Rodents infected with this virus may be a serious threat to public health.
Keywords: hepatitis E virus, viruses, rats, zoonosis, genotype 3, viral RNA, PCR, United States
Abstract
The role of rodents in the epidemiology of zoonotic hepatitis E virus (HEV) infection has been a subject of considerable debate. Seroprevalence studies suggest widespread HEV infection in commensal Rattus spp. rats, but experimental transmission has been largely unsuccessful and recovery of zoonotic genotype 3 HEV RNA from wild Rattus spp. rats has never been confirmed. We surveyed R. rattus and R. norvegicus rats from across the United States and several international populations by using a hemi-nested reverse transcription PCR approach. We isolated HEV RNA in liver tissues from 35 of 446 rats examined. All but 1 of these isolates was relegated to the zoonotic HEV genotype 3, and the remaining sequence represented the recently discovered rat genotype from the United States and Germany. HEV-positive rats were detected in urban and remote localities. Genetic analyses suggest all HEV genotype 3 isolates obtained from wild Rattus spp. rats were closely related.
Hepatitis E virus (HEV) is a major cause of acute hepatitis in developing countries, in which outbreaks arise most often through fecal contamination of drinking water or after flooding (1). Major outbreaks have been reported in India, Southeast Asia, Africa, and Mexico, and mortality rates are considerable (20%–30%) among pregnant women (1). In industrialized countries, HEV infections are reported sporadically and contamination of drinking water is an unlikely source, but cases are increasing as diagnostic tests are being performed more frequently (2). Moreover, zoonotic transmission of HEV through consumption of undercooked pork and deer meat has been confirmed (3,4), and detection of HEV in many mammalian hosts suggests the potential for multiple zoonotic sources of HEV infection in industrialized countries (5).
There are currently at least 4 genotypes of HEV known to infect humans. Genotypes 1 and 2 have been identified only from humans and are responsible for most outbreaks in developing countries (6). Genotypes 3 and 4 are believed to be involved in zoonotic transmission and have been isolated from swine (domesticated pig and wild boar), deer, mongoose, rabbits, cattle, and humans (5). Additional strains not known to infect humans have also been identified in rats and chickens, and the genetic diversity of HEV is only beginning to be understood.
Within the United States, HEV infections have been identified in travelers who have visited developing countries (7), and for several at-risk groups in the United States (i.e., swine veterinarians and farmers), the high number of reported seropositive persons is caused by swine–human contact (8,9). However, seroepidemiologic examinations of blood banks in the United States and other industrialized countries have shown high proportions of samples positive for antibodies against HEV (excluding persons who had traveled to HEV-endemic countries), but this finding was true in urban areas in which swine–human contact is absent (8,10,11).
HEV RNA has been detected in livers from commercially raised pigs (12) and represents an additional potential reservoir of infection. However, consumption of raw pork and wild game is uncommon in the United States, although it is a common practice in other industrialized nations in which high HEV seroprevalence has been reported (i.e., France) (13). This finding suggests that in addition to travel to HEV-endemic regions and swine–human contact, additional reservoirs of HEV infection exist in the United States, and evidence has accumulated indicating rodents as a potential HEV reservoir (14–18). In a survey of 26 rodent species in the United States, Favorov et al. (14) found 14 species of rodents seropositive for antibodies against HEV. Urban populations had ≈2× the proportion of seropositive rats relative to rural populations, and commensal Rattus spp. (R. rattus and R. norvegicus) rats had the highest proportion of seropositive animals (14).
The role of wild Rattus spp. rats as reservoirs in the epidemiology and transmission of HEV is unclear, but their ubiquity in urban environments and unparalleled propensity for carrying zoonotic pathogens makes them an obvious target of investigation. Multiple studies have reported finding IgG and IgM against HEV in R. norvegicus and R. rattus rat populations across the United States and Asia (14–18). Shukla et al. (19) successfully infected cell lines from Mus musculus mice, murid rodents closely related to Rattus spp. rats, with HEV genotype 3. In addition, Maneerat et al. (20) experimentally infected laboratory R. norvegicus rats with HEV isolated from infected humans, although the genotype of the infecting virus was unclear. After infection, the human virus strain effectively replicated in multiple tissues, and HEV RNA was detected in feces and serum for >30 days postexposure, suggesting that human strains of HEV can replicate in and be transmitted by R. norvegicus rats. However, recent discovery of a rat-specific strain of HEV not known to infect humans (21–23) suggests that high seroprevalence of antibodies against HEV may be caused by cross-reactivity rather than widespread infection with a human-infecting HEV genotype.
We used a reverse transcription PCR (RT-PCR) approach to survey R. rattus and R. norvegicus rats for HEV RNA. Our analysis detected HEV RNA in liver tissues from R. rattus and R. norvegicus rats at many localities across the United States. Sequencing of DNA from RT-PCR–positive samples indicated widespread infection with zoonotic HEV genotype 3: one rat in California was positive for the rat-specific strain. These findings suggest that wild Rattus spp. rats are competent hosts for genotype 3 HEV.
Materials and Methods
Rat Tissues
We obtained liver tissue samples from 446 R. rattus and R. norvegicus rats from museum collections (Technical Appendix) covering localities primarily in the United States (15 states) plus additional samples from China, Honduras, Madagascar, Mexico, Nicaragua, Peru, Russia, and Vietnam (Table). To maximize the likelihood of intact viral RNA, all liver samples selected were dissected from recently euthanized animals, immediately frozen, and maintained at −80°C until thawed for extraction.
Table. Rattus spp. rats tested for hepatitis E virus RNA*.
| Location | Species and sample size |
||
|---|---|---|---|
| R. norvegicus | R. rattus | No. positive | |
| United States | |||
| Aleutian Islands, Alaska | 18 | 7 | 6 |
| San Francisco Bay Area, California | 19 | 112 | 12 |
| Gainesville, Florida | NA | 21 | 4 |
| Oklahoma City, Oklahoma | 1 | NA | 1 |
| Memphis, Tennessee | 16 | NA | 6 |
| San Angelo, Texas | 2 | 11 | 2 |
| Little Rock, Arkansas | 2 | 6 | 0 |
| San Diego, California | 8 | 5 | 3 |
| Panama City, Florida | NA | 24 | 0 |
| Key Largo, Florida | NA | 5 | 0 |
| Spencer, Indiana | NA | 10 | 0 |
| Baton Rouge, Louisiana | NA | 12 | 0 |
| Prentiss, Mississippi | NA | 1 | 0 |
| Bernalillo, New Mexico | 2 | NA | 0 |
| Union County, Pennsylvania | 40 | NA | 1 |
| Corvallis, Oregon | 4 | NA | 0 |
| Houston, Texas | NA | 8 | 0 |
| Austin, Texas | NA | 14 | 0 |
| Kerns, West Virginia | 1 | NA | 0 |
| Seattle, Washington | 1 | 5 | 0 |
| Vietnam | NA | 18 | 0 |
| China | NA | 5 | 0 |
| Honduras | NA | 2 | 0 |
| Madagascar | NA | 5 | 0 |
| Mexico | 1 | 2 | 0 |
| Nicaragua | 1 | 11 | 0 |
| Peru | NA | 16 | 0 |
| Russia | 30 | NA | 0 |
*NA, no samples were available.
Hemi-nested RT-PCR
Total RNA was extracted from ≈30 mg of liver tissue by using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). We used a modification of the broad-spectrum RT-PCR approach of Johne et al. (22) to amplify a 334-bp fragment of HEV open reading frame 1 (ORF1). Primers were selected for their ability to amplify ORF1 from all known HEV genotypes, and all primer sequences are reported by Johne et al. (22). Attempts to amplify the ORF1 fragment from total extracted RNA resulted in amplification of a portion of an unidentified transcript in all R. rattus rat samples. When we sequenced the amplicon, it was clear that spurious amplification was caused by nonspecific binding of primer HEV-cas. To circumvent this problem, we used a hemi-nested approach, with the initial RT-PCR using the HEV-cs/HEV-casN primer combination and the nested PCR using the HEV-csN/HEV-casN primer combination. With the exception of the change in primer combinations, all other aspects of amplification followed the protocol of Johne et al. (22). Positive PCR amplicons (verified by agarose gel electrophoresis) were purified by using the Wizard SV Gel PCR Clean-Up System (Promega, Madison, WI, USA) and sequenced in both directions by using nested PCR primers (HEVcsN/HEVcasN).
Given the high sensitivity of a nested PCR approach, contamination can be a major issue and has been cited as problematic in investigations of HEV in rodents (24). Exceptional effort was made to ensure that no contamination occurred. All PCR steps were conducted in a sterile environment, under a laminar flow hood, and all surfaces, tubes, and equipment were UV irradiated between each PCR. This study was conducted in a newly constructed laboratory in which no HEV samples (or any other animal samples) had been handled, extractions were conducted in a room separate from that used for PCR amplifications, and all steps (extraction, RT-PCR, and nested PCR) included negative controls. In addition, a single HEV genotype 3 isolate was used as a positive control in PCRs, and we sequenced this isolate for the same locus targeted for the Rattus spp. rat samples. Any Rattus spp. rat HEV isolate exhibiting 100% nt identity to this positive control sequence was excluded as a contaminant.
Phylogenetic Analyses
In addition to the sequences we generated, we downloaded all complete HEV genome sequences from GenBank (accession numbers are shown in Figure A1) and extracted the ≈334-bp homologous portion of ORF1 from each genome. Total sequences were aligned by using the MAFFT aligner (25) implemented in Geneious version 5.5 (26). We conducted Bayesian, maximum-parsimony, and maximum-likelihood phylogenetic analyses on the combined alignment by using the avian HEV strain as an outgroup. For Bayesian analysis conducted in MrBayes version 3.2 (27), we partitioned the alignment by codon position and used a generalized time reversible + invariant sites + Γ substitution model, which Modeltest version 3.7 (28) indicated to be most appropriate. The analysis was run for 15,000,000 generations sampled every 1,000 generations, and burn-in values were determined empirically by evaluating likelihood scores. For maximum-parsimony analysis, we used tree bisection/reconnection branch swapping, 25 random additions of input taxa, and 1,000 bootstrap replicates to assess node support. For maximum-likelihood analysis, we used a generalized time reversible + invariant sites + Γ substitution model as indicated above, nearest-neighbor interchange branch-swapping, and 500 bootstrap replicates to assess node support. We generated a haplotype network for sequences generated in this study by using TCS software (29).
Results
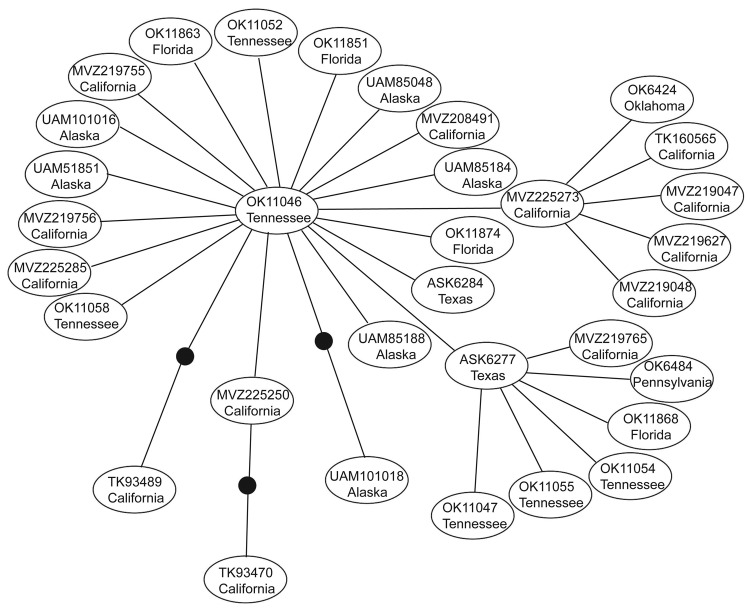
We excluded 7 isolates sequenced from 1 PCR batch that matched the positive control sequence. No subsequent matches with the positive control were detected, and no contamination was detected in negative controls. We identified 35 (7.85%) Rattus spp. rats positive for HEV by PCR from 446 rats examined. Most positive samples were from California (15 rats), but some were from rats in Tennessee, Florida, Oklahoma, Pennsylvania, Texas, and Alaska (Table). Phylogenetic analysis placed 34 of these positive rat samples in a closely related group within the HEV genotype 3 clade (Figure A1) termed subclade 3a by Lu et al. (6). This placement was supported in all analyses. Mean pairwise uncorrected genetic distances between HEV genotype 3 sequences and other known HEV genotypes were 36.19%, 24.12%, 24.91%, 24.05%, and 33.52% compared with the avian genotype, genotype 2, genotype 1, genotype 4, and rat genotype, respectively. Network analysis showed that HEV genotype 3 sequences from Rattus spp. rats formed a tight cluster (Figure), differed by only a few mutations, and represented a single strain. Mean pairwise sequence divergence within Rattus spp. rat HEV genotype 3 sequences was 0.51%. A single sequence (AF082843) isolated from an HEV-infected pig was also in this group.
The single sequence not nested within the genotype-3 clade was isolated from an R. norvegicus rat from the San Francisco Bay area of California. Phylogenetic analyses placed it in a strongly supported clade with 2 other sequences isolated from R. norvegicus rats in Germany (Figure A1). Uncorrected genetic distances indicated that the California rat HEV sequence is ≈2× as divergent from the 2 sequences isolated in Germany (California vs. GU345042 = 13.98%; California vs. GU345043 = 14.86%) as the 2 Germany sequences are from each other (GU345042 vs. GU345043 = 7.78%). These findings suggest a degree of distinction between rat HEV strains from the United States and Europe.
Discussion
A major conflict has surrounded the role of Rattus spp. rats (and other rodents) in HEV epidemiology since seroprevalence studies in the 1990s identified many species of rats positive for HEV antibodies in the United States and Asia (15–18). Maneerat et al. (20) infected 3 Wistar laboratory rats (R. norvegicus) with HEV (viral RNA was detected intermittently in feces for <30 days), but which genotype was used is unclear, and this result has not been duplicated (9). In addition, He et al. (24) isolated HEV genotype 1 from R. rattus and Bandicota bengalensis rats, but the study was later retracted because the authors were unable to rule out contamination as a source of detected viral RNA (30). More recently, Shukla et al. (19) successfully infected multiple M. musculus mouse cell cultures (in addition to infecting cow, rabbit, cat, dog, and chicken cultures) with HEV genotype 3, supporting the hypothesis that rodents may be competent hosts. However, there was substantial variation among different strains of HEV genotype 3 in the ability to infect cells derived from different hosts, including swine and human.
A recent attempt to infect adult Sprague-Dawley laboratory rats (R. norvegicus) with HEV genotypes 1, 2, and 3 failed (23). In this same study, infection of laboratory rats with the divergent rat genotype had limited success; only 25% of intravenously infected Sprague-Dawley rats and only 15.8% of nude rats seroconverted. This result is unexpected given that ≈80% of wild R. norvegicus rats from Los Angeles, California, where the study was conducted, were positive for IgG or IgM against HEV, suggesting that infection occurs in the wild (23). Johne et al. (22) also were unsuccessful in infecting rat liver cell lines with rat genotype HEV isolated from wild R. norvegicus rats from Germany. In contrast, we provide evidence of HEV genotype 3 infection in wild R. rattus and R. norvegicus rats.
Spread of HEV-positive rats indicates that infection in wild Rattus spp. rats is not restricted to any area of the United States or to urban areas. Our positive samples included both Rattus species of rat tested, and included the relatively remote Aleutian Islands in Alaska, and the urban San Francisco Bay area in California. Given the commensal nature of wild Rattus spp. rats and their ability to use human transportation vectors (i.e., commercial shipping) in dispersal, the prevalence of HEV in remote populations is not surprising. Recent work examining the genetic structure of R. rattus rats has shown that 2 mtDNA haplotypes have rapidly spread from their origin in India to every continent except Antarctica (31,32). Given the presence of HEV in domesticated animals (i.e., pigs) and human commensals (i.e., wild Rattus spp. rats), widespread domestication has likely enabled HEV to spread worldwide, potentially through interactions between humans, domesticated animals, and commensal rats. Furthermore, because R. rattus and R. norvegicus rats are sympatric over their contemporary range, lack of genetic distinction between strains infecting these 2 species is not unexpected (Figure A1, Figure).
In terms of infection rates, variation in handling of tissues from field-collected animals should be considered. Although we attempted to limit our analysis to the most well-preserved tissues, there is considerable variation among collection protocols and collectors in length of time between euthanasia and dissection, time between dissection and freezing, number of times tissues were thawed and frozen (i.e., in sorting, subsampling, shipping), and consistency of storage temperature. These factors can lead to nucleic acid degradation and negatively affect the ability to detect viral RNA. Therefore, our infection rate is likely not indicative of HEV infection rates in wild Rattus spp. rat populations.
Recent studies have reported major variation in diversity of competent mammalian hosts for various strains of HEV genotype 3 (19). Although seroprevalence studies have suggested infection rates ≤80% for HEV in US Rattus spp. rat populations (15,23), attempts to infect different laboratory strains of R. norvegicus rats with a genotype isolated from wild R. norvegicus rats have shown limited success; most attempts also failed in immunocompromised nude rats (22,23). These patterns, and the low genetic diversity of HEV-positive samples detected in this study (Figure), suggest that only a limited number of HEV genotype 3 strains may be capable of infecting Rattus spp. rats and other rodents (i.e., Mus spp.), possibly because of an HEV genotype 3/rat genotype recombinant.
Reduced genetic diversity of ORF1 sequences obtained from Rattus spp. rats requires further study, including sequencing genomes of these isolates to identify sequence diversity at other loci. Difficulty in transmitting virus from infected wild R. norvegicus rats into laboratory strains also indicates that certain life history or genetic characteristics may be essential for infection. Purcell et al. (23) reported a positive correlation between antibody prevalence and animal age in their study of seroprevalence of HEV in Rattus spp. rats, suggested that rats are readily infected in the wild, and that infection occurred in juvenile rats. This pattern is consistent with HEV infection in humans and swine (33,34), and suggests that infections should be attempted in wild and laboratory juvenile rats. Lending further support to this suggestion, the only report of major long-term infection (>30 days) of rats with HEV used weanling rats (20), and all other attempts we are aware of have used only adult rats (23). In addition, extreme variation in host specificity that Shukla et al. (19) observed among different HEV genotype 3 strains indicates the need for future transmission studies to include as many strains as possible.
Rattus species rats examined for hepatitis E virus.
Acknowledgments
We thank X.J. Meng for providing input on the study and manuscript and Chris Conroy, Eileen Lacey, Robert J. Baker, Link Olson, Loren Ammerman, Enrique Santoyo-Brito, Karen McBee, Joseph Cook, Sharon Birks, and Fred Sheldon for providing tissue samples.
This study was supported by the National Science Foundation (award no. DEB-1110806).
Biography
Mr Lack is a doctoral student in the Department of Zoology at Oklahoma State University. His research interests are evolutionary genetics, genomics, the biology of species invasions, and virus evolution.
Figure A1.
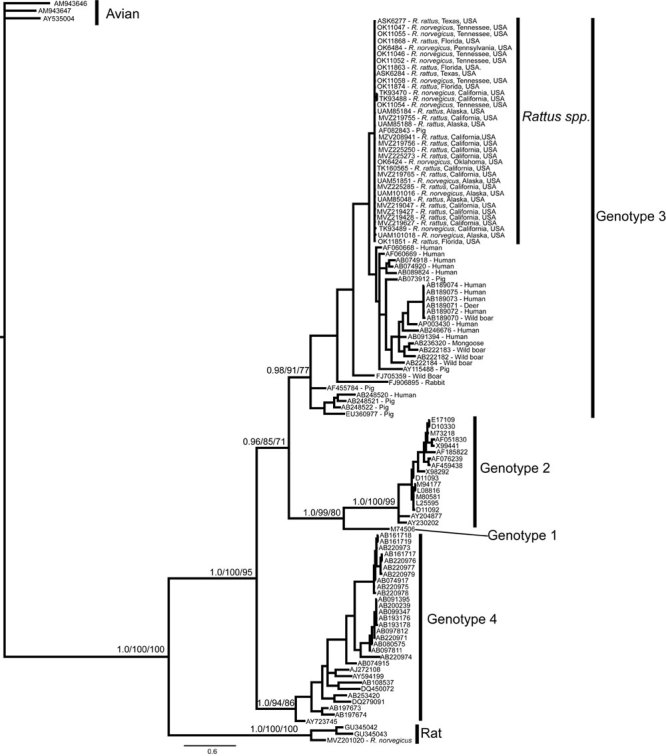
Bayesian phylogram resulting from analysis of a 334-bp fragment of hepatitis E virus (HEV) open reading frame 1. Node labels represent Bayesian posterior probabilities/maximum-likelihood bootstrap/maximum-parsimony bootstrap values, respectively, and are given only for nodes at the base of each genotype and nodes uniting the genotypes. For sequences obtained in this study from isolates from wild rats collected in the United States, terminal taxa labels correspond to tissue accession numbers shown in the Technical Appendix, followed by the species from which the tissue originated and the source population. For all other HEV genotype 3 sequences, the GenBank accession number is given, followed by the species from which the viral RNA was isolated. For all other sequences included, only the GenBank accession number is given. Scale bar indicates nucleotide substitutions per site.
Figure.
Genetic network showing the relationship among all hepatitis E virus genotype 3 sequences obtained in this study from isolates from wild rats collected in the United States. Each line represents a single mutational event and closed circles represent extinct or unsampled sequences. Each oval represents a single isolate, and the label corresponds to the tissue number shown in the Technical Appendix and the general sampling locality.
Footnotes
Suggested citation for this article: Lack JB, Volk K, Van Den Bussche RA. Hepatitis E virus genotype 3 in wild rats, United States. Emerg Infect Dis [serial on the Internet]. 2012 Aug [date cited]. http://dx.doi.org/10.3201/eid1808.120070
References
- 1.Aggarwal R. Hepatitis E: historical, contemporary and future perspectives. J Gastroenterol Hepatol. 2011;26:72–82. 10.1111/j.1440-1746.2010.06540.x [DOI] [PubMed] [Google Scholar]
- 2.Miyamura T. Hepatitis E virus infection in developed countries. Virus Res. 2011;161:40–6. 10.1016/j.virusres.2011.03.006 [DOI] [PubMed] [Google Scholar]
- 3.Tei S, Kitajima N, Takahashi K, Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–3. 10.1016/S0140-6736(03)14025-1 [DOI] [PubMed] [Google Scholar]
- 4.Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, et al. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol. 2003;84:2351–7. 10.1099/vir.0.19242-0 [DOI] [PubMed] [Google Scholar]
- 5.Meng XJ. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2010;140:256–65. 10.1016/j.vetmic.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. 10.1002/rmv.482 [DOI] [PubMed] [Google Scholar]
- 7.Bader TF, Krawczynski K, Polish LB, Favorov MO. Hepatitis E in a U.S. traveler to Mexico. N Engl J Med. 1991;325:1659. 10.1056/NEJM199112053252320 [DOI] [PubMed] [Google Scholar]
- 8.Meng XJ, Wiseman B, Elvinger F, Guenette DK, Toth TE, Engle RE, et al. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J Clin Microbiol. 2002;40:117–22. 10.1128/JCM.40.1.117-122.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011;161:23–30. 10.1016/j.virusres.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas DL, Yarbough PO, Vlahov D, Tsarev SA, Nelson KE, Saah AJ, et al. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J Clin Microbiol. 1997;35:1244–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mast EE, Kuramoto IK, Favorov MO, Schoening VR, Burkholder BT, Shapiro CN, et al. Prevalence of and risk factors for antibody to hepatitis E virus seroreactivity among blood donors in northern California. J Infect Dis. 1997;176:34–40. 10.1086/514037 [DOI] [PubMed] [Google Scholar]
- 12.Feagins AR, Opriessnig T, Guenette DK, Halbur PG, Meng XJ. Detection and characterization of infectious hepatitis E virus from commercial pig livers sold in local grocery stores in the USA. J Gen Virol. 2007;88:912–7. 10.1099/vir.0.82613-0 [DOI] [PubMed] [Google Scholar]
- 13.Mansuy JM, Bendall R, Legrand-Abravanel F, Saune K, Miedouge M, Ellis V, et al. Hepatitis E virus antibodies in blood donors, France. Emerg Infect Dis. 2011;17:2309–12. 10.3201/eid1712.110371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favorov MO, Kosoy MY, Tsarev SA, Childs JE, Margolis HS. Prevalence of antibody to hepatitis E virus among rodents in the United States. J Infect Dis. 2000;181:449–55. 10.1086/315273 [DOI] [PubMed] [Google Scholar]
- 15.Kabrane-Lazizi Y, Fine JB, Elm J, Glass GE, Higa H, Diwan A, et al. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am J Trop Med Hyg. 1999;61:331–5. [DOI] [PubMed] [Google Scholar]
- 16.Arankalle VA, Joshi MV, Kulkarni AM, Gandhe SS, Chobe LP, Rautmare SS, et al. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J Viral Hepat. 2001;8:223–7. 10.1046/j.1365-2893.2001.00290.x [DOI] [PubMed] [Google Scholar]
- 17.Hirano M, Ding X, Li TC, Takeda N, Kawabata H, Koizumi N, et al. Evidence for widespread infection of hepatitis E virus among wild rats in Japan. Hepatol Res. 2003;27:1–5. 10.1016/S1386-6346(03)00192-X [DOI] [PubMed] [Google Scholar]
- 18.Easterbrook JD, Kaplan JB, Vanasco NB, Reeves WK, Purcell RH, Kosoy MY, et al. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect. 2007;135:1192–9. 10.1017/S0950268806007746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, et al. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A. 2011;108:2438–43. 10.1073/pnas.1018878108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maneerat Y, Clayson ET, Myint KS, Young GD, Innis BL. Experimental infection of the laboratory rat with the hepatitis E virus. J Med Virol. 1996;48:121–8. [DOI] [PubMed] [Google Scholar]
- 21.Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, et al. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis. 2010;16:1452–5. 10.3201/eid1609.100444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johne R, Plenge-Bonig A, Hess M, Ulrich RG, Reetz J, Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol. 2010;91:750–8. 10.1099/vir.0.016584-0 [DOI] [PubMed] [Google Scholar]
- 23.Purcell RH, Engle RE, Rood MP, Kabrane-Lazizi Y, Nguyen HT, Govindarajan S, et al. Hepatitis E virus in rats, Los Angeles, California, USA. Emerg Infect Dis. 2011;17:2216–22. 10.3201/eid1712.110482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Innis BL, Shrestha MP, Clayson ET, Scott RM, Linthicum KJ, et al. Evidence that rodents are a reservoir of hepatitis E virus for humans in Nepal. J Clin Microbiol. 2002;40:4493–8. 10.1128/JCM.40.12.4493-4498.2002 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, et al. Geneious, version 5.0 [cited 2012 Apr 13]. http://www.geneious.com
- 27.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–5. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 28.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 29.Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–9. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- 30.He J, Innis BL, Shrestha MP, Clayson ET, Scott RM, Linthicum KJ, et al. Evidence that rodents are a reservoir of hepatitis E virus for humans in Nepal. Retraction of He J, Innis BL, Shresdtha MP, Clayton ET, Scott RM, et al. J Clin Microbiol. 2002;40:4493–8. J Clin Microbiol. 2006;44:1208. 10.1128/JCM.44.3.1208.2006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Aplin KP, Suzuki H, Chinen AA, Chesser RT, Have JT, Donnellan SC, et al. Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS ONE. 2011;6:e26357. 10.1371/journal.pone.0026357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lack JB, Greene D, Conroy C, Hamilton MJ, Braun JK, Mares MA, et al. Invasion facilitates extensive hybridization between three black rat species in the United States and Asia. Mol Ecol. 2012. In press. [DOI] [PubMed] [Google Scholar]
- 33.Meng XJ, Dea S, Engle RE, Friendship R, Lyoo YS, Sirinarumitr T, et al. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. J Med Virol. 1999;59:297–302. [DOI] [PubMed] [Google Scholar]
- 34.Arankalle VA, Tsarev SA, Chadha MS, Alling DW, Emerson SU, Banerjee K, et al. Age-specific prevalence of antibodies to hepatitis A and E viruses in Pune, India. J Infect Dis. 1995;171:447–50. 10.1093/infdis/171.2.447 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rattus species rats examined for hepatitis E virus.