Abstract
Epicardium-derived cells (EPDCs) invade the myocardium and differentiate into fibroblasts and vascular smooth muscle (SM) cells, which support the coronary vessels. The transcription factor Pod1 (Tcf21) is expressed in subpopulations of the epicardium and EPDCs in chicken and mouse embryonic hearts, and the transcription factors WT1, NFATC1, and Tbx18 are expressed in overlapping and distinct subsets of Pod1-expressing cells. Expression of Pod1 and WT1, but not Tbx18 or NFATC1, is activated with all-trans-retinoic acid (RA) treatment of isolated chick EPDCs in culture. In intact chicken hearts, RA inhibition leads to decreased Pod1 expression while RA treatment inhibits SM differentiation. The requirements for Pod1 in differentiation of EPDCs in the developing heart were examined in mice lacking Pod1. Loss of Pod1 in mice leads to epicardial blistering, increased SM differentiation on the surface of the heart, and a paucity of interstitial fibroblasts, with neonatal lethality. Epicardial epithelial-to-mesenchymal transition (EMT) and endothelial differentiation of coronary vessels are relatively unaffected. On the surface of the myocardium, expression of multiple SM markers is increased in Pod1-deficient EPDCs, demonstrating premature SM differentiation. Increased SM differentiation also is observed in Pod1-deficient lung mesenchyme. Together, these data demonstrate a critical role for Pod1 in controlling mesenchymal progenitor cell differentiation into SM and fibroblast lineages during cardiac development.
Keywords: Heart development, EPDC, Pod1, Tcf21, Chicken, Mouse
Introduction
During vertebrate embryonic development, epithelial cells from the proepicardium (PE), located at the venous pole of the primitive looped heart, migrate to the surface of the myocardium and form the epicardium (Gittenberger-de Groot et al., 2010). The mechanisms of epicardial formation and derivation of the coronary vasculature are conserved in vertebrates, including chicken and mouse embryos (Reese et al., 2002). A subset of epicardial cells undergoes EMT and invades the subepicardial space, thereby becoming EPDCs (Wessels and Perez-Pomares, 2004). Subepicardial EPDCs proliferate, invade the myocardium, and contribute to the fibrous matrix and coronary vasculature of the mature heart (Gittenberger-de Groot et al., 2010; Mikawa and Gourdie, 1996). EPDCs represent a multipotent progenitor population with the potential to differentiate into fibroblast and SM cell lineages, and also contribute to coronary endothelial cells and possibly cardiac myocytes at lower frequencies (Cai et al., 2008; Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Katz et al., 2012; Smart et al., 2011; Smart et al., 2009; Zhou et al., 2008). Multiple transcription factors (TFs), including Pod1 (Tcf21/Capsulin/Epicardin), Wilms’ Tumor 1 (WT1), NFATC1, and Tbx18, as well as signaling molecules, such as Retinoic Acid (RA), have been implicated in EPDC lineage development (Gittenberger-de Groot et al., 2010). The molecular interactions among specific signaling pathways and TFs in regulation of EPDC differentiation into particular cell lineages, including SM, are not well characterized.
Pod1 is a bHLH transcription factor expressed in the PE, epicardium, and EPDCs of embryonic chick and mouse hearts (Armstrong et al., 1993; Combs et al., 2011; Kraus et al., 2001; Quaggin et al., 1998; von Scheven et al., 2006). Loss of Pod1 leads to neonatal lethality with lung, kidney, and spleen defects, and mesenchymal Pod1 expression regulates lung and kidney epithelial morphogenesis (Lu et al., 2000; Quaggin et al., 1999). In kidney mesenchymal progenitors, Pod1 promotes myofibroblast differentiation (Plotkin and Mudunuri, 2008). However, the role of Pod1 in the heart has not been investigated in detail. In addition to Pod1, WT1, NFATC1, and Tbx18 also are expressed in the PE, epicardium, and EPDCs in chick and mouse embryos, and are required during heart morphogenesis (Armstrong et al., 1993; Combs et al., 2011; Kraus et al., 2001). WT1 regulates epicardial EMT, while epicardial NFATC1 promotes EPDC invasion of the myocardium (Combs et al., 2011; Kirschner et al., 2006; Moore et al., 1999; von Gise et al., 2011). Tbx18 contributes to formation of the sinus horn myocardium from pericardial mesenchyme (Bussen et al., 2004; Christoffels et al., 2006). However, the specific cellular functions and upstream regulatory mechanisms of these TFs, in particular Pod1, during EPDC development are relatively unknown.
Diverse signaling pathways regulate PE and EPDC development (Merki et al., 2005; Morabito et al., 2001; Schlueter et al., 2006; Zamora et al., 2007). In the epicardium and subepicardial EPDCs, Retinaldehyde Dehydrogenase-2 (RALDH2) is broadly expressed (Moss et al., 1998; Perez-Pomares et al., 2002). Upon invasion of the myocardium, EPDC expression of RALDH2 is downregulated, concurrent with EPDC differentiation into fibroblasts and SM cells. RA signaling in EPDCs is required to promote myocardial proliferation and to control coronary vasculogenesis, as determined by analysis of Retinoid X Receptor-α (RXRα) and RALDH2 null mouse models (Jenkins et al., 2005; Lin et al., 2010; Merki et al., 2005; Sucov et al., 1990). RA function in EPDCs, however, remains poorly understood.
In this study, we examine Pod1 expression relative to the expression patterns of other TFs in epicardium and EPDCs, as well as differential upstream regulation of Pod1 and other EPDC TFs. Pod1 function in EPDC differentiation in vivo also was examined in mice. Studies in chicken primary cell cultures and isolated hearts demonstrate that RA promotes Pod1 and WT1 expression, while also inhibiting SM differentiation, in EPDCs. Loss of Pod1 in mice leads to increased EPDC differentiation into SM and also in reduced numbers of interstitial fibroblasts in the developing heart.
Materials & Methods
Chick and mouse embryo collection
Fertilized white leghorn chicken eggs (Charles River Laboratories) were incubated at 38°C under high humidity, and embryos were sacrificed at E4 and E7. Pod1 heterozygous (Pod1+/−) mice, harboring one Pod1/LacZ knock-in allele, were obtained (Quaggin et al., 1999). The Pod1/LacZ loss-of-function allele contains a LacZ expression cassette in lieu of the Pod1 transcription initiation codon and basic helix-loop-helix (bHLH) domain; thus β-Galactosidase (βGal) is expressed instead of Pod1 from this locus (Quaggin et al., 1999). Pod1−/− mouse embryos were produced by timed mating of Pod1+/− animals with the presence of a copulation plug defined as embryonic day 0.5 (E0.5). Mouse embryos were collected from E11.5–E18.5. Wild type and Pod1+/− littermate embryos also were analyzed. Genotyping for the Pod1/LacZ allele was performed as previously described (Quaggin et al., 1999). All animal procedures were approved by the Cincinnati Children’s Hospital Medical Center Institutional Animal Care and Use Committee and performed following institutional guidelines.
Immunolocalization
Chick and mouse embryos were collected, fixed, dehydrated, and paraffin-embedded as previously described (Lincoln et al., 2004). Antibody labeling for immunofluorescence (IF), immunocytochemistry (ICC), and immunohistochemistry (IHC) was performed as previously described with modifications (Combs and Yutzey, 2009). Antigen retrieval was performed in boiling Citric Acid Based Antigen Unmasking Solution (1:100, Vector Laboratories) for 3–7 minutes under pressure. The following primary antibodies were used: Pod1 (1:100, Santa Cruz Biotechnology), Tbx18 (1:250, Santa Cruz), NFATC1 (1:100, Santa Cruz), WT1 (1:50, MyBioSource.com), NFATC1 (1:100, BD Pharmingen), ALDH1A2 (RALDH2) (1:100, Sigma Aldrich), WT1 (1:100, EMD Bioscience), Smooth Muscle Myosin (Myh11) (1:300, Biomedical Technologies), Calponin (1:100, Abcam), α-Smooth Muscle Actin (αSMA) (1:100, Sigma), Endomucin (Emcn) (1:250, eBioscience), E-Cadherin (1:150, Santa Cruz), SM22α (Transgelin)(1:100, Abcam), βGal (1:2000, Abcam), and Collagen Type I (Col1a1) (1:100, Millipore). Corresponding Alexa-donkey anti-rabbit-488, Alexa-donkey anti-mouse-568, Alexa-donkey anti-mouse-488, Alexa-donkey anti-rabbit-568, Alexa-goat anti-rabbit-488, Alexa-goat anti-mouse-555, Alexa-goat anti-mouse-488 (Invitrogen), or donkey anti-chicken-FITC (Abcam) conjugated secondary antibodies were applied as previously described (Combs and Yutzey, 2009). Alternatively, Renaissance Tyramide Signal Amplification Plus Fluorescein and Tetramethylrhodamine kits (Perkin Elmer) were used as described previously (Combs et al., 2011). For double IF experiments using two rabbit primary antibodies, Zenon Rabbit IgG Labeling Kit (Invitrogen) was used per manufacturer’s instructions. Nuclei were stained using 4’, 6-diamidino-2-phenylindole, dihydrochloride (DAPI) (1:10,000, Invitrogen).
For ICC, cultured EPDCs were fixed in 4% paraformaldehyde or cold 100% methanol (MeOH) for 1 hour at 4°C. Cells were washed in PBS and treated with 0.3% hydrogen peroxide for 30 minutes. ICC and IHC were performed using ImmunoPure ABC Ultra-Sensitive Peroxidase IgG Staining Kits (Fisher) or ImmunoCruz LSAB Staining Systems (Santa Cruz) per manufacturers’ instructions. After incubation, horseradish peroxidase detection with 3,3-Diaminobenzidine (DAB) Enhanced Metal Substrate Kit (Fisher) was performed per manufacturer’s instructions. Whole mouse heart IHC using anti-SM22α antibody was performed as previously described (Lincoln et al., 2004).
IF was detected using a Zeiss LSM 510 confocal microscope, and images were captured with Zeiss LSM version 3.2 SP2 software in parallel using identical confocal laser settings with constant PMT filters and integration levels. Alternatively, IF was detected using a Nikon A1-R LSM confocal microscope, and images were captured with NIS-Elements D 3.2 software in parallel using identical confocal laser settings, with constant PMT filters and integration levels.
Pictomicrographs of ICC and IHC tissue were obtained using either an Olympus BX51 microscope using NIS-Elements D 3.2 software, or using a Nikon SMZ1500 microscope, DXM1200F digital camera, and ACT-1 2.70 software.
Quantification of protein expression and colocalization
Images obtained by IF were used to quantify TF expression and colocalization in chick and mouse heart sections. The number of cells expressing each TF was quantified using Image J64 software. Single-channel images were converted to binary, a specific threshold value was set, and expression above this threshold value was used to quantify the number of cells expressing each TF, including Pod1, WT1, NFATC1, and Tbx18. Positive nuclei were counted in the epicardium and EPDCs. A Pod1 index was calculated by dividing the number of TF-positive (TF+) Pod1+ cells by the total number of Pod1+ cells, multiplied by 100%. Data were collected from three independent embryos (n=3) for each antibody combination, and approximately 730 cells were counted from 4–6 sections per embryo. Three independent experiments were performed in biological duplicate (n=3).
Quantification of the number of SM22α+ cells in the subepicardium and shallow myocardium, detected by IHC, per heart section was performed on E17.5 Pod1+/− and Pod1−/− tissue. SM22α expression was analyzed using pictomicrographs obtained at 600× magnification in comparable heart sections for each genotype. The number of SM22α-expressing cells was quantified in the ventricular subepicardium and shallow myocardium of the right and left free wall myocardium extending from the atrioventricular canal (AVC) to apex, exclusive of the interventricular septum. Three nonserial sections separated by at least 40 µm were quantified per embryo, and three embryos were analyzed per genotype (n=3). Comparable long-axis sections of Pod1−/− and Pod1+/− hearts were selected for analysis using the heart valves and septal structures as landmarks.
RNA probe generation and in situ hybridization
The chicken RALDH2 (Aldh1a2) sequence (835 bp) was amplified from E2.5 chick heart cDNA using forward 5’-GCT CGC CTT GCT TTT TCT CTG-3’ and reverse 5’-GTG GCC CTT GTT CTG TAG TTG G-3’ primers. The chicken Pod1 sequence (416 bp) was amplified from chick E3 PE cDNA using forward 5’-TTT GGC ATC TTC CAG ACC AT-3’ and reverse 5’-TTC AGG TCA CTC TCG GGT TT-3’ primers. The chicken Tbx18 sequence (1195 bp) was amplified from chick E6 limb cDNA using forward 5’-ACC AAG GCG GGC AGG CGC ATG TT-3’ and reverse 5’-TCG GCG AGG ACC CCA AGA AAC T-3’ primers. Sequences were amplified by polymerase chain reaction (PCR) and sub-cloned into pGEM-T vector (Promega). Identities were verified by sequencing. Antisense RNA probes were generated as described previously with modifications (Ehrman and Yutzey, 1999). The chicken RALDH2 digoxigenin-labeled riboprobe was synthesized using T3 polymerase from a plasmid linearized with XhoI. The chicken Pod1 probe was synthesized with SP6 polymerase from a plasmid linearized with NcoI. The chicken Tbx18 probe was synthesized with T7 polymerase from a plasmid linearized with NotI. Generation of the mouse Col1a1 riboprobe was described previously (Chakraborty et al., 2008).
In situ hybridization (ISH) was performed as previously described (Shelton and Yutzey, 2007) with the exception that 14 µm sections were treated with 20 µg/ml Proteinase K (Invitrogen) in PBS at 37°C for 6–18 minutes, depending on specimen’s age and species. Color reactions using 4-Nitro blue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl-phosphate solution (Roche Applied Science) were developed for 30–60 minutes. Images obtained by ISH were used to quantify the number of Col1a1-expressing interstitial cells within the ventricular myocardium of mouse embryos. Col1a1+ positive cells were counted in the right and left ventricular free wall myocardium of multiple comparable sections of Pod1+/− and Pod1−/− mouse E18.5 hearts as described above. Data were collected from three comparable sections each of three embryos (n=3) per genotype.
Chick PE and EPDC cultures
Aggregated PE tissue was dissected from chick E4 atrioventricular (AV) groove using tungsten needles (Ted Pella) as previously described (Combs et al., 2011). Six PE aggregates per culture were placed in 0.01% collagen-coated chamber slides (Fisher Scientific) containing PE culture media [M199 media (Cellgro Mediatech) with 10% Fetal Bovine Serum (FBS) (Fisher), 1% chick embryo extract (Sera Labs International), and 1% penicillin/streptomycin (Pen/Strep) (Invitrogen).]
Explants containing epicardium and EPDCs were dissected from the outer AVC regions of chick E7 hearts using tungsten needles. For each culture, twelve AVC explants from six hearts were minced into small pieces in complete culture media [M199 containing 10% FBS and 1% Pen/Strep] and cultured for two days, at which point the majority of beating myocardial clumps was removed using 10 µl Precision Barrier pipette tips (Denville Scientific).
PE and EPDC cultures were treated with all-trans-RA (RA) (1×10−6 M; Sigma) or MeOH as vehicle control (0.1%); diethylaminobenzaldehyde (DEAB), an aldehyde dehydrogenase inhibitor (1 µM; Sigma) or dimethyl sulfoxide (DMSO) as vehicle control (0.01%, Sigma); recombinant human BMP-2 (200 ng/ml; R&D Systems), recombinant mouse Noggin/Fc chimera (200 ng/ml; R&D Systems), recombinant mouse Wnt3a (150 ng/ml; R&D Systems), Wnt antagonist sFRP3 (150 ng/ml; R&D Systems), or bovine serum albumin (BSA) as vehicle control (1.5–2 µg/ml; Sigma), in complete culture media. Explanted cells were cultured for four days and then treated for four days, with media replenishment every two days, for a total of eight days. For examination of FGF signaling through the MAPK/ERK pathway, EPDC culture media was replaced after four days with EGM-2MV Microvascular Endothelial Cell Growth Medium-2 (Cambrex) with all SingleQuot additives, except hFGF-B, and incubated for one day, followed by treatment with bovine FGF basic (FGF2) (200 ng/ml; R&D Systems), Mitogen-Activated Protein Kinase Kinase (MEK) inhibitor U0126 (10 µM; Promega), or BSA+DMSO as vehicle controls, for four days, with replenishment of media after two days, for a total of nine days prior to RNA isolation. All culture experiments were performed at least three times in biological duplicate.
RNA isolation and real-time quantitative RT-PCR
Total RNA was isolated from chick PE and EPDC cultures as described previously (Combs and Yutzey, 2009). cDNA was generated from 500 ng total RNA from each culture using SuperScript II (Invitrogen) per manufacturer’s instructions. 1 µl cDNA in Power SybrGreen Master Mix (Applied Biosystems) was used for real-time quantitative reverse transcriptase PCR (qPCR) analysis (MJ Research, Opticon 2) of gene expression using the following primers: Pod1 5’-GGG TCC TTA GCA AAG CCT TC-3’ and 5’-TTT GCC GGC TAC CAT AAA AG-3’; WT1 5’-TCT AGG GGA CCA GCA GTA CTC-3’ and 5’-GAT GGG ACA GCT TGA AGT ATC G-3’; and Tbx18 5’-GCT TTG GTG GAG TCT TAC GC-3’ and 5’-TGT TGC GAC TGA GAT GGA AG-3’. Pod1, WT1, and Tbx18 PCR products were confirmed by sequencing. Primers and reaction conditions for NFATC1, Myh11, SM22α, GAPDH, β-actin, and RALDH2 were described previously (Combs and Yutzey, 2009; Landerholm et al., 1999; Lincoln et al., 2006b; Zheng et al., 2009). Samples were analyzed in triplicate and gene expression levels were determined as previously described (Lincoln et al., 2006a). The standard curve for each primer set was generated with a five-step 1:10 dilution series of chick E7 heart cDNA. All expression values were normalized to corresponding GAPDH expression levels, and consistent GAPDH expression was confirmed by normalization to α-actin. For each experiment, samples were collected in biological duplicates run in triplicate, and data were collected from at least three independent experiments for each condition (n=3–6).
Total RNA was collected from mouse E18.5 lungs using 800 µl Trizol reagent, and cDNA was generated from 1.2 µg RNA using SuperScript II. qPCR was performed using the Taqman gene expression assay (Applied Biosystems) for mouse Myh11 (Assay ID: Mm00443013_m1) on the StepOnePlus Real-Time PCR System (Applied Biosystems). Samples were analyzed in triplicate and gene expression values were calculated per manufacturer’s instructions based on the threshold cycle calibrated to a standard curve generated for each assay using a five-step 1:10 dilution series of wild type mouse E18.5 lung cDNA. Gene expression levels were normalized to corresponding β2-microglobulin (B2M) expression (Mm00437762_m1). Data were collected from individual Pod1−/− embryos and Pod1+/− littermates (n=4 per group).
Chick whole heart cultures and quantification of Pod1 and SM22α IF
Isolated E7 chick whole hearts were labeled with 25 µM carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) as described previously (Combs et al., 2011). Labeled hearts were cultured for 20 hours in complete culture media in 0.01% BSA-coated chamber slides and treated with MeOH+DMSO, RA, DEAB, or RA+DEAB as described above. Each treatment was performed in biological triplicate, and data were collected from three independent experiments for each treatment group (n=3). Hearts were fixed, processed, and sectioned (5 µm) as described above, with the exception that tissue and sections were cleared with d-Limonene (Hemo-De, Fisher). Pod1 and SM22α IF and quantification were performed as described above. Comparable long-axis sections were selected for quantification based on the position and morphology of AV valves, and pictomicrographs were obtained extending from the AVC to base of the left and right ventricular free walls. The number of Pod1+ EPDCs was quantified per microscopic field, and the average number of Pod1+ cells per field was calculated for each treatment group. SM22α expression was quantified similarly. Invasion of labeled EPDCs was quantified as described previously (Combs et al., 2011). The distance migrated by each cell from the epicardium into the subepicardium was quantified using ImageJ64 software, and the number of CFSE+ EPDCs in the subepicardium per microscopic field was determined. The average distance traveled by labeled EPDCs per microscopic field was calculated, as was the average number of invading EPDCs. Three independent experiments were performed in biological duplicate (n=6) for each condition.
β-Galactosidase Staining
E14.5 and E17.5 embryos were dissected, and hearts were stained, fixed, and paraffin-embedded as described previously (Lincoln et al., 2004; Sanes et al., 1986). 6 µm sections were cleared with xylene and mounted in Cytoseal (Fisher).
Statistical analysis
Statistical significance was determined by Student’s t-test with P•0.01 or P•0.05 as indicated. Data are reported as mean with standard error of the mean (s.e.m.).
Results
Epicardial cells and EPDCs demonstrate heterogeneity in TF expression
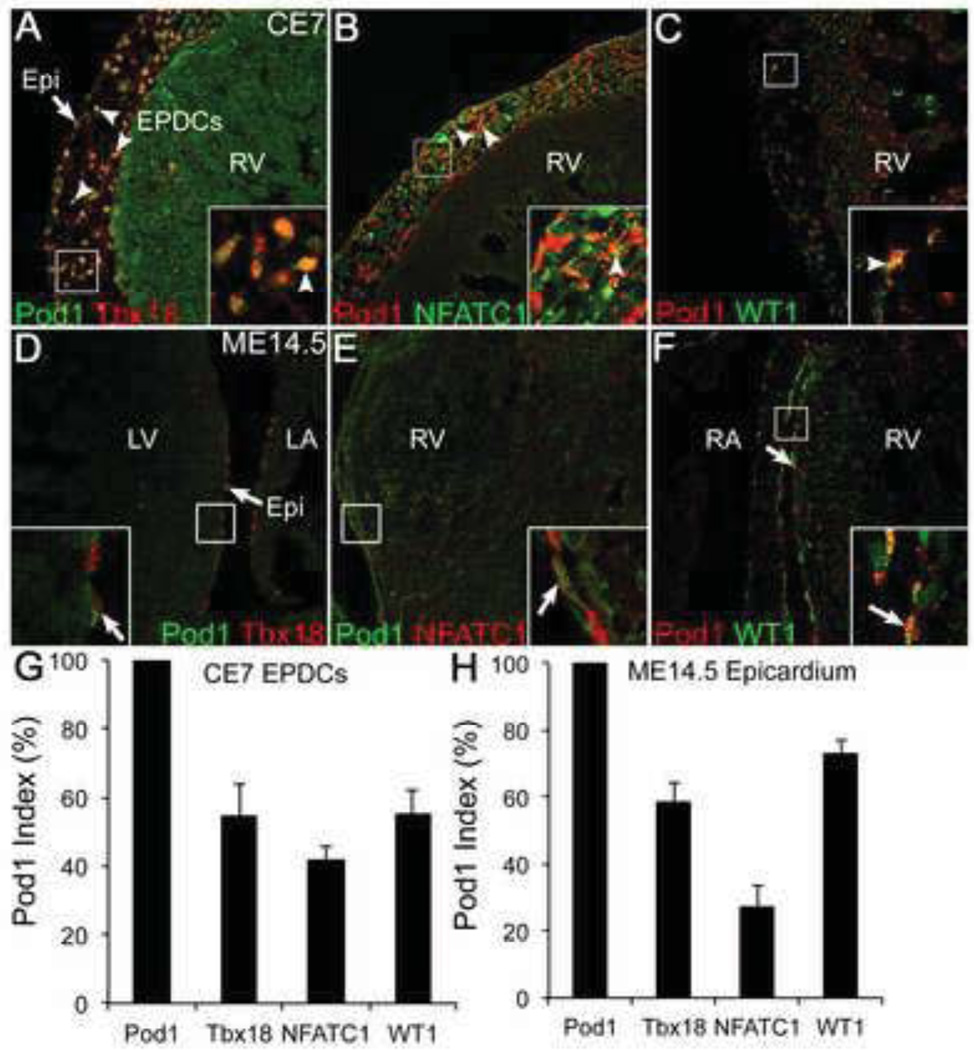
The bHLH TF Pod1 is expressed in the PE, epicardium, and EPDCs, but previous studies did not report if its expression is uniform or heterogeneous in these tissues (Cai et al., 2008; Combs et al., 2011; Ishii et al., 2007). Therefore, Pod1 expression in EPDCs in chick E7 and comparable mouse E14.5 heart sections was visualized in individual cells by IF with confocal laser scanning microscopy. In avian embryos, Pod1 is expressed in approximately 50% of EPDCs while WT1, NFATC1, and Tbx18 are expressed in comparable EPDC subpopulations (<50% of cells) (Fig. S1). Pod1 expression was defined further in terms of WT1, NFATC1, and Tbx18 co-expression in individual cells of the epicardium and EPDCs of chicken and mouse embryos (Fig. 1). Pod1 colocalization with WT1, NFATC1, or Tbx18 was visualized in individual cells by double IF with confocal laser scanning microscopy on chick E7 and mouse E14.5 heart sections. Corresponding representation of individual fluorescent channels is shown in Fig. S2. A Pod1 expression index was calculated for each TF by dividing the number of TF+ Pod1+ EPDCs by the total number of Pod1+ EPDCs, multiplied by 100%. Tbx18 is expressed in approximately 50% of Pod1+ EPDCs in chick and mouse embryos (Fig. 1A,D,G,H). NFATC1 is colocalized with Pod1 in a smaller fraction of EPDCs in chick and mouse (Fig. 1B,E,G,H), whereas WT1 is colocalized with Pod1 in approximately 50% of EPDCs in chick and 70% of epicardial cells in mouse (Fig. 1C,F,G,H). Together these data indicate that Pod1 is expressed in a subset of epicardial cells and EPDCs and that Pod1 is coexpressed with WT1, NFATC1, and Tbx18 in distinct and overlapping subpopulations of both mouse and chicken embryos.
Figure 1. Pod1, WT1, NFATC1, and Tbx18 are heterogeneously expressed in epicardium and EPDCs of chick E7 and mouse E14.5 hearts.
(A–F) Transcription factor (TF) expression in epicardium and EPDCs in chick E7 hearts (A–C) and mouse E14.5 hearts (D–F) was assessed by double immunofluorescence (IF) using the following antibodies: (A,D) anti-Pod1 (green) + anti-Tbx18 (red); (B,E) anti-Pod1 (red) + anti-NFATC1 (green); (C,F) anti-Pod1 (red) + anti-WT1 (green). Arrows and arrowheads indicate TF co-expression (yellow, insets) in epicardium and EPDCs, respectively. (G,H) TF heterogeneity was quantified by calculating a Pod1 index, which indicates the percentage of Pod1-positive (Pod1+) EPDCs that also express Tbx18, NFATC1, or WT1. The Pod1 index was calculated by dividing the number of TF+Pod1+ EPDCs by the total number of Pod1+ EPDCs, multiplied by 100%, per microscopic field. Error bars indicate standard error of the mean (s.e.m.). EPDC, epicardium-derived cell; Epi, epicardium; RV, right ventricle; LV, left ventricle; LA, left atrium; RA, right atrium.
RA signaling promotes Pod1 and WT1 expression in EPDCs
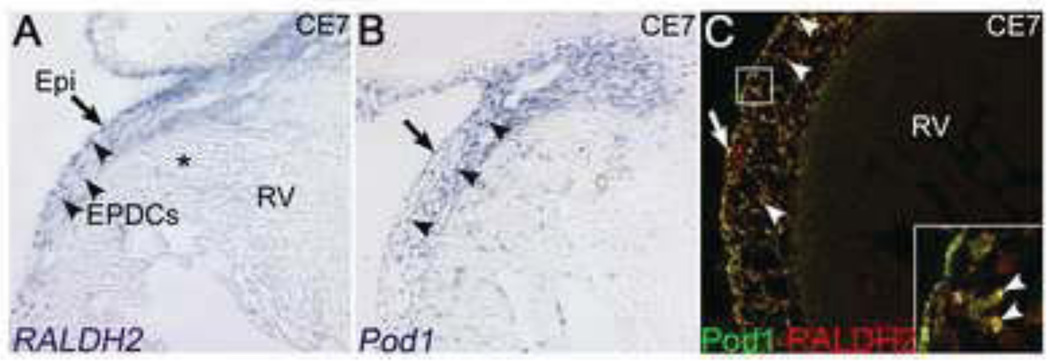
RA signaling is required for epicardial and EPDC development in the mouse embryo, as demonstrated by RXRα and RALDH2 gene ablation models (Jenkins et al., 2005; Lin et al., 2010). Expression of RALDH2 and Pod1 mRNA was examined in the chick E7 heart. RALDH2 is robustly expressed in chick E7 epicardium and EPDCs, as is Pod1 (Fig. 2A,B). In contrast, RALDH2 expression is downregulated, while Pod1 expression persists, in the myocardial interstitium that contains EPDCs (Fig. 2A). To determine if RALDH2 is expressed in Pod1+ EPDCs, RALDH2 and Pod1 co-expression was evaluated by double IF and confocal analysis. Pod1 and RALDH2 are co-expressed in a subset of EPDCs (inset, Fig. 2C) with approximately 80% of Pod1-expressing cells co-expressing RALDH2 (data not shown). Expression of RALDH2 is an indicator of RA biosynthesis and signaling (Rhinn and Dolle, 2012). Thus RA signaling is active in the majority of subepicardial EPDCs that express Pod1.
Figure 2. RALDH2 and Pod1 expression overlap in chick E7 EPDCs.
(A,B) As determined by RNA in situ hybridization (ISH), RALDH2 and Pod1 are robustly expressed in the chick E7 epicardium (arrows) and EPDCs (arrowheads). RALDH2 is downregulated in the myocardial interstitium (asterisk, A). (C) Double IF using anti-Pod1 (green) and anti-RALDH2 (red) antibodies show RALDH2 and Pod1 protein co-expression (yellow) in chick E7 epicardium (arrow) and EPDCs (arrowheads, inset).
The upstream regulation of TF expression in EPDCs was examined using an EPDC primary culture system. Cells were isolated from chick E7 epicardial AVC explants (boxed region in Fig. S3A) because this region is EPDC-rich, as indicated by the robust expression of Pod1 and Tbx18 (Figs. 2B, S3A). To verify that the isolated cells are EPDCs, TF expression was analyzed by ICC. Isolated cells express nuclear Pod1, Tbx18, and WT1 (Fig. S3B–D), which recapitulates TF expression seen in vivo (Fig. 1). Quantification of TF expression demonstrates that Pod1, Tbx18, and WT1 are individually expressed in 60–70% of isolated EPDCs, which is comparable to EPDCs in vivo (Figs. 1, S1). Together, these data indicate that isolated AVC cells express TFs characteristic of EPDCs.
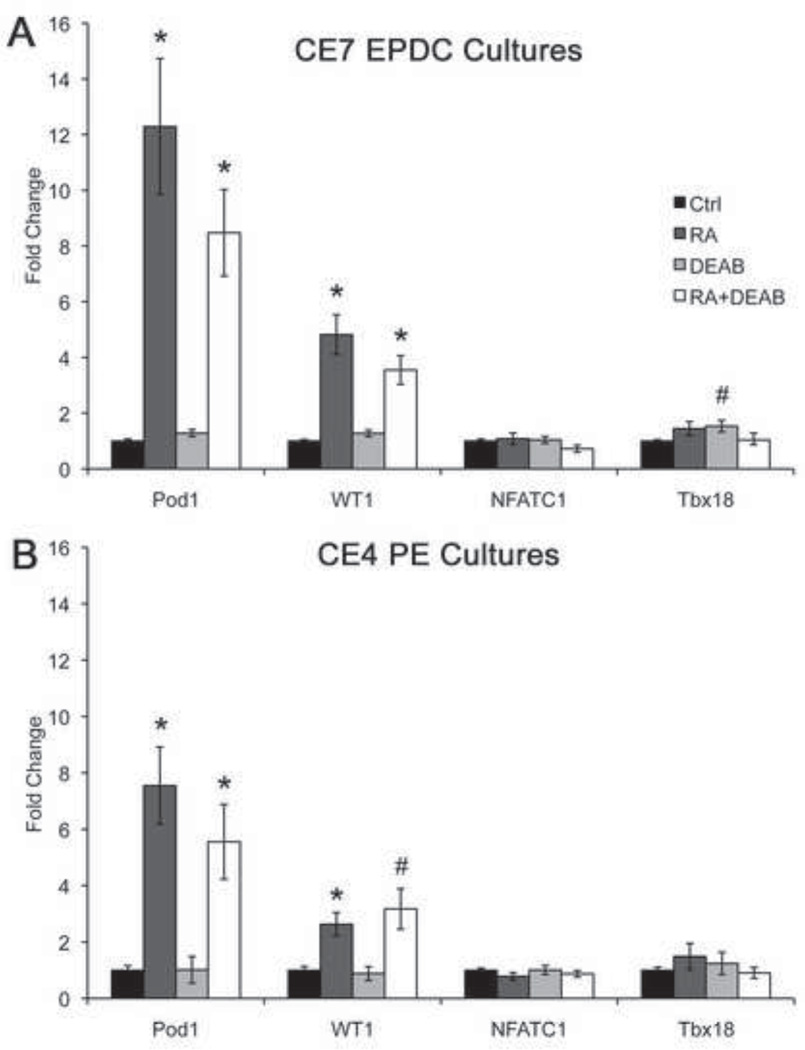
To determine if RA signaling differentially affects TF expression in EPDCs, EPDCs isolated from chick E7 AVC explants were grown in complete culture media for 4 days, followed by 4 days of treatment with RA, DEAB, and/or MeOH+DMSO as vehicle controls. Addition of RA to culture media results in 12.3-fold elevation in Pod1 mRNA expression, compared to MeOH+DMSO control, as detected by qPCR (Fig. 3A). RA also increases WT1 expression by 4.8-fold over control. RA treatment does not affect NFATC1 or Tbx18 expression in cultured EPDCs, which indicates that RA signaling preferentially promotes Pod1 and WT1 expression, without affecting NFATC1 and Tbx18. Addition of the RALDH inhibitor DEAB to culture media does not result in a significant change in TF gene expression relative to vehicle control and does not prevent induction by RA. The lack of inhibition of TF gene expression with DEAB treatment is likely due to the extremely low levels of RALDH2 expression in cultured EPDCs (3% of the GAPDH level, in contrast to 111% of the GAPDH level, in whole chick E7 hearts). Similar activation of Pod1 and WT1, but not NFATC1 and Tbx18, expression was observed in earlier PE cell aggregates isolated at E4 and treated with RA or RA+DEAB (Fig. 3B). Together these results indicate that RA specifically promotes Pod1 and WT1 expression, but not NFATC1 and Tbx18 expression, in isolated PE cells and EPDCs.
Figure 3. RA activates Pod1 and WT1, but not NFATC1 or Tbx18, gene expression in cultured chick proepicardial (PE) cells and EPDCs.
(A,B) TF gene expression was assessed in isolated chick E7 EPDCs (A) and E4 PE cells (B) treated with MeOH+DMSO (Ctrl) as vehicle controls, RA, the RALDH inhibitor DEAB, or RA+DEAB. Fold change in TF gene expression was quantified by qPCR relative to the control set to 1.0. Statistical significance of observed differences relative to control was determined by Student’s t-test (n=4–7). *P•0.01, #P•0.05.
The ability of BMP, FGF, or canonical Wnt signaling pathways to regulate TF expression in EPDCs also was examined (Fig. S4). EPDCs were isolated and cultured for 4 days as described above and treated for 4 days with BMP2 and/or Noggin (BMP inhibitor), FGF2 and/or U0126 (MEK inhibitor), Wnt3A and/or sFRP3 (soluble Wnt antagonist), or vehicle controls. Interestingly, FGF2 treatment specifically increases Pod1 expression in isolated EPDCs, and this effect is abrogated by the addition of the MEK inhibitor U0126 (Fig. S4B). However, manipulation of BMP or Wnt signaling pathways does not affect Pod1 or WT1 expression in EPDCs in vitro (Fig. S4A,C). The lack of response to BMP or Wnt treatments illustrates the specificity of Pod1 activation by RA and FGF2 in isolated EPDCs. Together these data show that FGF2, in addition to RA, promotes induction of Pod1 expression in cultured EPDCs.
RA promotes Pod1, while inhibiting SM22α expression, in cultured chick hearts
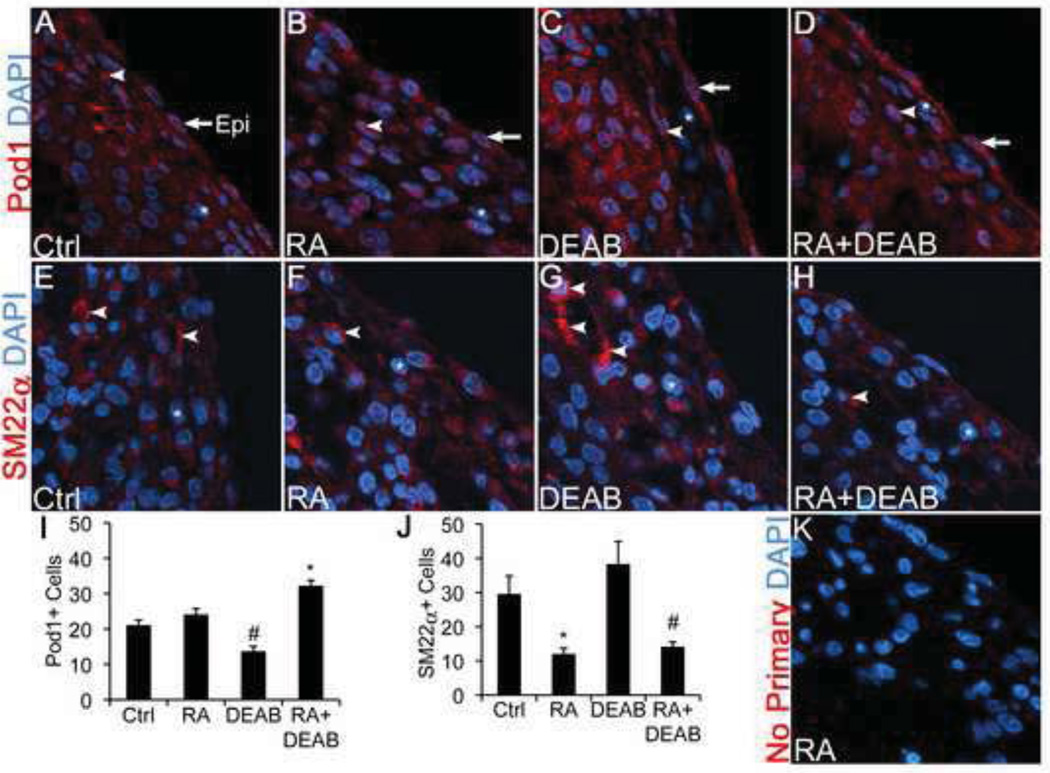
The effect of RA signaling on Pod1 expression was further examined in cultured whole chick hearts. Intact chick hearts were isolated at E7 and cultured in the presence of RA, DEAB, RA+DEAB, or MeOH+DMSO vehicle controls for 20 hours. Anti-Pod1 IF was visualized on sections of cultured hearts (Fig. 4), and the number of Pod1+ cells in the subepicardium per microscopic field was quantified (arrowheads, Fig. 4A–D,I). While RA treatment of chick hearts does not significantly affect the number of Pod1+ cells per field, compared to control hearts (Fig. 4A,B,I), addition of DEAB to culture media significantly decreases the number of Pod1+ cells per field, and this effect is abrogated by the addition of exogenous RA (Fig. 4C,D,I). Similar reduced Pod1 expression by RA signaling inhibition was observed upon quantification of anti-Pod1 by colorimetric IHC (black arrowheads, Fig. S5C,M). Thus RA signaling is necessary for full Pod1 expression in the chick subepicardium in vivo.
Figure 4. RA treatment increases Pod1 and decreases SM22α expression in intact chick E7 hearts.
(A–H) Chick E7 whole hearts were treated with vehicle controls (Ctrl) MeOH+DMSO (A,E), RA (B,F), DEAB (C,G), or RA+DEAB (D,H) in vitro. Expression of Pod1 and SM22α was detected by IF using anti-Pod1 antibody or anti-SM22α antibody, respectively. (A–D) Pod1 (red) is expressed in epicardium (arrows) and in EPDCs (arrowheads). (E–H) Expression of the smooth muscle (SM) marker SM22α (red) is indicated in the epicardium (arrows) and EPDCs (arrowheads). Nuclei are labeled with DAPI (blue). Asterisks indicate Pod1 or SM22α-negative cells. (I,J) Quantification of the average number of Pod1+ EPDCs (I) or SM22α+ EPDCs (J) per microscopic field is shown. Note that panels (A–H) are cropped and magnified regions of the microscopic fields. (K) A negative (no primary) control section has little background labeling. Statistical significance of observed differences relative to control was determined by Student’s t-test (n=6). *P•0.01, #P•0.05.
The effects of RA inhibition of EPDC differentiation into SM were examined in chick E7 whole heart cultures. Hearts were treated with RA, DEAB, RA+DEAB, or MeOH+DMSO, as described above. To determine if manipulation of RA signaling affects SM differentiation in whole hearts, anti-SM22α IF analysis was performed on cultured heart sections, and the number of SM22α+ cells in the subepicardium was quantified per microscopic field. RA treatment leads to significantly fewer SM22α+ cells per microscopic field (Fig. 4E,F,J). Therefore, RA signaling inhibits SM differentiation in EPDCs in chick hearts. DEAB treatment does not affect SM22α+ cell number, but addition of RA in the presence of DEAB inhibits SM differentiation (Fig. 4G,H,J). Similarly, DEAB treatment leads to increased Calponin expression, providing further evidence that RA signaling inhibits SM differentiation in the context of the whole heart (Fig. S5E–H,N). In contrast, RA signaling status does not affect EPDC migration into the subepicardium in whole heart cultures, as determined by quantification of CFSE-labeled EPDC subepicardial cell numbers or distance migrated in RA or DEAB-treated hearts (Fig. S6A–F). Together these results demonstrate that RA promotes Pod1 expression and inhibits SM differentiation, without affecting migration, in EPDCs.
Pod1/LacZ is expressed in mouse epicardium and EPDCs, and Pod1 is required during epicardial development in vivo
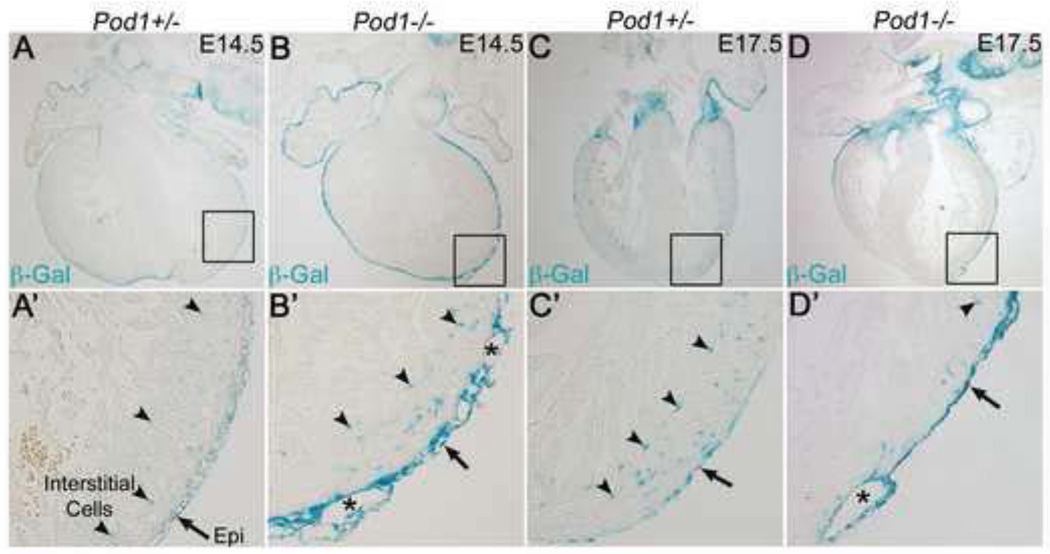
The studies in avian embryo cultures demonstrate that RA promotes Pod1 expression while inhibiting SM differentiation (Figs. 3, 4, S5). While previous studies have reported RA inhibition of SM differentiation (Azambuja et al., 2010), the role of Pod1 in regulating SM differentiation has not been previously reported. Therefore, Pod1 function in EPDCs in vivo was examined in mice lacking Pod1. To investigate Pod1 function in EPDCs in vivo, Pod1−/− mouse embryos were evaluated for defects in the epicardium or EPDCs. Pod1−/− mice are viable until birth, but a cardiac phenotype has not been described in depth for animals lacking Pod1 (Hidai et al., 1998; Lu et al., 2000; Quaggin et al., 1999). Since the Pod1 null allele contains a LacZ knock-in cassette (Quaggin et al., 1999), histological sections were examined by X-Gal staining to visualize βGal+ cells in the developing heart. At E14.5, Pod1+/− and Pod1−/− mouse embryos express βGal in the epicardium, subepicardial mesenchyme, and myocardial interstitial cells (arrowheads) (Fig. 5A’,B’). At E17.5, βGal expression persists in the epicardium and within the myocardial interstitium of Pod1+/− and Pod1−/− hearts (Fig. 5C’,D’). Thus, the Pod1/LacZ knock-in allele is expressed in epicardial progenitors and derivatives consistent with the observed expression of endogenous Pod1.
Figure 5. The epicardium detaches from the myocardium in Pod1−/− mice.
X-Gal staining was performed on E14.5 and E17.5 mouse heart sections. (A–B’) βGal expression from the Pod1 locus is detected in the epicardium (arrows) and EPDCs (arrowheads) of E14.5 Pod1+/− (A’) and Pod1−/− (B’) mouse hearts. (C–D’) βGal expression persists in the epicardium (arrows) and intramyocardial EPDCs (arrowheads) of Pod1+/− (C’) and Pod1−/− (D’) hearts at E17.5. (B’,D’) Epicardial blistering (asterisks) occurs in Pod1−/− hearts at E14.5 and E17.5.
In Pod1−/− embryos, the surface epicardium exhibits abnormal morphology and cellularity. Epicardial blistering is apparent at E14.5 and E17.5 (asterisks) (Fig. 5B’,D’) in regions where the epicardium has detached from the surface of the heart. In addition, hemopericardium, indicative of vascular rupture, is frequently observed upon harvest of Pod1−/− embryos, as previously described (Quaggin et al., 1999). The presence of an intact epithelial epicardium is evident by continuous E-Cadherin expression in the epicardium at E14.5 in Pod1−/− embryos, similar to Pod1+/− control (Fig. S7) (Batlle et al., 2000; Cano et al., 2000; Mahtab et al., 2008). In addition, epicardial EMT is apparent in βGal+ EPDCs observed in the space between the intact E-Cadherin+ epicardium and myocardium in the Pod1−/− embryos (arrow) (Fig. S7B’). To determine if coronary vessel endothelial cells are affected by loss of Pod1, IHC was performed using anti-Emcn antibody. At E18.5, Emcn expression is similar in Pod1+/− and Pod1−/− hearts, in which capillaries and coronary veins (arrows), but not arteries (arrowheads), contain Emcn+ endothelial cells (Fig. S8). Thus initial formation of the epicardium, EMT, and coronary endothelial differentiation all occur in the absence of Pod1. Epicardial maturation, however, is abnormal in Pod1−/− embryos.
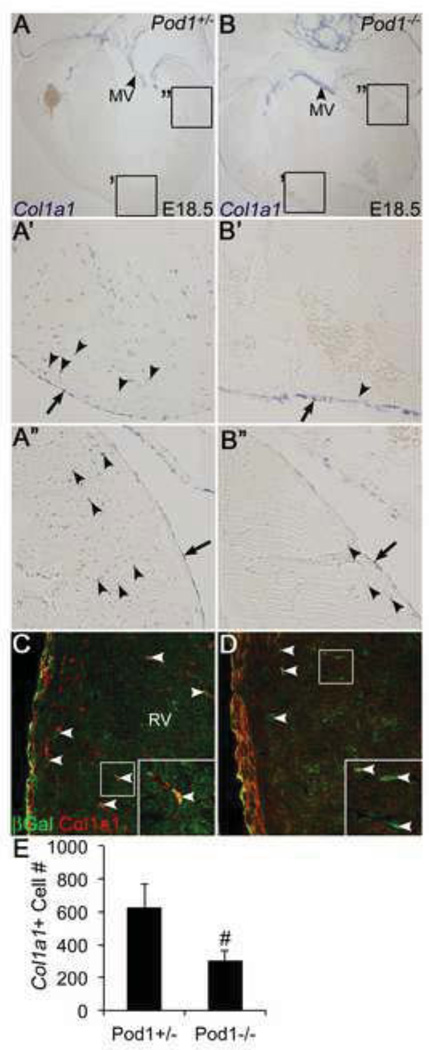
EPDC differentiation into fibroblasts was examined in Pod1+/− and Pod1−/− mouse embryos (Fig. 6). In order to determine if loss of Pod1 affects fibroblast lineage differentiation, expression of the fibroblast marker Col1a1 was evaluated by RNA ISH of E18.5 mouse heart sections, and the total number of Col1a1-expressing interstitial cells in the ventricular myocardium was quantified (Fig. 6E). Col1a1 is expressed in the epicardium of both Pod1−/− and Pod1+/− mouse hearts at E18.5 (arrows) (Fig. 6A’–B”). However, within the myocardium, the total number of Col1a1+ cells is decreased significantly in the Pod1−/− hearts, compared to heterozygous controls (arrowheads) (Fig. 6A’–B”,E). In Pod1+/− hearts, LacZ expression is maintained in differentiated fibroblasts as indicated by colocalized Col1a1 expression (Fig. 6C, inset). In Pod1−/− hearts, Pod1-deficient cells are present in the myocardial interstitium, as indicated by βgal expression (arrowheads). However they fail to activate Col1a1 expression (Fig. 6D, inset), indicating that Pod1 is necessary for fibroblast differentiation after EPDC migration into the myocardial interstitium. Together, these data indicate that Pod1 is required for Col1a1+ expression and fibroblast differentiation within the embryonic myocardium.
Figure 6. Cardiac interstitial Col1a1+ cells are rare in Pod1−/− mouse hearts.
(A–B”) RNA ISH was performed on E18.5 mouse heart sections using a Col1a1 mRNA probe. Col1a1 is expressed in the epicardium of Pod1+/− (A’,A”) and Pod1−/− (B’,B”) hearts (arrows). Interstitial Col1a1+ cells (arrowheads in A’–B”) are scarce within Pod1−/− myocardium (B’,B”). Normal Col1a1 expression is observed in mitral valves (MV, arrowheads) of Pod1+/− (A) and Pod1−/− (B) hearts. (C,D) Differentiation of Pod1-deficient EPDCs into fibroblasts was analyzed by double IF using anti-βGal (green) and anti-Col1a1 (red) antibodies. In E17.5 Pod1+/− mice, heterozygous for the Pod1/LacZ locus, βGal+ interstitial cells coexpress Col1a1 (yellow, arrowheads and inset). (D) In E17.5 Pod1−/− hearts, Pod1-deficient βGal+ interstitial cells (green) are negative for Col1a1 expression (arrowheads, inset). (E) Total Col1a1-expressing cells within the myocardial interstitium of Pod1+/− and Pod1−/− hearts were quantified (as in A–B”). Statistical significance of observed differences between Pod1+/− controls and Pod1−/− was determined by Student’s t-test (n=3). #P•0.05.
Loss of Pod1 leads to increased and premature SM expression in the heart
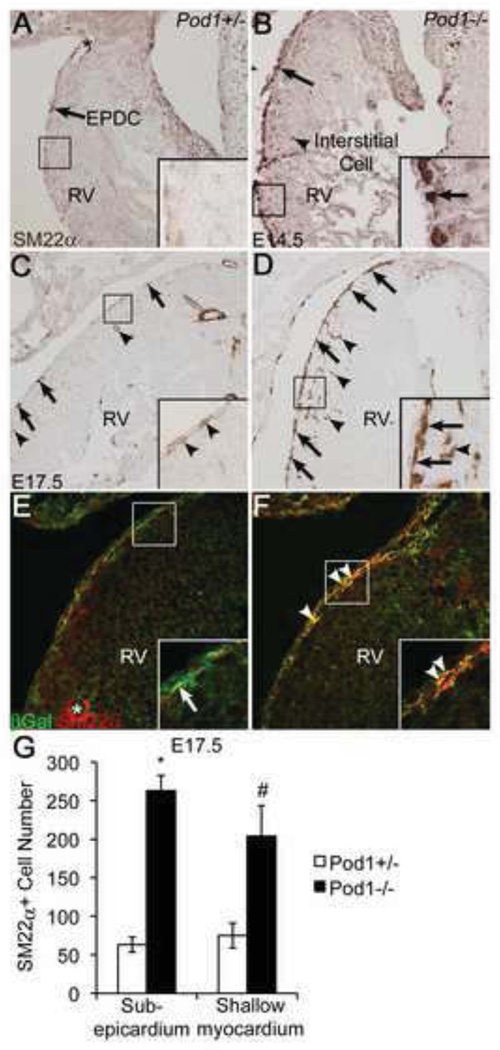
Differentiation of EPDC-derived SM in the myocardial interstitium is characterized by downregulation of RALDH2 and Pod1 (Fig. 2), consistent with a mechanism whereby these factors repress SM differentiation until EPDCs reach their final position surrounding the coronary vessels (Guadix et al., 2011; Perez-Pomares et al., 2002). To determine the effects of Pod1 deficiency on the timing and localization of SM differentiation, SM protein expression was analyzed by anti-SM22α IHC and IF in Pod1+/− and Pod1−/− embryonic hearts. At E14.5, little SM differentiation, as indicated by SM22α expression, is apparent in Pod1+/− embryos (Fig. 7A). In contrast, SM22α is robustly expressed in EPDCs on the heart surface, as well as in interstitial cells of the shallow myocardium, in Pod1−/− embryonic hearts (inset, Fig. 7B). At E17.5, intense SM22α expression is detected in the epicardium and in dispersed cells in the shallow myocardial interstitium of Pod1−/− embryos (inset, Fig. 7D), in contrast to localization of differentiated SM surrounding large coronary vessels in the Pod1+/− littermates (Fig. 7C). Quantification of these results demonstrates a 4.2-fold increase in the number of SM22α+ EPDCs and a 2.7-fold increase in the number of SM22α+ cells within the shallow myocardium of Pod1−/− heart sections at E17.5, compared to heterozygous controls (Fig. 7G). The total number of SM22α+ cells also is increased at E14.5 and E18.5 in Pod1−/− hearts, relative to controls (Fig. 7A,B; data not shown). Similarly, subepicardial activation of SM markers αSMA and Calponin also is increased in Pod1−/− embryos at E17.5 (Fig. S9). Likewise, whole mount IHC for SM22α demonstrates pervasive superficial SM22α expression over the surface of the heart in Pod1−/− hearts at E17.5, compared to the Pod1+/− control (Fig. S10). Thus, loss of Pod1 results in increased and aberrant EPDC differentiation into SM at the surface of the heart.
Figure 7. SM22α expression is increased in cells present in the subepicardium and shallow myocardium of Pod1−/− mouse hearts.
(A–D) Immunohistochemistry (IHC) was performed on E14.5 and E17.5 mouse heart sections using anti-SM22α antibody (brown). (A,C) SM22α is primarily expressed in coronary vascular SM (asterisks) of Pod1+/− controls at E14.5 (A) and E17.5 (C). SM22α is rarely expressed in the subepicardium (arrows, insets) of Pod1+/− hearts. (B,D) In Pod1−/− embryos, SM22α is expressed in EPDCs (arrows, insets) and in interstitial cells (arrowheads, insets) at E14.5 (B) and E17.5 (D). (E,F) Differentiation of Pod1-deficient EPDCs into SM was analyzed by double IF using anti-βGal (green) and anti-SM22α (red) antibodies. (E) In E17.5 Pod1+/− mice, heterozygous for the Pod1/LacZ locus, βGal+ subepicardial cells (green) are distinct from SM22α+ cells (red) in the subepicardium (arrow, inset) and in coronary vascular SM (asterisk). (F) In Pod1−/− hearts, Pod1-deficient βGal+ cells co-express SM22α (yellow) in the subepicardium (arrowheads, inset). (G) Total SM22α+ subepicardial and shallow intramyocardial cells per section at E17.5, labeled by IHC as in C–D, were quantified. Statistical significance of differences between Pod1+/− controls and Pod1−/− was determined by Student’s t-test (n=3). *P•0.01, #P•0.05.
The ability of Pod1-deficient EPDCs to differentiate into SM prior to deep myocardial invasion was assessed by colocalization of βGal, indicative of Pod1 locus expression, and SM22α in Pod1+/− and Pod1−/− E17.5 heart sections. In Pod1+/− embryos, SM22α is rarely expressed in Pod1+ surface EPDCs (arrow Fig. 7E inset). After EPDC migration into the ventricular interstitium, Pod1 is downregulated and SM22α is activated in SM cells surrounding coronary vessels (asterisk, Fig. 7E). Thus βGal is expressed in surface EPDCs that do not express SM22α but is not expressed in SM22α+ SM cells within the myocardium. In contrast, in the Pod1−/− heart, Pod1-deficient, βGal+ cells robustly co-express SM22α in the subepicardium (arrowheads, Fig. 7F inset). This result is consistent with premature differentiation of Pod1-deficient EPDCs into SM on the heart surface prior to invasion. However, differentiated SM22α+ SM is present in the coronary arteries within the myocardium of both Pod1−/− mice and Pod1+/− littermates at E17.5 (Fig. 7C,D), suggesting that Pod1 is not required for SM differentiation after myocardial invasion. Together, these data support a mechanism whereby Pod1 suppresses differentiation of subepicardial and intramyocardial SM progenitor cells when they are present at or near the surface of the heart.
Loss of Pod1 results in increased SM expression in the lungs
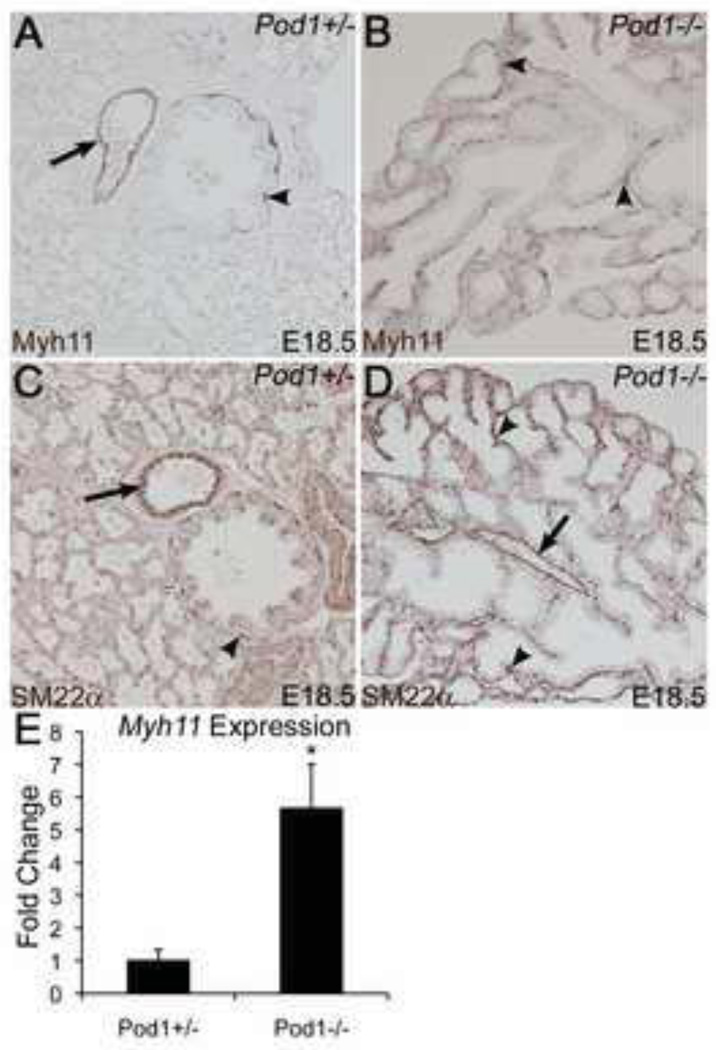
Pod1 is expressed in embryonic lung mesenchyme, and Pod1−/− mice die soon after birth with severe lung hypoplasia (Fernandes et al., 2004; Quaggin et al., 1999). To determine if loss of Pod1 affects differentiation of lung mesenchyme, expression of the SM markers Myh11 and SM22α was investigated by IHC in Pod1+/− and Pod1−/− lungs at E18.5. In Pod1+/− embryos expression of Myh11 and SM22α is predominant in differentiated SM of the large airways and arteries (Fig. 8A,C). In contrast, Myh11 and SM22α are widely expressed throughout the lung mesenchyme proximal to the airway epithelium in Pod1−/− littermates (Fig. 8B,D). In addition, Myh11 mRNA expression is increased in E18.5 Pod1−/− lungs relative to Pod1+/− littermates, as determined by qPCR (Fig. 8E). Thus loss of Pod1 leads to aberrant and pervasive SM differentiation in lung mesenchyme, consistent with increased SM differentiation in EPDCs in Pod1−/− hearts. Together these data provide evidence for a similar mechanism of Pod1 inhibition of mesenchymal cell differentiation into SM in embryonic heart and lungs.
Figure 8. Pod1−/− embryos have increased smooth muscle in the lung at E18.5.
IHC was performed on E18.5 mouse lung sections using anti-Myh11 (A,B) or anti-SM22α (C,D) antibodies (brown). (A,C) Myh11+ (A) and SM22α+ (C) SM cells surrounding a larger airway (arrowheads) and blood vessel (arrows) are indicated in Pod1+/− control lung. (B,D) Myh11+ (B) and SM22α+ (D) SM cells (arrowheads) surround hypoplastic airways of Pod1−/− lungs. A blood vessel is indicated (arrow, D). (E) Fold change in Myh11 mRNA expression in the lung was calculated by qPCR for four E18.5 Pod1+/− embryos and four E18.5 Pod1−/− embryos. Statistical significance was determined by Student’s t-test (n=4). *P•0.01.
Discussion
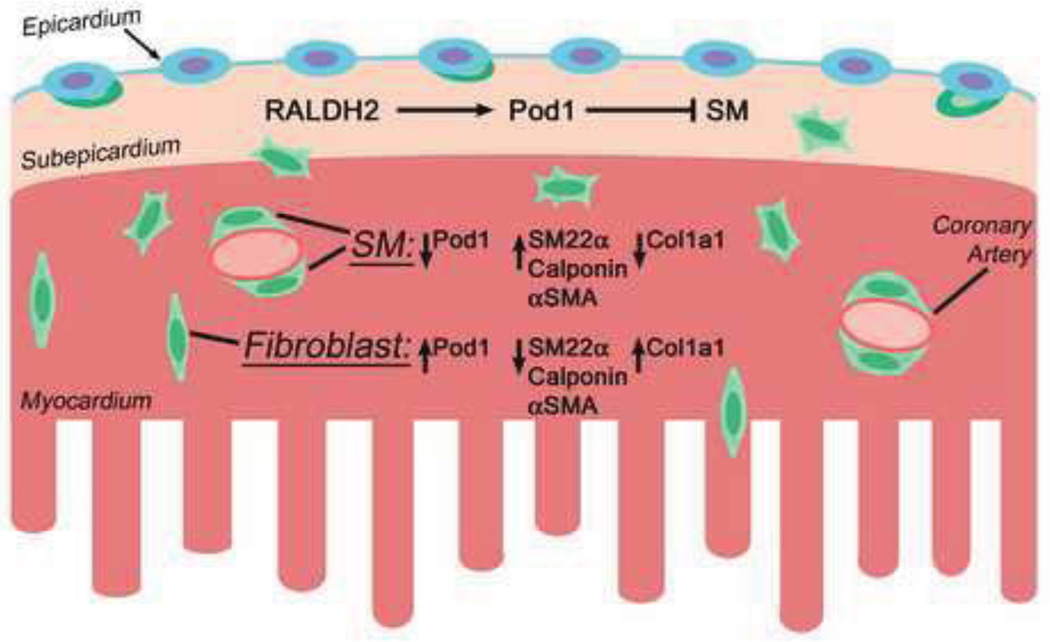
Here we demonstrate that Pod1-expressing epicardial cells and EPDCs differentially express WT1, NFATC1, and Tbx18 in overlapping and distinct subpopulations in chick and mouse embryos. RA differentially regulates epicardial TF gene expression by promoting Pod1 and WT1, but not NFATC1 or Tbx18, in PE cells and EPDCs. In addition, RA inhibits EPDC differentiation into SM in whole heart cultures. Loss of Pod1 in mice in vivo results in reduced presence of cardiac interstitial fibroblasts as well as aberrant increased SM differentiation in EPDCs and in lung mesenchyme of mouse embryos. These data support a model of regulation of EPDC differentiation (Fig. 9) in which RA signaling, evident in RALDH2 expression, induces Pod1 to inhibit SM differentiation in subepicardial EPDCs. After invading the heart, Pod1 expression is downregulated in the coronary vasculature consistent with an inhibitory role in SM differentiation, while Pod1 expression persists and is required for differentiation of interstitial fibroblasts.
Figure 9. Model of RA regulation of Pod1 and EPDC differentiation into smooth muscle and fibroblast lineages.
EPDCs (green cells) comprise a diverse group of cells with the potential to differentiate into epicardium-derived SM cells and fibroblasts (green cells) in the embryonic heart. In the epicardium (blue) and subepicardial mesenchyme (tan) overlying the ventricular myocardium (red), RA signaling, as indicated by RALDH2 expression, promotes Pod1 expression. In the subepicardial progenitor population, RA and Pod1 together inhibit cell differentiation into SM lineage, indicated by SM22α, Calponin, and αSMA expression. As EPDCs invade the myocardium, Pod1 expression is downregulated in some but not all EPDCs. Pod1 expression persists in cells that differentiate into fibroblasts, as indicated by Col1a1 expression. Thus downregulation of Pod1 allows EPDC differentiation into SM, while Pod1 is required for interstitial fibroblast differentiation.
Analysis of TF diversity indicates that Pod1+ epicardial cells and EPDC populations differentially express the TFs WT1, NFATC1, and Tbx18, implicated in epicardial cell lineage development. Pod1 is expressed in a subpopulation of EPDCs that can be further divided based on WT1, NFATC1, and Tbx18 expression in both chick E7 and mouse E14.5 hearts. Similar EPDC subpopulations are observed in both chicken and mouse embryonic hearts, supporting conservation of mechanisms regulating epicardial lineage development among vertebrate species (Reese et al., 2002). However, it is unclear if these Pod1-expressing subpopulations represent distinct EPDC progenitor lineages. The alterations in SM and fibroblasts, but not endothelial cells, with loss of Pod1 suggest that Pod1+ EPDCs are progenitors of those lineages. Additional evidence for an early separation of endothelial versus fibroblast/SM lineages in the PE and epicardium of chick and mouse embryos is that Scleraxis-lineage positive cells, including endothelial cells, are distinct from WT1/Tbx18-lineage positive cells in epicardial derivatives (Katz et al., 2012). It is unknown, however, if heterogeneity in Pod1, WT1, and Tbx18 expression represents diversified EPDC progenitors of coronary SM and fibroblast lineages. Alternatively, TF expression may oscillate, and a ‘negative’ cell may reactivate TF expression later during development. Further studies are necessary to define the molecular hierarchies and cell lineage relationships of EPDC progenitors and their subpopulations in the developing heart.
RA selectively activates WT1 and Pod1 expression and inhibits SM differentiation in isolated chick PE cells and EPDCs. RA activation of WT1 may be a direct mechanism, similar to that observed in zebrafish, in which RA directly activates WT1 gene expression via a Retinoic Acid Response Element (RARE) in the wt1a promoter (Bollig et al., 2009). However, conserved RARE sequences were not identified in mouse and chicken Pod1 proximal genomic sequences, as determined by rVISTA analysis (data not shown), suggesting that this regulatory interaction is indirect or may require more distal sequences. RA signaling in the epicardium is required to control coronary vascular morphogenesis (Dyson et al., 1995; Jenkins et al., 2005; Merki et al., 2005). RXRα−/− mouse embryos have a detached epicardium similar to that of Pod1−/− embryos (Jenkins et al., 2005), further supporting a mechanism by which RA induces Pod1 and maintains the undifferentiated epicardium. Additionally, RA inhibits SM differentiation in quail PE (Azambuja et al., 2010), which supports the mechanism presented here whereby RA signaling restricts EPDC differentiation into SM.
Here we demonstrate that Pod1 regulates the timing and localization of EPDC differentiation into SM and fibroblasts. These data are consistent with fate mapping analysis of the Pod1/Tcf21Cre lineage, which demonstrates that Pod1/Tcf21Cre+ derivatives contribute to coronary vascular SM and fibroblasts of the adult mouse heart (Acharya et al., 2011). The presence of Pod1-deficient cells in the interstitial myocardium suggests that loss of Pod1 does not prevent EPDC migration. However, fibroblast differentiation is severely reduced demonstrating that Pod1 is required for differentiation of this lineage in the myocardial interstitium. Strikingly, SM differentiation occurs prematurely and preferentially at the surface of the heart. Additional pathways implicated in SM differentiation include Notch, Transforming Growth Factor-β (TGFβ), and Platelet-Derived Growth Factor (PDGF) signaling pathways. Epicardial Notch signaling is required for SM differentiation (del Monte et al., 2011; Grieskamp et al., 2011), and TGFβ signaling promotes epicardial EMT and differentiation into SM (Austin et al., 2008; Compton et al., 2006). Likewise PDGF signaling through PDGFRα and PDGFRβ is required to direct EPDC differentiation into fibroblasts and SM cells, respectively (Mellgren et al., 2008; Smith et al., 2011). Together, these studies provide evidence that multiple signaling pathways, including RA activation of Pod1, contribute to EPDC lineage development and differentiation. However, little is known of the regulatory hierarchies of these pathways and specific downstream TF interactions.
Data presented here indicate that expression of multiple SM markers including SM22α, αSMA, and Calponin is increased in EPDCs and lung mesenchyme in the absence of Pod1. Myh11 expression is also increased in the E18.5 Pod1−/− lung mesenchyme and kidney (data not shown). Pod1 is a class II bHLH TF that negatively regulates gene expression by acting as a transcriptional repressor (Barnes and Firulli, 2009; Funato et al., 2003; Miyagishi et al., 2000a; Miyagishi et al., 2000b). In a multipotent mesenchymal cell line derived from adult mouse kidney, Pod1 binds to E-box DNA consensus sequences (CANNTG) within the SM22α and Calponin promoters, and overexpression of Pod1 inhibits SM22α and Calponin protein expression (Plotkin and Mudunuri, 2008). Together these data support a direct regulatory mechanism for Pod1 inhibition of SM differentiation in EPDC progenitor cells on the surface of the heart and in the myocardial interstitium prior to localization in the coronary vasculature. After migration into the myocardial interstitium, Pod1 expression is downregulated in a subset of EPDCs, and SM gene expression is induced in cells of the coronary vessels. A separate interstitial EPDC subpopulation maintains Pod1 expression and differentiates into fibroblasts, indicating that Pod1 likely has distinct regulatory functions in SM and fibroblast differentiation. Together these data provide evidence for Pod1 function in the regulation of the timing and localization of differentiation through direct repression of SM-specific gene expression in progenitor cell lineages in the developing heart and other organs.
Data presented here define a regulatory interaction between RA and Pod1 in the control of EPDC differentiation into SM. There is increasing evidence that developmental gene programs that control EPDC lineage specification and differentiation are reactivated with cardiac injury and repair (Smart et al., 2007; Zhou et al., 2011). In regenerating adult zebrafish hearts, EPDCs activate RALDH2 and Pod1/Tcf21 expression following ventricular resection (Kikuchi et al., 2011a; Kikuchi et al., 2011b; Lepilina et al., 2006). In adult mice, RALDH2 and WT1 are upregulated in EPDCs following myocardial infarction (Kikuchi et al., 2011b; Zhou et al., 2011). However, increased expression of Pod1 after myocardial injury has not been reported. If the developmental role of Pod1 is recapitulated with cardiac injury, then Pod1+ cells may represent a progenitor population that could support coronary revascularization. With the increasing emphasis on EPDCs as a source of cells in cardiac repair, it is possible that the RA/Pod1 regulatory interaction could be exploited to promote development of the SM lineage and enhance revascularization after myocardial injury.
Supplementary Material
Highlights.
Pod1/Tcf21, WT1, NFATC1, and Tbx18 are expressed in subsets of EPDCs in the embryo.
Retinoic acid selectively promotes Pod1 and WT1 expression in isolated chick EPDCs.
Pod1 is required to promote EPDC differentiation into fibroblasts in mouse embryos. (85)
Pod1−/− EPDCs prematurely differentiate into smooth muscle on the heart surface.
Pod1−/− lung mesenchymal cells have increased smooth muscle gene expression.
Acknowledgements
We are indebted to Asha Acharya and Michelle Tallquist for valuable reagents and discussion. This work was supported by NIH grants R01HL082716 and R01HL094319 (KEY) and American Heart Association-Great Rivers Pre-doctoral Fellowship Award 09PRE2080186 (CMB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests statement
The authors declare no competing financial interests.
References
- Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21cell lineages in the heart and kidney. Genesis. 2011;49:870–877. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JB. The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 1993;40:85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- Austin AF, Compton LA, Love JD, Brown CB, Barnett JV. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev. Dyn. 2008;237:366–376. doi: 10.1002/dvdy.21421. [DOI] [PubMed] [Google Scholar]
- Azambuja AP, Portillo-Sanchez V, Rodrigues MV, Omae SV, Schechtman D, Strauss BE, Costanzi-Strauss E, Krieger JE, Perez-Pomares JM, Xavier-Neto J. Retinoic acid and VEGF delay smooth muscle relative to endothelial differentiation to coordinate inner and outer coronary vessel wall morphogenesis. Circ. Res. 2010;107:204–216. doi: 10.1161/CIRCRESAHA.109.214650. [DOI] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int. J. Dev. Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bollig F, Perner B, Besenbeck B, Kothe S, Ebert C, Taudien S, Englert C. A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros. Development. 2009;136:2883–2892. doi: 10.1242/dev.031773. [DOI] [PubMed] [Google Scholar]
- Bussen M, Petry M, Schuster-Gossler K, Leitges M, Gossler A, Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18:1209–1221. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Cheek J, Sakthivel B, Aronow BJ, Yutzey KE. Shared gene expression profiles in developing heart valves and osteoblast progenitor cells. Physiol. Genomics. 2008;35:75–85. doi: 10.1152/physiolgenomics.90212.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- Combs MD, Braitsch CM, Lange AW, James JF, Yutzey KE. NFATC1 promotes epicardium-derived cell invasion into myocardium. Development. 2011;138:1747–1757. doi: 10.1242/dev.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. VEGF and RANKL regulation of NFATc1 in heart valve development. Circ. Res. 2009;105:565–574. doi: 10.1161/CIRCRESAHA.109.196469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton LA, Potash DA, Mundell NA, Barnett JV. Transforming growth factor-beta induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev. Dyn. 2006;235:82–93. doi: 10.1002/dvdy.20629. [DOI] [PubMed] [Google Scholar]
- del Monte G, Casanova JC, Guadix JA, MacGrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL. Differential Notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ. Res. 2011;108:824–836. doi: 10.1161/CIRCRESAHA.110.229062. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Dyson E, Sucov HM, Kubalak SW, Schmid-Schonbein GW, DeLano FA, Evans RM, Ross J, Jr, Chien KR. Atrial-like phenotype is associated with embryonic ventricular failure in retinoid X receptor alpha −/− mice. Proc. Natl. Acad. Sci. USA. 1995;92:7386–7390. doi: 10.1073/pnas.92.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrman LA, Yutzey KE. Lack of regulation in the heart forming region of avian embryos. Dev. Biol. 1999;207:163–175. doi: 10.1006/dbio.1998.9167. [DOI] [PubMed] [Google Scholar]
- Fernandes DJ, McConville JF, Stewart AG, Kalinichenko V, Solway J. Can we differentiate between airway and vascular smooth muscle? Clin. Exp. Pharmacol. Physiol. 2004;31:805–810. doi: 10.1111/j.1440-1681.2004.04084.x. [DOI] [PubMed] [Google Scholar]
- Funato N, Ohyama K, Kuroda T, Nakamura M. Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. J. Biol. Chem. 2003;278:7486–7493. doi: 10.1074/jbc.M212248200. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Winter EM, Poelmann RE. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J. Cell. Mol. Med. 2010;14:1056–1060. doi: 10.1111/j.1582-4934.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieskamp T, Rudat C, Ludtke TH, Norden J, Kispert A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ. Res. 2011;108:813–823. doi: 10.1161/CIRCRESAHA.110.228809. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Ruiz-Villalba A, Lettice L, Velecela V, Munoz-Chapuli R, Hastie ND, Perez-Pomares JM, Martinez-Estrada OM. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev. 1998;73:33–43. doi: 10.1016/s0925-4773(98)00031-8. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Langberg JD, Hurtado R, Lee S, Mikawa T. Induction of proepicardial marker gene expression by the liver bud. Development. 2007;134:3627–3637. doi: 10.1242/dev.005280. [DOI] [PubMed] [Google Scholar]
- Jenkins SJ, Hutson DR, Kubalak SW. Analysis of the proepicardium-epicardium transition during the malformation of the RXRalpha−/− epicardium. Dev. Dyn. 2005;233:1091–1101. doi: 10.1002/dvdy.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011a;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011b;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KM, Wagner N, Wagner KD, Wellmann S, Scholz H. The Wilms tumor suppressor Wt1 promotes cell adhesion through transcriptional activation of the alpha4integrin gene. J. Biol. Chem. 2006;281:31930–31939. doi: 10.1074/jbc.M602668200. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech. Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- Landerholm TE, Dong XR, Lu J, Belaguli NS, Schwartz RJ, Majesky MW. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development. 1999;126:2053–2062. doi: 10.1242/dev.126.10.2053. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lin SC, Dolle P, Ryckebusch L, Noseda M, Zaffran S, Schneider MD, Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Natl. Acad. Sci. USA. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev. Dyn. 2004;230:239–250. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. BMP and FGF regulatory pathways control cell lineage diversification of heart valve precursor cells. Dev. Biol. 2006a;292:292–302. doi: 10.1016/j.ydbio.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Lange AW, Yutzey KE. Hearts and bones: shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev. Biol. 2006b;294:292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:9525–9530. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtab EA, Wijffels MC, Van Den Akker NM, Hahurij ND, Lie-Venema H, Wisse LJ, Deruiter MC, Uhrin P, Zaujec J, Binder BR, Schalij MJ, Poelmann RE, Gittenberger-De Groot AC. Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: Correlation with abnormal epicardial development. Dev. Dyn. 2008;237:847–857. doi: 10.1002/dvdy.21463. [DOI] [PubMed] [Google Scholar]
- Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ. Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. USA. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Hatta M, Ohshima T, Ishida J, Fujii R, Nakajima T, Fukamizu A. Cell type-dependent transactivation or repression of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Mol. Cell. Biochem. 2000a;205:141–147. doi: 10.1023/a:1007057611868. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Nakajima T, Fukamizu A. Molecular characterization of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Int. J. Mol. Med. 2000b;5:27–31. doi: 10.3892/ijmm.5.1.27. [DOI] [PubMed] [Google Scholar]
- Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev. Biol. 2001;234:204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Drager UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev. Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev. Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- Plotkin M, Mudunuri V. Pod1 induces myofibroblast differentiation in mesenchymal progenitor cells from mouse kidney. J. Cell. Biochem. 2008;103:675–690. doi: 10.1002/jcb.21441. [DOI] [PubMed] [Google Scholar]
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- Quaggin SE, Vanden Heuvel GB, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech. Dev. 1998;71:37–48. doi: 10.1016/s0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ. Res. 2002;91:761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J, Manner J, Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev. Biol. 2006;295:546–558. doi: 10.1016/j.ydbio.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev. Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Dube KN, Riley PR. Coronary vessel development and insight towards neovascular therapy. Int. J. Exp. Pathol. 2009;90:262–283. doi: 10.1111/j.1365-2613.2009.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ. Res. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucov HM, Murakami KK, Evans RM. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc. Natl. Acad. Sci. USA. 1990;87:5392–5396. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Zhou B, Honor LB, Ma Q, Petryk A, Pu WT. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev. Biol. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheven G, Bothe I, Ahmed MU, Alvares LE, Dietrich S. Protein and genomic organisation of vertebrate MyoR and Capsulin genes and their expression during avian development. Gene Expr. Patterns. 2006;6:383–393. doi: 10.1016/j.modgep.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc. Natl. Acad. Sci. USA. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Zhang Y, Chen Y, Yang N, Wang XJ, Zhu D. Systematic identification of genes involved in divergent skeletal muscle growth rates of broiler and layer chickens. BMC Genomics. 2009;10:87. doi: 10.1186/1471-2164-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.