Abstract
Experimental studies in hemeproteins and model Tyr/Cys-containing peptides exposed to oxidizing and nitrating species suggest that intramolecular electron transfer (IET) between tyrosyl radicals (Tyr-O●) and Cys residues controls oxidative modification yields. The molecular basis of this IET process is not sufficiently understood with structural atomic detail. Herein, we analyzed using molecular dynamics and quantum mechanics-based computational calculations, mechanistic possibilities for the radical transfer reaction in Tyr/Cys-containing peptides in solution and correlated them with existing experimental data. Our results support that Tyr-O● to Cys radical transfer is mediated by an acid/base equilibrium that involves deprotonation of Cys to form the thiolate, followed by a likely rate-limiting transfer process to yield cysteinyl radical and a Tyr phenolate; proton uptake by Tyr completes the reaction. Both, the pKa values of the Tyr phenol and Cys thiol groups and the energetic and kinetics of the reversible IET are revealed as key physico-chemical factors. The proposed mechanism constitutes a case of sequential, acid/base equilibrium-dependent and solvent-mediated, proton-coupled electron transfer and explains the dependency of oxidative yields in Tyr/Cys peptides as a function of the number of alanine spacers. These findings contribute to explain oxidative modifications in proteins that contain sequence and/or spatially close Tyr-Cys residues.
Keywords: Electron Transfer, Tyrosyl Radical, Oxidation, Nitration, Computer Simulation
Introduction
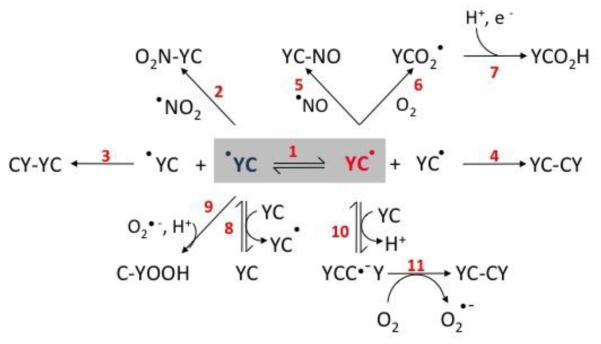
Tyrosine (Tyr) and cysteine (Cys) are key target residues in proteins for free radical-dependent post-translational modifications associated to oxidative stress conditions (1). Indeed, both amino acids can be oxidized by one-electron oxidants such as hydroxyl radical (●OH), nitrogen dioxide (●NO2), carbonate radical (CO3●−), peroxyl radicals (ROO●) and oxoferryl complexes to yield the corresponding amino acid-derived radicals (2-5), namely tyrosyl (Tyr-O●) and cysteinyl (Cys-S●) radicals, respectively. In proteins, Tyr-O● and Cys-S● are transient species and typically evolve to a variety of products including, 3,3′-dityrosine, 3-nitrotyrosine, cysteine disulfide, S-nitrosocysteine and cysteine sulfinic acid (1, 6) and/or participate in inter- or intra-molecular electron transfer (IET) processes (7-12) as schematically shown in Figure 1.
Figure 1. Electron transfer and oxidation reactions in Tyr-Cys-containing peptides.
The figure shows reactions involved in uni- and bimolecular processes related to radical-mediated oxidations in Tyr-Cys-containing peptides in the presence of oxygen, nitric oxide and nitrogen dioxide.The numbers in red indicate discrete reactions as follows. 1. Intramolecular electron transfer; 2. Combination reaction between tyrosyl radical and ●NO2 to yield 3-nitrotyrosine peptide; 3. Tyrosyl radical dimerization yielding 3,3′-di-tyrosine peptide; 4. Thiyl radical dimerization yielding disulfide peptide; 5. Combination reaction between cysteinyl radical and ●NO to yield S-nitrosocysteine peptide; 6-7. Thiyl radical reaction with oxygen to yield thiylperoxyl radical peptide (6) and subsequent rearrangement and reduction tosulfinic acid derivative (7); 8. Intermolecular electron transfer; 9. Reaction between tyrosyl radical and superoxide yielding tyrosine hydroperoxide; 10-11. Reaction of cysteinyl radical peptide with tyrosine-cysteine peptide to yield the disulfide radical anion (10), followed by the reduction of molecular oxygen to yield O2●- and the corresponding disulfide peptide (11).
In most cases only a few residues in proteins are oxidatively modified in vivo by free radical reactions, although the molecular bases of the selectivity are far from being understood (13, 14). In this regard, in spite of the initial sites of free radical attack in a protein, the formation and relative stability (or half-life) of Tyr-O● and Cys-S● seem to represent key factors controlling the final sites and yields of protein oxidative modifications. Particularly important is the fact that in peptides (8-10, 15, 16) and proteins (17-20) electron transfer processes (either inter or intramolecular) involving tyrosine and/or cysteine residues have been observed. Early work has shown that the redox potential for theTyr/Tyr-O● and Cys/Cys-S● couples are quite close: tyrosine has a one-electron redox potential value of 0.93-0.94 V at pH 7.0 and 25°C, while that of cysteine is slightly higher (i.e. ΔE°<+10 mV at pH 7.0) (10, 21-23). Similarly, the Keq for the one-electron oxidation reaction of GSH by Tyr-O● at pH 7.15 has been recently reported to be close to one (24). Nonetheless, the one-electron redox potentials of the amino acids such as Tyr and Cys within proteins can significantly differ, depending on factors that include local structure and solvent accessibility, among others (21, 25, 26).
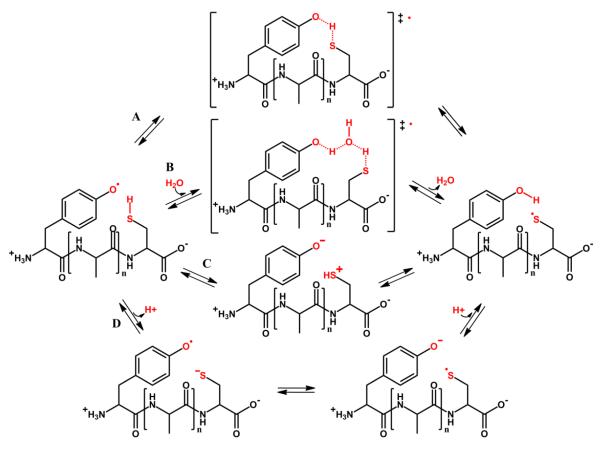
The presence of a Cys residue in the proximity of a Tyr residue has been reported to facilitate radical transfer in a way that Cys usually acts as a radical sink. For instance, hydrogen peroxide-mediated oxidation of myoglobin produces Cys100 thiyl radicals via an intermolecular electron transfer from Cys110 to Tyr103-phenoxyl radicals of another myoglobin molecule, ultimately yielding homodimers with disulfide bond formation (18, 19); similarly, peroxynitrite-mediated oxyhemoglobin oxidation results in the formation of thiyl radicals at Cys93 with the intermediacy of Tyr42 and Tyr24 phenoxyl radicals (19), and the formation of disulfide and dityrosine intermolecular cross-links (17). The role of Tyr-Cys IET in ribonucleotide reductase catalysis has been largely explored and established (20). Indeed, experimental studies performed in model peptides in aqueous solution, containing both Cys and Tyr residues suggest that rapid IET between the Tyr-O● and Cys residue controls the extent of Tyr nitration, with concomitant Cys oxidation and/or nitrosation, indicating the presence of a Cys-S● radical intermediate (16). These results were also related to the fact that many reported S-nitrosated mitochondrial proteins, share a Tyr and Cys sequence (27), highlighting the importance of the IET between both residues. A first insight into the modulation of tyrosine nitration by cysteine via IET mechanisms was determined by comparing the nitration yield of YAnC peptides with increasing amount of Ala spacers, which showed a decrease in the yield of nitrated peptides with increasing number of Ala spacers (16). The IET reaction between Tyr and Cys residues in the same peptide can potentially occur through different mechanisms which at the atomic level could be described as A, B, C and D in Figure 2.
Figure 2. Possible mechanisms for the intramolecular electron transfer (IET) reaction in the Tyr-Cys-containing peptides.
The figure shows four possible mechanisms studied in the present work. A. Involves a direct hydrogen- or time-concerted proton-coupled electron transfer (PCET) as described by reaction 2. B. Involves a water-bridged time concerted PCET as described by reaction 3. C. Consists of a first IET, i.e. Cys oxidation (reaction 4) followed by proton release and uptake steps (reaction 5) and D. involves the acid-base equilibrium of both residues and IET in the charged peptides in a solvent mediated non concerted PCET, as described by reactions 6, 7 and 8. The structures indicated in square brackets and the ‡symbol, correspond to proposed transition states.
Overall, the forward reaction consists of the transfer of the radical moiety from to the Cys residue, resulting in Cys-S● and an unmodified Tyr residue (Reaction 1). The general reaction could be described as either a hydrogen atom transfer (HAT), or a proton-coupled electron transfer (PCET) as defined in its broadest sense following Hammes-Shiffer and Soudackov́s work (28) (i.e. involving movement of a proton and an electron through different bonds in either a time-concerted or sequential manner):
| [1] |
Briefly, in Mechanism A the reaction is proposed to occur through a direct hydrogen-(●H), or time-concerted PCET directly from the Cys residue towards the Tyr-O●, which relies on the possible intramolecular hydrogen bond (HB) formation between Tyr and Cys residues to transfer the hydrogen or proton. Mechanism B involves a concerted IET, but with a proton transfer through a bridging water molecule, while in Mechanism C, IET occurs first, reducing the Tyr-O● radical to phenolate (Tyr-O−), with concomitant Cys oxidation, which later results in proton release. Finally, Mechanism D, involves the proton and IET steps significantly separated in time, resulting in an acid-base equilibrium-dependent three step reaction involving Cys deprotonation, IET in the charged peptide, and TyrO− protonation (Fig. 2, referred as Step 1D, 2D and 3D, respectively).
All the involved reactions can be reversible and display different thermodynamic and and kinetic properties that could be affected by the relative position of Tyr and Cys residues within the protein and their environment.
In this work we have used molecular-based computer simulation techniques (molecular dynamics and quantum mechanics-based calculations) to study the possible mechanisms of radical transfer (or IET) involving tyrosyl and cysteinyl residues in model peptides at the atomic level. To this end, we have analyzed each of the four mechanisms presented above in four peptides containing Tyr and Cys residues separated by different number of Ala spacers in explicit water, that allow to have a direct comparison with recent experimental data. Our results show that the radical transfer reaction is most likely to occur by Mechanism D, which involves an acid/base equilibrium, followed by a fast IET that can be described as a solvent-mediated, PCET reaction
Computational Methods
System setup, parameters and molecular dynamic (MD) simulations
For the present work we firstly built four different size peptides, in each of five possible charge or protonation states, corresponding to: TyrOH-(Ala)n-CysSH, TyrO●-(Ala)n-CysSH, TyrOH-(Ala)n-CysS●, TyrO●-(Ala)n-CysS− and TyrO−-(Ala)n-CysS● for n = 0, 1, 2 and 4. The amino terminal of all peptides was capped with an acetic acid and the carboxy terminal with a methylamine, through the corresponding peptide bonds. Classical molecular dynamics simulations for the peptides were performed in explicit solvent using the TIP3P water model. The simulated water box had a volume of ca. 65 nm3 and between 800 and 1,500 water molecules. All MD simulations were performed using the AMBER force field with the PARM99 set of parameters (29). The point charges for Tyr-O● and Tyr-O− were obtained using the restricted electrostatic potential (RESP) formalism (30) with the optimized structures computed with HF/6-31G* using the Gaussian-03 program (31). The same procedure was applied to determine point charges for Cys-S● and negatively charged Cys-S− residues.
All simulations were performed using the Periodic Boundary Conditions approximation and the Particle Mesh Ewald (PME) summation method with a grid spacing of 1 Å for treating long range electrostatic interactions, while a direct cut-off distance of 10 Å was used for direct interactions. The Shake method was used to constraint the H atoms at their equilibrium distance allowing the use of 2 fs time step. The Berendsen thermostat was used to keep the temperature constant at 300 K (32). All MD simulations were performed with the AMBER program package (33). The equilibration protocol for all peptides consisted of heating the optimized structures from 0 to 300 K gently in 0.12 ns, while the volume of the systems was kept constant. Then, constant isotropic pressure simulations were performed for 0.08 ns followed by other 12 ns of simulation at constant pressure.
Thermodynamic integration
Thermodynamic Integration (TI) scheme (32) as implemented in the AMBER packages (33) was used for computing the Gibbs free energy differences for the following reactions: i) The change in the solvation Gibbs free energy difference for the overall reaction (Reaction 1) and ii) The change in the solvation Gibbs free energy for the charge transfer reaction in the charged peptides (Step 2D). The method consists in slightly changing the system (in this case the peptide) as defined by the classical force field parameters from those corresponding to the initial, to the final state. The change is performed by varying the coupling parameter ζ in eleven discrete steps, from ζ =0 to ζ =1. For each reaction, 11 windows corresponding to ζ values of 0.01, 0.1, 0,2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 and 0.99 were simulated, and for each one at least a 2ns long MD was performed, or until convergence of the corresponding <δE/δζ> value for each window. The Gibbs free energy was then obtained by numerically integrating the <δE/δζ>vs ζ curve.
Quantum mechanics (QM)-based calculations
QM calculations were performed in vacuum and in implicit solvent with the B3LYP(34, 35) functional and 6-31G* basis set using the Gaussian-03 program (31). Compared to other density functional methods, B3LYP has shown good performance for the study of intramolecular addition of Cys-S● to Phe and Tyr residues (35). The different steps of the catalytic cycle of cytochrome oxidase, which include electron and proton or hydrogen transfer reactions have also been successfully studied using this level of theory (34, 36-38), while the experience using Density Functional Theory methods for PCET in metallo-enzymes was recently reviewed (39). The implicit solvation methods correspond to the Polarizable Continuum Model (PCM) as developed in (40). All QM calculations were performed only for the corresponding peptides in vacuum. For selected cases, QM calculations were also performed with the explicit inclusion of a small number of water molecules in the system. In all cases no significant differences were obtained in the observed trends. Given the known drawbacks (41) concerning the use of implicit solvation methods, especially when direct interactions with water molecules are present as in the case of charged peptides, for several reactions we also computed the change in the solvation Gibbs free energy along the reaction using classical explicit solvation methods and TI scheme as described above. These methodologies have the advantage of explicitly taking into account the solute solvent interaction and the entropy associated with water reorganization (42).
ET coupling matrix elements calculation using the Pathways algorithm
According to Marcus theory, the ET rate (kET) depends on the coupling matrix between donor and acceptor TDA, the reorganization energy (λ) and the reaction Gibbs free energy (ΔG°) as described by the following equation (43):
| (Equation 1) |
The electronic coupling matrix, usually referred as TDAbetween donor and acceptor for the TyrO●−(Ala)n-CysS- and TyrO−-(Ala)n-CysS● peptides were estimated using the pathways algorithm developed by Beratan et al. (44, 45) using the default parameters. The TDA is related to the probability of ET transfer and directly involved in the resulting ET rate, as described by Marcus theory. Briefly, the pathways method looks for the best possible path, the one with the highest TDA, that connects donor and acceptor selected orbitals. Orbitals are defined as either located on covalent bonds, or in free electron pairs as in oxygen atoms. This results, for example, in a water molecule displaying four orbitals with tetrahedral geometry, two corresponding to the O-H bonds and two corresponding to the free electron pairs. TDA for a given pathway is computed as the product of a number of steps, each with a given coupling value, that define the pathway. A step can either be through atom, when two connected orbitals share the central atom, which are assumed to have a coupling value of 0.6, or through space steps (or jumps) connecting orbitals separated by empty space, for which the coupling is assumed to decay exponentially with the orbital to orbital distance with a decay factor (β=−1.7). As a result of the different structures and ET paths, ET rates can vary as much as two orders of magnitude for the same donor acceptor distance. The TDA was computed between the Tyr-O and Cys-S located orbitals for a hundred snapshots taken from the corresponding peptide MD simulation. For each case, the average TDA along the whole MD is reported. Detailed visual inspection of the ET path allowed determination of the structural type of path involved.
Results
The results are organized as follows: first, the thermodynamics of the overall radical transfer reaction is studied in vacuum and in water (Reaction 1), for each of the different Ala spacer-containing peptides. Then, all steps for each possible mechanism are comparatively studied for all peptides in aqueous solution.
Overall reaction
We began studying the overall reaction (Reaction 1) using both QM and MD simulations. We divided the reaction in two parts. Firstly, the intrinsic (or internal) energy of the ET reaction for isolated Tyr or Cys (N and O acetyl capped) residues in vacuum and in implicit water (using PCM approximation) were computed using the B3LYP method, as a measure of the intrinsic radical stabilization capacity of each residue. This reaction would then correspond to an intermolecular reaction analyzed for comparative purposes. The results show that the reaction is only slightly exergonic (−0.1 and −1.6 kcal/mol for vacuum and implicit water respectively) showing that the radical is similarly stabilized in both residues with a slight preference for isolated Cys-S● in water.1
Afterwards, we computed the overall reaction in vacuum and in implicit water, starting from four random selected snapshots (corresponding to different conformations), two for each TyrO●-(Ala)n-CysSH and two for each TyrOH-(Ala)n-CysS● (n=0, 1, 2, 4) peptide (obtained from explicit water MD simulations) which therefore corresponds to the intramolecular reaction. The data in Table I for the averaged peptides/snapshots show that the reaction is also exergonic (with a mean of ≈ −6.2 and −6.4 kcal/mol in vacuum and in implicit water, respectively), further supporting the higher stabilization of the radical in the Cys residue. No significant dependence on the number of Ala spacers was observed. Moreover, the reactions in vacuum and in implicit water did not show significant differences among them. Interestingly, the reaction was more favorable in the peptides (intramolecular) than for the isolated amino acid residues (intermolecular), probably reflecting that in the peptides the radical located on the Cys thiol group, is stabilized by the larger size of the system.
Table I. Reaction 1 associated energy changes (kcal/mol) computed for four randomly selected snapshots/conformations of each peptide in vacuum and in implicit water.
| n = 0 | n = 1 | n = 2 | n = 4 | |
|---|---|---|---|---|
| Vacuum | −6.4 ± 6.0 | −4.4 ± 2.2 | −7.1 ± 4.0 | −6.8 ± 2.7 |
| Implicit Water | −7.1 ± 6.6 | −4.8 ± 3.4 | −6.4 ± 2.1 | −7.2 ± 2.8 |
In a second step, we computed the change in the solvation Gibbs free energy difference (ΔGSV) between both states (TyrO●-(Ala)n-CysSH andTyrOH-(Ala)n-CysS●), for all peptides using explicit water MD simulations, which provide a better description of the specific water solute interactions as compared to the implicit method. The corresponding value was computed using a thermodynamic integration (TI) scheme (See Methods). Briefly, the method consists in slightly changing the peptide parameters from those corresponding to the TyrO●-(Ala)n-CysSH state, to those for the TyrOH-(Ala)n-CysS● state. In this case the change corresponds to switching the corresponding partial charges of the Tyr and Cys residues, and removing the Cys-SH hydrogen Lennard-Jones parameters, which are slowly created in the position corresponding to that of Tyr-OH. The calculation yielded values of −9 ± 3 kcal/mol for all the peptides, showing that solvation contribution to the reaction is exergonic. No significant differences were observed for the different peptides.
Mechanism A
We began our mechanistic analysis by looking at the dynamic structure of theTyrO●-(Ala)n-CysSH and TyrOH-(Ala)n-CysS● (n=0, 1, 2 and 4) peptides in explicit water from a 10 ns long MD simulations of each peptide. The general observation is that all the studied peptides show no appreciable structure, and move forming and breaking intramolecular and peptide-water hydrogen bonds (HB) contacts during the time scale of the simulation. In order to analyze the likeliness of HB formation between the Tyr and Cys residues, we measured the distance probability functions between the Tyr-O● oxygen atom and the Cys thiol hydrogen and that between tyrosine phenol hydrogen and the Cys-S● sulfur atoms in each peptide (the corresponding plots are shown in Supplementary Material, Fig. S1). We also computed the amount of time that a HB interaction is present between the above-described atoms (Table II). An HB was considered established whenever the O-S distance was less than 3.5 Å and the O-H-S angle larger than 140 degrees.
Table II. Internal HBoccupancy(in %) between the TyrOH/O● and CysSH/S● groups for all Tyr-Cys containing peptides.
| Peptide | n = 0 | n = 1 | n = 2 | n = 4 |
|---|---|---|---|---|
| TyrO●-(Ala)n-CysSH | 0.00 | 0.26 | 0.06 | 0.02 |
| TyrOH-(Ala)n-CysS● | 0.00 | 0.02 | 0.02 | 0.00 |
The analysis of the distance probability plots show that only for the peptide containing one Ala spacer the Tyr phenolic hydrogen (Tyr-HOH) or Cys sulfur hydrogen (Cys-HSH) can be found relatively close (less than 3Å) to the radical atoms although with very low probability (and even less probability for the two Ala spacer). In the YC dipeptide (i.e. without Ala spacers) the hydrogen and radical atoms cannot come closer than 3Å, while for the four Ala peptides they are always further than 5Å. This behavior is further reflected in the data from Table II, showing that conformations in which an internal HB is established are extremely unlikely to be found.
To get some insight into the kinetics of a possible hydrogen- or time-concerted PCET reaction for the most likely structures, we selected one snapshot for each peptide where Tyr and Cys residues are establishing an HB or are as close as possible, and computed the corresponding reaction energy profile with the scan of the HB donor hydrogen atom difference distance to both the Tyr-O●and Cys-S●atoms as the reaction coordinate. The results for the overall reaction (ΔE) and activation (E#) energies are summarized in Table III.
Table III. Energy difference (ΔE, (kcal/mol)) and activation energy (E‡, (kcal/mol)) for the PCET reaction in vacuum. The forward reaction corresponds to PCET from CysSH to TyrO●, and the backward reaction from TyrOH to CysS●.
| Forward reaction | Backward reaction | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| n | 0 | 1 | 2 | 4 | 0 | 1 | 2 | 4 |
| ΔE (kcal/mol) | −3.27 | 1.3 | −2.37 | n.c | 3.21 | −2.07 | 0.68 | 0.36 |
| E‡(kcal/mol) | 17.7 | 10.7 | 10.53 | n.c | 24.09 | 9.88 | 10.92 | 11.00 |
The results show that the ΔE for reaction 2 depends considerably on the selected snapshot, but is always smaller that the value obtained for overall computed reaction 1, described previously. This possibly reflects the fact that selected snapshots are biased for structures with HB between donor and acceptor atoms and therefore the hydrogen is similarly stabilized by both residues. Moreover, the computed barriers for Hydrogen/PCET were quite high (> 10 kcal/mol) and no trend is observed for increasing number of alanine spacer residues. The same results were obtained when including one or two waters molecules explicitly in the QM calculation, showing that the barriers are not affected by the presence of the solvent (data not shown). Taken together the fact that almost no structure is found with Tyr-O● and Cys-SH groups (or the Tyr-OH and Cys-S● groups) close or forming an HB, and even for the HB establishing conformations, the relative high barrier for hydrogen or concerted and direct PCET transfer, argue against this mechanism as being operative in the studied peptides.
A note should be made concerning the computed barrier for hydrogen-transfer or PCET. The barriers computed in the present work do not consider the quantum nature of the transferring atom. Protons and hydrogen atoms could also be transferred by quantum tunneling. Many theoretical studies have been devoted to this issue including calculations in enzymes (28, 46-48). However, analysis of the emblematic cases in comparison to the structures of the peptides show that for the present case donor and acceptor atoms are never found close enough (e.g. < 2 Å) to allow such type of transfer to be significant.
Mechanism B
This possibility involves a concerted IET, with a proton transfer through a bridging water molecule, i.e the proton is transferred from the Cys to a water molecule that transfers its proton to Tyr, while ET occurs simultaneously. To analyze this possibility for the above-described peptides, we studied the probability of finding water molecules bridging the Tyr-O and Cys-S interaction in the TyrO●-(Ala)n-CysSH and TyrOH-(Ala)n-CysS● peptides. For this sake we computed the probability that one, two or three water molecules were found to be simultaneously HB Tyr-O to Cys-HS and Tyr-HOH and Cys-S atoms, and considering always the non radical residue as being the HB donor. No structure containing water molecules bridging both reactants was found (not shown), suggesting that water-mediated PCET is highly unlikely.
Mechanism C
In this case IET is proposed to occur first, reducing the Tyr-O● radical to phenolate (Tyr-O-), with concomitant Cys oxidation.Afterwards, the proton is released from the resulting cysteine cation radical (Cys-SH●+) and uptaken by the Tyr-O-, either intramolecularly or mediated by the water solvent. To analyze this possibility, we computed the energy associated with the proposed IET. The results obatined from QM calculations for the isolated residues in vacuum and in implicit solvent, yield ΔE values of 135.7 and 50.6 kcal/mol, respectively. This value strongly suggests that IET occurring before Cys deprotonation is extremely unlikely, and therefore this mechanism is unlikely to be relevant in the present case.
Mechanism D
In this case the proton and IET steps are proposed to be significantly separated in time, resulting in an acid-base equilibrium three step dependent reaction. First, the Cys side chain acid-base equilibrium allows formation of the negative thiolate moiety (Cys-S−) (Step 1D). In the second step (Step 2D), the negatively charged radical peptide undergoes IET, which in fact corresponds to internal charge redistribution, resulting in Cys-S● and a Tyr-O−. Finally, the Tyr-O− must capture a proton from the solvent in a reaction that corresponds to Tyr acid base equilibrium referred as step 3D. Key processes for the forward and back reactions are the Cys and Tyr acid-base reactions governed by the corresponding pKa values, which are 8.37 and 10.46 (49) for isolated Cys and Tyr residues in water, respectively. In a first approximation we can expect that the corresponding pKa values will remain similar for all peptides, since each residue is completely exposed to the solvent as shown by previously described MD results. Therefore, significant amount of charged Cys can be found for the forward reaction, while once the charge is transferred to the Tyr residue the equilibrium is considerably displaced to the neutral state.
The kinetics of acid-base phenomena in aqueous solution is determined by the thermodynamics, namely, spontaneous processes are barrierless (50), while those non spontaneous exhibit kinetic rate constants that can be predicted from the Ka values. In this case, using Eyring equation (51)2, the proton transfer rate constant from Cys to water (kCys-) is estimated to be 2.4 × 104 s−1, which is similar or even faster than the estimates for the overall rate reaction in tyrosine-containing peptides k=103-104 s−1 (9, 15, 52) suggesting that ET (Step 2D) being at least partially rate-limiting. On the other hand, tyrosine protonation is spontaneous, and so, it is expected to be diffusion-controlled.
We now turn our attention to the IET in the charged peptides, corresponding to Step 2D. The reaction corresponds to a change in the negative charge localization from the Cys to the Tyr residue or vice-versa. For two isolated Cys and Tyr peptides in vacuum this results in an energy change of −7.0 kcal/mol, showing a preferred intrinsic stabilization of the negative charge in the Tyr residue. Now, in order to estimate the change in the solvation Gibbs free energy for the charge transfer reaction, the Gibbs free energy change for all the peptides during the charge process in explicit water was computed using TI. In this case the TI protocols consisted in changing the partial charges of the Tyr and Cys atoms, from those corresponding to having a Tyr-O● and Cy-S− to those corresponding to Tyr-O− and Cys-S●. The corresponding free energy change for all the peptides was about 4 kcal/mol positive, showing that solvation favors localization of the radical in the Tyr residue, while preferentially stabilizing the charge in the Cys residue, consistent with the above-described observation for the QM (PCM) results. The higher stabilization of the charged state in the Cys residue is consistent with the lower pKa for Cys compared to that of Tyr. More interestingly, no significant change in the solvation free energy is obtained for the peptides containing increasing number of alanine residues. Finally, combining the results for the intrinsic and solvation effects overall charge transfer from TyrO− to Cys-S●is only slightly endergonic (ca. +3 kcal/mol).
ET kinetics
Having analyzed the thermodynamics of Step 2D, we now turn our attention to possible influence on ET kinetics in the different peptides. In this context, and to analyze ET rates in a comparative way for the TyrO●-(Ala)n-CysS− and TyrO−-(Ala)n-CysS● peptides, we first computed the ET coupling matrix (TDA) between Tyr-O and Cys-S located orbitals using the Pathways algorithm (see Methods), using a hundred snapshots taken from the MD simulation of the corresponding peptide. The results are presented in Table IV; although the method is not expected to yield quantitative coupling values, the results are reliable when analyzed as qualitative comparative trends (53).
Table IV.
Results for the ET coupling matrix (TDA) calculations using the pathway algorithm. <TDA> and SD are the average and standard deviation of the obtained coupling values in eV along the MD simulation. Through bonds TDA is the predicted coupling if only through bonds ET is allowed. Relative <TDA> are the relative average coupling values with respect to Tyr-Cys-containing peptides. In the first column Y and C represent Tyr andCys residues, respectively.
| Starting Peptide |
Through bonds TDA | <TDA> | SD | Relative <TDA> |
Relative Cys-Cys peptide amounta MPO, H2O2+NaNO2 |
|
|---|---|---|---|---|---|---|
| Y●C− | 6.1−10−3 (10 bonds) | 6.1×10−3 | 1×10−4 | 1 | 1 | 1 |
| Y−C● | 6.1×10−3 (10 bonds) | 6.5×10−3 | 2.4×10−3 | 1 | ||
| Y●AC− | 1.3×10−3 (13 bonds) | 1.7×10−3 | 1.3×10−3 | 0.28 | 0.64 | 0.81 |
| Y−AC● | 1.3×10− (13 bonds) | 7.0×10−3 | 12×10−3 | 1.08 | ||
| Y●A2C− | 2.8×10−4 (16 bonds) | 2.4×10−3 | 5.7×10−3 | 0.39 | 0.41 | 0.37 |
| Y−A2C● | 2.8×10−4 (16 bonds) | 1.6×10−3 | 3×10−3 | 0.25 | ||
| Y●A4C− | 1.3×10−5 (22 bonds) | 1.5×10−3 | 4.8×10−3 | 0.25 | 0.20 | 0.19 |
| Y−A4C● | 1.3×10−5 (22 bonds) | 1.6×10−4 | 3×10−4 | 0.02 | ||
RelativeCys-Cys (disulfide) peptide amounts were taken from Table II of the work of Zhang et al. (16) as determined by HPLC separation and UV detection, after reactions of the corresponding peptides with myeloperoxidase (MPO) plus H2O2 in the absence (MPO, H2O2) or presence (+NaNO2) of sodium nitrite.
As expected if only through bonds ET pathways are considered, the more Ala are inserted between Tyr and Cys residues, the lower predicted coupling values, with relative values of 1, 0.16, 0.046 and 0.002 for peptides containing zero, one, two and four Ala respectively (as determined by the values shown in the 2nd column of Table IV). When looking at the average predicted coupling values (3rd and 5th column) a similar trend is observed but the differences are much smaller. The reason for the smaller differences is due to the fact that, for longer peptides more and more IET pathways are predicted that shortcut between Tyr-O and Cys-S atoms, either through a space jump or through two consecutive jumps involving a bridging water molecule. This trend is so far the only observed difference in computed parameters between different peptides. Moreover, the trend is consistent with the experimentally observed amount of Cys disulfide (Cys-Cys) peptides after tyrosyl radical formation (Table IV); indeed, the reaction yields depend on the nature and extent of radical transfer reactions and are consistent with a relevant role of the IET rate, as determined by the electronic coupling in the present reaction.
As shown in Equation 1, the ET rate not only depends on the coupling, but also on the reorganization energy, which is defined as the energy needed to distort or move the structure along the reactant surface, to the optimal structure corresponding to that of the products. For the corresponding reaction, possibly most of the reorganization energy will come from the change in peptide solvent interactions as the charge distribution is altered due to the ET process, i.e. how much energy is needed to drive solvent structure to that of the product charge distribution with the distribution corresponding to the reactant state. As a first attempt to estimate the reorganization energy in a comparative manner, we computed the total non bonded energy of the system (intermolecular interactions) for both charge distributions (TyrO●-(Ala)n-CysS− and TyrO--(Ala)n-CysS●) along the TI reaction coordinate (i.e. from ζ=0 to ζ=1) for step 2D which corresponds to the process of changing the charge distribution form one state to the other. The non-bonded energy is expected to capture most of solvation energy, since water molecules have no internal energy in the TIP3P model, and peptide internal energy (bonds and angles) are expected to be approximately the same along the whole reaction. The resulting plots are shown in Supplementary Material (Fig. S2A). The results show that moving forward in the RC (which corresponds to changing ζ from 0 to 1) results in an increase in the TyrO●-(Ala)n-CysS− energy, and a decrease in the TyrO−-(Ala)n-CysS● energy as the solvent structure adopts optimal conformation for solvating the TyrO−-(Ala)n-CysS● charge distribution (corresponding to ζ=1). Interestingly, for all peptides a similar trend for the energy changes are observed and therefore very similar reorganization energy values can be expected.
To have another estimation of the change in the solvation Gibbs free energy, and therefore the main component of λ (the reorganization energy), and given that main changes in solvation occur for the Tyr-O and Cys-S atoms, we also computed the water charge/radical interaction Gibbs free energy (J(r)) for each TI window as determined by the S/O centered water radial distribution function (g(r)) using the following equation:
| (Equation 2) |
Where J(r) is the Gibbs free energy of the water molecule which interacts with the TyrO and/or CysS atom at distance r, k is the Boltzmann constant and T the temperature. This equation allows the determination of the Gibbs free energy of solvation for the corresponding CysS and TyrO atoms corresponding to each water molecule from the first solvation shell by computing the difference in J(r) between the value in the bulk solvent (set to zero) and the peak in the g(r) function. The resulting plots (see Supplementary Material, Fig. S2B) and the estimated reorganization energy indicate that the differences are much smaller compared to previously computed non-bonded energy. The contribution to the reorganization energy, thus estimated as the end point value for the original charge distribution. (i.e. the value at ζ=1, for the charge distribution of the TyrO●-(Ala)n-CysS− state and vice versa) is predicted to be between 2-4 kcal/mol and similar for all peptides.
In summary, taking into account the estimation of the coupling and reorganization energies for each peptide, our data indicate that although reorganization energy estimates were similar for all peptides, the coupling and therefore the ET rate significantly decreased with the number of Ala spacers, a fact that is consistent with the experimental data from Zhang et al. (16).
Discussion
Previous results on ET between Tyr and Cys residues
The results presented in the present manuscript can be related to previous studies performed by Prütz et al. where the redox potential of isolated Tyr and Cys amino acids were established at different pH values (10), which show that at pH<8 (i.e. Tyr and Cys are neutral species) the difference in redox potential is ca. zero, implying a minimal change in Gibbs free energy for the intermolecular ET reaction. Interestingly, at pH > 10, where both Cys and Tyr residues are deprotonated the difference in the redox potential for intermolecular ET from Tyr to Cys is increased, and results in a ca. 3.7 kcal/mol for the ΔG of the reaction. Also, more recent studies by Folkes et al. (24) showed that the equilibrium constant for reaction between tyrosyl radicals and glutathione at pH 7.15, is close to one. Our results for isolated neutral aminoacid residues, yielded values which are only slightly exergonic (−1.6 kcal/mol) which, given the approximations involved in the calculations, are in reasonable agreement with the mentioned above experimental values. For the peptides, however, our results (Table I) clearly show that the intramolecular electron transfer process indicated in reaction 1 is exergonic, a fact consistent with the experimental observation by Zhang et al. (16). where the process was actually shown to occur3. Therefore, at least for the neutral peptide there seems to be significant differences in the reaction energetics between the inter- and intra-molecular electron transfer reactions. Also, noteworthy is the fact that our results of the ET reaction in the charged peptides yields a value of +3 kcal/mol (combining the in vacuum intrinsic −7 kcal/mol and the +4 kcal/mol corresponding to solvent contribution), a result that is analogous to the data obtained by Prutz et al. at pH >10 (10).
How does the radical transfer reaction occur in Tyr/Cys-containing peptides in aqueous solution?
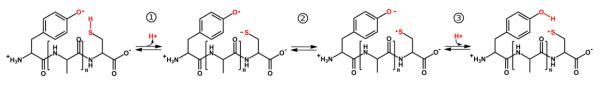
The present study was partially motivated by previous results of 3-nitrotyrosine and disulfide (Cys-Cys) yields of different Tyr and Cys-containing peptides in water exposed to the oxidizing/nitrating system of myeloperoxidase/H2O2/nitrite (16). In the corresponding work the amount of Cys-Cys product after Tyr-O● formation (which is an indicative of intramolecular radical transfer occurrence corresponding to the overall reaction) showed a clear decrease in the Cys-Cys product with an increasing number of Ala spacers (Table IV); this observation is also consistent with the notion that aliphatic amino acids such as alanine can not act as “stepping stones” or “relay” amino acids during IET processes (7). Taken together, our results strongly suggest that Mechanism D is the most likely. The choice is based on the following reasons. First, mechanism A, B and C are discarded on the following basis. Mechanism A, requires the establishment of a strong HB between Tyr and Cys residues, a conformation which is present less than 1% of the time, as determined by the explicit water MD simulations. Moreover, even for these structures the barrier for HAT or PCET are larger than 10 kcal/mol. Mechanism B is discarded, since the required water bridged structure/conformation is never obtained. Finally, for mechanism C the ab-initio calculations yield a possible barrier value of more than 50 kcal/mol, clearly showing that the mechanism C is extremely unlikely. Second, and more important, for mechanisms A, B and C no difference in any of the computed parameters is obtained for the different Ala-containing peptides. The only significant difference among the different peptides was obtained for the electronic coupling term that is expected to control the ET rate in mechanism D, since also both estimations of the reorganization energy yielded no significant differences among the peptides. The predicted relative ET rates according to estimated couplings (presented in Table IV), are consistent with the observed experimental trend in the amount of inter-peptide disulfide formation. The observed consistency makes a strong point for Mechanism D as the responsible for the radical transfer mechanism in the system. The overall proposed mechanism, which involves three steps (Step 1, 2 and 3D), is shown in Figure 3.
Figure 3. Proposed mechanism of intramolecular electron transfer in tyrosine-cysteine-containing peptides.
The mechanism involves three discrete steps that may occur significantly separated in time: (1) deprotonation of the cysteine thiol; (2) intramolecular electron transfer from cysteine to tyrosine which determines KIET; and (3) protonation of the tyrosine phenolate.
It is important to note, that under the proposed mechanism, the effective overall observed reaction rate, as experimentally determined, will result from a combination of the ET rate and pKa equilibrium. For example, at pH 7.4 (16), there is already a significant amount of deprotonated Cys (ca. 10 %), which undergoes oxidation via reactions step 2D. For this subpopulation the rate is directly the ET rate, and final reprotonation of TyrO- is expected to be very fast as mentioned previously. For the protonated Cys species the estimated rates for Step 1D (based on pKa value) are similar to the electron migration rates in the peptide unit as reported by (9, 15, 52). On the other hand, the rate of this step is expected to be similar in the studied peptides of different length. Conversely, the trends in the computed electronic coupling terms, which are related to the electron transfer rate constants through Marcus equation (Step 2D), correlate very well with the experimental trends (Table IV), indicating that this a key rate-controlling step in the overall radical transfer process.
Our QM and MD results show that although intrinsically the Cys to Tyr charge transfer is exergonic, the solvent tends to localize the charge on the Cys residue. Our best estimation, combining QM value for Cys-S− to Tyr-O● electron transfer in vacuo (−7 kcal/mol) and the +4 kcal/mol corresponding to the change in solvation Gibbs free energy determined from the TI calculation, yield a value of ca. −3 kcal/molfor the charge transfer reaction in all of the studied peptides. Given the approximations, the relative small value obtained suggests that once deprotonated radical states localized in either Tyr or Cys residues in each peptide maybe significantly populated (i.e. the equilibrium constant for IET, KIET, must be not very far from unity). Therefore, the outcome of the oxidation process yielding Tyr nitration, Cys nitrosation or any of the other routes shown in Figure 1, will depend on the presence of the secondary reactants such as ●NO2, ●NO and oxygen.
It should be noted however, that in proteins the local environment of each residue may stabilize/destabilize either the radical or charged state of the residue therefore, favoring a specific modification. The fact that radical transfer mechanism occurs through a deprotonation/protonation coupled with internal ET mechanism, may explain why Cys are able to inhibit Tyr nitration but neighboring Met are not, as experimentally observed for model peptides (16). Also important is the fact that for the overall radical transfer reaction from the Tyr to Cys residue (Reaction 1), our results show a gained energy both in the gas phase (QM) and in water (MD), suggesting that the overall intramolecular reaction in small peptides is thermodynamically favored by about −15 kcal/mol, as estimated by adding the intrinsic energy difference from Table I and the change in the solvation Gibbs free energy estimated with the TI scheme. This result is consistent with the experimental observation that nearby Cys residues are able to reduce Tyr-O● yielding the more stable radical Cys-S● resulting in the formation of Cys-Cys bridges or Cys S-nitrosation (16).
Finally, our results suggest that most likely mechanism involves acid/base equilibrium and IET in the charged peptides. On the other hand, direct hydrogen, direct concerted PCET or even water-mediated transfer are very unlikely to occur, since during the simulation, hydrogen (or proton) donor and acceptor atoms never are close enough for establishing proper HB interactions. Moreover, even for the best possible structures the instantaneous reaction, which usually corresponds to a PCET, has a quite high barrier. Nevertheless, the lack of intramolecular HB could be just a particular property of the present studied model peptides. In proteins, the local structure may impose direct or another residue-mediated HB between Tyr and Cys residues, therefore making possible a direct concerted PCET mechanism (20, 39, 54-57).
Conclusions
In the present work we have analyzed several mechanistic possibilities at the atomic level for a radical transfer reaction in Tyr/Cys-containing peptides in aqueous solution. Our results show that tyrosine radical (Tyr-O●) to cysteine transfer is most likely to be mediated by an acid/base equilibrium that involves deprotonation of cysteine residue to form thiolate, followed by a fast (e.g. k≅103-104 s−1 in tyrosine-containing peptides (9, 15, 52)), yet rate limiting, internal electron transfer (i.e. charge redistribution) resulting in cysteine radical and tyrosine phenolate. Finally, proton uptake by the Tyr residue completes the reaction. Therefore, the reaction can be described as a sequential, acid base equilibrium-dependent and solvent-mediated PCET reaction (Figure 3); within this general mechanism, electrons could be physically transferred via alternative pathways depending on the nature of the peptide and/or polypeptide chain sequence and structure and the characteristics of the milieu (Figure 4). While in short peptides the through-bond path (Type A) predominates, as the peptide chain lengthens, the jumping pathways of type B and C shown in Figure 4 become more likely. For one alanine spacer through space jumping paths (Type B) correspond to ca. 10%, while water-mediated paths are rare. For the two and four alanine spacers jumping paths are between 20 to 35% and equally distributed between types B and C. The resulting Cys-S● can evolve to secondary end products, which may vary depending on reaction conditions (16) (as shown in Figure 1), including cysteine disulfide (Table IV). The overall mechanism fully explains the dependency of tyrosine nitration and cysteine disulfide yields as a function of the number of Ala spacers observed experimentally previously (16). Importantly, while the capacity of ET will be highly influenced by the pKa of the corresponding amino acids, the intramolecular ET process will be in competition with direct bimolecular reaction of the primary radicals (e.g. Cys●) with reactants such as oxygen, nitric oxide (●NO), nitrogen dioxide (●NO2), and other radical species or even spin traps (see Figure 1). The pKa of Cys and Tyr will be influenced both by neighboring amino acids as well as by the degree of hydrophobicity of where they are located. Indeed, hydrophobicity will increase the pKa value (58) and therefore possibly hinder the ET process. Our study has focused on model peptides with an in silico approach and taking into consideration previous experimental data on Tyr-Cys-containing peptides (16) and protein oxidation (17, 18). These findings provide new elements to mechanistically and kinetically explain more probable oxidative modifications in proteins that contain Tyr-Cys sequences and may assist in the understanding of the cytoprotective actions of tyrosine- and/or cysteine-containing peptides during oxidative stress conditions in vitro and in vivo (59, 60).
Figure 4. Selected snapshots for alternative charge redistribution pathways in the Tyr-(Ala4)-Cys peptide.
The figure shows the three paths for ET as predicted with the pathways algorithm and taken from the explicit water MD simulation (see Methods for details). The peptide is shown as bold sticks. (A) Through-bond path, (B) through-space jump and (C) through two consecutive jumps involving a bridging water molecule. The predicted ET paths are shown in orange arrows. Surrounding waters were omitted for clarity. The relative contribution of each pathway (A, B or C) to intramolecular electron transfer in the shown studied peptides for one, two and four alanine spacers, are indicated in the text. The percent contribution for each path type in each peptide was computed by visually inspecting and counting the corresponding path type in one hundred snapshots.
Supplementary Material
Molecular basis of the tyrosine to cysteine radical transfer in peptides
Acid/base-dependent solvent-mediated proton-coupled electron transfer in peptides
Electron transfer as rate limiting step in cysteine-mediated tyrosine radical reduction
Through bond- and through space-mediated intramolecular electron transfer
Mechanisms of cysteine-mediated tyrosine nitration inhibition
Acknowledgements
This work was supported by grants PICT-2010-0416, UBA CyT 2010/2012 and PIP 12-200801-01207 to MAM; the Howard Hughes Medical Institute, Agencia Nacional de Investigación e Innovación (ANII)/Fondo Clemente Estable (FCE_2486) and Comisión Sectorial de Investigación Científica to RR, National Institutes of Health to BK and RR (2 RO1Hl063119-05) and Agencia Nacional de Investigación e Innovación (ANII)/Fondo Clemente Estable to SB (FCE_362). Computer power was gently provided by the Ce CAR at FCEN-Universidad de Buenos Aires, Argentina and Sim One at INSIBIO, CONICET-Universidad Nacional de Tucumán, Argentina.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The calculations performed with the whole zwiterionic species, i.e including the charged carboxylate and amino groups, yielded a value of −0.3 kcal/mol for the reaction in vacuum. No significant difference was observed when either the whole amino acid or just the side chain (i.e para methyl phenol and ethyl thiol) were considered.
In this case, the Eyring equation becomes, kCys-=(kT/h) × Ka
This implies a KIET >1 for reaction 1. Direct pulse radiolysis determinations for kf and krfor reaction 1 are underway.
References
- 1.Dean RT, Fu S, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324(Pt 1):1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartesaghi S, Wenzel J, Trujillo M, Lopez M, Joseph J, Kalyanaraman B, Radi R. Lipid peroxyl radicals mediate tyrosine dimerization and nitration in membranes. Chem Res Toxicol. 2010;23:821–835. doi: 10.1021/tx900446r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonini MG, Augusto O. Carbon dioxide stimulates the production of thiyl, sulfinyl, and disulfide radical anion from thiol oxidation by peroxynitrite. J Biol Chem. 2001;276:9749–9754. doi: 10.1074/jbc.M008456200. [DOI] [PubMed] [Google Scholar]
- 4.Quijano C, Alvarez B, Gatti RM, Augusto O, Radi R. Pathways of peroxynitrite oxidation of thiol groups. Biochem J. 1997;322(Pt 1):167–173. doi: 10.1042/bj3220167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian SY, Chen YR, Deterding LJ, Fann YC, Chignell CF, Tomer KB, Mason RP. Identification of protein-derived tyrosyl radical in the reaction of cytochrome c and hydrogen peroxide: characterization by ESR spin-trapping, HPLC and MS. Biochem J. 2002;363:281–288. doi: 10.1042/0264-6021:3630281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichorner H, Metodiewa D, Winterbourn CC. Generation of superoxide and tyrosine peroxide as a result of tyrosyl radical scavenging by glutathione. Arch Biochem Biophys. 1995;323:429–437. doi: 10.1006/abbi.1995.0064. [DOI] [PubMed] [Google Scholar]
- 7.Cordes M, Kottgen A, Jasper C, Jacques O, Boudebous H, Giese B. Influence of amino acid side chains on long-distance electron transfer in peptides: electron hopping via “stepping stones”. Angew Chem Int Ed Engl. 2008;47:3461–3463. doi: 10.1002/anie.200705588. [DOI] [PubMed] [Google Scholar]
- 8.Prutz WA, Butler J, Land EJ. Phenol coupling initiated by one-electron oxidation of tyrosine units in peptides and histone. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;44:183–196. doi: 10.1080/09553008314550981. [DOI] [PubMed] [Google Scholar]
- 9.Prutz WA, Butler J, Land EJ, Swallow AJ. Unpaired electron migration between aromatic and sulfur peptide units. Free Radic Res Commun. 1986;2:69–75. doi: 10.3109/10715768609088056. [DOI] [PubMed] [Google Scholar]
- 10.Prutz WA, Butler J, Land EJ, Swallow AJ. The role of sulphur peptide functions in free radical transfer: a pulse radiolysis study. Int J Radiat Biol. 1989;55:539–556. doi: 10.1080/09553008914550591. [DOI] [PubMed] [Google Scholar]
- 11.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Electron transfer reactions of fluorotyrosyl radicals. J Am Chem Soc. 2006;128:13654–13655. doi: 10.1021/ja0636688. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Gao J, Muller P, Giese B. Electron transfer in peptides with cysteine and methionine as relay amino acids. Angew Chem Int Ed Engl. 2009;48:4232–4234. doi: 10.1002/anie.200900827. [DOI] [PubMed] [Google Scholar]
- 13.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration--functional alteration or just a biomarker? Free Radic Biol Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobrowski K, Wierzchowski KL, Holcman J, Ciurak M. Pulse radiolysis studies of intramolecular electron transfer in model peptides and proteins. IV. Met/S:.Br -->Tyr/O. radical transformation in aqueous solution of H-Tyr-(Pro)n-Met-OH peptides. Int J Radiat Biol. 1992;62:507–516. doi: 10.1080/09553009214552431. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Xu Y, Joseph J, Kalyanaraman B. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides with MPO, H2O2 and NO2-: EPR spin trapping studies. J Biol Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- 17.Romero N, Radi R, Linares E, Augusto O, Detweiler CD, Mason RP, Denicola A. Reaction of human hemoglobin with peroxynitrite. Isomerization to nitrate and secondary formation of protein radicals. J Biol Chem. 2003;278:44049–44057. doi: 10.1074/jbc.M305895200. [DOI] [PubMed] [Google Scholar]
- 18.Witting PK, Mauk AG. Reaction of human myoglobin and H2O2. Electron transfer between tyrosine 103 phenoxyl radical and cysteine 110 yields a protein-thiyl radical. J Biol Chem. 2001;276:16540–16547. doi: 10.1074/jbc.M011707200. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J Am Chem Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- 20.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc Lond B Biol Sci. 2006;361:1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeFelippis M, Murthy C, Broitman F, Weinraub D, Faraggi M, Klapper M. Electrochemical Properties of Tyrosine Phenoxy and Tryptophan Indoyl Radicals in Peptides and Amino Acid Analogues. J. Phys. Chem. 1991;95:3416–3419. [Google Scholar]
- 22.DeFelippis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: the tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 23.Harriman A. Further Comments on the Redox Potentials of Tryptophan and Tyrsosine. J Phys Chem. 1987;91:6102–6104. [Google Scholar]
- 24.Folkes LK, Trujillo M, Bartesaghi S, Radi R, Wardman P. Kinetics of reduction of tyrosine phenoxyl radicals by glutathione. Arch Biochem Biophys. 2011;506:242–249. doi: 10.1016/j.abb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 26.Vass I, Styring S. pH-dependent charge equilibria between tyrosine-D and the S states in photosystem II. Estimation of relative midpoint redox potentials. Biochemistry. 1991;30:830–839. doi: 10.1021/bi00217a037. [DOI] [PubMed] [Google Scholar]
- 27.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 28.Hammes-Schiffer S, Soudackov AV. Proton-coupled electron transfer in solution, proteins, and electrochemistry. J Phys Chem B. 2008;112:14108–14123. doi: 10.1021/jp805876e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheatham TE, 3rd, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J Comp Chem. 2000;21:1049–1074. [Google Scholar]
- 31.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR. Gaussian 03. Gaussian; Pittsburgh, PA: 2003. revision A.I. [Google Scholar]
- 32.Leach A. Molecular Modelling: Principles and Applications. 2001 [Google Scholar]
- 33.Case DA, Darden TA, Cheartham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang W, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco, CA: 2006. [Google Scholar]
- 34.Mitani M, Inoue M, Yoshioka Y. A B3LYP study on the mechanism of second H2O formation in a fully reduced cytochrome c oxidase. Chemical Physics Letters. 2007;440:296–301. [Google Scholar]
- 35.Naumov S, Schoneich C. Intramolecular addition of cysteine thiyl radical to phenylalanine and tyrosine in model peptides, Phe (CysS*) and Tyr(CysS*): a computational study. J Phys Chem A. 2009;113:3560–3565. doi: 10.1021/jp8076358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshioka Y, Kawai H, Yamaguchi K. Theoretical study of role of H2O molecule on initial stage of reduction of O2 molecule in active site of cytochrome c oxidase. Chemical Physics Letters. 2003;374:45–52. [Google Scholar]
- 37.Yoshioka Y, Mitani M. B3LYP study on reduction mechanisms from O2 to H2O at the catalytic sites of fully reduced and mixed-valence bovine cytochrome c oxidases. Bioinorganic Chemistry and Applications 2010. 2010;ID 182804:18. doi: 10.1155/2010/182804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshioka Y, Satoh H, Mitani M. Theoretical study on electronic structures of FeOO, FeOOH, FeO(H2O), and FeO in hemes: As intermediate models of dioxygen reduction in cytochrome c oxidase. Journal of Inorganic Biochemistry. 2007;101:1410–1427. doi: 10.1016/j.jinorgbio.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 39.Siegbahn PEM, Blomberg MRA. Quantum Chemical Studies of Proton-Coupled Electron Transfer in Metalloenzymes. Chemical Reviews. 2010;110:7040–7061. doi: 10.1021/cr100070p. [DOI] [PubMed] [Google Scholar]
- 40.Cossi M, Scalmani G, Rega N, Barone V. New developments in the polarizable continuum model for quantum mechanical and classical calculations on molecules in solution. Journal of Chemical Physics. 2002;117:43–54. [Google Scholar]
- 41.Klamt A, Mennucci B, Tomasi J, Barone V, Curutchet C, Orozco M, Luque FJ. On the performance of continuum solvation methods. A comment on “Universal approaches to solvation modeling”. Acc Chem Res. 2009;42:489–492. doi: 10.1021/ar800187p. discussion 493-487. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Lazaridis T. Water at biomolecular binding interfaces. Phys Chem Chem Phys. 2007;9:573–581. doi: 10.1039/b612449f. [DOI] [PubMed] [Google Scholar]
- 43.Marcus RA. On the Theory of Oxidation-Reduction Reactions Involving Electron Transfer I. J Chem Phys. 1956;24:966–978. [Google Scholar]
- 44.Beratan DN, Onuchic JN, Betts JN, Bowler BE, Gray HB. Electron tunneling pathways in ruthenated proteins. 1990;112:7915–7921. [Google Scholar]
- 45.Beratan DN, Onuchic JN, Winkler JR, Gray HB. Electron-tunneling pathways in proteins. Science. 1992;258:1740–1741. doi: 10.1126/science.1334572. [DOI] [PubMed] [Google Scholar]
- 46.Carra C, Iordanova N, Hammes-Schiffer S. Proton-coupled electron transfer in a model for tyrosine oxidation in photosystem II. Journal of the American Chemical Society (2003) 2008;125:10429–10436. doi: 10.1021/ja035588z. [DOI] [PubMed] [Google Scholar]
- 47.Edwards SJ, Soudackov AV, Hammes-Schiffer S. Analysis of kinetic isotope effects for proton-coupled electron transfer reactions. Journal of Physical Chemistry A. 2009;113:2117–2126. doi: 10.1021/jp809122y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammes-Schiffer S, Hatcher E, Ishikita H, Skone JH, Soudackov AV. Theoretical Studies of Proton-Coupled Electron Transfer: Models and Concepts Relevant to Bioenergetics. Coord Chem Rev. 2008;252:384–394. doi: 10.1016/j.ccr.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dawson RMC, Elliot DC, Elliot WH, Jones KM. Data for Biochemical Research. 3rd ed New York: 1986. [Google Scholar]
- 50.Eigen M. Protonenübertragung, Säure-Base-Katalyse und enzymatische Hydrolyse. Teil I: Elementarvorgänge, Angewandte Chemie. 1963;75:489–508. [Google Scholar]
- 51.Eyring H. The Activated Complex in Chemical Reactions. J. Chem. Phys. 1935;3:107–115. [Google Scholar]
- 52.Bobrowski K, Poznanski J, Holcman J, Wierzchowski K. Pulse Radiolysis Studies of Intramolecular Electron Transfer in Model Peptides and Proteins. 8 Trp[NH.+] -->Tyr[O.] Radical Transformation in H-Trp-(Pro)n-Tyr-OH, n)= 3-5, Series of Peptides. J. Phys. Chem. B. 1999:10316–10324. [Google Scholar]
- 53.Alvarez-Paggi D, Martin DF, DeBiase PM, Hildebrandt P, Marti MA, Murgida DH. Molecular basis of coupled protein and electron transfer dynamics of cytochrome c in biomimetic complexes. J Am Chem Soc. 132:5769–5778. doi: 10.1021/ja910707r. [DOI] [PubMed] [Google Scholar]
- 54.Himo F. C-C bond formation and cleavage in radical enzymes, a theoretical perspective. Biochimica et Biophysica Acta – Bioenergetics. 2005;1707:24–33. doi: 10.1016/j.bbabio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Himo F, Siegbahn PEM. Quantum chemical studies of radical-containing enzymes. Chemical Reviews. 2003;103:2421–2456. doi: 10.1021/cr020436s. [DOI] [PubMed] [Google Scholar]
- 56.Siegbahn PEM, Eriksson L, Himo F, Pavlov M. Hydrogen atom transfer in ribonucleotide reductase (RNR) Journal of Physical Chemistry B. 1998;102:10622–10629. [Google Scholar]
- 57.Stubbe J, Van Der Donk WA. Protein radicals in enzyme catalysis. Chemical Reviews. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 58.Yokoyama K, Uhlin U, Stubbe J. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J Am Chem Soc. 132:8385–8397. doi: 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szeto HH, Schiller PW. Novel therapies targeting inner mitochondrial membrane--from discovery to clinical development. Pharm Res. 2011;28:2669–2679. doi: 10.1007/s11095-011-0476-8. [DOI] [PubMed] [Google Scholar]
- 60.Ye Y, Quijano C, Robinson KM, Ricart KC, Strayer AL, Sahawneh MA, Shacka JJ, Kirk M, Barnes S, Accavitti-Loper MA, Radi R, Beckman JS, Estevez AG. Prevention of peroxynitrite-induced apoptosis of motor neurons and PC12 cells by tyrosine-containing peptides. J Biol Chem. 2007;282:6324–6337. doi: 10.1074/jbc.M610800200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.