This clone is a major public health problem because it limits antimicrobial drug therapy.
Keywords: Pseudomonas aeruginosa, bacteria, antimicrobial resistance, multidrug-resistant, outbreak, metallo-β-lactamase, ST175, bacteria, Spain, pandemic, clone, VIM-2 β-lactamase, IMP-22 β-lactamase
Abstract
A total of 183 patients were colonized or infected with multidrug-resistant Pseudomonas aeruginosa isolates at a hospital in Spain during 2007–2010; prevalence increased over this period from 2.8% to 15.3%. To characterize these isolates, we performed molecular epidemiologic and drug resistance analysis. Genotyping showed that 104 (56.8%) isolates belonged to a single major clone (clone B), which was identified by multilocus sequence typing as sequence type (ST) 175. This clone was initially isolated from 5 patients in 2008, and then isolated from 23 patients in 2009 and 76 patients in 2010. PCR analysis of clone B isolates identified the blaVIM-2 gene in all but 1 isolate, which harbored blaIMP-22. ST175 isolates were susceptible to only amikacin (75%) and colistin (100%). Emergence of the ST175 clone represents a major health problem because it compromises therapy for treatment of P. aeruginosa nosocomial infections.
Members of the bacterial genus Pseudomonas, especially P. aeruginosa, are among the major nosocomial pathogens because of their ubiquitous nature and ability to colonize and survive in hospital reservoirs and because of their role in causing infections in immunocompromised and critically ill patients (1). P. aeruginosa shows a high level of intrinsic resistance to antimicrobial drugs and an ability to become even more drug resistant. These characteristics are caused by selective pressure of mutations in chromosomal genes that lead to ampC hyperexpression, repression or inactivation of oprD, and overexpression of efflux pumps (2). In addition, P. aeruginosa is able to acquire other drug-resistance determinants by horizontal transfer of mobile genetic elements coding for class B carbapenemases (also called metallo-β-lactamases [MBLs]), which hydrolyze all β-lactams except aztreonam (3).
Because they can be disseminated horizontally through transfer of resistance determinants, MBLs have become a serious concern in hospitals worldwide over the past decade. Such acquired MBLs include the IMP and VIM types SPM-1, GIM-1, SIM-1, AIM-1, KHM-1, NDM-1, and SID-1 (4,5). MBL genes are normally encoded in class 1 integrons along with other resistance determinants, such as the aminoglycoside-modifying enzymes. The integrons are frequently located in plasmids or transposons, the dissemination of which contributes to the global spread of this resistance mechanism (6,7). The versatility and ability of P. aeruginosa to combine different resistance mechanisms has led to emergence of strains that are resistant to multiple antimicrobial drugs, which severely limits therapeutic options for treating infections (8,9). Interim definitions defining multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant bacteria, including P. aeruginosa, have been recently reported (10).
Although the prevalence of P. aeruginosa strains producing carbapenemases in Spain was considered low (0.4% of carbapenem-resistant isolates) (11) compared with prevalence in other countries in Europe, such as Italy (12.6% of carbapenem-resistant isolates) (12), detection of these isolates is no longer sporadic (13,14). Recent evidence from multicenter studies indicates an ≈10-fold increase in prevalence of these isolates in the past 5 years (15). During 2007 and 2008, a polyclonal outbreak of VIM-2–producing P. aeruginosa was detected in a hospital in Spain. At the same time, another outbreak in the hematology department of a hospital in Spain was caused by a P. aeruginosa ST235 clone, which produced GES-1 and GES-5, 2 extended-spectrum β-lactamases (16). We have observed a sharp increase in infections with drug-resistant P. aeruginosa that produces carbapenemase. Thus, we conducted a study to determine the clinical and molecular epidemiologic characteristics of drug-resistant P. aeruginosa isolates detected at a major hospital during 2007–2010.
Materials and Methods
Study Population
We conducted a retrospective study of all non–cystic fibrosis adult patients who were colonized or infected with P. aeruginosa isolates during January 2007–December 2010 at the Hospital Universitario 12 de Octubre in Madrid. This hospital is a 1,300-bed tertiary-care facility serving a population of 600,000 persons (≈42,000 admissions/year). We classified resistance patterns in P. aeruginosa according to recently published proposed interim definitions (10). An isolate was defined as MDR if it was resistant to >1 drug in >3 categories of drugs and XDR if it was resistant to >1 drug in <2 drug categories. The drugs on which our categorization was based included antipseudomonal cephalosporins (ceftazidime [CAZ], cefepime [FEP]), carbapenems (imipenem [IMP], meropenem [MER]), piperacillin/tazobactam (PIP-TZ), aztreonem, fluoroquinolones (ciprofloxacin [CIP]), and aminoglycosides (gentamicin [GEN], tobramycin [TOB], amikacin [AMK]).
Unique (nonduplicate) clinical isolates from colonized or infected patients were collected during the study. A case was defined as nosocomial if infection or colonization was detected in a person >48 hours after admission or if a person had documented evidence of hospitalization within the previous 12 months. Colonization was defined as isolation of P. aeruginosa from >1 clinical specimens in the absence of clinical signs consistent with infection. Medical charts were reviewed and demographic, clinical, and microbiological data were collected.
Antimicrobial Drug Susceptibility Testing
Identification and antimicrobial drug susceptibility testing of P. aeruginosa isolates included in this study were performed by using semi-automated microdilution panels (Soria, Melguizo, Spain) (17). The antimicrobial drugs tested were PIP-TZ, CAZ, FEP, ATM, IMP, MER, CIP, GEN, TOB, AMK and colistin. Break points were applied according to Clinical and Laboratory Standards Institute (CLSI) guidelines (18).
Genotyping Analysis
Epidemiologic relatedness of isolates was studied by using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). PFGE was conducted by macrorestriction of chromosomal DNA with SpeI and separation of restriction fragments by using a CHEF DRIII PFGE system (Bio-Rad Laboratories, Hercules, CA, USA). Migration of DNA fragments was normalized by using an appropriate mass marker, and computer-assisted analysis of PFGE patterns was conducted by using Bionumerics software (Applied Maths, St-Martens-Latem, Belgium). PFGE types were defined on the basis of DNA banding patterns in accordance with criteria defined by Tenover et al. (19).
MLST was performed on selected isolates according to published protocols (20). Standard DNA amplification and sequencing of 7 housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were performed. Isolates were assigned a sequence type (ST) number according to the allelic profiles available in the MLST database (http://pubmlst.org/paeruginosa).
Characterization of Acquired MBLs and Integron Analysis
The presence of horizontally acquired β-lactamases was determined by using phenotypic and genetic approaches. Phenotypic tests included analysis with Etest MBL strips (AB Biodisk, Solna, Sweden) for detection of class B carbapenemases. On the basis of positive results from preliminary phenotypic tests, the potential presence of genes encoding acquired metallo-β-lactamases was explored by using PCR amplification and DNA sequence analysis.
Described primers and conditions were used to amplify genes encoding VIM and IMP type β-lactamases (11,13). After PCR amplification, sequencing reactions were performed by using the BigDye Terminator Kit (PE Applied Biosystems, Foster City, CA, USA), and sequences were analyzed by using an ABI prism 3100 DNA Sequencer (PE Applied Biosystems). Resulting sequences were compared with those available in GenBank (www.ncbi.nih.gov/BLAST). Integrons harboring MBL-encoding genes were characterized by PCR and DNA sequencing by using specific primers to amplify the IntI1 and qacEΔ1 markers, the DNA region located between intI1 and qacEΔ1, and the corresponding MBL-encoding gene (11,13).
Statistical Analysis
Univariate analysis was performed by using the t test for continuous variables and the χ2 or Fisher exact tests for categorical variables. A p value <0.05 was considered significant. Data were stored and analyzed by using SPSS version 17.0 for Windows software (Analytical Software, St. Paul, MN USA).
Results
General Characteristics
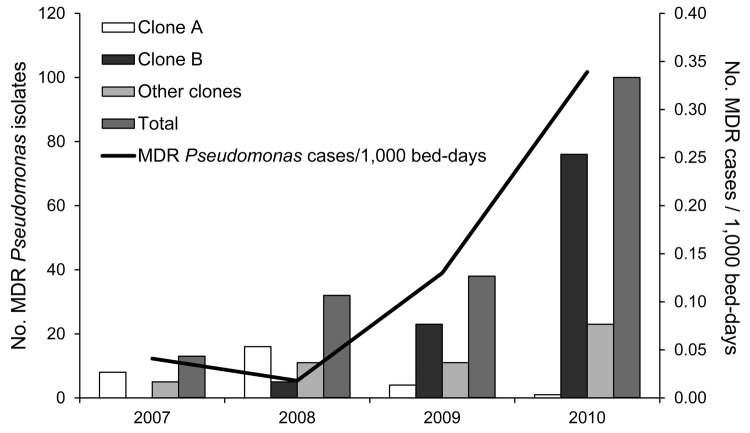
During the 4-year study, of 2,145 patients who were infected or colonized with Pseudomonas spp., 183 harbored MDR or XDR isolates: 13 (2.8%) of 460 in 2007, 32 (6.2%) of 517 in 2008, 38 (7.4%) of 514 in 2009, and 100 of (15.3%) 654 in 2010. The estimated annual incidence of MDR Pseudomonas–infected persons increased from 0.04/1,000 bed-days in 2007 to 0.34/1,000 bed-days in 2010 (Figure 1). Of 183 isolates, 177 were identified as P. aeruginosa and 6 as P. putida.
Figure 1.
Annual incidence of infections/colonizations by multidrug-resistant (MDR) Pseudomonas spp. and temporal distribution of human cases according to clonal type, Spain.
The mean ± SD age of patients was 65.1 ± 15.7 years, and 70.5% were men. Most (95%) cases were nosocomially acquired. The main clinical wards in which drug-resistant bacteria were isolated were internal medicine (57 cases, 31.1%), surgery (32, 17.5%), intensive care (24, 13.1%), pulmonology (21, 11.5%), and hematology (15, (8.2%). Of 183 patients, 143 (78.1%) were considered infected, including 36 (19.7%) with lower respiratory tract infection, 30 (16.4%) with urinary tract infection, 28 (15.3%) with bacteremia, and 22 (12%) with intraabdominal infection. A total of 42 (23%) patients died during hospitalization (Table).
Molecular Typing of an MDR P. aeruginosa Clone
Genotyping analysis of clinical isolates by PFGE showed that 104 isolates belonged to a single major clone (clone B), 29 belonged to a second clone (clone A), and 2 belonged to 2 clones (clones C and D). The remaining isolates, including the 6 P. putida isolates, showed unique PFGE patterns.
MLST analysis was performed on 9 isolates in clone B to determine their relationship to other strains that had been described. All isolates that we examined exhibited the same allelic profile (acsA [28], aroE [22], guaA [5], mutL [3], nuoD [3], ppsA [14], and trpE [19]), and were identified as ST175 according to the MLST database. MLST analysis of isolates in clone A had already been performed, and these isolates were classified as ST235 (16).
Clone B was initially isolated in February 2008 from a patient admitted to the pulmonology ward with a diagnosis of lower tract respiratory infection. Subsequently, 4 additional clone B isolates were detected in 2008, 23 in 2009, and 76 in 2010 (Figure 1). Twelve patients infected or colonized with this clone were referred to Hospital Universitario 12 de Octubre from 4 other hospitals in Madrid and from a fifth hospital in the Canary Islands. From 5 of these patients, the MDR P. aeruginosa isolate was recovered at the time of admission to Hospital Universitario 12 de Octubre.
Comparison of clinical characteristics for patients infected or colonized with clone B and other MDR P. aeruginosa clones did not show major differences, but patients infected with clone B were slightly older (mean age 67.5 vs. 61.9 years; p = 0.016) and they were more frequently admitted to the internal medicine (37.5% vs. 22.8%; p = 0.033) and pulmonology wards (16.3% vs. 5.1%; p = 0.018). Patients infected with clone B had more frequent underlying respiratory disease such as chronic obstructive pulmonary disease (36.5% vs. 22.8%; p = 0.046) (Table) than patients infected with other clones. We also found similar differences between patients infected with clone A and those infected with other MDR Pseudomonas spp. clones as reported (16).
Antimicrobial Drug Susceptibility
Antimicrobial drug–resistance patterns of 183 Pseudomonas spp. isolates were CAZ (100% resistant), FEP (100%), IMP (100%), MER (100%), GEN (97.3%), TOB (96.7%), and CIP (94%), and resistance to PIP-TZ (50.3%), AMK (42.1%), and ATM (41%) was more variable (Table A1). When break points recommended by the European Committee on Susceptibility Testing (EUCAST; www.eucast.org) were applied, the percentage of PIP-TZ–resistant and ATM-resistant isolates increased to 98.4% and 97.8%, respectively. There were no differences in MIC ranges for the other drugs tested when either CLSI or EUCAST break points were applied. All isolates were susceptible to COL.
Resistance profiles of MDR isolates in clone B were CAZ (100% resistant), FEP (100%), IMP (100%), MER (100%), CIP (100%), GEN (100%), and TOB (100%), and some isolates were also resistant to ATM (13.5%) and AMK (25%). The percentage of isolates resistant to PIP/TZ according to break points recommended by CLSI and EUCAST were 20.2% and 97.1%, respectively (Table A1).
Resistance profiles of isolates in clone A were CAZ (100% resistant), FEP (100%), IMP (100%), MER (100%), CIP (96.5%), GEN (100%), TOB (89.6%), AMK (86.2%) and ATM (86.2%). As a result, this clone was classified as XDR, as were clones C and D (Table A1).
Detection of MBL Genes
All isolates in clone B were positive by phenotypic methods for MBL. PCR analysis and sequencing identified MBL VIM-2 in 103 isolates and IMP-22 in 1 isolate (Table A1). VIM-2 was also detected in isolates in clone C, in 4 unique clones of P. aeruginosa, and in 5 of 6 isolates of P. putida. In 1 P. putida isolate, VIM-1 and VIM-2 MBLs were detected. A VIM-1-type MBL was also found in 1 clone of P. aeruginosa (Table A1). No MBL genes were detected among isolates in clone A, which demonstrated that these isolates were GES-1/GES-5 extended-spectrum β-lactamase/class A carbapenemase producers (16). Overall, the percentage of MBL-producing MDR or XDR Pseudomonas spp. isolates during the study was 63.9%. Moreover, if one considers the clone producing GES-1-GES-5, the overall prevalence of isolates producing acquired carbapenemases reaches 79.8%.
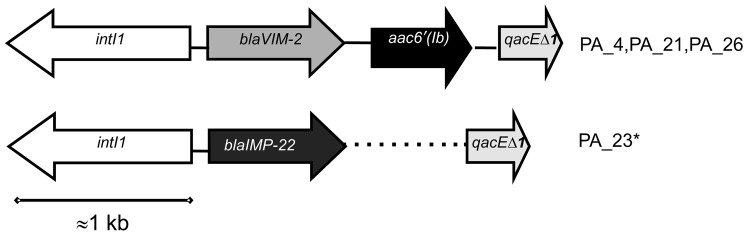
To investigate the genetic content of integrons from clone B isolates, we conducted PCR mapping of blaVIM-2 and blaIMP-22 genes for 4 strains. The structure of integrons from isolates producing VIM-2 was IntlI-VIM2-aac6'Ib-qacEΔ1 (Figure 2) in all isolates. IMP-22 was also found to be encoded in a class 1 integron, but we were unable to amplify the fragment between blaIMP-22 and qacEΔ1 by PCR, perhaps because of the large size of this DNA fragment.
Figure 2.
Structure of the class I integron detected in 4 representative isolates of the Pseudomonas spp. epidemic multidrug-resistant sequence type 175 clone, Spain. PA, P. aeruginosa. *Dotted line indicates undetermined part not amplified by PCR.
Discussion
Outbreaks by MBL-producing P. aeruginosa have been documented in hospitals in several countries, and VIM-is the most dominant MBL variant in Spain and worldwide (14,21–27). We report a large outbreak of VIM-2–producing MDR P. aeruginosa. The predominant clone belonged to ST175, a strain that was first detected on February 2008, and it has become increasingly common in Hospital Universitario 12 de Octubre, affecting 104 patients. This MDR P. aeruginosa clone was found in in several wards of the hospital, although it was more frequently associated with patients in the internal medicine and pulmonology wards.
The spread of the ST175 clone should be considered an emerging pandemic. It was first identified in 2005 in the United Kingdom and Canada (http://pubmlst.org/paeruginosa) and has been reported in Hungary, the Czech Republic, Poland, Spain, the Unites States, and China (15,28–31). Our study also identified this clone in patients referred to Hospital Universitario 12 de Octubre from 5 other hospitals in Spain, supporting the notion that this clone has disseminated nationwide (28). ST175 has been associated with multidrug-resistant isolates and acquisition of different β-lactamases, mostly located on mobile elements such as integrons (15,31,32).
In our study, all MDR P. aeruginosa isolates belonging to ST175 producedVIM-2 type MBLs, except for 1 that produced IMP-22. PCR mapping showed that VIM-2 was inserted in a class 1 integron with an aminoglycoside-modifying enzyme (aac6′-1b). The IMP-type MBLs are most common among Pseudomonas spp. isolates in Asia, although they have been reported less frequently in some countries in Europe such as Italy and Austria (33,34). We report a lineage of ST175 MDR P. aeruginosa that produces IMP-22, which adds this MBL to the list of acquired β-lactamases associated with this epidemic clone. A thorough understanding of the genetic mechanism involved and horizontal and longitudinal dissemination is necessary, particularly for those carrying integron- and plasmid-borne MBLs, given their additional capacity for intraspecies and interspecies spread of multidrug resistance.
Although the origin of the ST175-VIM-2 MDR P. aeruginosa strain is unknown, this clone emerged in Hospital Universitario 12 de Octubre in February 2008, seven months after the first detection of VIM-2 type MBLs in a strain of P. putida. This species and other Pseudomonas species might play a major role as potential reservoirs for MDR determinants by enhancing their transfer to P. aeruginosa clones (3).
The ST175 clone was able to persist in Hospital Universitario 12 de Octubre for >34 months and has disseminated widely in spite of control measures that have been implemented, such as strict isolation of patients, active surveillance of patients at the time of entry into intensive care units, and environment investigation of possible sources of colonization. The spread of this clone among patients admitted to different sections of the hospital and high selective pressure for antimicrobial drug resistance may encourage its persistence. The design and implementation of infection control strategies in these hyperendemic situations is challenging. We recently faced a similar situation in Hospital Universitario 12 de Octubre with a large outbreak of MDR Acinetobacter baumannii that persisted for >30 months but that was finally controlled (35).
This study also detected emergence of multiple strains of Pseudomonas spp. that produced VIM-2- and VIM-1-type MBLs, including >6 P. aeruginosa and 6 P. putida clones. The polyclonal nature of MBL-based resistance might have major epidemiologic implications because sporadically isolated strains may eventually spread in the hospital environment or act as a reservoir for horizontal transfer of resistance determinants.
The emergence of MBL-producing MDR P. aeruginosa is a major health problem because it leaves the clinician with almost no therapeutic options for treating nosocomial infections caused by P. aeruginosa. Our results showed that isolates belonging to ST175 had susceptibility only to PIP/TZ (79.8%), ATM (86.5%), AK (75%), and colistin (100%). The difference in PIP/TZ susceptibility depending on the break point criteria applied is remarkable. When the EUCAST susceptibility testing criteria were applied, only 2.9% isolates were susceptible to PIP/TZ compared with ≈80% when criteria recommended by CLSI were applied. A recent report concluded that in P. aeruginosa bacteremia caused by isolates with reduced PIP/TZ susceptibility (32/4 µg/mL or 64/4 µg/mL), empirically prescribed PIP/TZ therapy was associated with increased patient deaths (36). In our study, most isolates had PIP/TZ susceptibility in this range. Fortunately, the 2012 CLSI PIP/TZ break point (37), which was implemented during the review of this report, has been reported as 16/4 µg/mL, thus agreeing with the break point established by EUCAST. Confluence of susceptibility testing criteria among agency standards are useful for optimizing strategies to treat severe MDR P. aeruginosa infections.
In summary, we report a large outbreak of infections caused by a VIM-2–producing ST175 MDR P. aeruginosa strain that was responsible for 76% of infections or colonizations by MDR P. aeruginosa in 2010 at Hospital Universitario 12 de Octubre, and >50% of infections or colonizations during the study period. This epidemic clone is also circulating in other hospitals in Spain and other countries. The underlying reasons for the widespread success of this clone still need to be fully elucidated fully, including the potential for an enhanced ability to acquire MDR determinants that facilitate persistence under conditions of antimicrobial drug selective pressure encountered in the hospital environment (21,22). Deciphering the epidemiologic and molecular aspects driving the emergence and spread of such strains is crucial to the implementation of efficient measures to control their dissemination.
Acknowledgment
We thank Tobin Hellyer for reviewing the manuscript.
This study was supported by Spanish Network for the Research in Infectious Diseases (RD06/0008) from the Instituto de Salud Carlos III, Spain.
Biography
Dr Viedma is a clinical microbiologist at Hospital Universitario 12 de Octubre, Madrid, Spain. Her research interest is the molecular epidemiology of multidrug-resistant microorganisms.
Table A1. Clonal distribution, type of carbapenemase, and drug resistance patterns of MDR Pseudomonas isolates, Spain, January 2007–December 2010 *.
| Isolate | No. isolates/y |
|
Carbapenemase type (no. isolates) | ST | No. (%) resistant isolates |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2007 | 2008 | 2009 | 2010 | PIP/TZ | CAZ | FEP | IMP | MER | ATM | GEN | TOB | AMK | CIP | ||||
| All MDR isolates (183) | 13 | 32 | 38 | 100 | VIM-1, VIM-2, IMP-22, GES1–5 (146) | NA | 92 (50.3)/ 180 (98.4) | 183 (100) | 183 (100) | 183 (100) | 183 (100) | 75 (41.0)/ 179 (97.8)† | 178 (97.3) | 177 (96.7) | 77 (42.1) | 172 (94) | |
| P. aeruginosa (clonal types) | |||||||||||||||||
| B (104) | 0 | 5 | 23 | 76 | VIM-2 (103), IMP-22 (1) | 175, 175 | 21 (20.2)/ 101 (97.1)† | 104 (100) | 104 (100) | 104 (100) | 104 (100) | 14 (13.5)/ 100 (95.2)† | 104 (100) | 104 (100) | 26 (25) | 104 (100) | |
| A (29)‡ | 8 | 16 | 4 | 1 | GES 1/5 (29) | 235 | 29 (100) | 29 (100) | 29 (100) | 29 (100) | 29 (100) | 25 (86.2) | 29 (100) | 26 (89.6) | 25 (86.2) | 28 (96.5) | |
| C (2) | 0 | 0 | 1 | 1 | VIM-2 (2) | NA | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | |
| D (2) | 0 | 0 | 0 | 2 | Noncarbapenemase (2) | NA | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 0 | 2 (100) | 2 (100) | 2 (100) | 2 (100) | |
| Unique clones (40) | 4 | 10 | 8 | 18 | Noncarbapenemase (35) | NA | 31 (88.6)/ 35 (100)† | 35 (100) | 35 (100) | 35 (100) | 35 (100) | 26 (74.3) | 30 (85.7) | 32 (65.7) | 17 (48.3) | 25 (71.4) | |
| 0 | 1 | 0 | 0 | PA_5 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 1 | 0 | PA_10 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | |||||||
| 0 | 0 | 1 | 0 | PA_12 VIM-1 | NA | 4 (80)/ 5 (100)† | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) | 3 (60) | 5 (100) | ||
| 0 | 0 | 1 | 0 | PA_13 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 0 | 0 | 1 | PA_28 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| P. putida (clonal types) | |||||||||||||||||
| Unique clones (6) | 1 | 1 | 2 | 2 | 0 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 0 | 0 | 0 | PP_1 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 1 | 0 | 0 | PP_11 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 0 | 1 | 0 | PP_14 VIM-2 | NA | 3 (50)/ 6 (100)† | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 4 (66.6) | 6 (100) | 6 (100) | 2 (33.3) | 6 (100) | ||
| 0 | 0 | 1 | 0 | PP_19 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 0 | 1 | 0 | PP25_VIM-1/VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 0 | 0 | 1 | 0 | PP_27 VIM-2 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
*MDR, multidrug resistant; ST, sequence type; PIP/TZ, piperacillin-tazobactam; CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; MER, meropenem; ATM, aztreonam; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; CIP, ciprofloxacin; NA, not available. None of the isolates showed resistance to colistin. †Percentage drug-resistant isolates when the European Committee on Susceptibility Testing break point was applied. ‡Viedma et al. (16).
Table. Characteristics of 183 patients colonized or infected with clone B or nonclone B multidrug-resistant Pseudomonas isolates, Spain, January 2007–December 2010*.
| Characteristic | Patients with clone B, n = 104 | Patients with non–clone B, n = 79 | p value |
|---|---|---|---|
| Mean ± SD age, y | 67.5 ± 15.4 | 61.9 ± 15.7 | 0.016 |
| Male | 74 (71.1) | 55 (69.6) | 0.822 |
| Hospital location | |||
| Surgical ward | 15 (14.4) | 17 (21.5) | 0.211 |
| Intensive care unit | 11 (10.6) | 13 (16.4) | 0.243 |
| Internal medicine ward | 39 (37.5) | 18 (22.8) | 0.033 |
| Hematology ward | 4 (3.8) | 11 (13.9) | 0.014 |
| Pulmonology ward | 17 (16.3) | 4 (5.1) | 0.018 |
| Hospitalization within previous 3 mo | 48 (46.1) | 37 (46.8) | 0.927 |
| Surgical procedure | 29 (27.9) | 32 (40.5) | 0.073 |
| Transplant | 4 (3.8) | 8 (10.1) | 0.089 |
| Hematologic malignancy | 8 (7.7) | 11(13.9) | 0.171 |
| Solid tumor | 21 (20.2) | 13 (16.4) | 0.520 |
| Diabetes mellitus | 29 (27.9) | 21 (26.9) | 0.845 |
| Chronic obstructive pulmonary disease | 38 (36.5) | 18 (22.8) | 0.046 |
| Central catheter | 83 (79.8) | 59 (76.7) | 0.410 |
| Urinary catheter | 60(57.7) | 44(55.7) | 0.441 |
| Infections | 82 (78.8) | 61 (77.2) | 0.791 |
| Respiratory tract | 25 (24) | 11 (13.9) | 0.088 |
| Intraabdominal | 7 (6.7) | 15 (19) | 0.012 |
| Wound | 4 (3.8) | 8 (10.1) | 0.089 |
| Urinary tract | 19 (18.3) | 11 (13.9) | 0.432 |
| Bacteremia | 12 (11.5) | 16 (20.2) | 0.105 |
| Died during hospitalization | 20 (19.2) | 22 (27.8) | 0.170 |
*Values are no. (%) unless otherwise indicated.
Footnotes
Suggested citation for this article: Viedma E, Juan C, Villa J, Barrado L, Orellana MÁ, Sanz F, et al. VIM-2–producing multidrug-resistant Pseudomonas aeruginosa clone ST175, Spain. Emerg Infect Dis [serial on the Internet]. 2012 Aug [date cited]. http://dx.doi.org/10.3201/eid1808.111234
References
- 1.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50:43–8. 10.1128/AAC.50.1.43-48.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34:634–40. 10.1086/338782 [DOI] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–58. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–93. 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. 10.1128/CMR.18.2.306-325.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–7. 10.1128/AAC.44.4.891-897.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riccio ML, Pallecchi L, Fontana R, Rossolini GM. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob Agents Chemother. 2001;45:1249–53. 10.1128/AAC.45.4.1249-1253.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–10. 10.1128/AAC.48.12.4606-4610.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitliks SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998;244:379–86. 10.1046/j.1365-2796.1998.00379.x [DOI] [PubMed] [Google Scholar]
- 10.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 11.Gutiérrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51:4329–35. 10.1128/AAC.00810-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossolini GM, Luzzaro F, Migliavacca R, Mugnaioli C, Pini B, De Luca F, et al. First countrywide survey of acquired metallo-β-lactamase in gram-negative pathogens in Italy. Antimicrob Agents Chemother. 2008;52:4023–9. 10.1128/AAC.00707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juan C, Beceiro A, Gutiérrez O, Alberti S, Garau M, Pérez JL, et al. Characterization of the new metallo-β-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. Antimicrob Agents Chemother. 2008;52:3589–96. 10.1128/AAC.00465-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peña C, Suárez C, Tubau F, Gutiérrez O, Domínguez A, Oliver A, et al. Nosocomial spread of Pseudomonas aeruginosa producing the metallo-β-lactamase VIM-2 in a Spanish hospital: clinical and epidemiological implications. Clin Microbiol Infect. 2007;13:1026–9. 10.1111/j.1469-0691.2007.01784.x [DOI] [PubMed] [Google Scholar]
- 15.Riera E, Cabot G, Mulet X, García-Castillo M, Del Campo R, Juan C, et al. Pseudomonas aeruginosa carbapenem resistance mechanism in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother. 2011;66:2022–7. 10.1093/jac/dkr232 [DOI] [PubMed] [Google Scholar]
- 16.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, et al. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum b-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother. 2009;53:4930–3. 10.1128/AAC.00900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantón R, Perez-Vazquez M, Oliver A, Sanchez Del Saz B, Gutiérrez MO, Martinez-Ferrer M, et al. Evaluation of the Wider system, a new computer-assisted image-processing device for bacterial identification and susceptibility testing. J Clin Microbiol. 2000;38:1339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, vol. 26, no. 3. 16th informational supplement. M100–S16. Wayne (PA): The Institute; 2006. [Google Scholar]
- 19.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42:5644–9. 10.1128/JCM.42.12.5644-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pournaras S, Maniati M, Petinaki E, Tzouvelekis LS, Tsakris A, Legakis NJ, et al. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-beta-lactamase gene variants blaVIM-2 and blaVIM-4. J Antimicrob Chemother. 2003;51:1409–14. 10.1093/jac/dkg239 [DOI] [PubMed] [Google Scholar]
- 22.Gibb AP, Tribuddharat C, Moore RA, Louie TJ, Krulicki W, Livermore DM, et al. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new bla(IMP) allele, bla(IMP-7). Antimicrob Agents Chemother. 2002;46:255–8. 10.1128/AAC.46.1.255-258.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crespo MP, Woodford N, Sinclair A, Kaufmann ME, Turton J, Glover J, et al. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-beta-lactamase, in a tertiary care center in Cali, Colombia. J Clin Microbiol. 2004;42:5094–101. 10.1128/JCM.42.11.5094-5101.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryoo NH, Lee K, Lim JB, Lee YH, Bae IK, Jeong SH. Outbreak by meropenem-resistant Pseudomonas aeruginosa producing IMP-6 metallo-beta-lactamase in a Korean hospital. Diagn Microbiol Infect Dis. 2009;63:115–7. 10.1016/j.diagmicrobio.2008.08.019 [DOI] [PubMed] [Google Scholar]
- 25.Deplano A, Rodriguez-Villalobos H, Glupczynski Y, Bogaerts P, Allemeersch D, Grimmelprez A, et al. Emergence and dissemination of multidrug resistant clones of Pseudomonas aeruginosa producing VIM-2 metallo-beta-lactamase in Belgium. Euro Surveill. 2007;12:E070118.2. [DOI] [PubMed] [Google Scholar]
- 26.Lolans K, Queenan AM, Bush K, Sahud A, Quinn JP. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-beta-lactamase (VIM-2) in the United States. Antimicrob Agents Chemother. 2005;49:3538–40. 10.1128/AAC.49.8.3538-3540.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castanheira M, Bell JM, Turnidge JD, Mathai D, Jones RN. Carbapenem resistance among Pseudomonas aeruginosa strains from India: evidence for nationwide endemicity of multiple metallo-beta-lactamase clones (VIM-2, −5, −6, and −11 and the newly characterized VIM-18). Antimicrob Agents Chemother. 2009;53:1225–7. 10.1128/AAC.01011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Castillo M, Del Campo R, Morosini MI, Riera E, Cabot G, Willems R, et al. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J Clin Microbiol. 2011;49:2905–10. 10.1128/JCM.00753-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol. 2011;49:2578–83. 10.1128/JCM.00102-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemec A, Krizova L, Maixnerova M, Musilek M. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res Microbiol. 2010;161:234–42. 10.1016/j.resmic.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 31.Libisch B, Balogh B, Füzi M. Identification of two multidrug-resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr Microbiol. 2009;58:111–6. 10.1007/s00284-008-9285-7 [DOI] [PubMed] [Google Scholar]
- 32.Elias J, Schoen C, Heinze G, Valenza G, Gerhaz E, Riedmiller H, et al. Nosocomial outbreak of VIM-2 metallo-β-lactamase producing Pseudomonas aeruginosa associated with retrograde urography. Clin Microbiol Infect. 2010;16:1494–500. 10.1111/j.1469-0691.2010.03146.x [DOI] [PubMed] [Google Scholar]
- 33.Pagani L, Colinon C, Migliavacca R, Labonia M, Docquier JD, Nucleo E, et al. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-beta-lactamase. J Clin Microbiol. 2005;43:3824–8. 10.1128/JCM.43.8.3824-3828.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duljasz W, Gniadkowski M, Sitter S, Wojna A, Jebelean C. First organisms with acquired metallo-beta-lactamases (IMP-13, IMP-22, and VIM-2) reported in Austria. Antimicrob Agents Chemother. 2009;53:2221–2. 10.1128/AAC.01573-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acosta J, Merino L, Viedma E, Poza M, Sanz F, Otero JR, et al. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg Infect Dis. 2011;17:1064–7. 10.3201/eid1706.091866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam VH, Gamez EA, Weston JS, Gerard LN, Larocco MT, Caeiro JP, et al. Outcomes of bacteremia due to Pseudomonas aeruginosa with reduced susceptibility to piperacillin-tazobactam: implications on the appropriateness of the resistance breakpoint. Clin Infect Dis. 2008;46:862–7. 10.1086/528712 [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 22nd informational supplement. M100–S22. Wayne (PA): The Institute; 2012. [Google Scholar]