Abstract
A chemically dynamic spermine-based polymer: polyspermine imidazole-4, 5-amide (PSIA, Mw > 7 kDa) was designed, synthesized, and evaluated in terms of its ability to deliver nucleic acids. This polymer was made from an endogenous monomer professionally condensing genes in sperms, spermine, and a known safety drug metabolite, imidazole-4, 5-dicarboxylic acid, through a bis-amide bond conjugated with the imidazole ring. This polymer can condense pDNA at a W/W ratio above 10 to form polyplexes (100–200 nm in diameter), which is consistent with the observation by transmission electron microscopy (TEM), and the zeta potential was in the range of 10–20 mV. The pDNA packaged polymer was stable in phosphate buffer solution (PBS) at pH 7.4 (simulated body fluid) while the polyplexes were releasing pDNA into the solution at pH 5.8 (simulated endo-lysosomes) due to the degradation of the bis-amide linkages in response to changes in pH values. PSIA-polyplexes were able to achieve efficient cellular uptake and luciferase gene silencing by co-transfection of pDNA and siRNA in COS-7 cells and HepG2 cells with negligible cytotoxicity. Biodistribution of Rhodamine B-labeled PSIA-polyplexes after being systemically injected in BALB/c nude-mice showed that the polyplexes circulated throughout the body, accumulated mainly in the kidney at 4 hours of sample administration, and moved to the liver and spleen after 24 hours. All the results suggested that PSIA offered a promising example to balance the transfection efficiency and toxicity of a synthetic carrier system for the delivery of therapeutic nucleic acids.
Keywords: gene delivery, polyspermine, cytotoxicity, transfection efficiency, biodistribution
Introduction
A variety of nucleic acids such as plasmid DNA (pDNA) and small interfering RNA (siRNA) are used as effective therapeutic drugs in gene therapy, vaccination, and tissue regeneration by regulating the relevant gene on (expression) or off (silencing).1–3 However, many biological barriers need to be conquered to achieve the safe and efficient delivery of nucleic acids to the nuclei or cytoplasm of target cells. Among the carrier systems for delivering polynucleotides (pDNA and siRNA), synthetic non-viral delivery systems possess a series of advantages over viral vectors, including freedom from immunity and viral mutation, ability to package multiple genes or siRNA of choice into particulates via a single mechanism, and adaptability for simple and costefficient manufacturing processes.4–6
To deliver gene materials to targeted inter- and intra-cellular sites effectively using non-viral systems, the synthetic gene carriers must accomplish five tasks simultaneously: (A) packing gene materials to nano-particulate forms to avoid prephagocytosis degradation, (B) attaching to targeted cells selectively, (C) facilitating endosomal escape of gene materials, (D) releasing gene materials in cytoplasm, and (E) metabolizing themselves to nontoxic species.2 While there have been numerous cationic polymers reported to date for gene delivery, such as polyethyleneimine (PEI),7–11 poly-L-Lysine (PLL),12 and poly (amide amine) (PAMAM),13–15 none have the ability to meet all these criteria.
Recently, spermine, an endogenous multi-amino monomer professionally condensing genes in the body, has been widely studied for nucleic acid delivery as the basic building block to form gene carriers. Previous studies focus mainly on the construction of spermine-based degradable16–20 or non-degradable carrier systems,21 some interesting results of more efficient gene transfection with less cytotoxicity were obtained in the last decade, but these carriers couldn’t degrade back to their original state because of their non-degradable or non-directly connected degradable linkages. Therefore, the accumulation of degradation products with unknown safety from the delivery systems would be a major concern for the clinical application of the vehicles.
In this work, we designed and synthesized a chemically dynamic polyspermine with bis-amide linkages conjugated with an imidazole ring (PSIA), which was formed from an endogenous gene condensing monomer, spermine, and a drug metabolite, imidazole-4, 5-dicarboxylic acid with established safety.22 Then, gene condensation, pH-responsive degradation, and cytotoxicity were evaluated. Furthermore, cellular uptake, gene transfection as well as in vivo biodistribution of PSIA-polyplexes were investigated. It was supposed that PSIA-polyplexes may be further coated with a self-assembled surface to neutralize its positive charges and to immobilize cell-targeting moieties with optimized surface population. Electrostatic adsorption and phase-guided assembly are currently being used in our laboratory to form a rationally designed surface and to accomplish task (B) from above.
Materials and methods
Materials
Polyethyleneimine 25 kDa (PEI 25 kDa), spermine, N,N′-dicyclohexylcarbodiimide (DCC), N-hydroxysuccinimide (NHS), 4, 5-imidazoledicarboxylic acid, trifluoroacetic acid (TFA), dimethyl sulfoxide BioReagent (DMSO, for molecular biology), and ethidium bromide (EB) were purchased from Sigma-Aldrich (St Louis, MO). N,N- dimethylformamide (DMF, 99.8%, extra dry over molecular sieves) was purchased from Acros Organics (Geel, Belgium). 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazoliumbromide (MTT) and penicillin-streptomycin solution were purchased from Solarbio Science and Technology Co, Ltd (Beijing, China). A poly(ethylene glycol) standards kit ( ranging from 106 to 20100 in molecular weight) was purchased from Polymer Standards Service GmbH (Mainz, Germany). Trypan blue solution (0.4%) was purchased from Amresco (Solon, OH). A Micro BCA™ protein assay kit was purchased from Thermo Scientific Pierce (Rockford, IL). A luciferase assay kit was purchased from Promega ( Madison, WI). Water was purified using a Milli-Q instrument (Millipore, Billerica, MA). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum gold (FBS), and trypsin-EDTA solution were obtained from PAA Laboratories (Pasching, Austria). OPTIMEM-I reduced serum medium and Lyso-Tracker™ green DND-26 were purchased from Invitrogen (Carlsbad, CA). Plasmid DNA (pDNA) encoding firefly luciferase pGL3-control (Promega) was amplified using EndoFree™ Plasmid Maxi (Qiagen, Venlo, The Netherlands). The DNA concentration was determined by measuring the absorbance at 260 nm. pGL3 luciferase gene siRNA and Allstars Negative Control siRNA (Cat No 1027280) were purchased from Qiagen. All the materials used for siRNA experiments were RNase-free. All other chemicals were of analytical grade.
Cell culture and animals
The cell lines COS-7 (African green monkey kidney fibroblast) and HepG2 (Human hepatocellular liver carcinoma) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium containing 10% FBS, 100 units/mL penicillin G sodium, and 100 μg/mL streptomycin. Cells were maintained at 37°C in a humidified incubator (5% CO2). Male BALB/c nude-mice aged 6 weeks (20–25 g) were provided by the Laboratory Animal Center of the Chinese Academy of Sciences and maintained under standard housing conditions. The animal experiments were carried out in accordance with the guidelines approved by the Regulations for the Administration of Affairs Concerning Laboratory Animals and The National Regulation of China for Care and Use of Laboratory Animals.
Synthesis of PSIA
4, 5-imidazoledicarboxylic acid (100 mg, 0.64 mmol), DCC (528 mg, 2.56 mmol), NHS (295 mg, 2.56 mmol), and TFA (1 mL, 13.5 mmol) were dissolved in DMF (5 mL) and stirred at 25°C for 12 hours. The mixture was filtrated and added dropwise to a DMF solution (5 mL) of spermine (130 mg, 0.64 mmol) slowly over 6 hours under a high-purity nitrogen atmosphere, stirring at 25°C for another 24 hours. The resulting solution was then filtrated in vacuum and evaporated to remove the solvent. The viscous residue was dissolved again in water and dialyzed through a cellulose membrane with a molecular weight cutoff of 7000 kDa (Thermo Scientific, Waltham, MA) for 24 hours to remove the small fragments. The dialysate was stored at −80°C for 4 hours and freeze-dried (Christ Alpha 1–2 LD Freezedryer, Osterode, Germany) to obtain PSIA (40 mg, yield of 17.4%).
Characterization of PSIA
The chemical structure and average molecular weight of PSIA was confirmed by various characterization methods, including 1H-nuclear magnetic resonance (1H-NMR), Fourier transform infrared spectrometry (FT-IR) and size exclusion chromatography (SEC). 1H-NMR spectrum was obtained in DMSO-d6 with 0.03% (v/v) tetramethylsilane (TMS) as the internal standard using a Varian Mercury Plus 400 MHz spectrometer (Palo Alto, CA). The FT-IR spectrum was recorded in a KBr pellet using a Bruker Optics FT-IR spectrometer (Billerica, MA) in the range of 400–4000 cm−1 with 4 cm−1 resolution and 16 scans per spectrum. The molecular weight (Mw) of PSIA at a concentration of 2 mg/mL in water was measured by SEC with calibrated PEG standards and PEI 25 kDa using a Waters HPLC system (Milford, MA) equipped with a model 2414 refractive index detector, a model 1525 binary HPLC pump, and a series of Ultrahydrogel ™ columns (120 and 250) at 40°C. As an eluent, 0.05% NaN3 aqueous solution was used at a flow rate of 1.0 mL/minute.
Preparation and characterization of polyplexes made from PSIA and pDNA
Polyplexes were formed by mixing PSIA with the pDNA solution at various polymer to gene ratios (W/W), followed by incubating for 30 minutes at room temperature. The formed polyplexes were loaded on a 1.0% agarose gel in 1 × Tris-Acetate-EDTA (TAE) buffer containing 0.5 μg/mL ethidium bromide with 6 × loading buffer and subjected to electrophoresis for 45 minutes at 110 V (DNA marker: DS™ 5000, Dongsheng Biotech Co, Ltd, Guangzhou, China). The retardation of pDNA electrophoresis was visualized using a UV illuminator. The morphology of the polyplexes formed at a polymer to gene ratio (W/W) of 20 was imaged using a transmission electron microscopic (TEM; JEM 2010 system JEOL, Tokyo, Japan). The particle size and zeta potential of the polyplexes at various polymer to gene ratios (W/W) were measured in water using a Brookhaven particle size analyzer (90 Plus; Upton, NY). The mean values of the particle size and zeta potential (±standard deviation) were calculated from three experiments.
Degradation of PSIA under different pH
PSIA was dissolved in phosphate buffer to a concentration of 2.0 mg/mL at pH 7.4 and 5.8 for simulating body fluid and endo-lysosomes. The polymers to be characterized (Mw) were incubated under the respective pH at 37°C, and sampled for SEC measurement. The degradation rates of the polymers were determined by the changes of molecular weight measured by SEC.
Cellular uptake experiment
COS-7 cells and HepG2 cells were seeded in a 6-well plate at a density of 2 × 105 cells per well in 1 mL DMEM medium and incubated for 24 hours. PSIA was fluorescently labeled with Rhodamine B by mixing PSIA and Rhodamine B succinimidyl ester at a polymer to dye molecule ratio (W/W) of 2 and stirring under the catalysis of TFA for 24 hours at room temperature in the dark. The polyplexes made from Rhodamine B-labeled PSIA and pDNA at a polymer to gene ratio (W/W) of 20 were prepared as described above and added into the cells at a pDNA concentration of 2.5 μg/well for 4 hours at 37°C, followed by adding Lyso-Tracker Green DND-26 (50 nmol/mL) to the cells and incubating at 37°C for another 60 minutes. The extracellular fluorescence was quenched with 0.4% trypan blue (500 μL/well) for 2 minutes. After the medium was removed, cells were washed three times with PBS and visualized using a fluorescence microscope (Nikon Eclipse TS-100 Inverted Fluorescent Microscope, Japan) to follow the internalization of the polyplexes.
Transfection efficiency of polyplexes made from PSIA and pDNA
COS-7 cells and HepG2 cells were seeded in a 48-well plate at a density of 5 × 104 cells per well to reach approximately 90% confluence at the time of transfection and incubated for 24 hours. pDNA (0.3 nM) was added into a PSIA solution at various polymer to gene W/W ratios to form polyplexes. Spermine and PEI 25 kDa were used as references at W/W ratios of 10 and 2, respectively. The cells were treated with the carrier-gene complexes in OPTIMEM-I Reduced Serum Medium for 4 hours. The transfection medium was then replaced with fresh medium and incubated for an additional 48 hours. Finally, the transfected cells were washed with PBS solution and lysed with 1 × cell lysis buffer (Promega), followed by centrifugation at 12000 rpm for 3 minutes ( Eppendorf 5810 R Centrifuge, Hamburg, Germany). To determine relative gene expression, 20 μL supernatant was mixed with 20 μL substrate (Luciferase Assay System, Promega) and the luminescence was measured using a single tube luminometer (Sirius-Single Tube Luminometer, Berthold Detection Systems GmbH, Pforzheim, Germany). Gene transfection activities of PSIA and the two references were defined as the luminescence per unit protein, the amount of which was determined using a Micro BCA™ Assay Kit. The value of the luminescence per unit protein (±standard deviation) was calculated from the mean of three experiments.
Co-transfection efficiency of pDNA and siRNA using PSIA
COS-7 cells and HepG2 cells were seeded in a 48-well plate at a density of 5 × 104 cells per well to reach approximately 90% confluence at the time of transfection and incubated for 24 hours. Polyplexes formed from pDNA (0.09 nM) and siRNA (22.5 nM) formulated with PSIA at various polymer to nucleic acid W/W ratios were transfected into the cells for 4 hours in the OPTIMEM-I Reduced Serum Medium. The transfection medium was replaced with fresh culture medium and further incubated for 48 hours. To determine the extent of luciferase gene expression, the transfected cells were washed with PBS solution and lysed with 1 × cell lysis buffer followed by centrifugation at 12000 rpm for 3 minutes. The supernatant (20 μL) was mixed with 20 μL of substrate and the luminescence was measured using a single tube luminometer. The protein concentrations of the samples were determined using a Micro BCA™ Protein Assay Kit. Luciferase activity of a sample was expressed as relative light units (RLUs) normalized on protein concentration. The value of the luminescence per unit protein (±standard deviations) was calculated from the mean of three experiments.
Cytotoxicity assay
Cytotoxicity of PSIA in comparison with PEI 25 kDa was evaluated by percent viability of COS-7 cells and HepG2 cells at various polymer concentrations. Cells were seeded in a 96-well plate at a density of 1 × 104 cells per well and incubated for 24 hours, followed by treatment with the polymer solutions at the desired concentrations. After an additional 4 hours of incubation, 112.5 μL fresh culture medium and 12.5 μL MTT solution at a concentration of 5 mg/mL in PBS buffer were added into the cells for 6 hours. Viable cells were determined by measuring the absorbance of the samples at 570 nm (with 630 nm as the reference) using a SpectraMax M3 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA). Cell viabilities were calculated by taking the non-treated cells (normal cells) as 100%. The data were expressed as the mean values (±standard deviations) of six experiments.
Biodistribution in BLAB/c nude-mice
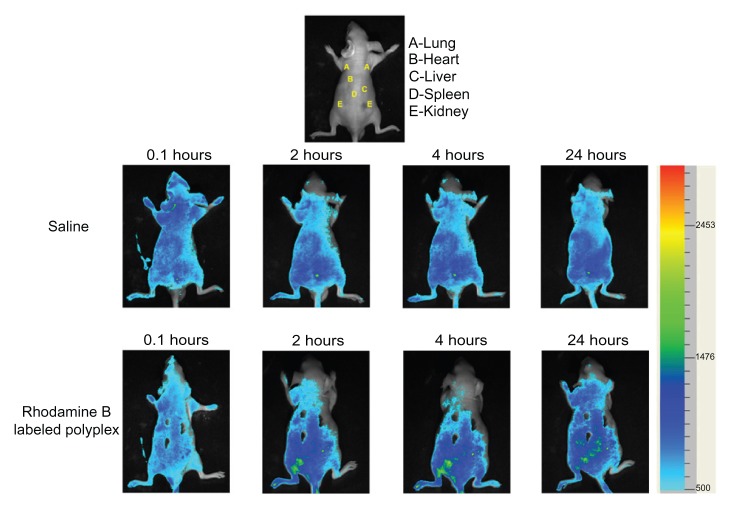
The polyplexes formed from Rhodamine B-labeled PSIA and pDNA at a polymer to gene ratio (W/W) of 20 were prepared as described above (saline as a control), and then BLAB/c nude-mice were intravenously injected with the polyplexes at a dose of 5 μg pDNA per mouse (in 200 μL saline) via the tail vein. After systematic injection of the polyplexes at 0.1 hours, 2 hours, 4 hours, and 24 hours, the mice were anesthetized with an intraperitoneal injection of 20% urethane and observed using Small Animal Fluorescense Imaging Facility (Zhongke Kaisheng Medical Technology Co, Ltd, Guangzhou, China).
Statistical analysis
Data were expressed as the mean values (±standard deviations) of independent measurements. Statistical analysis was performed using an independent samples t-test. Statistical significance was assigned for P-values less than 0.05.
Results and discussion
Synthesis and characterization of PSIA
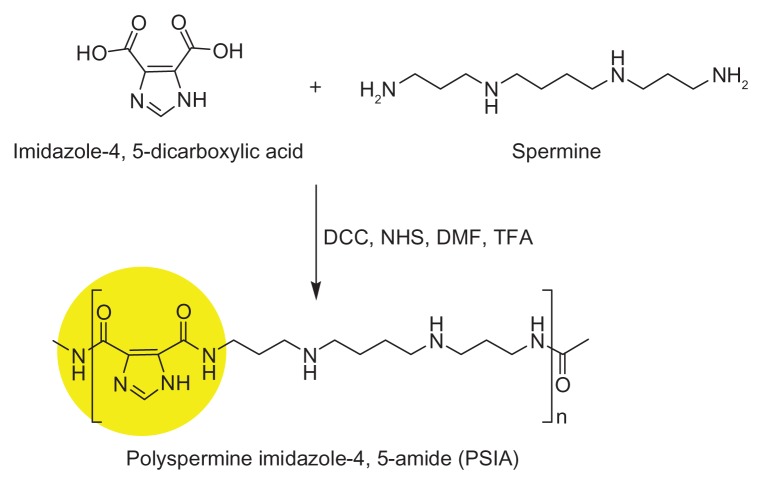
PSIA, containing bis-amide linkages conjugated with the imidazole ring, was synthesized via a one-step condensation reaction between spermine and carboxyl activation of imidazole bis-carboxylic acid, in the presence of a catalytic amount of TFA (Figure 1). The polymerization was performed at room temperature under stirring for 24 hours and further dialyzed in water through a molecular weight cut-off cellulose membrane of 7 kDa. The expected product was lyophilized to a water-soluble white powder prior to storage at −80°C.
Figure 1.
Chemical structure and polymerization scheme of PSIA.
Abbreviations: DCC, N,N′-dicyclohexylcarbodiimide; DMF, N,N-dimethylformamide; NHS, N-hydroxysuccinimide; TFA, trifluoroacetic acid.
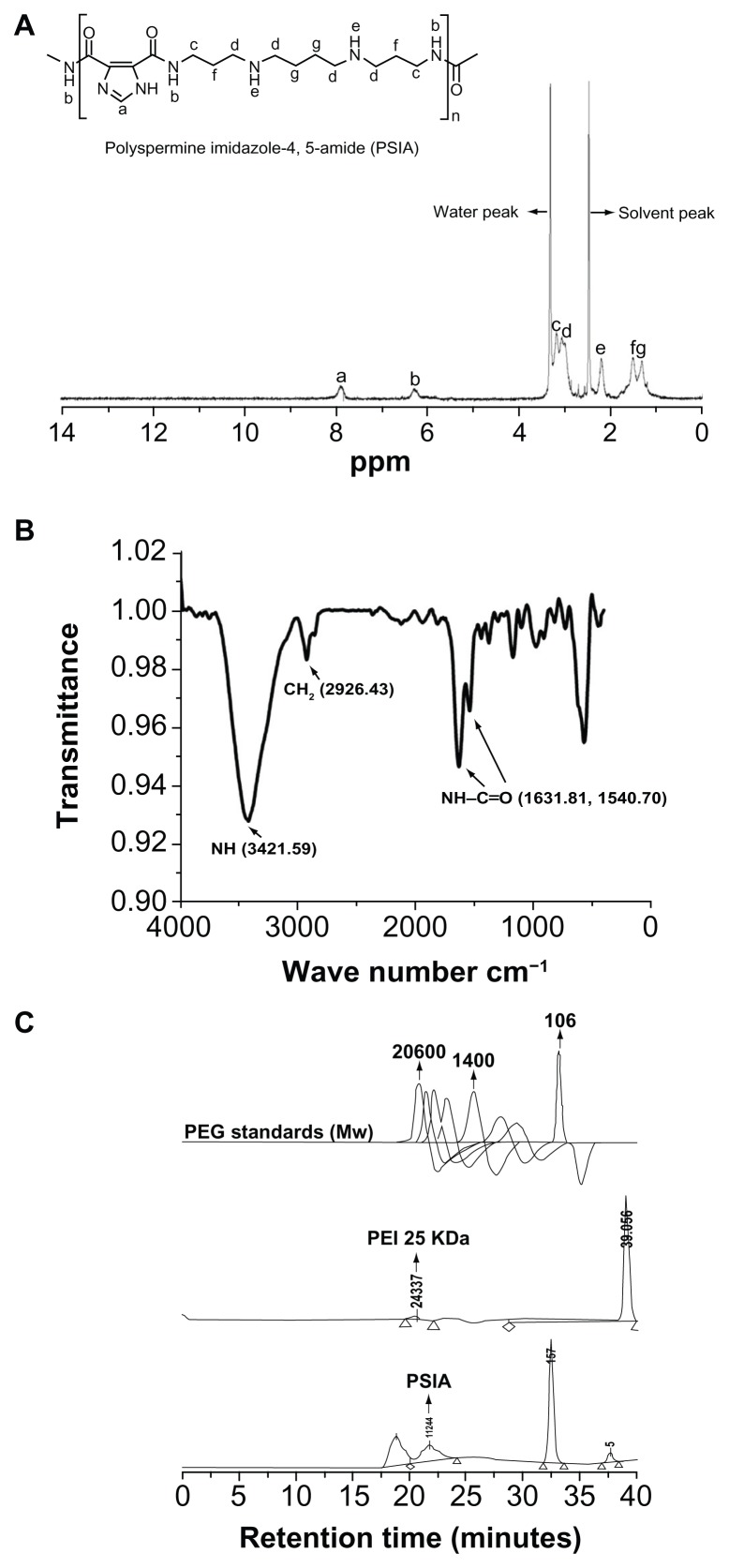
The chemical structure and average molecular weight (Mw) of PSIA were demonstrated by 1H-NMR, FT-IR, and SEC. The 1H-NMR spectrum of PSIA in DMSO-d6 was in accordance with the desired polymer (Figure 2A). The methylene protons close to nitrogen atoms were present at δ = 2.98–3.20 ppm. The presence of the remaining methyl groups were confirmed by a signal at δ = 1.29–1.53 ppm indicating methylene protons and δ = 7.80–8.10 ppm indicating methine protons of the imidazole ring, respectively. The signal at δ = 2.12–2.33 ppm indicates the amino protons of the spermine skeleton. Also, due to the generation of new bis-amide linkages (O=C–NH) in the polymer chain, the signal at δ = 6.14–6.50 ppm appeared, which was assigned to the amide protons, indicating that the polymer was built by bis-amide linkages. In addition, we observed that the intense absorptions at 1631.81 cm−1 and 1540.70 cm−1 were shown in the FT-IR spectrum (Figure 2B), which was attributed to the stretching vibration of the newly formed bis-amide bonds (as amide I and II peaks, respectively) rather than the original bis-carboxyl bonds, further demonstrating the synthetic process of the polymer. Moreover, the average molecular weight (Mw) of PSIA measured by SEC was 14.2 kDa, with polydispersity to be 1.37 by eluting a solution of the polymer (2 mg/mL in water) through a series of SEC Ultrahydrogel columns with PEG as relative standards. PEI 25 kDa was used as a calibration standard to establish a reliable method for the determination of the molecular weight distribution of the polymer (Figure 2C). These results confirm the successful formation of PSIA.
Figure 2.
Chemical structure and molecular weight characterization of PSIA: (A) 1H-NMR spectrum (DMSO-d6); (B) FT-IR spectrum (within a KBr disk); (C) SEC charts (with PEG as standards and PEI 25 kDa as a calibration standard).
Abbreviations: PSIA, polyspermine imidazole-4, 5-amide; 1H-NMR, proton nuclear magnetic resonance; FT-IR, fourier transform infrared; SEC, size exclusion chromatography; PEG, polyethylene glycol; PEI, polyethyleneimine.
Physicochemical characterization of polyplexes made from PSIA and pDNA
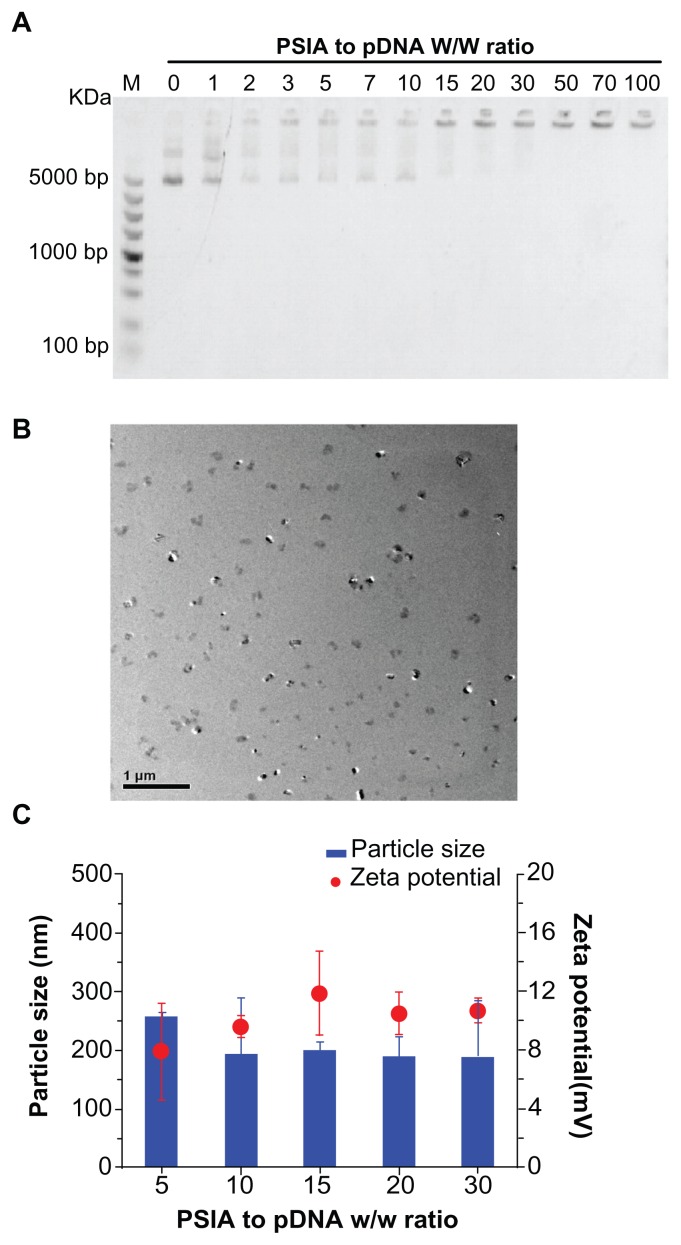
The ability of PSIA gene condensation was examined using agarose gel electrophoresis, TEM, dynamic light scattering (DLS), as well as zeta potential measurement. As shown in Figure 3A, the complete retardation of electrophoresis of pDNA-polymer complexes when the polymer to nucleic acid ratio (W/W) was over 10, indicated that the polymer was capable of impeding the mobility of pDNA and condensing pDNA into nano-particulates. The morphology of PSIA-polyplexes was observed by TEM in a dehydrated condition, and confirmed the formation of the polyplexes, which were spherical in shape and 100–200 nm in average diameter (Figure 3B). Furthermore, the particle size and zeta potential of the polyplexes in water were determined by DLS and plotted against PSIA to pDNA ratio (W/W) in Figure 3C. The particle sizes varied in the range between 100 to 200 nm, which is consistent with the measurement by TEM. The zeta potential of the polyplexes was in the range of 7.93–11.87 mV and remained relatively constant over those ranges. Electrophoresis, TEM, and DLS all suggest that PSIA has the ability to pack pDNA into polyplexes.
Figure 3.
Physicochemical characterization of polyplexes made of PSIA and pDNA at various polymer to gene ratios (w/w) in water: (A) agarose gel electrophoresis of polyplexes; (B) TEM image of polyplexes at the polymer to gene ratio (w/w) of 20 (scale bar: 200 nm); (C) particle size distribution and zeta potential measurement of polyplexes.
Abbreviations: PSIA, polyspermine imidazole-4, 5-amide; TEM, transmission electron microscopy.
Degradation of PSIA
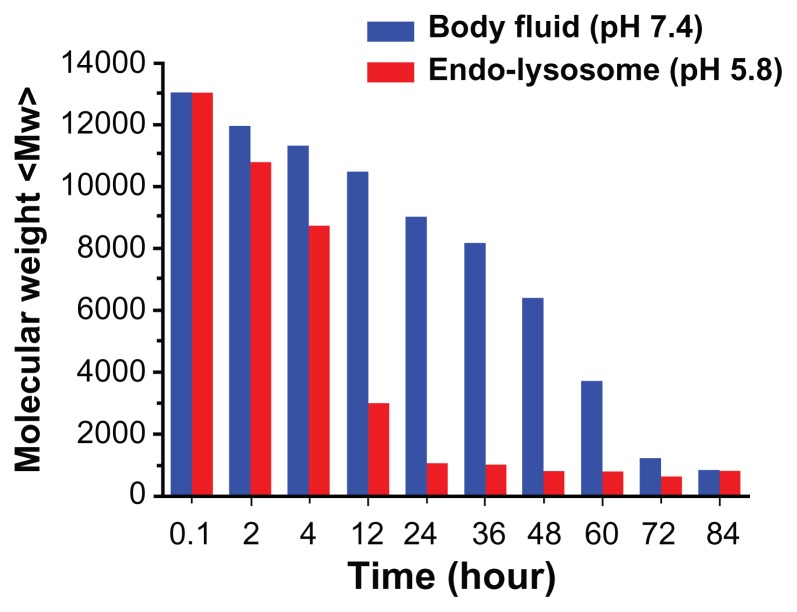
On the basis of the biological pathways for the intracellular delivery of nucleic acids, a pH-responsive degradable gene carrier was used to release pDNA into the cytoplasm, to minimize cytotoxicity of the carrier and to generate free amino groups to facilitate endosomal escape. The degradability of PSIA at pH values simulating both body fluid (pH 7.4) and endo-lysosomes (pH 5.8) was analyzed by SEC method and plotted as a function of time. Incubating this polyelectrolyte in PBS, at pH 7.4 and pH 5.8 at 37°C, the average molecular weight of PSIA degraded versus time at significantly different rates responsive to different buffers. As shown in Figure 4, PSIA exhibited slow degradation of the molecular weight at pH 7.4, until small fragments (smaller than 1 kDa) relative to its original molecular weight were detected at the time of 84 hours. However, the time taken for the polymer to reach the same distribution of molecular weight (no more than 1 kDa) at pH 5.8 was remarkably decreased between 12 to 20 hours, indicating that the bis-amide linkages of the polymer chain upon degradation were capable of generating two free amino groups to enhance the proton-sponging effect,23,24 and further improving the biocompatibility with reduced cytotoxicity.
Figure 4.
The degradation curves of PSIA in PBS, at pH 7.4 and pH 5.8 at 37°C (relative to the original molecular weight of PSIA).
Abbreviations: PSIA, polyspermine imidazole-4, 5-amide; PBS, phosphate buffer solution.
Cellular uptake experiment
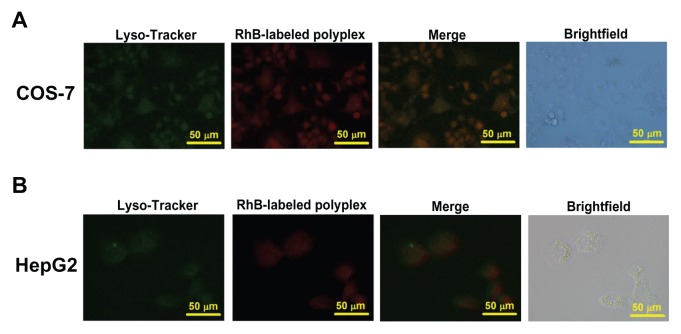
The cellular uptake of the polyplexes formed from PSIA and pDNA was performed on COS-7 cells and HepG2 cells. PSIA was pre-treated with Rhodamine B succinimidyl ester to form a fluorescently labeled polymer, which was able to pack gene into Rhodamine B-labeled polyplexes. Lyso-Tracker Green DND-26 was used to stain the lysosomes. The effect of extracellular fluorescence on the cellular uptake assay was ruled out by adding trypan blue to quench the non-internalization of the fluorescent molecules. We observed that in COS-7 cells treated with Rhodamine B-labeled polyplexes for 4 hours, a large number of red fluorescent polyplexes were visualized around the green fluorescent lysosomes in the cytoplasmic compartment. Meanwhile, in terms of the intracellular fluorescence of HepG2 cells incubated with the polyplexes under the same treatment as described above, red fluorescence was also detected in the cytoplasm (Figure 5). However, the fluorescent intensity of the polyplexes in HepG2 cells was not comparable to that in COS-7 cells, revealing that the internalization of the nano-particulates depended on the absorption properties of different cell types.
Figure 5.
Cellular uptake of Rhodamine B-labeled polyplexes formed from PSIA and pDNA at the polymer to gene ratio (w/w) of 20 in COS-7 cells (A) and HepG2 cells (B) scale bar 50 μm.
Abbreviation: PSIA, polyspermine imidazole-4, 5-amide.
In vitro transfection and cell viability assay
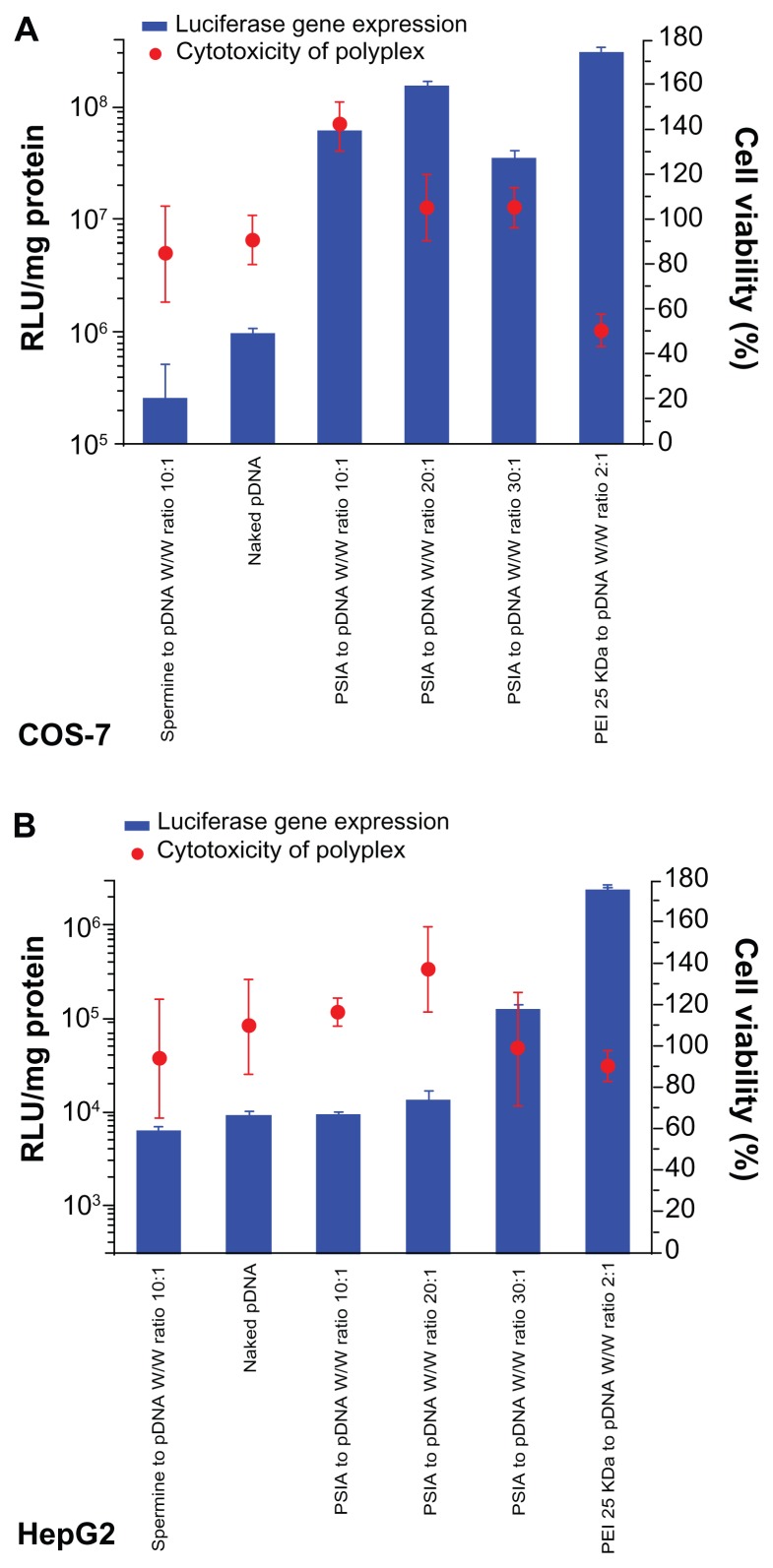
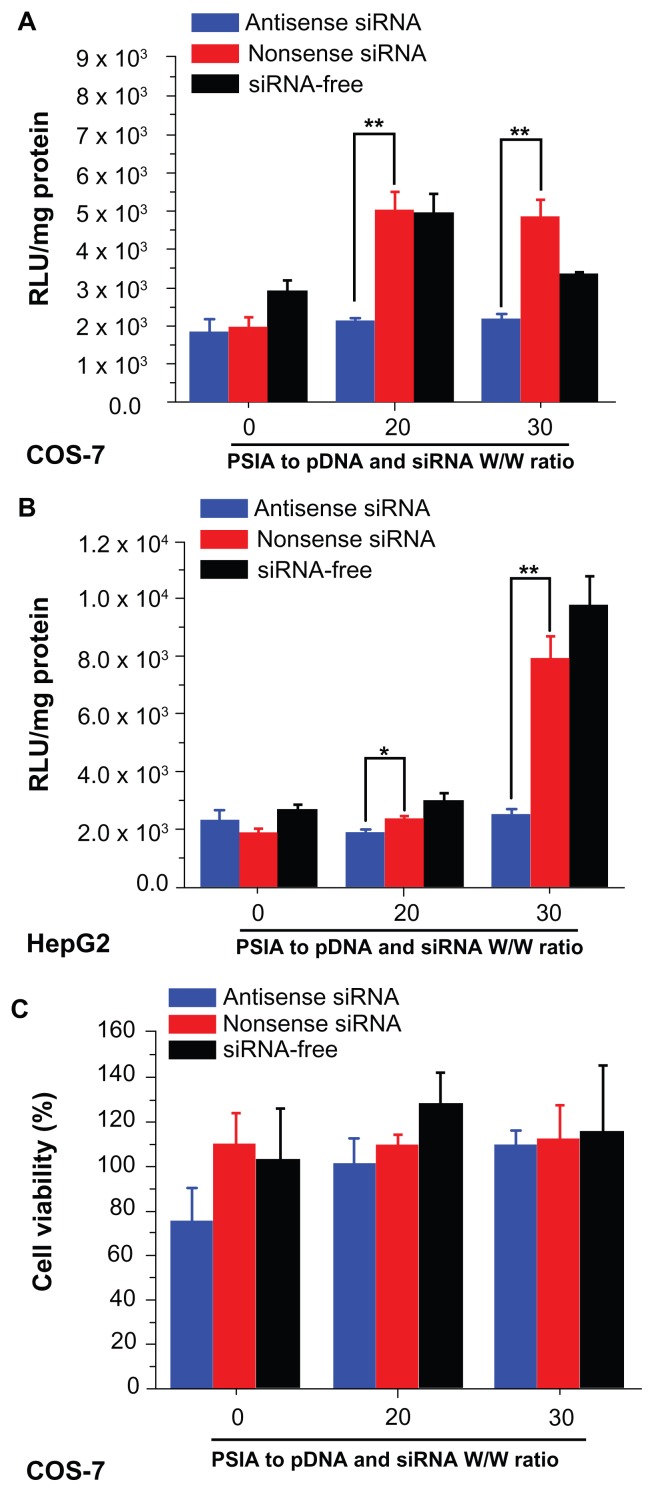
The capability of PSIA to deliver pDNA or co-deliver pDNA and siRNA into cells was determined through the expression of luciferase gene in COS-7 cells and HepG2 cells in vitro. For the transfection of pDNA, the cells were treated with the polyplexes formed from PSIA and pDNA. For the co- transfection of pDNA and siRNA, the cells were treated with the luciferase gene, combined with antisense and nonsense siRNA in naked and PSIA-packed forms, respectively. As shown in Figure 6A, the best luciferase expression achieved by PSIA-pDNA complexes at a polymer to gene ratio (W/W) of 20, was comparable to that of PEI 25 kDa at the optimal mass ratio in COS-7 cells. Conversely, almost no gene expression was detected for polymer-free pDNA with the protection of small-molecular-weight spermine added into the cells directly, due to the electrostatic repulsion between the nucleic acids and the cytomembrane, which are both negatively charged on the surface, and thus impede the penetration of the nucleic acid into the cytoplasm. HepG2 cells transfected with the polyplexes formed from PSIA and pDNA as a function of the polymer to gene ratios (W/W) showed that the transient expression of luciferase gene was not high when compared to PEI 25 kDa, but the improvement in transfection efficiency was achieved by PSIA compared to naked pDNA (Figure 6B). These results were in accordance with the cellular uptake assay. After COS-7 and HepG2 cells were treated with the polyplexes made of PSIA, luciferase gene, antisense siRNA, and nonsense siRNA at various polymer to nucleic acid W/W ratios, significantly different expression rates of luciferase gene were found (Figure 7). For both the cells incubated with naked luciferase gene and naked siRNA, nonsense or antisense, the level of luciferase expression defined as the substrate luminescence per mg protein was the same as for the cells that received no treatment, indicating a baseline. However, when PSIA was added to luciferase gene and siRNA in both the cells, at the W/W ratios of 20 and 30, the samples co-transfected with polyplexes loaded with nonsense siRNA and luciferase gene showed drastically elevated luciferase expression (the red bars in Figure 7A and B). On the contrary, for the samples co-transfected with polyplexes of antisense siRNA and luciferase gene, the level of luciferase expression remained at baseline, indicating complete silencing of the gene. Furthermore, the cytotoxicity effect of the polyplexes involving the carrier molecules on the cell transfection assays was ruled out by MTT assay (Figure 7C and D).
Figure 6.
Luciferase activity and cytotoxicity of polyplexes formed from PSIA and pDNA in transfecting luciferase gene in COS-7 cells (A) and HepG2 cells (B) at various polymer to gene ratios (W/W).
Abbreviations: PEI, polyethylenimine; PSIA, polyspermine imidazole-4, 5-amide.
Figure 7.
Expression of luciferase gene by co-transfecting pDNA and siRNA (blue: antisense; red: nonsense; black: siRNA-free) into COS-7 cells (A) and HepG2 cells (B) using PSIA; cytotoxicity of polyplexes formed from PSIA-pDNA-antisense siRNA complex (blue bar), PSIA-pDNA-nonsense siRNA complex (red bar), and PSIA-pDNA complex (black bar) at various polymer to gene ratios (W/W) in COS- 7 cells (C) and HepG2 cells (D); the value ‘0’ on the x-axis refers to naked pDNA and siRNA solution or naked pDNA solution.
Notes: *P < 0.05; **P < 0.01.
Abbreviations: PSIA, polyspermine imidazole-4, 5-amide; RLU, relative light unit.
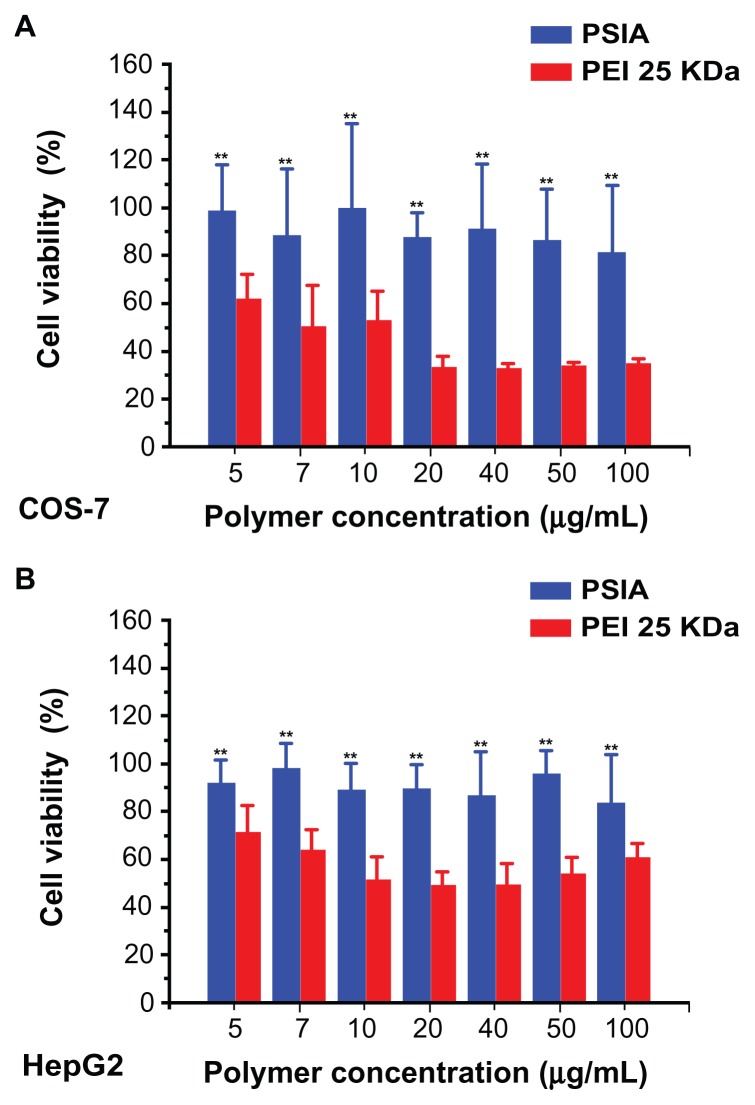
Likely due to the degradation of the polymer shown in Figure 4, cell viability assays showed a negligible cytotoxicity profile using culturing of PSIA with COS-7 and HepG2 cells as a function of polymer concentration, which was much better than that of PEI 25 kDa with the same treatment for MTT assay. As shown in Figure 8, the viability of both the cell lines treated with PEI 25 kDa dropped dramatically to 30% for COS-7 and 50% for HepG2 of the original when the polymer concentration reached 20 μg/mL, but those treated with PSIA remained unchanged during the entire experiment (up to 100 μg/mL in polymer concentration). Hence, the cytotoxicity-free characteristics of PSIA give it potential for clinical application.
Figure 8.
Viability of COS-7 cells (A) and HepG2 cells (B) treated with PSIA and PEI 25 kDa at various polymer concentrations.
Note: **P < 0.01 vs PEI 25 kDa.
Abbreviations: PSIA, polyspermine imidazole-4, 5-amide; PEI, polyethyleneimine.
Biodistribution in BALB/c nude-mice
The biodistribution of Rhodamine B-labeled PSIA-polyplexes after being systemically injected in BALB/c nude-mice at different time points is presented in Figure 9. All the sampling mice survived during the experiment (in the case of the systemic injection of PEI 25 kDa-polyplexes at the same concentration as pDNA, sudden death occurred in the mice due to the significant cytotoxicity of PEI 25 kDa). No fluorescence could be detected at the initial time (0.1 hours) of sample administration. After 4 hours, it was found that the fluorescent polyplexes circulated throughout the body and accumulated gradually in the spleen and kidney. Only a small amount of the polyplexes was observed in the lung, heart, and kidney, and was mainly distributed to the liver and spleen at 24 hours. Therefore, the biodistribution of the polyplexes in vivo was conducive to the exploration of new strategies by passive targeting in the treatment of genetic-related diseases.
Figure 9.
Biodistribution of Rhodamine B-labeled polyplexes formed from PSIA and pDNA at the polymer to gene ratio of 20 after being systemically injected in BALB/c nude-mice (saline as a control).
Abbreviation: PSIA, polyspermine imidazole-4, 5-amide.
Conclusion
An imidazole-conjugated polyspermine with bis-amide linkages as chemically dynamic moieties (PSIA) was synthesized and investigated for intracellular delivery of nucleic acids. PSIA could pack pDNA into nano-particulates with positive surface charges. Also, PSIA showed pH-responsive degradability in acidic conditions and further enhanced the so-called proton-sponge mechanism. The desired cellular uptake and luciferase gene expression by transfection or silencing by co-transfection in both cell lines (COS-7 and HepG2) with high cell viability achieved by PSIA-nucleic acid polyplexes confirmed the capability of PSIA as a gene delivery system. In vivo biodistribution of PSIA-polyplexes indicated that the polyplexes initally concentrate in the kidneys, and are distributed to the liver and spleen after 24 hours. Therefore, these experimental results suggest that PSIA could be a potential alternative as a non-viral vector for safe and efficient nucleic acid delivery.
Acknowledgments
The Ministry of Science and Technology (2009ZX09310-007) of the People’s Republic of China and Technology Committee of Science and Technology Special Nanotechnology Grant (No 1052nm03900) are gratefully acknowledged for financial support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mulligan RC. The basic science of gene therapy. Science. 1993;260(5110):926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 2.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 5.Wolff JA. The “grand” problem of synthetic delivery. Nat Biotechnol. 2002;20(8):768–769. doi: 10.1038/nbt0802-768. [DOI] [PubMed] [Google Scholar]
- 6.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Therapy. 2008;15(16):1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 7.Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Nat Acad Sci U S A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boletta A, Benigni A, Lutz J, Remuzzi G, Soria MR, Monaco L. Nonviral gene delivery to the rat kidney with polyethylenimine. Hum Gene Ther. 1997;8(10):1243–1251. doi: 10.1089/hum.1997.8.10-1243. [DOI] [PubMed] [Google Scholar]
- 9.Zanta MA, Boussif O, Adib A, Behr JP. In vitro gene delivery to hepatocytes with galactosylated polyethylenimine. Bioconjug Chem. 1997;8(6):839–844. doi: 10.1021/bc970098f. [DOI] [PubMed] [Google Scholar]
- 10.Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 1998;5(9):1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 11.Goula D, Remy JS, Erbacher P, et al. Size, diffusibility and transfection performance of linear PEI/DNA complexes in the mouse central nervous system. Gene Ther. 1998;5(5):712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 12.Wagner E, Ogris M, Zauner W. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv Drug Deliv Rev. 1998;30(1–3):97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 13.Tomalia DA, Naylor AM, Goddard WA. Starburst Dendrimers: Molecular-Level Control of Size, Shape, Surface Chemistry, Topology, and Flexibility from Atoms to Macroscopic Matter. Angew Chem Int Edit. 1990;29(2):138–175. [Google Scholar]
- 14.Haensler J, Szoka FC., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug Chem. 1993;4(5):372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka S, Iwai M, Harada Y, et al. Targeted killing of carcinoembryonic antigen (CEA)-producing cholangiocarcinoma cells by polyamidoamine dendrimer-mediated transfer of an Epstein-Barr virus (EBV)- based plasmid vector carrying the CEA promoter. Cancer Gene Ther. 2000;7(9):1241–1250. doi: 10.1038/sj.cgt.7700219. [DOI] [PubMed] [Google Scholar]
- 16.Maslov MA, Kabilova TO, Petukhov IA, et al. Novel cholesterol spermine conjugates provide efficient cellular delivery of plasmid DNA and small interfering RNA. J Control Release. 2012;160(2):182–193. doi: 10.1016/j.jconrel.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Gagnon KT, Watts JK, Pendergraff HM, et al. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide- oligospermine conjugates. J Am Chem Soc. 2011;133(22):8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim MS, Kwon YJ. Dual mode polyspermine with tunable degradability for plasmid DNA and siRNA delivery. Biomaterials. 2011;32(16):4009–4020. doi: 10.1016/j.biomaterials.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Vinogradov SV. Short biodegradable polyamines for gene delivery and transfection of brain capillary endothelial cells. J Control Release. 2010;143(3):359–366. doi: 10.1016/j.jconrel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu RZ, Lu ZR. Design, synthesis and evaluation of spermine-based pH-sensitive amphiphilic gene delivery systems: Multifunctional nonviral gene carriers. Sci Chin Chem. 2011;54(2):359–368. [Google Scholar]
- 21.Ghonaim HM, Soltan MK, Blagbrough IS. Heterocyclic N-4, N-9- diacyl spermines: nanoparticle lipopolyamine vectors for efficient pDNA and siRNA delivery. J Pharm Pharmacol. 2010;62(10):1276–1277. [Google Scholar]
- 22.Novak E, Lakings DB, Paxton LM. Tolerance and disposition of cefpimizole in normal human volunteers after intramuscular administration. Antimicrob Agents Chemother. 1987;31(11):1706–1710. doi: 10.1128/aac.31.11.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behr JP. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997;51(1–2):34–36. [Google Scholar]
- 24.Xu D, Hu A, Su J, Wu F, Yuan W. Micro and Nanotechnology for Intracellular Delivery Therapy Protein. Nano-Micro Lett. 2012;4(2):118–123. [Google Scholar]