Abstract
Metazoan cells are exposed to a multitude of signals, which they integrate to determine appropriate developmental or physiological responses. Although the Hippo pathway was only discovered recently, and our knowledge of Hippo signal transduction is far from complete, a wealth of interconnections amongst Hippo and other signaling pathways have already been identified. Hippo signaling is particularly important for growth control, and I describe how integration of Hippo and other pathways contributes to regulation of organ growth. Molecular links between Hippo signaling and other signal transduction pathways are summarized. Different types of mechanisms for signal integration are described, and examples of how the complex interconnections between pathways are used to guide developmental and physiological growth responses are discussed. Features of Hippo signaling appear to make it particularly well suited to signal integration, including its responsiveness to cell-cell contact and the mediation of its transcriptional output by transcriptional co-activator proteins that can interact with transcription factors of other pathways.
Keywords: Hippo, Signaling, Yorkie, Morphogen, Regeneration
1.1 Introduction: Integration of intercellular signaling
Virtually all signaling pathways are integrated at some level, and biologists increasingly realize that we must consider webs of interconnected signaling networks rather than simple linear pathways. The interconnection of signaling pathways enables cells to integrate the different kinds of information they receive, such as relative position, developmental stage, and nutritional status. This integration can take a simple hierarchical form, for example when one pathway regulates the production of a ligand for another, or can involve deeper and more complex interconnections.
Interconnection with other pathways appears to be a particularly prominent feature of the Hippo pathway. One important role for this is to enable cells to integrate the information provided by Hippo signaling, which is often related to cell contact or cell polarity, together with that provided by other pathways, such as positional information from BMP signaling. However, interconnections between Hippo and other pathways have been identified at many different levels, and they serve to modulate an ever-increasing variety of developmental and physiological responses. As these interconnections have been most intensively studied in Drosophila, where Hippo signaling was first discovered, I focus on this model system, but also include examples from other organisms.
1.2 Hippo, a conserved pathway for growth control
At its most basic, the Hippo pathway is, like many other pathways, a signal transduction pathway that conveys signals perceived at the plasma membrane to a transcriptional response in the nucleus (Figure 1). In many (though not all) cases, the transcriptional output of the pathway appears to be directed towards the regulation of organ growth, and consequently Hippo signaling has major roles in developmental growth control, and its dysregulation has been linked to a number of cancers. As these roles of the Hippo pathway, together with its basic mechanics, have been discussed extensively in recent reviews [1–3], and in other articles of this issue, only a brief outline of the pathway is provided here.
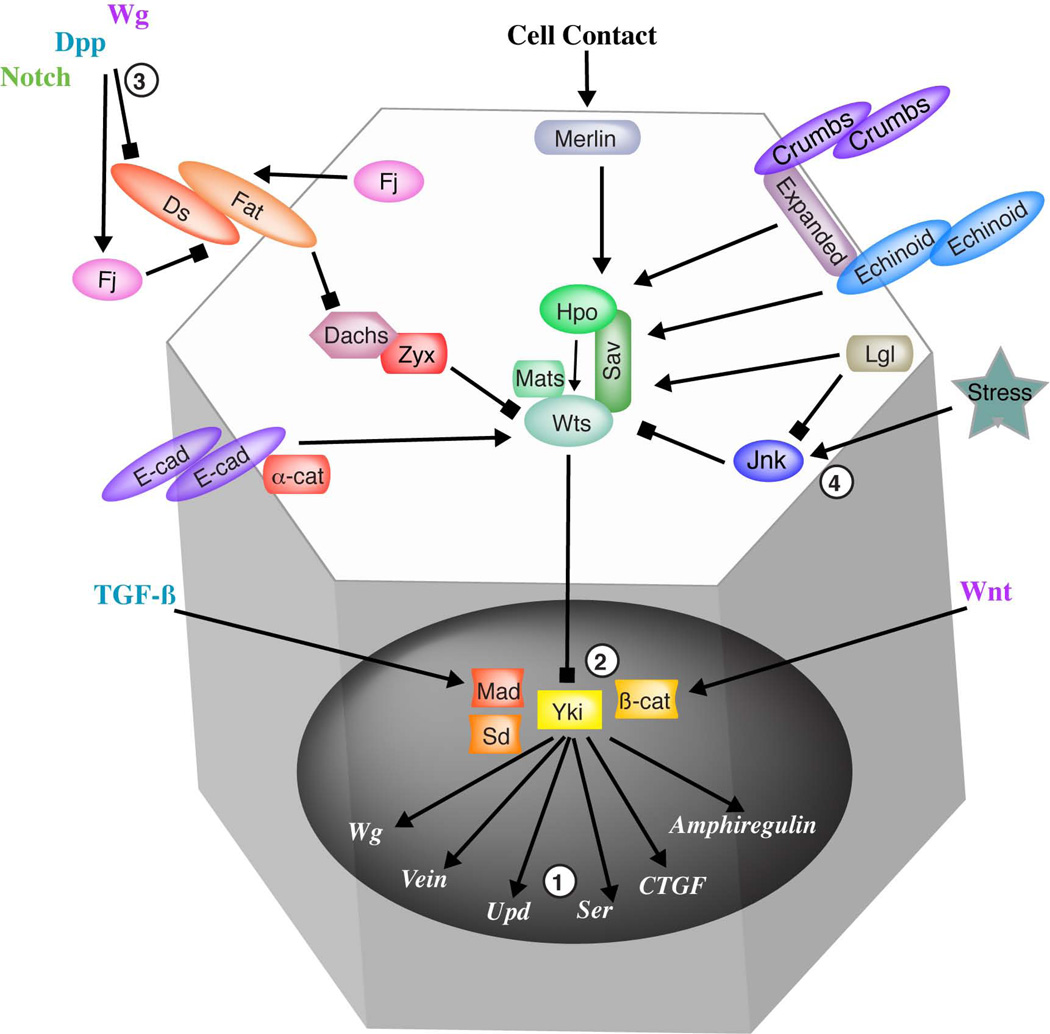
Figure 1. Hippo signaling and some of its connections with other pathways.
The schematic depicts regulatory some of connections within the Hippo pathway, and between Hippo signaling and other pathways discussed in the text. The Drosophila names of pathway components are used, and the schematic is over-simplified in that many Hippo signaling pathway components are not shown. Interactions with other pathways depicted include 1) Within the nucleus, Yki/Yap/Taz promotes the expression of ligands for other signaling pathways. 2) Yki/Yap/Taz interacts with Smad proteins (Mad) and β-catenin, which are transcription factors of TGF-β and Wnt pathways. When Yki/Yap/Taz is cytoplasmic this interaction inhibits transcriptional activation by these pathways, but when its nuclear they cooperate to promote transcription of downstream genes. 3) Fj and Ds expression are regulated by the Notch, Dpp, and Wg signlaing pathways in the Drosophila wing, at least some of this regulation is mediated through Vg (not shown). 4) Wounding, apoptosis, or infection (“stress”) can activate the Jnk pathway, which can lead to Yki activation.
Some individual components of Hippo signaling were first discovered years (e.g. Yap, Warts) [4–6], or even decades (e.g. Fat, Dachs) [7, 8] before the term Hippo signaling was coined, but it is only within the last decade that the interconnectedness of these components into a conserved intercellular signaling pathway has become appreciated [1–3]. At the core of both the discovery of the pathway and its signal transduction mechanism is a conserved kinase cascade, in which the protein kinase Hippo (Mst1,2 in vertebrates) promotes activation of the protein kinase Warts (Lats1,2 in vertebrates). This is achieved by Hippo directly phosphorylating Warts, and two additional components of the core kinase cassette: Mob as tumor suppressor (Mats, Mobkl1a,b in vertebrates) and Salvador (Sav, WW45 in vertebrates) [9, 10]. Warts kinase can then negatively regulate a transcriptional co-activator protein, Yorkie (Yki, Yap and Taz in vertabrates) [11], mainly by phosphorylating it to promote its cytoplasmic retention through 14-3-3 binding [12–17].
Upstream and downstream of this conserved kinase cassette, the pathway becomes more complex, as there is a considerable diversity of upstream regulatory inputs. In Drosophila, three transmembrane receptor proteins have so far been identified as Hippo pathway receptors: Fat, Crumbs, and Echinoid [18–26]. Other upstream inputs whose regulation is less well understood have also been described in Drosophila, including regulation dependent upon Merlin, Lethal giant larvae, and Jun kinase (Jnk) [19, 23, 27–30]. There is also a surprising diversity amongst the upstream regulatory inputs into the pathway between phyla. Merlin is a conserved regulator between Drosophila and vertebrates, which at least in vertebrates plays a crucial role in contact-dependent inhibition of cell proliferation through affects on Hippo signaling [13, 31–33]. However, in vertebrates components of the E-cadherin/alpha-catenin complex are Hippo pathway regulators [34, 35], and an equivalent role in Drosophila has not yet been described. Conversely, components of the Fat branch of the Hippo pathway have been identified in vertebrates, but don’t appear to have significant effects on the Hippo pathway in these species, at least in most tissues [36–38]. Nonetheless, a common theme has emerged in which most regulators of Hippo signaling are associated with cell-cell junctions, and thus well positioned to inform cells about cell polarity and cell density, which thus have major influences on Hippo activity.
One feature of Hippo signaling that provides significant potential for signal integration concerns the nature of the transcriptional output of the pathway. This is provided by a transcriptional co-activator protein (Yorkie in Drosophila, Yap and Taz in vertebrates) rather than a DNA-binding protein [11]. Thus, interaction with other proteins is intrinsically required for regulation of downstream transcriptional targets. Moreover, Yki/Yap/Taz co-activators have been found to be able to associate with multiple, distinct DNA-binding partners [39, 40]. The major partner for Yki/Yap/Taz appears to be Scalloped in Drosophila, and its homologues the Tead/TEF proteins in vertebrates [16, 41–45]. However, a number of additional DNA-binding partners have also been identified, including some that are regulated by other signaling pathways, such as the Smad proteins [46–49], which are transcription factors of TGF-β related pathways [50].
1.3 Regulation of other pathways by Hippo signaling
While integration between signaling pathways can occur at almost any level, conceptually one of the simplest forms is when one pathway regulates the amount of signal perceived by another, for example, by regulating the production of a ligand for that pathway. In most contexts, the main targets of Yki/Yap/Taz activity appear to be promoters of organ growth. These include a number of direct, autonomous promoters of cellular growth. However, a significant part of the growth regulating activity of Hippo appears to derive from regulation of secreted growth factors that activate other pathways.
An early example of this was the regulation of the Drosophila Wnt ligand Wingless (Wg) in the proximal part of the developing wing [19, 51]. Wg acts as a mitogen here, promoting proximal wing growth [52], and this regulation of Wg contributes significantly to upregulation of growth associated with certain Hippo pathway mutants [51]. Another example from Drosophila is the upregulation of Serrate, a ligand for the Notch pathway, which promotes leg growth [53, 54]. Drosophila Hippo signaling has also been linked to regulation of glypicans, which play a prominent role in modulating signaling by several classes of secreted growth factors [55]. Finally, a striking recent example of growth promotion by Drosophila Hippo signaling through regulation of other pathways has come from studies of the adult intestine.
The Drosophila intestine is maintained by intestinal stem cells (ISCs) and has a simple architecture comprising four basic cell types: ISCs, undifferentiated progenitors, and two types of differentiated cells [56, 57]. The differentiated cells are continuously lost at a low rate, but are replaced from the ISCs, which are the only cells that proliferate. In response to infection or injury, ISC proliferation is increased to facilitate repair [58–60]. This increased proliferation is mediated through the Hippo pathway, but the effect of Hippo signaling is at least mostly non-autonomous: infection or damage of the non-proliferating differentiated cells leads to activation of Yki, which then promotes the expression of cytokines that are secreted from these cells, and which stimulate the proliferation of nearby ISCs [28, 30, 61, 62]. These cytokines include multiple ligands for the Jak-Stat pathway (Unpaired proteins), and also ligands for the EGFR pathway. The characterization of the central role of non-autonomous effects of Hippo signaling in controlling ISC proliferation emphasizes the importance of regulation of other pathways to growth control by Hippo signaling.
Activation of secreted ligands for other pathways has also been linked to growth regulation by vertebrate Hippo pathways. One prominent example is the regulation of amphiregulin, a mammalian EGFR ligand, by Yap within breast epithelial and adenocarcinoma cells [63, 64]. This upregulation of amphiregulin is essential to the ability of Yap to transform these cells. Another major growth factor target of Hippo signaling in mammalian cells is Connective Tissue Growth Factor (CTGF, also known as CCN2) [15, 43]. Regulation of CTGF by Yap/Taz appears to be widespread amongst many different cell types, and consequently its expression is often now used as a marker of Hippo pathway activity. CTGF has diverse roles, including links to tissue repair and carcinogenesis [65], which could contribute to Hippo pathway influences on growth in vertebrates.
Other examples of Hippo regulating other pathways have been identified, but the mechanisms by which they are connected are more complex or less well defined. For example, multiple links between Hippo and Notch signaling have been identified during Drosophila oogenesis. During early oogenesis, when Notch is required for polar cell specification [66], Yki promotes Notch activity, and thereby influences polar cell fate [67]. Later in oogenesis, when Notch influences gene expression within main body follicle cells, and their transition from a mitotic to an endoreplicative cell cycle, Yki inhibits Notch activity; consequently mutations in upstream negative regulators of Yki like warts result in phenotypes that resemble those of Notch mutations [68–70]. Yki affects endocytic trafficking and the levels of Notch protein at the apical membrane in these cells, but whether this accounts for the observed effects on Notch signaling is unknown. Hippo pathway mutations can also affect levels of apical membrane proteins in imaginal discs, including several transmembrane receptors [71], and moreover can influence the size of the apical membrane, but this effect appears to be distinct from its effects on growth [72, 73].
1.4 Regulation of Hippo signaling by other pathways
1.4.1 Regulation of Fat-Hippo signaling by morphogen gradients
Hippo pathway regulation is complex, and a number of distinct upstream inputs have been identified. Many of these upstream inputs are related to cell-cell contact, and not directly affected by other pathways. However, one prominent exception to this occurs within the developing Drosophila wing, where the Fat branch of the Hippo pathway is regulated downstream of signaling pathways that organize wing patterning and growth. Fat is a large (over 5000 amino acids) cadherin protein that functions as a receptor for both Hippo signaling, and a genetically separable planar cell polarity (PCP) pathway [reviewed in 74, 75]. Fat activity is regulated by its ligand, Dachsous (Ds, another large cadherin molecule) and a Golgi-localized kinase, Four-jointed (Fj), which modulates binding between Ds and Fat [19, 51, 76–82]. A defining feature of Fj and Ds expression is that they are always expressed in complementary gradients. These gradients are then interpreted in a unique fashion: rather than responding simply to the absolute amount of Ds or Fj, the vector of the Ds and Fj expression gradients directs the Fat-PCP pathway [75], whereas the relative slope of the Ds and Fj expression gradients can influence Fat-Hippo signaling [78, 79, 83]. How these gradients are interpreted is not fully understood, but it appears at least in part to involve the polarized membrane distribution of the myosin protein Dachs [53, 78], which facilitates interactions between Zyxin and Warts [84].
Since Fat-Hippo pathway activity depends upon the Fj and Ds gradients, it ultimately depends upon the mechanisms that establish their graded expression. In the wing, Fj and Ds expression are influenced by wing patterning signals including Dpp, Wg, and Notch [78], although at least some of this regulation is mediated through Vestigial (Vg), a transcription factor that is itself regulated by Dpp, Wg, and Notch [85–90], and which promotes Fj expression whilst inhibiting Ds expression [51, 83, 91, 92]. However, as Vg is a wing-specific transcription factor [93, 94], the gradients of Fj and Ds expression that are established in other organs must be established independently of Vg.
In addition to influencing growth within the developing wing, an alternative mechanism by which the graded expression of Fj and Ds could drive wing growth involves recruitment of neighboring, non-wing cells into the developing wing [83, 89]. This can occur through induction of Vg, whose expression defines the future wing blade [93, 94]. It relies on the steep gradients of Fj and Ds expression that occur at the edge of the developing wing, which result in strong activation of Yki through the Fat-Hippo pathway. This Yki activation can act in conjunction with Wg signaling to induce Vg expression in neighboring cells, thereby re-specifying them as wing blade cells [83].
Whether morphogen gradients influence growth through the Fat-Hippo pathway in other organs and organisms has not been as well characterized, but Fat pathway components do have significant affects on Drosophila leg growth [53, 95]. Moreover, the cricket leg has been established as a model for insect leg regeneration, and a requirement for Fat and Hippo pathway genes has been demonstrated in this system by RNAi [96]. The expression of Fat pathway components, and the results of the authors’ manipulations, can be interpreted by a model in which leg regeneration is influenced by the steepness of expression gradients of Fat pathway regulators [96].
1.4.2 Regulation of Hippo signaling by Jnk and other pathways in regeneration and tumorigenesis
Hippo signaling is also important for regeneration in other contexts. In mammals, Yap activity contributes to intestinal regeneration in a mouse model [97]; how Yap activity is regulated in this case is not known. Hippo signaling has also been studied in Drosophila for its influence on regenerative growth after tissue damage [28–30, 61, 62, 98]. In both imaginal discs and in the adult intestine the Jnk pathway plays a crucial role in regulating Yki activity during regeneration, as blocking Jnk activity can prevent Yki activation after tissue damage, and directly activating Jnk is sufficient to stimulate Yki activation [28–30]. The role of Jnk is complex however, as it also has a pro-apoptotic role [99], and it’s not yet clear how these opposing roles are balanced.
Jnk signaling also regulates Yki activity in certain classes of tumors, but again its effect is complex. Yki is activated within tumors associated with mutation of lethal giant larvae (lgl) [23, 29, 100], and at least in the wing this requires Jnk activity [29]. However, within tumors associated with mutation of scribble, Jnk has an anti-proliferative, pro-apoptotic role [101], and down-regulation of Jnk actually increases Yki activation [102]. Presumably, some of the effects of Jnk are indirect, and a better biochemical understanding of how Jnk affects Yki activity is needed.
Other pathways also regulate Yap during tumor formation. For example, Sonic hedgehog (Shh) signaling has been linked to medulloblastoma, and the influence of Shh on neural progenitor proliferation in medulloblastoma depends on Yap [103], but how Yap is upregulated by Shh is not yet known. In colorectal carcinoma cells, Wnt signaling upregulates Yap levels [104]. This upregulation is not through the Hippo pathway, but instead involves a direct transcriptional upregulation of Yap by transcription factors of the Wnt pathway (β-catenin and TCF4) [104].
Integration of Hippo signaling with other pathways
In addition to the upstream-downstream connections between pathways described above, some pathways are even more closely integrated with Hippo signaling. TGF-β signaling pathways are intimately connected to Hippo pathways through direct binding of their transcription factors. TGF-β signaling influences transcription by regulating the sub-cellular localization of DNA-binding transcription factors of the Smad protein family [50]. Smad proteins can be partners for Yki/Yap/Taz proteins [46–49], and, in some cases, such as in the Drosophila wing, contribute to growth regulation by Hippo signaling [49]. The interaction between Yap/Taz proteins and Smad proteins has also been implicated in TGF-β signaling functions in some contexts, including mammalian ES cell renewal [47]. Notably, these proteins interact not only in the nucleus, where they can co-regulate downstream genes, but also in the cytoplasm. This cytoplasmic interaction, which has so far only been characterized in mammalian cells, enables Hippo signaling to exert another layer of control on TGF-β signaling. When Taz/Yap are cytoplasmic, their interaction with Smads in the cytoplasm restrains TGF-β signaling [105]. This results in a cell density-dependent influence on TGF-β signaling [105], as cell density influences Yap/Taz localization through the Hippo pathway [13, 105].
Hippo also exhibits multiple layers of interaction with Wnt signaling pathways beyond the transcriptional regulation described above. As in TGF-β pathways, crucial cytoplasmic interactions between pathway components have been identified. Yap or Taz can interact in the cytoplasm with the transcription factor of canonical Wnt pathways, β-catenin, and thereby retain it in the cytoplasm [106]. Taz has also been reported to inhibit Wnt signaling by interacting in the cytoplasm with Dvl, another key component of Wnt pathways [107]. Conversely, under conditions where the Hippo pathway is inactivated, and Yap is able to translocate to the nucleus, Yap can interact with β-catenin within the nucleus to promote the expression of Wnt target genes [108]. In the developing heart, this led to elevated cardiomyocyte proliferation [108]. Yki and Wnt also synergize to co-regulate Vg in the Drosophila wing [83], but the mechanism by which they synergize here has not been determined.
Summary: Integration of intercellular signaling in the Hippo pathway
Although interconnections occur between many signaling pathways, integration with other pathways appears to be a particularly prominent feature of Hippo signaling. This can occur at many different levels, and a number of examples of upstream-downstream integration have been described (Figure 1). Many of these play important roles in the growth control functions associated with the Hippo pathway. In addition, in some instance, such as for Wnt and TGF-β pathways, even tighter integration has been observed, through interactions between the respective transcription factors of these pathways. Biologically, this integration could make sense because these pathways provide different kinds of information. Hippo signaling is often regulated by cell contact or cell density, whereas pathways it interacts with often provide positional information based on the concentrations of secreted protein ligands.
Highlights.
Hippo signaling is integrated with other signal transduction pathways
This integration can occur at many different levels of Hippo signaling
Regulation of other pathways contributes to growth regulation by Hippo signaling
Hippo signaling is tightly integrated with TGF-β and Wnt signaling
ACKNOWLEDGEMENTS
We thank Cordelia Rauskolb for comments of the manuscript. Research in K.D.I.’s lab is supported by the Howard Hughes Medical Institute, NIH grant 2R01 GM078620, and HFSP grant RGP0016/2010.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 5.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 6.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 7.Bridges CB, Morgan TH. Contributions to the genetics of Drosophila melanogaster.II. The second-chromosome group of mutant characters. Publs Carnegie Instn. 1919;278:123–304. [Google Scholar]
- 8.Waddington CH. The genetic control of wing development in Drosophila. J Genet. 1940;41:75–139. [Google Scholar]
- 9.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 10.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett FC, Harvey KF. Fat Cadherin Modulates Organ Size in Drosophila via the Salvador/Warts/Hippo Signaling Pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 19.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 20.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The Tumor-Suppressor Gene fat Controls Tissue Growth Upstream of Expanded in the Hippo Signaling Pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The Fat Cadherin Acts through the Hippo Tumor-Suppressor Pathway to Regulate Tissue Size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 24.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue T, Tian A, Jiang J. The Cell Adhesion Molecule Echinoid Functions as a Tumor Suppressor and Upstream Regulator of the Hippo Signaling Pathway. Developmental cell. 2012 doi: 10.1016/j.devcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 28.Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Current biology : CB. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental biology. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau Y-KI, Murray LB, Houshmandi SS, Xu Y, Gutmann DH, Yu Q. Merlin is a potent inhibitor of glioma growth. Cancer Research. 2008;68:5733–5742. doi: 10.1158/0008-5472.CAN-08-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama T, Osada H, Murakami H, Tatematsu Y, Taniguchi T, Kondo Y, et al. YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis. 2008;29:2139–2146. doi: 10.1093/carcin/bgn200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Developmental cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, et al. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet. 2008;40:1010–1015. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- 37.Mao Y, Mulvaney J, Zakaria S, Yu T, Morgan KM, Allen S, et al. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development. 2011;138:947–957. doi: 10.1242/dev.057166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Hateren NJ, Das RM, Hautbergue GM, Borycki AG, Placzek M, Wilson SA. FatJ acts via the Hippo mediator Yap1 to restrict the size of neural progenitor cell pools. Development. 2011;138:1893–1902. doi: 10.1242/dev.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends in Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 41.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 45.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 48.Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, et al. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 49.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padgett RW, Das P, Krishna S. TGF-beta signaling, Smads, and tumor suppressors. Bioessays. 1998;20:382–390. doi: 10.1002/(SICI)1521-1878(199805)20:5<382::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 51.Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- 52.Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development. 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- 53.Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, et al. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- 54.Buckles GR, Rauskolb C, Villano JL, Katz FN. four-jointed interacts with dachs, abelson and enabled and feeds back onto the Notch pathway to affect growth and segmentation in the Drosophila leg. Development. 2001;128:3533–3542. doi: 10.1242/dev.128.18.3533. [DOI] [PubMed] [Google Scholar]
- 55.Baena-Lopez LA, Rodriguez I, Baonza A. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci U S A. 2008;105:9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 57.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 58.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell stem cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong A, Gupta A, Pai RK, Tun M, Lowe AW. The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through Hippo pathway co-activator YAP1 activation. The Journal of biological chemistry. 2011;286:18301–18310. doi: 10.1074/jbc.M110.215707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine & growth factor reviews. 2008;19:133–144. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Grammont M, Irvine KD. fringe and Notch specify polar cell fate during Drosophila oogenesis. Development. 2001;128:2243–2253. doi: 10.1242/dev.128.12.2243. [DOI] [PubMed] [Google Scholar]
- 67.Chen HJ, Wang CM, Wang TW, Liaw GJ, Hsu TH, Lin TH, et al. The Hippo pathway controls polar cell fate through Notch signaling during Drosophila oogenesis. Developmental biology. 2011;357:370–379. doi: 10.1016/j.ydbio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Yu J, Poulton J, Huang YC, Deng WM. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS ONE. 2008;3:e1761. doi: 10.1371/journal.pone.0001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Polesello C, Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 70.Meignin C, Alvarez-Garcia I, Davis I, Palacios IM. The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 72.Hamaratoglu F, Gajewski K, Sansores-Garcia L, Morrison C, Tao C, Halder G. The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J Cell Sci. 2009;122:2351–2359. doi: 10.1242/jcs.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, et al. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 75.Thomas C, Strutt D. The roles of the cadherins Fat and Dachsous in planar polarity specification in Drosophila. Developmental dynamics : an official publication of the American Association of Anatomists. 2011 doi: 10.1002/dvdy.22736. [DOI] [PubMed] [Google Scholar]
- 76.Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- 77.Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 78.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brittle AL, Repiso A, Casal J, Lawrence PA, Strutt D. Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat. Curr Biol. 2010;20:803–810. doi: 10.1016/j.cub.2010.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simon MA, Xu A, Ishikawa HO, Irvine KD. Modulation of Fat:Dachsous binding by the cadherin domain kinase four-jointed. Curr Biol. 2010;20:811–817. doi: 10.1016/j.cub.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishikawa HO, Takeuchi H, Haltiwanger RS, Irvine KD. Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science. 2008;321:401–404. doi: 10.1126/science.1158159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000386. e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS biology. 2011;9 doi: 10.1371/journal.pbio.1000624. e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neumann CJ, Cohen SM. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- 86.Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 87.Neumann CJ, Cohen SM. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 1996;122:3477–3485. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- 88.Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, et al. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 89.Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development. 2007;134:3001–3010. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- 90.Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- 91.Baena-Lopez LA, Garcia-Bellido A. Control of growth and positional information by the graded vestigial expression pattern in the wing of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13734–13739. doi: 10.1073/pnas.0606092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwank G, Tauriello G, Yagi R, Kranz E, Koumoutsakos P, Basler K. Antagonistic growth regulation by Dpp and Fat drives uniform cell proliferation. Developmental cell. 2011;20:123–130. doi: 10.1016/j.devcel.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 93.Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 94.Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- 95.Waddington CH. The development of some 'leg genes' in Drosophila. DrosophilaJGenet. 1943;45:29–43. [Google Scholar]
- 96.Bando T, Mito T, Maeda Y, Nakamura T, Ito F, Watanabe T, et al. Regulation of leg size and shape by the Dachsous/Fat signalling pathway during regeneration. Development. 2009;136:2235–2245. doi: 10.1242/dev.035204. [DOI] [PubMed] [Google Scholar]
- 97.Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes & development. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Developmental biology. 2011;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 99.Kanda H, Miura M. Regulatory roles of JNK in programmed cell death. J Biochem. 2004;136:1–6. doi: 10.1093/jb/mvh098. [DOI] [PubMed] [Google Scholar]
- 100.Menendez J, Perez-Garijo A, Calleja M, Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14651–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 102.Chen CL, Schroeder MC, Kango-Singh M, Tao C, Halder G. Tumor suppression by cell competition through regulation of the Hippo pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:484–489. doi: 10.1073/pnas.1113882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Konsavage WM, Jr, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/beta-Catenin Signaling Regulates Yes-associated Protein (YAP) Gene Expression in Colorectal Carcinoma Cells. The Journal of biological chemistry. 2012;287:11730–11739. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta- SMAD pathway. Developmental cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 106.Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. The EMBO journal. 2012 doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 108.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]