A quick glance at the recent scientific literature might give the impression that nucleic acid polymerases are suffering from an identity crisis. The last few years have brought us “E. coli DNA polymerase I as a reverse transcriptase” (1), “A mutant T7 RNA polymerase as a DNA polymerase” (2), and, in a recent issue of the Proceedings, “Conferring RNA polymerase activity to a DNA polymerase: A single residue in reverse transcriptase controls substrate selection” (3).
Because polymerases have traditionally been divided into four classes based on their substrate specificities [the use of DNA or RNA templates and deoxyribonucleotides (dNTPs) or ribonucleotides (rNTPs)], the blurring of the distinction between classes might give the impression that an important biological barrier has been breached. That this is not a correct perception is suggested by earlier reports of circumstances in which wild-type polymerases do not discriminate strictly between deoxyribo and ribo substrates. Thus the template specificity of DNA polymerases shows a range of stringency (4), from polymerases with a fairly strict requirement for a DNA template, to reverse transcriptases, which need to use both RNA and DNA templates during the retroviral replication cycle. The rather relaxed specificity of Escherichia coli DNA polymerase I (1) can be thought of as intermediate between these two extremes. In an analogous way, the discrimination by wild-type polymerases against nucleotide substrates with the “wrong” sugar structure is not absolute, and is further relaxed when Mn2+ replaces Mg2+ as the metal cofactor (2, 5–7).
Structural studies support the view that the similarities between polymerases transcend arbitrary divisions into classes on the basis of the use of deoxyribo or ribo templates and nucleotides (for recent reviews, see refs. 8–10). The inescapable conclusion from the six published polymerase structures (11–19), together with a handful of additional structures presented at recent scientific meetings, is that the majority of polymerases belong to a polymerase superfamily and have closely related active sites similarly positioned within a polymerase cleft whose shape has been compared with that of a half open right hand (Fig. 1). Thus far, the only exception to this generalization is mammalian DNA polymerase β, which is now recognized to be more appropriately assigned to a related but distinct family of nucleotidyl transferases (22, 23). Thus it appears that there is a generic polymerase module that provides the active site architecture to carry out the phosphoryl transfer reaction, and that subtle modifications to this module achieve the substrate specificities characteristic of each polymerase class.
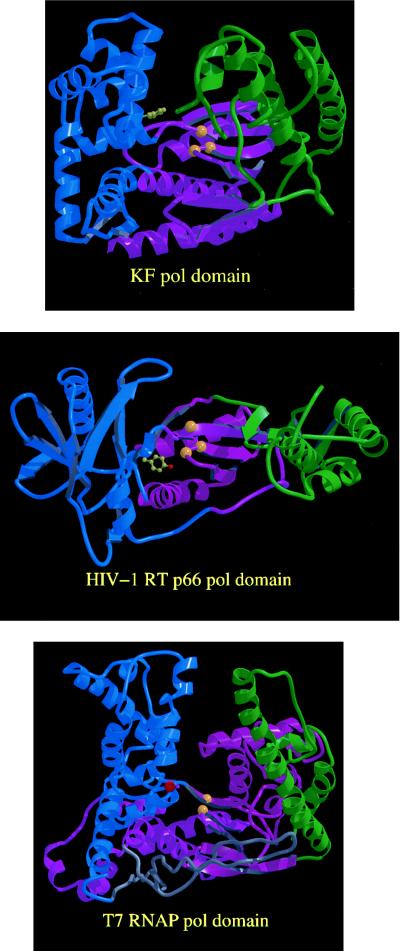
Figure 1.
The backbone crystal structures of the polymerase domains of Klenow fragment, HIV-1 reverse transcriptase (RT), and T7 RNA polymerase. In each case the palm subdomain is colored purple, the thumb green, and the fingers blue. The three structures are positioned such that the active site regions of the palm subdomains (with yellow spheres showing the α-carbon positions of the active-site carboxylates) have similar orientations. The locations of residues that influence recognition of the sugar portion of the incoming nucleotide are indicated. Phe-762 of Klenow fragment and Tyr-115 of HIV-1 reverse transcriptase [the homolog of Phe-155 in Moloney murine leukemia virus (MoMLV) reverse transcriptase] are shown in ball-and-stick representation; in T7 RNA polymerase the α-carbon of Tyr-639 is marked by a red sphere. This figure was created by Jimin Wang (Yale University) using raster3d (20, 21).
Although the determinants of template specificity in polymerases remain unclear, as does the mechanism by which Mn2+ perturbs nucleotide specificity, three recent studies have provided new insights into the way in which polymerases select a nucleotide substrate with the appropriate sugar structure. In each case a polymerase mutant that affects nucleotide specificity has been obtained. The paper by Gao et al. (3) in a recent issue of the Proceedings describes a reverse transcriptase mutant that can use rNTPs; previous papers identified a mutation that allows the incorporation of dNTPs by an RNA polymerase (2), and a side chain that influences the incorporation of dideoxyNTPs (ddNTPs) by DNA polymerases of the polymerase I (pol I) family (24). Together, these papers define a region on the polymerase cleft that monitors the 2′ and 3′ substituents on the sugar of the incoming nucleotide. In each case the phenotype of the mutant protein is a loss of specificity and not a complete specificity switch. This is what would be expected if the mutation removed a discriminator mechanism. It is also important to realize that the mutations under consideration cause a relaxation in stringency in the nucleotide incorporation step only. Neither in reverse transcriptase nor in RNA polymerase does the mutant enzyme work particularly well when synthesizing a product composed entirely of the “wrong” substrate. Presumably additional barriers are encountered when adding nucleotides to a primer terminus having the wrong sugar structure or when trying to accommodate a product duplex that is not of the expected kind. Thus, these mutant proteins could be thought of as the first step toward polymerases having novel specificities.
It is not hard to imagine a plausible mechanism that would allow DNA polymerases to select dNTPs in preference to rNTPs; since the unwanted substrate has a larger substituent at the 2′ position (OH vs. H), a simple steric “gate” will suffice. This is borne out by the reverse transcriptase study of Gao et al. (3), which examines a hypothesis proposed on the basis of the three-dimensional structure of an active fragment of the polymerase domain of the MoMLV reverse transcriptase (19). Model-building of substrates onto this structure, using information from the complex of HIV-1 reverse transcriptase with DNA (13) and from the ternary complex of DNA polymerase β with DNA and nucleotide substrates (25), identified a side chain, Phe-155, positioned such that it might clash with the 2′OH of a ribonucleotide substrate. Gao et al. (3) showed that replacement of Phe-155 with a smaller side chain (Val) does indeed have the predicted effect; dNTPs are preferred over rNTPs by 10,000-fold in the wild-type enzyme, but by only 30-fold in the F155V mutant enzyme (taking the ratio of Vmax/Km values reported for each enzyme).
Phe-155 of MoMLV reverse transcriptase is within a highly conserved region of the protein sequence, just five residues C-terminal to the invariant active-site aspartate of “motif A” in the unified polymerase alignment of Delarue et al. (26). Only Phe or Tyr are found at this position in reverse transcriptases (27) (Fig. 2). In HIV-1 reverse transcriptase, the corresponding residue, Tyr-115, has also been mutated to valine (31); however the mutant was not characterized in sufficient detail to establish whether dNTP/rNTP discrimination is affected. Modeling a dNTP into the HIV-1 reverse transcriptase–DNA complex has led Arnold and his colleagues (32, 33) to propose that Tyr-115 may interact directly with the base of the incoming dNTP and may also form part of the binding site for the sugar moiety.
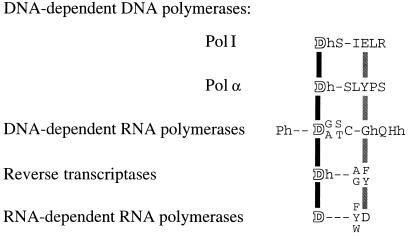
Figure 2.
Alignment of the conserved sequence motif A (26) of nucleic acid polymerases. The consensus motifs for each polymerase family are based on published compilations for the pol I (26, 28), and pol α (26, 29) families, for the single subunit DNA-dependent RNA polymerases (30), and for RNA-dependent polymerases (27). The motif is aligned on the invariant aspartate residue (white outlined letters, connected by a black vertical bar). The positions that align with Phe-155 of MoMLV reverse transcriptase are connected by a shaded vertical bar. Positions that are almost invariably occupied by a hydrophobic amino acid are designated “h;” hyphens indicate nonconserved positions.
Examination of the motif A sequences in other polymerase classes reveals some fascinating coincidences (Fig. 2). An obvious caveat is that it is risky to equate sequence position with structural location; nevertheless, the rather extended conformation of motif A in the known polymerase structures may make this a reasonable initial approximation. If Phe-155 does indeed provide a steric gate that blocks the entry of rNTPs, then one would expect to find residues fulfilling a similar function in other DNA polymerases. In the pol I family of DNA polymerases, the analogous position in motif A is occupied by an invariant glutamate; moreover, in Klenow fragment, replacement of this Glu by a smaller side chain likewise decreases discrimination against ribonucleotides (M. Astatke and C.M.J., unpublished work). In the pol α family, the corresponding position is an invariant tyrosine. Mutations at this position, Tyr-865, in human DNA pol α affect polymerase fidelity and sensitivity to inhibitors directed to the dNTP binding site, implicating this side chain in some kind of interaction with the incoming dNTP (34). A final prediction that can be made is that RNA polymerases should lack an analogous steric gate. Although this is hard to assess in the absence of structural information, the sequence data could be construed as consistent with this prediction. The RNA replicase sequences in particular show an interesting distinction from the reverse transcriptase sequences. Although the majority have an aromatic side chain (Phe, Tyr, or Trp) within motif A, it is only four residues C-terminal to the aspartate, compared with five in reverse transcriptases.
The other two papers mentioned above (2, 24) deal with discrimination by a polymerase against a nucleotide that is smaller than the normal substrate (rNTP vs. dNTP, and dNTP vs. ddNTP, respectively). In these cases steric exclusion is clearly not an option, and the most reasonable hypothesis is that the normal substrate should be able to make an interaction that is not possible for a smaller molecule. A precedent for this type of model is provided by the work of Blow and Fersht and their colleagues (35, 36) on tyrosyl-tRNA synthetase, where the specific interactions that are thought to ensure the selection of tyrosine over phenylalanine can be seen in the three-dimensional structure. This example is particularly apt for the present discussion of polymerase specificity since here, too, the discrimination is between OH and H groups.
Sousa and Padilla (2) have described a mutation, Y639F, in T7 RNA polymerase that allows the incorporation of dNTPs as well as rNTPs. An obvious initial hypothesis is that the hydroxyl of Tyr-639 could provide the discriminator interaction with the ribose 2′ hydroxyl, and that the phenylalanine substitution, by removing this interaction, results in the observed loss of specificity. However, a closer examination of the kinetic data suggests that the actual situation may be more complex; the Y639F mutation lowers specificity for the ribonucleotide by making the dNTP a better substrate, rather than by making the rNTP a worse substrate (as would be expected if Y639F were a simple loss-of-contact mutation). Another curious circumstance is that all DNA-dependent polymerases, regardless of whether their nucleotide substrate is dNTP or rNTP, have a tyrosine in the sequence position corresponding to Tyr-639 of T7 RNA polymerase (26). In Klenow fragment, the homologous residue, Tyr-766, which occupies a structurally equivalent position at the C terminus of a long helix in the fingers subdomain (11, 14), is implicated in various aspects of the enzyme–dNTP interaction, including dNTP insertion fidelity (37–39), though the available evidence suggests that this may be via an interaction with the DNA template (40–42). Presumably the immediate structural environment of these homologous tyrosine residues must be sufficiently different in DNA and RNA polymerases, so that the tyrosine functions as a discriminator against deoxynucleotides in RNA polymerases only.
In the pol I family of polymerases, a neighboring residue (Phe-762 in Klenow fragment), on the same α-helix as the invariant tyrosine discussed above, is also implicated in sugar selectivity, this time at the 3′ position of the ribose ring. Almost all the polymerases in the pol I family have either a phenylalanine or a tyrosine at this position (40), and, in every case that has been studied, polymerases with a Phe discriminate strongly against dideoxynucleotides, while those with a Tyr are comparatively permissive. Moreover, the specificity of a polymerase can be switched simply by mutating this residue (24, 43). As in the RNA polymerase case discussed above, the natural substrate has the larger side chain (3′OH vs. H); however the effect of changes in the protein side chain is the opposite, in that it is Tyr, not Phe, that causes a relaxation in specificity. A simple loss-of-contact explanation cannot apply here since it is hard to imagine any contact made by Phe that cannot also be made by Tyr. A plausible model invoked by Tabor and Richardson (24) to explain the data is that the dNTP 3′OH normally interacts with another group, “X,” within the ternary complex, and that this interaction can be satisfied by the phenolic OH of the Tyr side chain if there is no 3′OH on the nucleotide substrate. Tabor and Richardson propose that “X” may be one of the active site metal ions, though other possibilities are not excluded by the available data.
The three papers discussed above address the question of nucleotide recognition at the 2′ and 3′ positions of the ribose ring. Fig. 1 shows that the side chains implicated in this process in three polymerases from different subfamilies all lie within the same small portion of the polymerase cleft, the junction between fingers and palm close to the catalytic site. This supports existing data implicating this junction region in nucleotide binding (44), and now identifies a small part of this region as a possible sugar recognition determinant. An interesting aspect of the results is that the discriminators appear to be structurally rather simple, since changes in a single side chain virtually abolished specificity in each of the three polymerases studied. Further studies, including structure determinations of appropriate enzyme–substrate complexes, will be necessary to understand the details of the recognition process. The problem of sugar recognition by polymerases also serves to focus attention on the more general question of how enzymes distinguish between competing substrates, especially in the situation when the unwanted substrate is smaller than the natural substrate. The two examples discussed above certainly suggest that the process of excluding a smaller substrate may be more complex than simple models would lead us to expect.
Acknowledgments
I am very grateful to Nigel Grindley and Tom Steitz for helpful discussions, and to Jimin Wang for preparing Fig. 1.
References
- 1.Ricchetti M, Buc H. EMBO J. 1993;12:387–396. doi: 10.1002/j.1460-2075.1993.tb05670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa R, Padilla R. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornberg A, Baker T A. DNA Replication. San Francisco: Freeman; 1992. [Google Scholar]
- 5.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van de Sande J H, Loewen P C, Khorana H G. J Biol Chem. 1972;247:6140–6148. [PubMed] [Google Scholar]
- 7.Ide H, Yagi R, Yamaoka T, Kimura Y. Nucleic Acids Res Symp Ser. 1993;29:133–134. [PubMed] [Google Scholar]
- 8.Arnold E, Ding J, Hughes S H, Hostomsky Z. Curr Opin Struct Biol. 1995;5:27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- 9.Joyce C M, Steitz T A. J Bacteriol. 1995;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa R. Trends Biochem Sci. 1996;21:186–190. [PubMed] [Google Scholar]
- 11.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 12.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 13.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sousa R, Chung Y J, Rose J P, Wang B C. Nature (London) 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 15.Davies J F, Almassy R J, Hostomska Z, Ferre R A, Hostomsky Z. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 16.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Eom S H, Wang J, Lee D-S, Suh S W, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 18.Korolev S, Nayal M, Barnes W M, Di Cera E, Waksman G. Proc Natl Acad Sci USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure (London) 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 20.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 21.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 23.Martin G, Keller W. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 24.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 26.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 27.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K-I, Korn D, Hunkapiller M W, Wang T S-F. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAllister W T, Raskin C A. Mol Microbiol. 1993;10:1–6. doi: 10.1111/j.1365-2958.1993.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 31.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 32.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 33.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D, Jr, Raag R, Nanni R G, Hughes S H, Arnold E. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 34.Dong Q, Copeland W C, Wang T S-F. J Biol Chem. 1993;268:24163–24174. [PubMed] [Google Scholar]
- 35.Brick P, Blow D M. J Mol Biol. 1987;194:287–297. doi: 10.1016/0022-2836(87)90376-7. [DOI] [PubMed] [Google Scholar]
- 36.Fersht A R, Shi J-P, Knill-Jones J, Lowe D M, Wilkinson A J, Blow D M, Brick P, Carter P, Waye M M Y, Winter G. Nature (London) 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 37.Polesky A H, Steitz T A, Grindley N D F, Joyce C M. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 38.Carroll S S, Cowart M, Benkovic S J. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 39.Bell, J. B., Eckert, K. A., Joyce, C. M. & Kunkel, T. A. (1997) J. Biol. Chem. 272, in press. [DOI] [PubMed]
- 40.Astatke M, Grindley N D F, Joyce C M. J Biol Chem. 1995;270:1945–1954. doi: 10.1074/jbc.270.4.1945. [DOI] [PubMed] [Google Scholar]
- 41.Beese L S, Derbyshire V, Steitz T A. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 42.Eom S H, Wang J, Steitz T A. Nature (London) 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 43.Mizrahi V, Huberts P. Nucleic Acids Res. 1996;24:4845–4852. doi: 10.1093/nar/24.24.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]