Abstract
In this study, a novel redox-responsive hyperbranched poly(amido amine) (named PCD) was synthesized and used as a cationic polymer to form a ternary complex with small interfering RNA (siRNA) and hyaluronic acid (HA) for siRNA delivery. Here, it is hypothesized that different mixing orders result in different assembly structures, which may affect the siRNA delivery efficiency. To investigate the effects of mixing orders on siRNA delivery efficiency in two human breast cancer cell lines, three ternary complexes with different mixing orders of siRNA/PCD/HA were prepared and characterized: mixing order I (initially prepared siRNA/PCD binary complex further coated by negatively charged HA), mixing order II ( initially prepared HA/PCD binary complex further incubated with siRNA), and mixing order III ( initially prepared siRNA/HA mixture further electrostatically compacted by positively charged PCD). With an optimized siRNA/PCD/HA charge ratio of 1/20/16, the particle sizes and zeta potentials of these ternary complexes were 124.8 nm and 27.3 mV (mixing order I), 147.5 nm and 29.9 mV (mixing order II), and 128.8 nm and 19.4 mV (mixing order III). Also, the effects on stability, cellular uptake, and gene silencing efficiency of siRNA formulated in ternary complexes with different mixing orders were investigated. The results showed that mixing orders I and III displayed better siRNA transfection and protection than mixing order II in human breast cancer MCF-7 and MDA-MB-231 cells. More interesting, at the siRNA/PCD/HA charge ratio of 1/20/16, the gene silencing effects on vascular endothelial growth factor expression in MDA-MB- 231 cells were as follows: mixing order III > mixing order I > mixing order II. Based on these results, a likely explanation for the difference in functionality dependent on mixing orders is the formation of different assembly structures. These results may help future optimization of siRNA ternary complexes for achieving better delivery efficiencies, especially for target-specific siRNA delivery to cells with HA receptor overexpression.
Keywords: small interfering RNA, poly(amido amine)s, siRNA ternary complex, mixing order, hyaluronic acid, siRNA delivery
Introduction
Since small interfering RNA (siRNA) was shown to achieve sequence-specific gene knockdown in mammalian cells,1 and has been considered a potential agent for the treatment of many diseases, including cancer, viral infections, and hereditary genetic disorders.2,3 However, the clinical application of siRNA in disease therapy has been hindered by delivery problems. Degradation by endogenous enzymes, poor cellular uptake, and several intracellular barriers such as endosomal escape and unpacking of siRNA/polycation complex must be circumvented when developing new, safe, and efficient siRNA delivery vectors.4–6
Nonviral vectors appear a viable alternative to viral vectors for their low cost, easy manufacturing, large cargo capacity, and nonimmunogenicity.7–9 Among the nonviral vectors designed to overcome these barriers,10–12 redox-responsive poly(amido amine)s (rPAAs) have gained particular attention because of their good endosomal escape capacity (proton sponge effect) and good unpacking ability. 13–15 Importantly, these rPAAs/siRNA complexes are stable in the predominantly oxidizing extracellular environment, but they rapidly disassemble to release siRNA because of the cleavage of disulfide bonds in the reducing intracellular compartment.16 Additionally, rPAAs are peptidomimetic and water-soluble in nature, making them attractive candidates for siRNA delivery in pharmaceuticals.17 In addition, it has been reported that cationic polymers with branched structures, especially hyperbranched ones, tend to result in better gene delivery efficiencies than their linear counterparts.18–23
Although siRNA/polycation binary complexes have many advantageous properties, these complexes are prone to aggregation with serum proteins and can induce systemic toxicity during intravenous administration. Recently, polyanion-coating24–27 has become an effective solution to this problem. Among these polyanion-coating polymers, hyaluronic acid (HA) is a biodegradable, biocompatible, and nontoxic natural polysaccharide,28 and is approved for injections by the US Food and Drug Administration.29 It has many desirable properties for use in vector optimization and is used for targeting HA receptors (such as CD44), which are overexpressed in many malignant cells.30–33 Recent studies have demonstrated that HA-coated electrostatic complexes display a great number of attractive properties, including preparation of very small vesicles,34 better transfection efficacy,25 and better target specificity.35
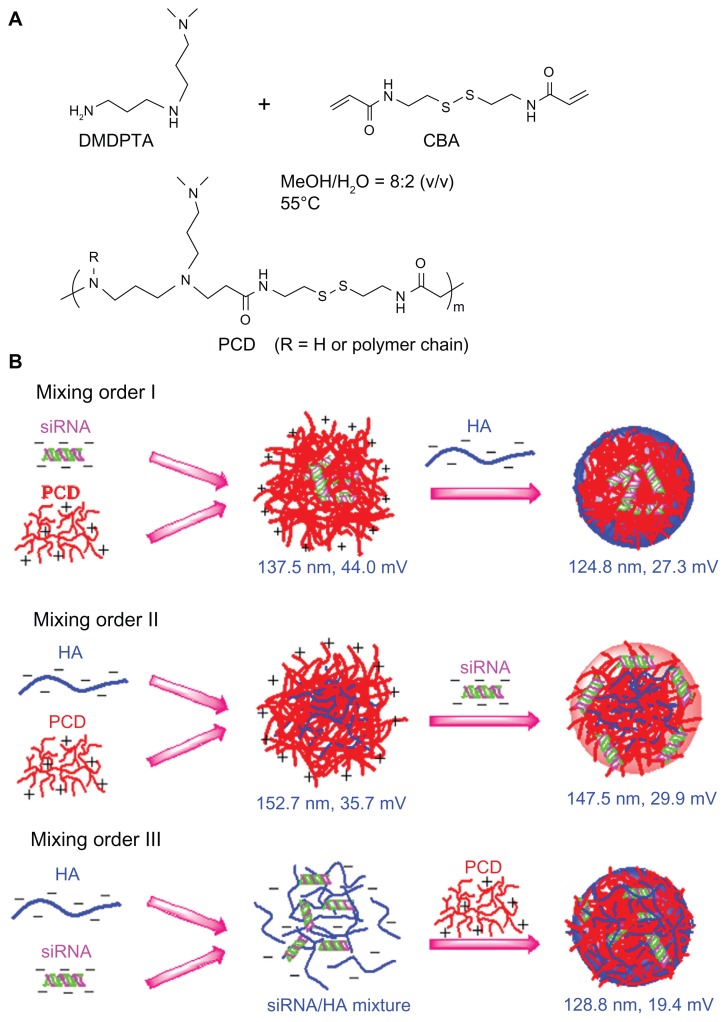
In this study, the authors synthesized a type of redox-responsive hyperbranched poly(amido amine)s (named PCD) to form siRNA/PCD/HA ternary complexes with three different mixing orders (Figure 1B). As these ternary complexes form spontaneously upon mixing, the authors hypothesized that different mixing orders result in different assembly structures, which may affect the siRNA delivery efficiency. To address this hypothesis, the effects of mixing orders on siRNA delivery efficiency were systemically investigated in two human breast cancer cell lines, MCF-7 (low CD44 expression) and MDA-MB-231 (high CD44 expression) cells.
Figure 1.
Schematic illustrations showing (A) the synthesis of a novel redox-responsive hyperbranched poly(amido amine) (named PCD) and (B) the formation of small interfering RNA (siRNA) ternary complexes with three different mixing orders.
Note: The ternary complexes were prepared in an optimized siRNA/PCD/hyaluronic acid (HA) charge ratio of 1/20/16.
Abbreviations: CBA, N, N′-cystamine bisacrylamide; DMDPTA, N, N′-dimethyldipropylenetriamine.
Materials and methods
N, N′-cystamine bisacrylamide (CBA) was purchased from Alfa Aesar China (Tianjin) Co, Ltd (Tianjin, China). Branched polyethylenimine (PEI) (molecular weight, 25 kDa), dithiothreitol (DTT), and N, N′-dimethyldipropylenetriamine (DMDPTA) were purchased from Sigma-Aldrich Shanghai Trading Co, Ltd (Shanghai, China). Hyaluronic acid sodium salt (HA) (molecular weight, 125 kDa) was obtained from Shandong Freda Biochem Co, Ltd (Shandong, China). Vascular endothelial growth factor (VEGF)–directed siRNA (sense: 5′-GGA GUA CCC UGA UGA GAU CdTdT-3′; antisense: 5′-GAU CUC AUC AGG GUA CUC CdTdT-3′) and fluorescein-labeled siRNA (FAM-siRNA) (sense: 5′-UUC UCC GAA CGU GUC ACG UTT-3′; antisense: 5′-ACG UGA CAC GUU CGG AGA ATT-3′) were both synthesized and purified with high-performance liquid chromatography by Shanghai GenePharma Co, Ltd (Shanghai, China).
Human breast cancer cell lines MCF-7 and MDA-MB-231 were purchased from the Institute of Basic Medical Science, Chinese Academy of Medical Sciences (Beijing, China). RPMI-1640 medium and Leibovitz’s L-15 medium were obtained from Macgene Biotech Co, Ltd (Beijing, China). Opti-MEM® medium was purchased from Invitrogen (Grand Island, NY). Human VEGF ELISA Kit was purchased from RayBiotech, Inc (Norcross, GA).
Synthesis and characterization of PCD
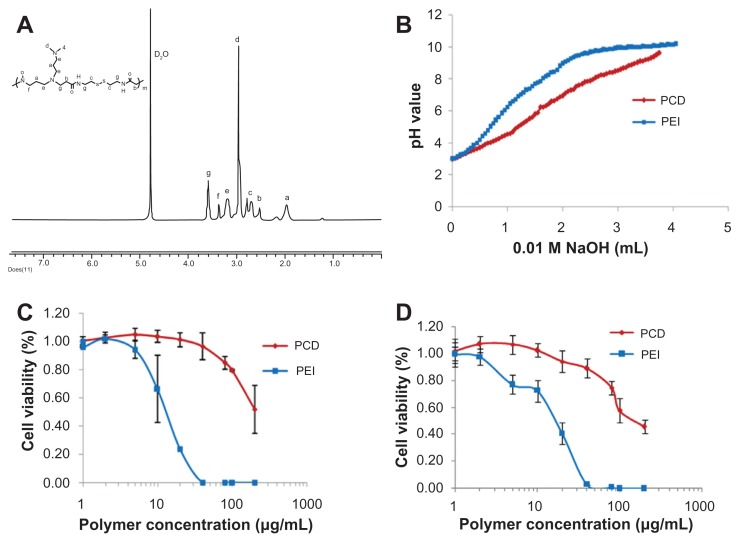
The synthetic scheme of PCD is illustrated in Figure 1A. PCD synthesis was done by Michael-type addition polymerization as previously reported with some minor modifications.36 In brief, equimolar CBA (2 mmol) and DMDPTA (2 mmol) were added to a light-protected flask using methanol/water (8:2, v/v) mixture as a solvent. The reaction was conducted under an argon atmosphere at 55°C in order to produce a hyperbranched structure.37 After 5 days, an excess amount of DMDPTA (about 10% mol) was added to consume any unreacted acryloyl functional groups and was allowed to proceed for 2 days at 55°C. The resulting solution was diluted with distilled water and acidified to approximately pH 4 with hydrochloric acid. The polymer was obtained by lyophilization after extensive dialysis against distilled water through a dialysis membrane (molecular weight cutoff, 3.5 kDa) for 3 days. The chemical structure of the synthesized PCD was characterized by proton nuclear magnetic resonance (1H-NMR) (AVANCE III, 400 MHz, deuterium oxide; Bruker, Billerica, MA).
Buffer capacity of PCD
The buffer capacity of PCD was determined by acid-base titration, as previously reported, with some modifications.21 In brief, 5 mL polymer solution with 0.025 mmol of amine groups was adjusted to pH 3.0 with 0.1 M hydrochloric acid. The solution was then titrated with 50 μL aliquots of 0.01 M sodium hydroxide (NaOH), and the pH value was measured after each addition. As a reference, the same procedure was also applied to branched PEI (25 kDa). The buffer capacity is defined as the percentage of amine groups becoming protonated from pH 7.4 to pH 5.1 and can be calculated according to the following equation (where ΔVNaOH is the volume of 0.01 M NaOH solution used to bring the pH value of the polymer solution from 5.1 to 7.4, and N mol [0.025 mmol] is the total molar amount of protonable amine groups in PCD or PEI):
Cytotoxicity assay
The cytotoxicity of PCD and PEI on both MCF-7 and MDA-MB- 231 human breast cancer cells was measured by the sulforhodamine assay. MCF-7 (or MDA-MB-231) cells were seeded into a 96-well plate at a population of 4 × 103 per well and were maintained at 37°C in a humidified atmosphere of 5% carbon dioxide. After 24 hours’ incubation, the medium was replaced with fresh Opti-MEM medium containing various concentrations of PEI and PCD. After an additional 4 hours’ incubation, the Opti-MEM medium was removed and replaced with fresh RPMI-1640 medium (Leibovitz’s L-15 medium for MDA-MB-231 cells) containing 10% fetal bovine serum (FBS). Cell viability was measured at 48 hours via the sulforhodamine assay, and the absorbance was read on a microplate reader at 540 nm. The cell viability (%) was calculated according to the following formula (where OD540 (sample) is the optical density from the wells treated with samples and OD540 (control) is from the wells treated with fresh medium without PCD or PEI):
Preparation of siRNA complexes
The siRNA binary complexes were prepared by mixing an siRNA solution with a solution of PEI or PCD followed by incubation at room temperature for at least 15 minutes. For siRNA/PCD/HA ternary complexes, preparation was completed by the following three methods (Figure 1B): (1) mixing order I – siRNA/PCD binary complexes were firstly prepared, and a solution of HA was added to these binary complexes to form the ternary complexes; (2) mixing order II – HA/PCD binary complexes were prepared by mixing HA solution and PCD solution followed by incubation at room temperature for at least 15 minutes, and then a solution of siRNA was added to the HA/PCD binary complexes to form the ternary complexes; (3) mixing order III – siRNA solution was mixed with the HA solution and incubated at room temperature for at least 15 minutes, and then the siRNA/HA mixture was further electrostatically compacted by PCD. Similarly, the siRNA/PEI/HA ternary complexes were also prepared in three different mixing orders as previously described. All the ternary complexes in this study were prepared in the charge ratio of siRNA/polycation/HA (P/N/C) unless otherwise specified.
Particle size and zeta potential of siRNA complexes
The particle size and zeta potential of various siRNA complexes were measured at 25°C using dynamic light scattering (Malvern Zetasizer Nano ZS; Malvern Instruments Ltd, Malvern, Worcestershire, UK). All siRNA complexes were diluted with 5% glucose solution prior to the measurement. All results are presented as means plus or minus standard deviations of three or more independent experiments.
Evaluation of FAM-siRNA fluorescence quenching
The FAM-siRNA fluorescence quenching was evaluated as reported.35,38 FAM-siRNA/PCD/HA ternary complexes (P/N/C charge ratio = 1/20/16) were prepared by different mixing orders using the same siRNA concentration. Then, relative fluorescence intensity of FAM-siRNA in each ternary complex was determined using a spectrofluorophotometer (Cary-Eclipse; Varian Medical Systems, Inc, Palo Alto, CA) at an excitation wavelength of 480 nm and an emission wavelength of 520 nm. To evaluate the release of siRNA from ternary complexes under a reducing environment, the complexes were preincubated with 10 mM DTT for 4 hours prior to determination of fluorescence intensity.
Gel retardation assay of siRNA complexes
The gel retardation assay of various siRNA ternary complexes was evaluated using agarose gel electrophoresis. The siRNA/PCD/HA ternary complexes were prepared by mixing order I, as described earlier. As a reference, siRNA/PEI/HA ternary complexes were also prepared by mixing order I. The siRNA complexes were loaded onto 1% agarose gel containing 0.5 μg/mL of ethidium bromide. Electrophoresis was carried out at 80 V for 3 minutes and then at 100 V for 10 minutes in TBE (45 mM tris, 45 mM boric acid, and 1 mM ethylenediaminetetraacetic acid) buffer solution. The retardation of the complexes was visualized using an EC3 Imaging System (UVP, LLC, Upland, CA). To evaluate the release of siRNA from ternary complexes under a reducing environment, the complexes were preincubated with 10 mM DTT for 4 hours prior to electrophoresis.
Serum stability of siRNA complexes
Free siRNA and three siRNA ternary complexes were separately incubated with 10% FBS at 37°C. An aliquot of siRNA complex was collected at 2, 4, 8, 12, 16, 24, and 36 hours after incubation, followed by quenching the enzyme activity with addition of ethylenediaminetetraacetic acid buffer. The samples were then mixed with a solution containing 10% sodium dodecyl sulfate and 20 mM DTT. After incubation at 37°C for an additional 30 minutes, the samples were loaded onto 1% agarose gel for electrophoresis.
Cellular uptake of siRNA complexes
MCF-7 (or MDA-MB-231) cells were seeded into a 6-well plate at a density of 3 × 105 cells per well, and cultured for 24 hours in RPMI-1640 medium (Leibovitz’s L-15 medium for MDA-MB- 231 cells) supplemented with 10% FBS and antibiotics (penicillin 100 units/mL and streptomycin 0.1 mg/mL). After 24 hours’ incubation, the cells were washed with phosphate-buffered saline (PBS) twice and fresh Opti-MEM medium containing various FAM-siRNA complexes were separately added to each well for transfection. After incubation at 37°C for 4 hours, the cells were washed with PBS and detached with trypsin. After that, the cells were washed an additional four times and the mean fluorescence intensity of intracellular FAM-siRNA per 10,000 cells was measured by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA). The relative fluorescence intensity is defined as the ratio of fluorescence intensity in cells treated with siRNA complex to untreated cells. The untreated cells were used as the control.
Intracellular distribution of FAM-siRNA
For confocal microscopy analysis, MDA-MB-231 human breast cancer cells at a density of 3 × 105 cells per dish were cultured into glass-based dishes and incubated for 24 hours to allow for adhesion. After 24 hours’ incubation, the cells were washed with PBS twice and then fresh Opti-MEM medium containing FAM-siRNA/PEI binary complex, FAM-siRNA/PCD binary complex, and FAM-siRNA/PCD/HA ternary complexes prepared in different mixing orders were separately added to each dish for transfection. After 2 hours’ incubation, cells were incubated with Hoechst 33258 (Sigma-Aldrich, St Louis, USA) for nucleus labeling. To observe the intracellular distribution of the FAM-siRNA, cells were incubated with LysoTracker® Red (Invitrogen, Carlsbad, CA) for 15 minutes at the end of uptake study for lysosome labeling. After a total incubation of 4 hours, the cells were washed and observed with a Leica TCS SP5 confocal fluorescence microscope (Leica Microsystems Heidelberg GmbH, Heidelberg, Germany).
Evaluation of VEGF-directed siRNA gene silencing efficiency
MDA-MB-231 human breast cancer cells were seeded into a 6-well plate at a density of 3 × 105 cells per well and were allowed to grow for 24 hours in Leibovitz’s L-15 medium. After 24 hours’ incubation, the cells were washed with PBS twice and fresh Opti-MEM medium containing various VEGF-directed siRNA complexes were separately added to each well for transfection. After incubation at 37°C for 4 hours, the transfection medium was replaced with Leibovitz’s L-15 medium containing 10% FBS. The incubation was continued for an additional 8 hours and the medium was replaced with fresh Leibovitz’s L-15 medium containing 10% FBS and heparin (20 μg/mL).39,40 Finally, the medium was collected after 18 hours’ incubation, and the amount of VEGF secreted from cells in the medium was analyzed using a human VEGF immunoassay kit (RayBiotech) according to the manufacturer’s instructions.
Statistical analysis
For statistical analysis between two groups, Student’s t-test for independent means was applied. The differences between any two of several groups were analyzed by one-way analysis of variance followed by least significant difference multiple comparisons. Statistical analysis was performed with SPSS software (v 13.0; SPSS Inc, Chicago, IL). A value of P < 0.05 was considered statistically significant.
Results and discussion
Characterization of ternary complexes prepared in mixing order I
The PCD was successfully synthesized by Michael addition polymerization between acryloyl group of CBA and amine group of DMDPTA (Figure 1A). The synthesized PCD was hygroscopic and readily dissolved in water. According to GPC measurement, the molecular weight of PCD was around 20.3 kDa with a polydispersity index of 1.78. The chemical structure of PCD was validated by 1H NMR spectrum, as shown in Figure 2A. The synthesized PCD displayed better buffer capacity (about 42%) than PEI (about 30%), indicating PCD had better endosomal escape capacity based on the so-called “proton sponge” effect (Figure 2B). In addition, PCD displayed lower cytotoxicity than PEI in both MCF-7 and MDA-MB-231 cells (Figure 2C and D). The lower cytotoxicity may be explained by the rapid fragmentation of the synthesized PCD in the reductive cytosol, resulting in small polycation fragments with negligible cytotoxicity.
Figure 2.
Characterization of the synthesized novel redox-responsive hyperbranched poly(amido amine) (named PCD): (A) proton nuclear magnetic resonance spectrum of PCD, (B) acid-base titration curves of PCD and polyethylenimine (PEI), (C) cytotoxicity of PCD and PEI on MCF-7 human breast cancer cells, and (D) cytotoxicity of PCD and PEI on MDA-MB-231 human breast cancer cells.
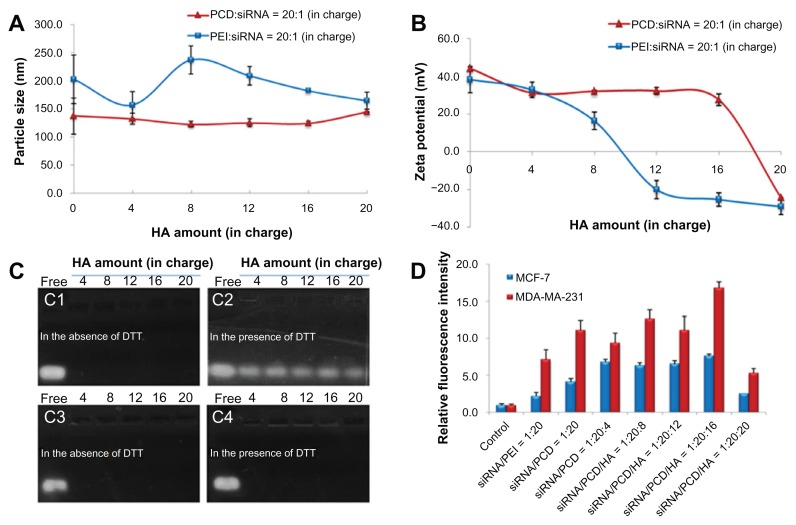
The binding capacity of siRNA with PCD was examined using the gel retardation assay. The result showed that as N/P charge ratio increased to 20/1, the migration of siRNA was retarded efficiently. Therefore, preparation of the siRNA/PCD complex was carried out at the N/P charge ratio of 20/1 in this study. The particle size and zeta potential of siRNA/PCD binary complex were about 140 nm and 40 mV, respectively (Figure 3A and B). To optimize the charge ratio of P/N/C in ternary complex, the preparation method of siRNA complex was firstly fixed at mixing order I (Figure 1B). After optimization of the P/N/C charge ratio, the effects of ternary complexes with different mixing orders on siRNA delivery would be further investigated. Figure 3A and B show the particle size and surface potential of various siRNA complexes with the different charge ratios of siRNA/polycation/HA. With an N/P charge ratio of 20/1, the particle size of PCD ternary complexes was not significantly changed with the increased amount of HA from 4 to 20 (in charge), while the particle size of PEI ternary complexes could be affected by the amount of HA (Figure 3A). Compared with PEI ternary complexes, PCD ternary complexes had relatively smaller vesicles. These results demonstrated that more protonated amine groups existed in PCD (hydrochloride salt), thus forming more compact complexes than that of PEI. It is likely because of more intense electrostatic interactions between positively charged PCD and negatively charged HA or siRNA. In some cases, these differences of protonated amine groups may explain the different surface potentials between PEI and PCD ternary complexes. For example, at an N/P/C charge ratio of 20/1/12, the surface potential of PCD ternary complexes was positive, whereas PEI ternary complexes displayed negative charge (Figure 3B). When excess HA was added (eg, at the ratio of 20), both PEI and PCD ternary complexes displayed negative charge.
Figure 3.
Characterization of small interfering RNA (siRNA) ternary complexes prepared in mixing order I: particle size (A) and zeta potential (B) of siRNA complexes with different amounts of hyaluronic acid (HA) (in charge); (C) gel retardation assay of siRNA/novel redox-responsive hyperbranched poly(amido amine) (named PCD)/HA (C1 and C2) and siRNA/polyethylenimine (PEI)/HA (C3 and C4) ternary complexes with different amounts of HA (from 4 to 20 in charge) in the absence of dithiothreitol (DTT) (C1 and C3) and in the presence of DTT (C2 and C4), the charge ratio of siRNA/polycation is equal to 1/20; (D) cellular uptake of fluorescein-labeled siRNA transfected by various siRNA formulations after incubation at 37°C for 4 hours in MCF-7 and MDA-MB-231 human breast cancer cells.
Note: The untreated cells were used as the control.
To evaluate the binding stability of siRNA in the complexes, the gel retardation assay was conducted (Figure 3C). In the absence of DTT, there is no siRNA release from both PEI and PCD ternary complexes with HA amounts from 4 to 20 (in charge) when the N/P charge ratio is 20/1, suggesting good binding stability of siRNA in PCD (Figure 3C1) or PEI (Figure 3C3) ternary complexes. However, in the presence of DTT (mimicking the reductive intracellular environment), siRNA was evidently released from PCD ternary complexes (Figure 3C2), indicating PCD ternary complexes could exhibit a redox-responsive release of siRNA in the reducing environment. On the contrary, there was almost no siRNA released from PEI ternary complexes in the presence of DTT (Figure 3C4), suggesting that PEI ternary complexes were not readily unpacked in the reductive conditions.
It is known that good cellular uptake is an essential prerequisite for efficient siRNA delivery. Cellular uptake of ternary complexes with different HA amounts in MCF-7 and MDA-MB-231 cells are shown in Figure 3D. For PCD ternary complexes, when the amount of HA reached 16 (in charge), ternary complex showed the best cellular uptake in both cells. However, as the amount of HA increased to 20 (in charge), the cellular uptake in both cells dramatically reduced, possibly because of the negative charge in ternary complexes (Figure 3D). Normally, cells do not easily take up anionic complexes, as they tend to be electrostatically repelled by the negatively charged cell membranes. Therefore, the ternary complex containing HA developed in the study should have a little positive charge so as to enhance the cellular uptake of ternary complexes. As seen in Figure 3D, the relative fluorescence intensity of the complexes without HA (siRNA/PCD = 1/20) showed values comparable with the complexes with HA (siRNA/PCD/HA = 1/20/4, 8, and 12) in MDA-MB-231 cell line, and only siRNA/PCD/HA with the ratio of 1/20/16 showed a higher fluorescence value. On the contrary, for the MCF-7 cell line, the complexes with HA (siRNA/PCD/HA = 1/20/4, 8, 12, and 16) showed higher fluorescence values than the complexes without HA (siRNA/PCD = 1/20). Importantly, the fluorescence intensity was much higher in MDA-MB-231 than that of MCF-7 with respect to the complexes without HA (siRNA/PCD = 1/20) and with HA (siRNA/PCD/HA = 1/20/4, 8, 12, and 16). These results suggest that the different fluorescence intensities between MDA-MB-231 and MCF-7 cells after treatment with the complexes were most likely due to the biological characters of different cell species. Compared with complexes without HA, the ternary complexes with HA (especially for siRNA/PCD/HA = 1/20/16) improved intracellular fluorescence intensity in MDA-MB-231 or MCF-7 cells to some extent, suggesting that HA receptor- mediated endocytosis partly contributed to the improvement of cellular uptake,41 although siRNA cell transfection here was mainly based on the significant positive-charge effects. Therefore, the ternary complexes with HA (siRNA/PCD/HA = 1/20/16) and the MDA-MB-231 cell line were used for the evaluation of siRNA gene silencing efficacy.
Effects of mixing orders on particle size and zeta potential
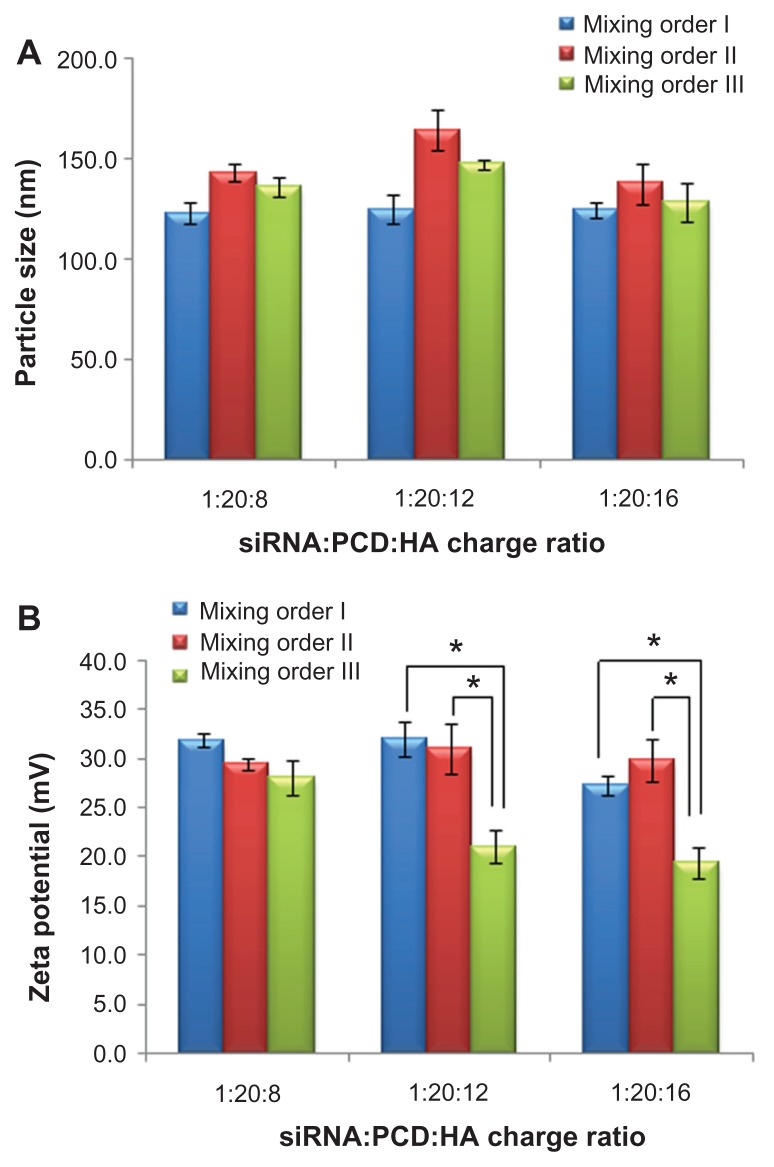
As shown in Figure 4A, the different mixing orders did not greatly influence the hydrodynamic size (from about 130 to about 140 nm) of siRNA ternary complexes. However, the zeta potential of vesicles (Figure 4B) was affected by the different mixing orders, depending on the charge ratio of P/N/C. With a 1/20/8 charge ratio, similar zeta potentials (approximately 30 mV) were observed in the siRNA ternary complexes prepared in different mixing orders. When the P/N/C charge ratio was 1/20/12 or 1/20/16, the mixing order III showed significantly lower zeta potential (approximately 20 mV) than the other two mixing orders (approximately 30 mV). The different zeta potentials (observed in different ternary complexes at the P/N/C charge ratio of 1/20/16) are more likely due to the different surface densities of positive charges existing in assembly structures of ternary complexes with different mixing orders, although similar hydrodynamic particle sizes were observed among these three complexes.
Figure 4.
Particle size (A) and zeta potential (B) of small interfering RNA (siRNA) ternary complexes prepared by three mixing orders at different charge ratios of siRNA/novel redox-responsive hyperbranched poly(amido amine) (named PCD)/hyaluronic acid (HA).
Note: *P < 0.01, significant difference between the two groups.
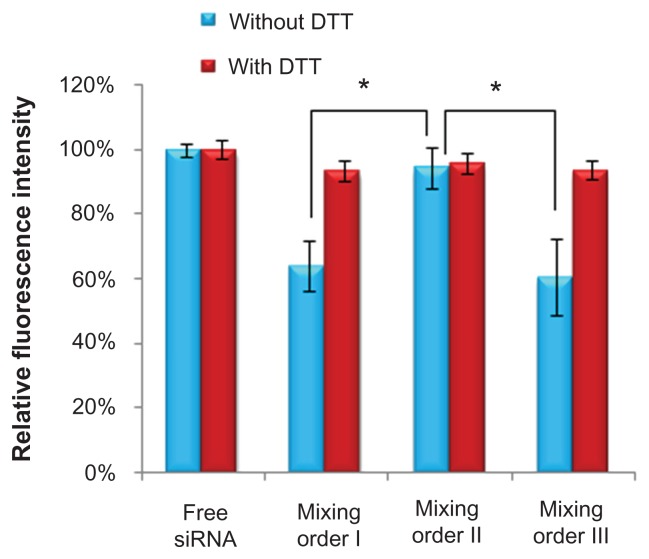
Effects of mixing orders on FAM-siRNA fluorescence quenching
Recently, many studies have shown that tight compaction of FAM-siRNA increased the fluorescence quenching because of close spatial proximity.35,38,42 As shown in Figure 5, the mixing order II resulted in higher fluorescence intensity than the other two mixing orders without addition of DDT, indicating most of FAM-siRNA molecules were adsorbed with a relatively loose state on the surface layer of ternary complexes by electrostatic interaction when prepared in mixing order II (Figure 1B).
Figure 5.
Relative fluorescence intensity of fluorescein-labeled small interfering RNA (siRNA) in various siRNA/novel redox-responsive hyperbranched poly(amido amine) (named PCD)/hyaluronic acid ternary complexes (siRNA/polycation/hyaluronic acid charge ratio of 1/20/16) prepared by different mixing orders.
Note: *P < 0.01, significant difference between the two groups.
Abbreviation: DTT, dithiothreitol.
For ternary complexes prepared in mixing orders I and III, obvious fluorescence quenching was observed under the condition without DTT, whereas significantly increased fluorescence intensity was detected in the presence of DTT (Figure 5). These results indicate most of FAM-siRNA were compressed in the vesicles and thus resulted in decrease of fluorescence intensity. After incubation with DTT, siRNA was released from these complexes by the cleavage of disulfide bonds in PCD.
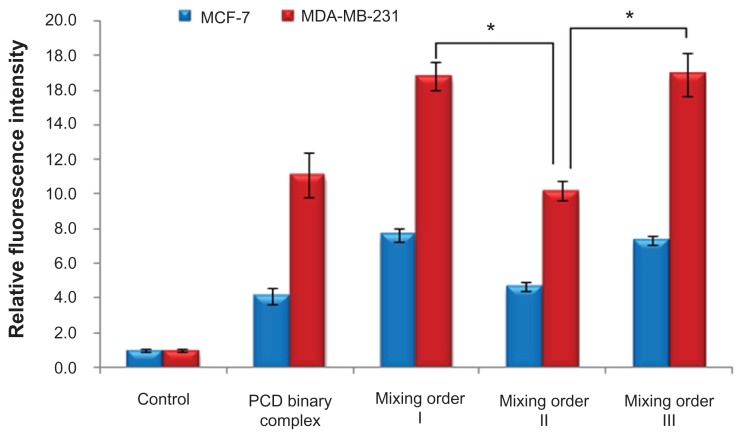
Effects of mixing orders on cellular uptake
Ideal siRNA vectors capable of protecting siRNA molecules from degradation in the biological milieu, steering them to target cells, and facilitating their cellular entry, are practically desired. In this study, the addition of HA was expected to decrease the zeta potential of nano-sized siRNA complexes and enhance targeting efficiency of siRNA delivery into the cells with HA receptor (CD44) overexpression. To further confirm this hypothesis, the effect of mixing orders on cellular uptake was investigated. As seen in Figure 6, the relative fluorescence intensity of the ternary complexes with HA (siRNA/PCD/HA = 1/20/16, prepared in mixing order II) showed values comparable with the binary complexes without HA (siRNA/PCD = 1/20) in MDA-MB-231 and MCF-7 cell lines, but significantly lower than the other two ternary complexes with HA (siRNA/PCD/HA = 1/20/16, prepared in mixing orders I and III). Moreover, similar fluorescence intensity was observed between mixing orders I and III. These results demonstrate that the ternary complexes prepared in mixing orders I and III (siRNA/PCD/HA = 1/20/1) could exhibit better cell uptake in MDA-MB-231 and MCF-7 cells than that prepared in mixing order II. Considering the result of fluorescence quenching of FAM-siRNA (Figure 5), it is easy to understand that most of FAM-siRNA molecules were adsorbed with a relatively loose state on the surface layer of ternary complexes by electrostatic interaction when prepared in mixing order II. On the contrary, the ternary complexes prepared in mixing orders I and III achieved a relatively higher cellular uptake than that prepared in mixing order II, which more likely correlated with receptor-mediated endocytosis due to the more exposed HA on the surface of vesicles prepared in mixing orders I and III. Also, the reason for similar cellular uptake between the mixing orders III and I is more likely to be similar surface density of exposed HA in these two complexes. A more recent study confirmed that HA was coated on the outer layer of the vesicles, as determined by Raman microscopy, when ternary complexes were prepared by mixing order I.34 As for the mixing order III, it is mainly because of the similar surface density of HA existing in the outer layer of ternary complexes, which is sufficient to perform the receptor-mediated endocytosis at the P/N/C charge ratio of 1/20/16. Accordingly, the ternary complex prepared in mixing orders I or III not only ensures better cellular uptake to the target cells but also provides better protective effect of siRNA from degradation.
Figure 6.
Cellular uptake (4 hours) of fluorescein-labeled small interfering RNA (siRNA) in MCF-7 and MDA-MB-231 human breast cancer cells after incubation with novel redox-responsive hyperbranched poly(amido amine) (named PCD)/siRNA binary complex and various siRNA/PCD/hyaluronic acid ternary complexes (siRNA/polycation/hyaluronic acid charge ratio of 1/20/16) prepared by different mixing orders.
Notes: The untreated cells were used as the control. *P < 0.01, significant difference between the two groups.
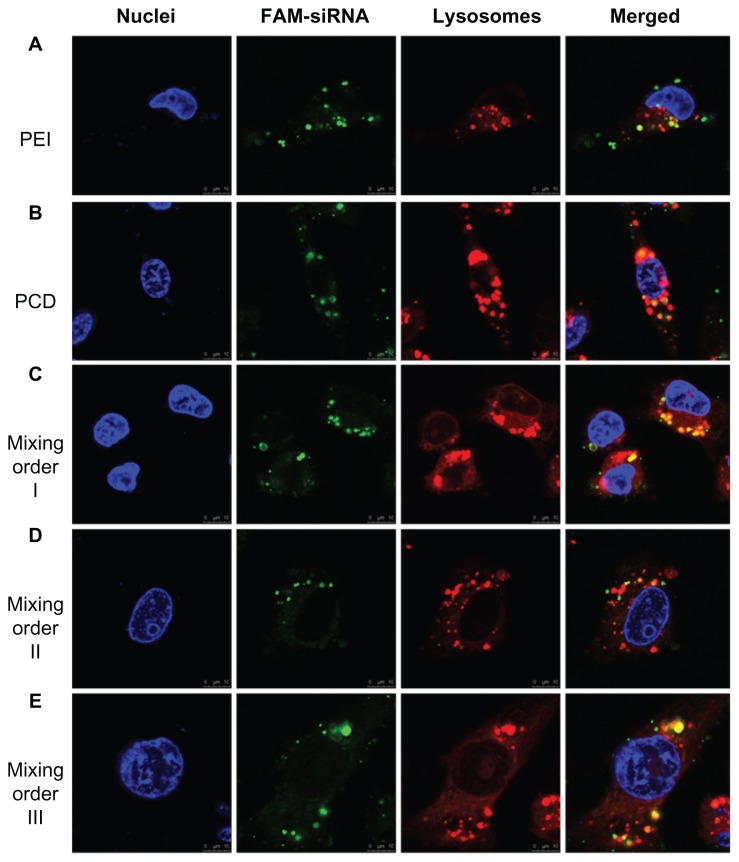
The confocal microscopy observations were also used to confirm intracellular distribution of siRNA after treatment with various ternary complexes prepared in different assembly sequences (Figure 7). In MDA-MB-231 cells, agglomerate distribution of green fluorescence was observed in the cells after treatment with PEI/siRNA binary complexes, indicating most siRNA was still tightly bound to PEI after cell internalization and not prone to disassembly and release from the complexes in the reductive cytosol (Figure 7A). In contrast, siRNA was in a relatively loose state for siRNA/PCD binary complexes, suggesting that the complexes could be unpacked to release siRNA by the cleavage of disulfide bonds in the reductive cytosol (Figure 7B). In addition, a certain amount of siRNA (diffused green fluorescence) was detached from the PCD ternary complexes (mixing orders I, II and III) in the reductive cytosol (Figure 7C–E), similar to the previous report.24 These results were consistent with the flow cytometry analysis shown in Figure 6. Therefore, it was demonstrated that the mixing orders I and III displayed more effective cell internalization and better targeting efficiency than mixing order II. Based on better cellular internalization and intracellular distribution of siRNA, gene silencing efficiency of these ternary complexes should be further evaluated in MDA-MB- 231 cells with higher HA receptor overexpression, so as to obtain an optimal siRNA delivery vector.
Figure 7.
Confocal microscopic images of MDA-MB-231 human breast cancer cells after incubation for 4 hours with (A) fluorescein-labeled small interfering RNA (FAM-siRNA)/polyethylenimine (PEI) binary complex, (B) FAM-siRNA/novel redox-responsive hyperbranched poly(amido amine) (named PCD) binary complex, and FAM-siRNA/PCD/hyaluronic acid ternary complexes prepared by (C) mixing order I, (D) mixing order II, and (E) mixing order III.
Notes: The siRNA/polycation charge ratio is equal to 1/20 for binary complexes, and siRNA/polycation/hyaluronic acid charge ratio is equal to 1/20/16 for ternary complexes. Nuclei and lysosomes were stained with Hoechst 33258 (blue) and LysoTracker® (red), respectively. The scale bars represent 10 μm.
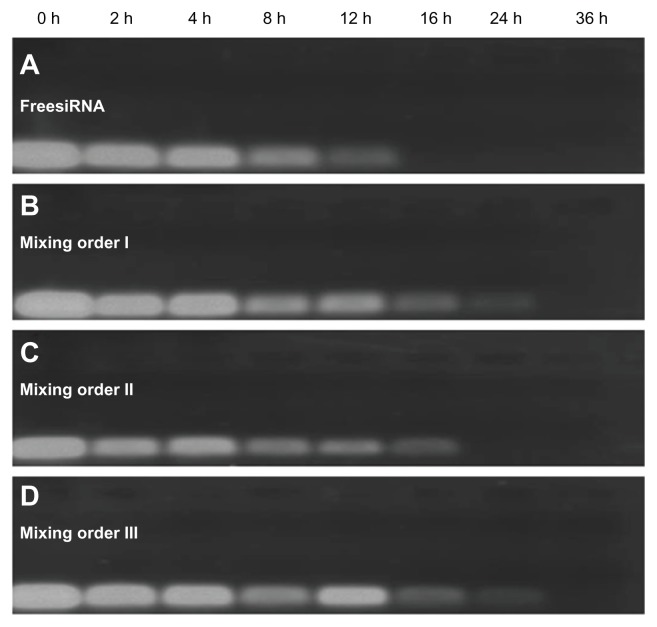
Effects of mixing orders on serum stability
Considering the importance of serum stability of the ternary complex in vivo condition, the effects of mixing order on serum stability were also studied in the presence of serum ( Figure 8). Free siRNA was not stable and completely degraded within 16 hours in the presence of serum (Figure 8A). In a comparison of serum stability of ternary complexes prepared in different mixing orders, a detectable siRNA band could be seen in mixing orders I and III at the 24-hour point, but no detectable siRNA band was seen in mixing order II at the same time point (Figure 8B–D). It is possible that more siRNA was located in the inner layer of the vesicles (or the siRNA was more tightly compacted) when ternary complexes were prepared in mixing orders I and III, thus providing better protection of siRNA from enzyme-mediated digestion. Furthermore, more siRNA is possibly located in the outer layer of vesicles (or siRNA was compacted in loose state) when ternary complexes were prepared in mixing order II, thus the siRNA is prone to subjected degradation in the presence of serum. From the results of fluorescence quenching and serum stability tests, it is demonstrated that significantly different condensation states of siRNA were formed in different assembly structures when ternary complexes were prepared in different mixing orders and may therefore affect siRNA delivery efficiency.
Figure 8.
Serum stability of free small interfering RNA (siRNA) and siRNA ternary complexes – siRNA/novel redox-responsive hyperbranched poly(amido amine) (named PCD)/hyaluronic acid charge ratio of 1/20/16 – in the presence of 10% fetal bovine serum: (A) free siRNA, (B) mixing order I, (C) mixing order II, and (D) mixing order III.
Abbreviation: h, hours.
Effects of mixing orders on VEGF-siRNA gene silencing efficacy
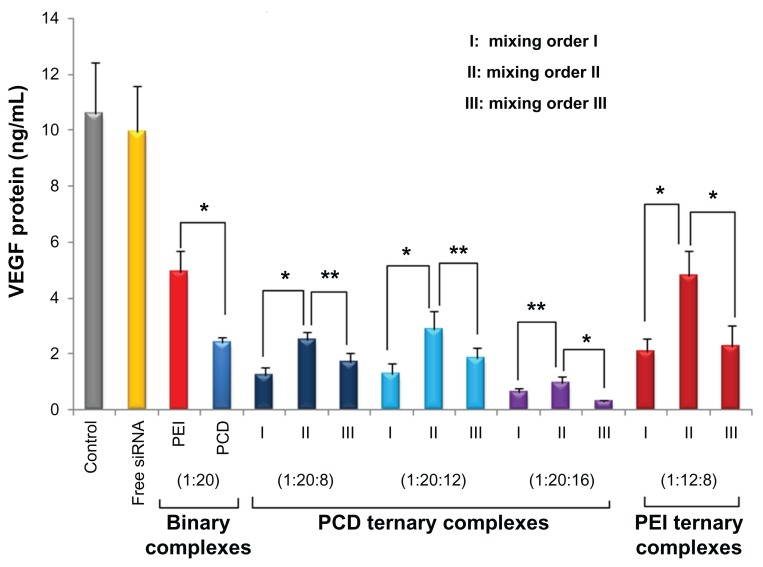
To investigate siRNA delivery efficiency of ternary complexes prepared in different mixing orders, the level of VEGF-directed protein after gene silencing was measured by enzyme-linked immunosorbent assay. When formulated with VEGF-directed siRNA to form binary complexes, PCD/siRNA complexes showed significantly higher inhibition of VEGF (approximately 75% inhibition) than branched PEI (approximately 50% inhibition) in MDA-MA- 231 cells ( Figure 9). At the P/N/C charge ratio of 1/20/8, the gene silencing effects achieved by PCD ternary complexes were superior to that of PEI ternary complexes prepared by the same method. An earlier study indicated that vector unpacking to release nucleic acid had been considered as a limiting step for efficient gene delivery.43 In the present study, the better gene silencing effects achieved by PCD ternary complex possibly resulted from their better unpacking capacities in the reductive condition, which suggests that unpacking and releasing of siRNA into the cytoplasm is essential for maximum gene silencing activity of the siRNA.44
Figure 9.
Vascular endothelial growth factor (VEGF) gene silencing activities of various VEGF-directed small interfering RNA (siRNA) complexes in MDA-MB-231 human breast cancer cells. The siRNA ternary complexes were prepared in different charge ratios of siRNA/polycation/hyaluronic acid. For binary complexes, the siRNA/polycation charge ratio is equal to 1/20. The untreated cells were used as the control.
Notes: Results presented as means and standard deviations of three experiments; *P < 0.01, **P < 0.05 between the two groups.
Abbreviations: PCD, novel redox-responsive hyperbranched poly(amido amine); PEI, polyethylenimine.
For ternary complexes, either with PEI or PCD, the results demonstrated that mixing order II exhibited worse transfection efficiency of siRNA than mixing orders I and III. For PCD ternary complexes, when HA charge ratio was 8 or 12, mixing order II showed a similar gene silencing effect to that for PCD binary complexes (Figure 9). Similarly, the gene silencing effect achieved by PEI binary complexes (P/N charge ratio = 1/20) was close to that of PEI ternary complexes prepared by mixing order II (P/N/C charge ratio = 1/20/8). Therefore, it is apparent that mixing order II should not be encouraged during the preparation of ternary complexes.
In contrast, siRNA/PCD/HA ternary complexes prepared by mixing orders I and III generally exhibited a similarly higher gene silencing efficiency than siRNA/PCD binary complexes (P < 0.05). When the P/N/C charge ratio was 1/20/8 or 1/20/12, ternary complexes prepared in mixing order I exhibited a higher gene silencing effect than mixing order III. As the P/N/C charge ratio increased to 1/20/16, ternary complexes prepared in mixing order I exhibited a lower gene silencing effect than that of mixing order III. Therefore, whether the gene silencing effect of ternary complexes (prepared by mixing order I) is better than that of ternary complexes (prepared by mixing order III) may depend on the charge ratio of P/N/C. It is important to note that the differences in siRNA gene silencing efficiency among these three ternary complexes are due to the difference of surface exposed density of HA and condensation state of siRNA in different assembly structures. Taken together, it can be concluded that the siRNA/PCD/HA ternary complexes prepared by mixing orders I and III are the promising vectors for siRNA delivery but that mixing order II is not encouraged to be used for siRNA delivery.
Conclusion
To the authors’ knowledge, this is the first comparative study of three ternary complexes prepared by different mixing orders. In a comparison of three positively charged siRNA/PCD/HA ternary complexes, the mixing orders I and III displayed better siRNA delivery efficiencies than mixing order II in MDA-MA-231 human breast cancer cells. With an optimized siRNA/PCD/HA charge ratio of 1/20/16, the gene silencing effects on VEGF expression in MDA-MA- 231 cells were ranked as follows: mixing order III > mixing order I > mixing order II. These results could benefit the development of an efficient vector for siRNA delivery. Knowledge of the assembly structures and the uptake mechanisms of these ternary complexes should be further studied in future.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program Nos 2007CB935800 and 2009CB930300), the Program for New Drug R&D (No 2009ZX09310-001), National Natural Science Foundation of China projects (No 81072597), Beijing Natural Science Foundation project (No 7112089), and Programs from the Ministry of Education (Nos BMU20110268 and BMU20110263).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 3.Tebes SJ, Kruk PA. The genesis of RNA interference, its potential clinical applications, and implications in gynecologic cancer. Gynecol Oncol. 2005;99(3):736–741. doi: 10.1016/j.ygyno.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 6.Schiffelers RM, Ansari A, Xu J, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavazzana-Calvo M, Thrasher A, Mavilio F. The future of gene therapy. Nature. 2004;427(6977):779–781. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- 8.Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 10.Juliano R, Bauman J, Kang H, Ming X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol Pharm. 2009;6(3):686–695. doi: 10.1021/mp900093r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 12.Jeong JH, Kim SW, Park TG. Molecular design of functional polymers for gene therapy. Prog Polym Sci. 2007;32(11):1239–1274. [Google Scholar]
- 13.Bauhuber S, Hozsa C, Breunig M, Göpferich A. Delivery of nucleic acids via disulfide-based carrier systems. Adv Mater. 2009;21(32–33):3286–3306. doi: 10.1002/adma.200802453. [DOI] [PubMed] [Google Scholar]
- 14.Meng F, Hennink WE, Zhong Z. Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials. 2009;30(12):2180–2198. doi: 10.1016/j.biomaterials.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Jere D, Kim JE, Arote R, et al. Akt1 silencing efficiencies in lung cancer cells by sh/si/ssiRNA transfection using a reductable polyspermine carrier. Biomaterials. 2009;30(8):1635–1647. doi: 10.1016/j.biomaterials.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Piest M, Lin C, Mateos-Timoneda MA, et al. Novel poly(amido amine)s with bioreducible disulfide linkages in their diamino-units: structure effects and in vitro gene transfer properties. J Control Release. 2008;130(1):38–45. doi: 10.1016/j.jconrel.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Christensen LV, Chang CW, Kim WJ, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17(5):1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 18.Breunig M, Hozsa C, Lungwitz U, et al. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130(1):57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30(29):5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo H, Jin GW, Kang H, et al. Biodegradable branched poly(ethylenimine sulfide) for gene delivery. Biomaterials. 2010;31(5):988–997. doi: 10.1016/j.biomaterials.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Lin C, Blaauboer CJ, Timoneda MM, et al. Bioreducible poly(amido amine)s with oligoamine side chains: synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126(2):166–174. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Son S, Singha K, Kim WJ. Bioreducible BPEI-SS-PEG-cNGR polymer as a tumor targeted nonviral gene carrier. Biomaterials. 2010;31(24):6344–6354. doi: 10.1016/j.biomaterials.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Zhou L, Zhou Y, et al. Synthesis and gene delivery of poly(amido amine)s with different branched architecture. Biomacromolecules. 2010;11(2):489–495. doi: 10.1021/bm901215s. [DOI] [PubMed] [Google Scholar]
- 24.Kurosaki T, Kitahara T, Fumoto S, et al. Ternary complexes of pDNA, polyethylenimine, and gamma-polyglutamic acid for gene delivery systems. Biomaterials. 2009;30(14):2846–2853. doi: 10.1016/j.biomaterials.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 25.Duceppe N, Tabrizian M. Factors influencing the transfection efficiency of ultra low molecular weight chitosan/hyaluronic acid nanoparticles. Biomaterials. 2009;30(13):2625–2631. doi: 10.1016/j.biomaterials.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Yoshihara C, Hamada K, Koyama Y. DNA/polyethyleneimine/hyaluronic acid small complex particles and tumor suppression in mice. Biomaterials. 2010;31(10):2912–2918. doi: 10.1016/j.biomaterials.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 27.Xu P, Quick GK, Yeo Y. Gene delivery through the use of a hyaluronate-associated intracellularly degradable crosslinked polyethyleneimine. Biomaterials. 2009;30(29):5834–5843. doi: 10.1016/j.biomaterials.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh EJ, Park K, Kim KS, et al. Target specific and long-acting delivery of protein, peptide, and nucleotide therapeutics using hyaluronic acid derivatives. J Control Release. 2010;141(1):2–12. doi: 10.1016/j.jconrel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Yao J, Fan Y, Du R, et al. Amphoteric hyaluronic acid derivative for targeting gene delivery. Biomaterials. 2010;31(35):9357–9365. doi: 10.1016/j.biomaterials.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 30.Götte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66(21):10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 31.Draffin JE, McFarlane S, Hill A, Johnston PG, Waugh DJ. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 2004;64(16):5702–5711. doi: 10.1158/0008-5472.CAN-04-0389. [DOI] [PubMed] [Google Scholar]
- 32.Sheridan C, Kishimoto H, Fuchs RK, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eliaz RE, Szoka FC., Jr Liposome-encapsulated doxorubicin targeted to CD44: a strategy to kill CD44-overexpressing tumor cells. Cancer Res. 2001;61(6):2592–2601. [PubMed] [Google Scholar]
- 34.Ito T, Koyama Y, Otsuka M. Analysis of the surface structure of DNA/polycation/hyaluronic acid ternary complex by Raman microscopy. J Pharm Biomed Anal. 2010;51(1):268–272. doi: 10.1016/j.jpba.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Ito T, Iida-Tanaka N, Niidome T, et al. Hyaluronic acid and its derivative as a multi-functional gene expression enhancer: protection from non-specific interactions, adhesion to targeted cells, and transcriptional activation. J Control Release. 2006;112(3):382–388. doi: 10.1016/j.jconrel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Wu C, Oupický D. Bioreducible hyperbranched poly(amido amine)s for gene delivery. Biomacromolecules. 2009;10(10):2921–2927. doi: 10.1021/bm900724c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong CY, You YZ, Wu DC, Liu Y, Pan CY. Thermal control over the topology of cleavable polymers: from linear to hyperbranched structures. J Am Chem Soc. 2007;129(17):5354–5355. doi: 10.1021/ja070871+. [DOI] [PubMed] [Google Scholar]
- 38.Vader P, van der Aa LJ, Engbersen JF, Storm G, Schiffelers RM. A method for quantifying cellular uptake of fluorescently labeled siRNA. J Control Release. 2010;148(1):106–109. doi: 10.1016/j.jconrel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64(10):3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 40.Kim SH, Jeong JH, Kim TI, Kim SW, Bull DA. VEGF siRNA delivery system using arginine-grafted bioreducible poly(disulfide amine) Mol Pharm. 2009;6(3):718–726. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim EK, Kim HO, Jang E, et al. Hyaluronan-modified magnetic nanoclusters for detection of CD44-overexpressing breast cancer by MR imaging. Biomaterials. 2011;32(31):7941–7950. doi: 10.1016/j.biomaterials.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 42.Merkel OM, Beyerle A, Librizzi D, et al. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol Pharm. 2009;6(4):1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 43.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 44.Hoon Jeong J, Christensen LV, Yockman JW, et al. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]