also fell into this general category, in that the direction of the genotype-phenotype association was consistent across all ethnically distinct individual populations in which the variant was common (MAF>5%) and present (not monomorphic). It is acknowledged that the meta-analysis methodology itself is designed to favor identification of “consistent-direction” SNPs among the top signals.
Figure 6. Examples of SNPs identified in the meta-analysis.
(A) SNP rs8101143 (P=1.9×10−7; ranked #1 in the meta-analysis) illustrates genotype-phenotype associations in the same direction across all six individually evaluated populations; (B) SNP rs576523, which is only polymorphic in the YRI population and, interestingly, was still the second most significant SNP in the overall meta-analysis (P=2.3×10−7) illustrates a SNP with a strong signal in the meta-analysis despite having a genotype-phenotype association only present in one population because the variant was monomorphic in all other populations; (C) SNP rs6971109 illustrates a top hit in the meta-analysis despite the fact that the consensus direction of the genotype-phenotype association for the SNP was opposite in one or more of the ethnic populations. The circular symbol indicates that the directionality of the association in that population was opposite that of the other populations (which are all similarly denoted with diamonds). The dashed vertical line in all panels identifies a P value of 0.05.
Another category was genotype-phenotype associations present only in one ethnically-restricted population because the variant was monomorphic or rare in all other populations. An example is shown for SNP rs576523 (Fig. 6B). rs576523 is only polymorphic in the YRI population and, interestingly, is the second-most significant SNP in the meta-analysis (P=2.3×10−7). In fact, three of the top 10 SNPs in Table 1 are considered polymorphic only in one population (rs576523 in YRI, rs2863344 in CEU, and rs6771019 in YRI). These results include a number of the “surviving” SNPs depicted in Fig. 5, and they likely represent some of the strongest findings given the strengths of the associations despite the fact that they are population-restricted.
The third group includes SNPs in which the consensus direction of the genotype-phenotype association for the SNP was opposite for one or more of the ethnic populations, yet the strength of the association in the consensus direction was robust enough to achieve a meta-analysis P value reaching top significance (e.g., an association was positive for CEU, YRI, and ASW, but opposite for ASN, yet meta-analysis P value achieved <10−4). An example of this is shown in Fig. 6C, for SNP rs6971109. Such SNPs might have relevance for general testing in most individuals with the knowledge that in a single ethnic population, the association might not be relevant. Only 20 of the top 321 SNPs fit this group, and in none of these 20 was the opposite direction-outlier population’s association statistically significant.
Potential Functional Role of Top Meta-Analysis SNPs
Of the 321 top SNPs with P<10−4, most (162 SNPs) are located in uncharacterized regions of the genome (i.e., not annotated to any known gene based on location), a finding that has been consistent in our previous studies using an unbiased genome-wide approach to chemotherapy susceptibility pharmacogenomics. Many others are located in introns (145 SNPs), although none are at known splice sites. Eight (8) SNPs are located either near known genes (rs263003 with PARL; rs3106134 with SMARCAD1; and rs972249 with KRT40) or in 3′ or 5′ untranslated (UTR) gene regions (rs7448390 and rs17101607 located in the 3′UTR of YIPF5; rs11635570 located in the 3′UTR of MTFMT; rs17039288 located in the 5′UTR of MYT1L; and rs3738414 located in the 5′UTR of VTCN1). One SNP (rs10907177) was a synonymous coding variant in a poorly-characterized gene region (C1ORF159).
Perhaps most interesting was a missense SNP (rs11722476) within SMARCAD1 (Fig. 7A). This SNP had a meta-analysis P value of 6.7×10−5. The G to A DNA change results in a serine to asparagine amino acid change in the SMARCAD1 (SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin) protein. The genotype-phenotype association for this SNP in the n=503 individuals is shown in Fig. 7B.
Figure 7. Meta-analysis of the cross-population GWAS identified rs11722476, a missense SNP within SMARCAD1.
(A) Zoom-in view of the genomic region on chromosome 4 around this SNP. The fact that a number of other signals within SMARCAD1 were found is illustrated by the striking number of SNPs which were found at P values smaller than the arbitrary cutoff of P<10−4; (B) Genotype-phenotype association for this SNP in the n=503 individuals.
Effect of Thymidine Phosphorylase
Because inactive 5′DFUR requires activation to 5-FU via a final anabolizing enzyme, thymidine phosphorylase, and because thymidine phosphorylase levels can be affected by the relative expression of the thymidine phosphorylase gene (TYMP) in various human tissues32, we lastly interrogated whether TYMP levels within LCLs correlated with 5′DFUR susceptibility in our study. Using our broad gene expression data in HapMap CEU1 and YRI119, we found a significant relationship between 5′DFUR AUC and TYMP expression in both CEU1 (P=1.5×10−4) and YRI1 (P=2.4×10−6). In both populations, the direction of the relationship indicated that higher TYMP expression correlated with lower AUC (β=−3.97 for CEU1 and -5.10 for YRI1), as might be hypothesized. However, the overall proportion of AUC variation explained by TYMP was only 0.15 for CEU1, and 0.22 for YRI1, supporting our above GWAS findings that there are other important sources of genetic variability in determining capecitabine (5′DFUR) sensitivity.
Discussion
We have herein described a novel human cell-based approach to identifying germline pharmacogenomic polymorphisms governing susceptibility to the widely-used chemotherapy drug capecitabine. By utilizing the inherent powerful genetic information encapsulated within the HapMap and a high-throughput cell-based chemotherapy susceptibility testing method, we were able to perform the largest known GWAS for capecitabine susceptibility pharmacogenomics, in over 500 individual samples. This comprehensive, “across-populations” chemotherapy study is the first of its kind—distinguished from previous cell-based pharmacogenomic approaches by the novel idea of conducting a meta-analysis across multiple ethnic populations. This exercise permitted a built-in method for statistical veracity testing of resulting SNPs, since the meta-analysis assimilated raw P values from individual population-based GWA studies and only the top SNPs showing strong association after consideration of all individual populations achieved robust meta-analysis P values. In this sense, many of our top SNPs from this meta-analysis might be therefore considered clinically testable for replication in the ethnically diverse human populations that are typically encountered in true clinical practice and in clinical research settings.
Our two most compelling genetic findings deserve particular discussion. SNP rs4702484 on chromosome 5 was identified among the top three SNPs in our meta-analysis in addition to being identified by our CEU intra-population GWAS (at a P value reaching genome-wide significance). While this SNP is intronic within ADCY2, close analysis of the genomic region around this SNP demonstrates that the SNP is quite proximal to the methionine-folate pathway gene MTRR (see Fig. 2B). We therefore hypothesize that upstream regulation of MTRR (via polymorphism of its extended promoter region) may be likely, a mechanism which would fit with the purported site of activity of 5-FU related compounds like capecitabine within the folate metabolism/nucleotide biosynthesis pathway. While other capecitabine/5-FU pathway candidate genes (e.g., TYMS, MTHFR, DPD) have been well studied previously33–35, we could find no prior positive reports implicating MTRR pharmacogenetic variation with 5-FU-related phenotypes (one recent study reported a negative finding with a different MTRR polymorphism36). This increases the potential novelty of our finding. Perhaps most interestingly, if this relationship is indeed confirmed by ongoing functional studies of MTRR, this genetic relationship would be an example of a GWAS approach identifying a de facto “candidate gene” that has been largely previously ignored by classic candidate gene methods.
Separately, while a number of our other top meta-analysis SNPs are interesting for possible functional and clinical importance, the identification of SNP rs11722476 (a missense SNP within SMARCAD1) was especially intriguing. First, there were a noticeably large (and disproportionate) number of repetitive, strong signals within SMARCAD1 among our top 321 meta-analysis SNPs (see Fig. 7A), and two signals within SMARCAD1 (potentially in linkage disequilibrium [LD]) among the most significant (P<10−5) overall SNPs (Table 1). These findings, along with the identification of the above missense SNP in this region, compositely suggest importance of this gene for capecitabine. SMARCAD1, a member of a helicase superfamily including proteins essential to genome replication, repair, and expression37, has interestingly been previously mentioned (although only tangentially) in reports that would be consistent with a 5-FU-related importance. One study, using deletion mapping of chromosome 4q22–35, showed that SMARCAD1 was frequently deleted in head and neck cancers38, which are often treated successfully by 5-FU. This might imply that SMARCAD1 gene dosage effects could potentially underlie one mechanism of 5-FU susceptibility for these tumors. A second unrelated study found (via expression analysis) that SMARCAD1 has particularly high levels in endocrine tissues39. Breast tissue would be considered highly endocrine-responsive (estrogen/progesterone receptors) and therefore, while still speculative, this could begin to suggest a role for SMARCAD1 polymorphism in explaining the sensitivity of breast cancers to capecitabine. Of course, such hypotheses would need to be confirmed by formal molecular studies, highlighting the fact that GWA studies are often excellent for permitting new hypothesis generation.
None of the typically-studied capecitabine/5-FU pathway candidate genes themselves (TYMS, TYMP, MTHFR, DPYD, among others) were identified among the top SNP signals in our study. This could be due, in part, to tissue-restricted down regulation of some of these genes in LCLs. However, it also illustrates the idea that a combined approach (genome-wide plus candidate gene methods) may ultimately yield the most comprehensive approach to drug susceptibility pharmacogenomics, perhaps especially in oncology. Additionally, there may be differences in the pharmacogenomics of capecitabine compared to 5-FU (just as there are differences in the toxicity profiles for these two drugs), and there has been relatively much less clinical investigation into the pharmacogenomics of capecitabine. One recent study implicated a role for TYMS in patients with colon cancer receiving a regimen containing capecitabine40. A second prior study had also previously found the strongest evidence for TYMS among the typically-studied capecitabine candidate genes41.
Our study has recognized limitations. While SNP rs4702484 did achieve genome-wide statistical significance in the CEU-only GWAS, none of the meta-analysis results achieved P values below the generally-accepted GWA cutoff of ~5×10−8 30. In this sense, the overall statistical power gained by combining six panels in the meta-analysis was probably less than we expected. This may be due to different underlying LD structures between the individual populations42,43. At the same time, it has been argued that additional factors in the comprehensive evaluation of GWAS findings need to be considered beyond just the P value threshold44, and we would suggest that SNPs in our meta-analysis with P values of ~10−6 or smaller—especially when the association directionality is consistent across all 6 individually-tested panels—have a higher likelihood of true importance. The fact that 4 SNPs indeed achieved P values of ~10−7 (approaching traditional genome-wide significance) despite our sample size (~500 individuals) which would typically be considered too small for conducting a GWAS may emphasize the potential relevance of these findings. We believe it also simultaneously validates the utility of the meta-analysis approach. Secondly, the majority of the top identified SNPs are located in regions of the genome without obvious apparent functional explanation. This could reflect two possibilities: either this simply reflects the greater statistical probability of more commonly identifying variants in non-coding regions of the genome since those regions inherently comprise a vastly greater total percentage of the genome; or this signals the novelty and the advantage of GWAS studies like this one for interrogating chemotherapeutic pharmacogenomic traits, where an unbiased approach may be exactly what is desired, since candidate-gene or single-gene methods have often fallen short18. Thirdly, although the genetic information in this study is from human individuals in the HapMap project, and although meta-analysis approaches when conducted properly may obviate the need for additional replication in a separate population, the phenotypes were derived in a cell-line model and therefore the results require validation in clinical populations of patients taking capecitabine.
It should lastly be mentioned that while our cross-population method offers the advantage of identifying relatively common SNPs testable in all or most individuals regardless of ethnic background, we did find allelic heterogeneity among the top SNPs (and often one or more populations were monomorphic at a given identified locus) and, despite the value of a cross population meta-analysis, one of our strongest results was found via a single-population analysis, underscoring the value of individual population analyses as well.
In summary, we conducted a large, cell-based meta-analysis of genome-wide association findings for capecitabine chemotherapy susceptibility across multiple divergent human populations. The resulting list of novel SNPs and related genes, along with SNPs in previously-identified capecitabine/5-FU pathway candidate genes, deserve study in clinical settings toward the goal of identifying underlying genetic factors influencing toxicity from, and perhaps response to, this commonly-used cancer agent. The ongoing multi-center clinical study examining comprehensive capecitabine toxicity pharmacogenomics (www.clinicaltrials.gov study identifier NCT00977119) indeed plans to utilize these data for specifically that purpose.
Acknowledgments
Supported by The University of Chicago Breast Cancer SPORE P50 CA125183 (MED), NIH/NIGMS UO1 GM61393 Pharmacogenomics of Anticancer Agents Research Group (MED; NJC), NIH/NCI F32 CA136123 (PHO), NIH TL1 RR25001 (ALS), and NIH/NIGMS K08 GM089941 (RSH).
Footnotes
There are no relevant financial relationships to disclose for any authors.
References
- 1.Efferth T, Volm M. Pharmacogenetics for individualized cancer chemotherapy. Pharmacology & Therapeutics. 2005;107:155–176. doi: 10.1016/j.pharmthera.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy J, Saltz L, Twelves C, et al. Efficacy of capecitabine versus 5-fluorouracil in colorectal and gastric cancers: A meta-analysis of individual data from 6171 patients. Ann Oncol. 2011 doi: 10.1093/annonc/mdr031. [DOI] [PubMed] [Google Scholar]
- 4.Walko CM, Lindley C. Capecitabine: A review. Clin Ther. 2005;27:23–44. doi: 10.1016/j.clinthera.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Aprile G, Mazzer M, Moroso S, Puglisi F. Pharmacology and therapeutic efficacy of capecitabine: Focus on breast and colorectal cancer. Anticancer Drugs. 2009;20:217–229. doi: 10.1097/CAD.0b013e3283293fd4. [DOI] [PubMed] [Google Scholar]
- 6.Biganzoli L, Martin M, Twelves C. Moving forward with capecitabine: A glimpse of the future. Oncologist. 2002;7 (Suppl 6):29–35. [PubMed] [Google Scholar]
- 7.Clemons M, Joy AA, Abdulnabi R, et al. Phase ii, double-blind, randomized trial of capecitabine plus enzastaurin versus capecitabine plus placebo in patients with metastatic or recurrent breast cancer after prior anthracycline and taxane therapy. Breast Cancer Res Treat. 2010;124:177–186. doi: 10.1007/s10549-010-1152-0. [DOI] [PubMed] [Google Scholar]
- 8.Huang RS, Duan S, Bleibel WK, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartford CM, Duan S, Delaney SM, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113:2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang RS, Duan S, Kistner EO, et al. Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res. 2008;68:3161–3168. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bleibel WK, Duan S, Huang RS, et al. Identification of genomic regions contributing to etoposide-induced cytotoxicity. Hum Genet. 2009;125:173–180. doi: 10.1007/s00439-008-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donnell PH, Gamazon E, Zhang W, et al. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenet Genomics. 2010;20(5):327–337. doi: 10.1097/FPC.0b013e3283396c4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla SJ, Duan S, Wu X, Badner JA, Kasza K, Dolan ME. Whole-genome approach implicates cd44 in cellular resistance to carboplatin. Human Genomics. 2009;3:128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from africans. Mol Cancer Ther. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welsh M, Mangravite L, Medina MW, et al. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziliak D, O’Donnell PH, Im HK, et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl Res. 2011;157:265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell PH, Dolan ME. Cancer pharmacoethnicity: Ethnic differences in susceptibility to the effects of chemotherapy. Clin Cancer Res. 2009;15:4806–4814. doi: 10.1158/1078-0432.CCR-09-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Duan S, Kistner EO, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark AL, Zhang W, Mi S, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10:505–512. doi: 10.1038/tpj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark AL, Zhang W, Zhou T, et al. Population differences in the rate of proliferation of international hapmap cell lines. Am J Hum Genet. 2010;87:829–833. doi: 10.1016/j.ajhg.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgerson DG, Boyko AR, Hernandez RD, et al. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009;5:e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wettergren Y, Odin E, Carlsson G, Gustavsson B. Mthfr, mtr, and mtrr polymorphisms in relation to p16ink4a hypermethylation in mucosa of patients with colorectal cancer. Mol Med. 2010;16:425–432. doi: 10.2119/molmed.2009.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bono JS, Twelves CJ. The oral fluorinated pyrimidines. Invest New Drugs. 2001;19:41–59. doi: 10.1023/a:1006404701008. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RC, Nelson GW, Troyer JL, et al. Accounting for multiple comparisons in a genome-wide association study (gwas) BMC Genomics. 2010;11:724. doi: 10.1186/1471-2164-11-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. American Journal of Human Genetics. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pentheroudakis G, Twelves C. Capecitabine (xeloda): From the laboratory to the patient’s home. Clin Colorectal Cancer. 2002;2:16–23. doi: 10.3816/CCC.2002.n.007. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Diasio RB. Regulation of human dihydropyrimidine dehydrogenase: Implications in the pharmacogenetics of 5-fu-based chemotherapy. Pharmacogenomics. 2007;8:257–265. doi: 10.2217/14622416.8.3.257. [DOI] [PubMed] [Google Scholar]
- 34.Salgado J, Zabalegui N, Gil C, Monreal I, Rodriguez J, Garcia-Foncillas J. Polymorphisms in the thymidylate synthase and dihydropyrimidine dehydrogenase genes predict response and toxicity to capecitabine-raltitrexed in colorectal cancer. Oncol Rep. 2007;17:325–328. [PubMed] [Google Scholar]
- 35.Etienne-Grimaldi MC, Francoual M, Formento JL, Milano G. Methylenetetrahydrofolate reductase (mthfr) variants and fluorouracil-based treatments in colorectal cancer. Pharmacogenomics. 2007;8:1561–1566. doi: 10.2217/14622416.8.11.1561. [DOI] [PubMed] [Google Scholar]
- 36.Pardini B, Kumar R, Naccarati A, et al. 5-fluorouracil-based chemotherapy for colorectal cancer and mthfr/mtrr genotypes. Br J Clin Pharmacol. 2010 doi: 10.1111/j.1365-2125.2010.03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki N, Ikeda S, Ohara R, et al. The novel protein complex with smarcad1/kiaa1122 binds to the vicinity of tss. J Mol Biol. 2008;382:257–265. doi: 10.1016/j.jmb.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 38.Cetin E, Cengiz B, Gunduz E, et al. Deletion mapping of chromosome 4q22–35 and identification of four frequently deleted regions in head and neck cancers. Neoplasma. 2008;55:299–304. [PubMed] [Google Scholar]
- 39.Adra CN, Donato JL, Badovinac R, et al. Smarcad1, a novel human helicase family-defining member associated with genetic instability: Cloning, expression, and mapping to 4q22-q23, a band rich in breakpoints and deletion mutants involved in several human diseases. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- 40.Pander J, Wessels JA, Gelderblom H, van der Straaten T, Punt CJ, Guchelaar HJ. Pharmacogenetic interaction analysis for the efficacy of systemic treatment in metastatic colorectal cancer. Ann Oncol. 2010 doi: 10.1093/annonc/mdq572. [DOI] [PubMed] [Google Scholar]
- 41.Largillier R, Etienne-Grimaldi MC, Formento JL, et al. Pharmacogenetics of capecitabine in advanced breast cancer patients. Clin Cancer Res. 2006;12:5496–5502. doi: 10.1158/1078-0432.CCR-06-0320. [DOI] [PubMed] [Google Scholar]
- 42.Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]